Abstract
Autophagy influences cell survival through maintenance of cell bioenergetics and clearance of protein aggregates and damaged organelles. Several lines of evidence indicate that autophagy is a multifaceted regulator of cell death, but controversy exists over whether autophagy alone can drive cell death under physiologically relevant circumstances. Here we review the role of autophagy in cell death and examine how autophagy interfaces with other forms of cell death including apoptosis and necrosis.
Cell death occurs by multiple mechanisms, but original descriptions were based on morphological features and how dead cells and their fragments are cleared. Morphologically, three broad classifications of cell death occur under physiological conditions during development: apoptosis, autophagic cell death, and necrosis1.
Apoptosis is characterized by cell shrinkage, chromatin condensation, nuclear fragmentation, plasma membrane blebbing, and debris clearance by neighboring phagocytes1, 2. Two molecular mechanisms control apoptosis, the intrinsic pathway initiated by intracellular stimuli such as DNA damage, and the extrinsic pathway initiated by extracellular death receptors; both pathways depend upon caspase activation2–4.
Cell death that depends on autophagy is distinguished by cytoplasmic vacuolization, autophagosome formation, and clearance of material via the lysosome1, 3. During macroautophagy (herein referred to as autophagy), cytoplasmic materials and organelles are sequestered and engulfed by a double membraned structure termed the autophagosome, and delivered to the lysosome for degradation and recycling3, 5. Necrosis results in organelle swelling and lysis, inflammation, and release of intracellular components. In contrast to apoptosis and autophagic cell death, necrosis generally occurs when cells are exposed to pathological conditions3, 6.
The molecular machinery underlying these different forms of cell death are promiscuous, and recent evidence indicates a complex interplay among them, making it difficult to unambiguously distinguish one form of cell death from another7. The mechanism by which a cell dies is often specific to the cell type, stimulus, context and environment. The recent explosion in discoveries of cell death subtypes highlights the many ways a cell can die3. In this Review, we address the crosstalk between autophagy, apoptosis and necrosis, highlighting the emerging and context-specific roles of autophagy during cell death.
Defining autophagic cell death
Autophagy (“self eating”) sequesters, degrades and recycles cellular material8. Autophagy is necessary and beneficial to the cell and organism because it prevents buildup of toxic protein aggregates, removes damaged organelles, and provides the cell and organism with bioenergetic substrates needed to survive8. Initially identified through genetic screens in yeast9, 10, the core autophagy machinery is well characterized and conserved in mammals8. The Atg1/Ulk1 protein kinase complex initiates autophagy by recruiting and activating the Vacuolar protein sorting 34 (VPS34) containing class III PI3K complex necessary for PI(3)P production and subsequent recruitment of autophagy factors to the autophagosomal membrane11. Autophagosome elongation and maturation occur through the coordinated activity of two ubiquitin-like conjugation systems that facilitate Atg8 (LC3/GABARAP in mammals) lipidation with phosphatidylethanolamine12–18. Insertion of lipidated Atg8into the autophagosomal membrane drives autophagosome maturation14. Autophagosomes fuse with lysosomes to form autolysosomes that degrade the cytoplasmic material sequestered within autophagosomes, and recycle it to generate amino acids, fatty acids, and nucleotides19, 20. During starvation AMP-activated protein kinase (AMPK) phosphorylates Tuberous Sclerosis Complex 2 (TSC2) leading to inactivation of mechanistic Target of Rapamycin Complex (mTORC), a negative regulator of autophagy, and autophagy induction to promote cellular and organismal survival21. In contrast to its role in cell survival, the molecular mechanisms underlying autophagic cell death are poorly understood.
The term “autophagic cell death” is controversial; it is important to clarify what can be considered autophagic cell death versus cell death accompanied by autophagy, and what criteria define a process as autophagic cell death3. Whereas many dying cells initiate autophagy, this does not imply that autophagy is driving cell death. Dying cells can initiate autophagy as a pro-survival attempt22, 23. Autophagic vesicles can accumulate in cells due to impaired autophagic flux rather than increased autophagy levels24. This can be detrimental and elicit necrotic or apoptotic cell death25, 26. Thus, it is important to determine if autophagy is altering the dynamics of an alternative cell death mechanism or if cell death is occurring by autophagy.
Autophagic cell death was initially identified based on morphological observations and defined as cell death in the presence of lysosomes27. To be termed autophagic cell death, death should be suppressed by genetic inhibition of at least two autophagy pathway components and autophagy must mediate death independent of other cell death pathways3. However, autophagy often drives other forms of cell death and although autophagy may not be the final mechanism resulting in death, it is often differentially required for cell death to occur, reflecting biological complexity28.
Whether autophagy results in cell survival or demise is highly contextual and it is unclear what determines a given outcome29–33. Confounding this paradox, many of the core autophagy genes have autophagy-independent functions34–37. Not all core autophagy genes are required for autophagy in every context, making it difficult to distinguish between an autophagy-dependent or -independent function38, 39. Below we discuss how autophagy functions during developmental and pathological cell death.
Autophagy and programmed cell death
Autophagy and apoptosis are tightly regulated processes underlying cell and tissue homeostasis, development, and disease40. Programmed Cell Death (PCD) occurs in all metazoans, frequently through caspase-dependent apoptosis40. Compelling and physiologically relevant evidence of autophagy as a cell death mechanism stems from its role in developmental PCD7, 31, 32, 41. Autophagic cell death can occur in combination with caspases or in rare cases alone to drive developmental PCD29.
Significant evidence for a role of autophagy during developmental PCD comes from Drosophila studies. During the transformation from larval to adult stage, pulses of the steroid ecdysone trigger the PCD of the larval salivary gland and midgut42–45. Salivary gland degradation exhibits high levels of autophagy and caspases43, 46–49 (Fig. 1). Mutations in either single autophagy genes or impairment of apoptotic caspase proteases partially block salivary gland degradation and clearance46 43, 49. Combined inhibition of both processes results in a further delay in clearance indicating these pathways act in parallel46. The delay in salivary gland clearance, as opposed to a complete block, resulting from combined inhibition of apoptosis and autophagy raises the possibility that other cell death pathways may be involved. It is also possiblean alternative form of cell death is used in the absence of apoptosis and/or autophagy.
Figure 1. Autophagic cell death in development.
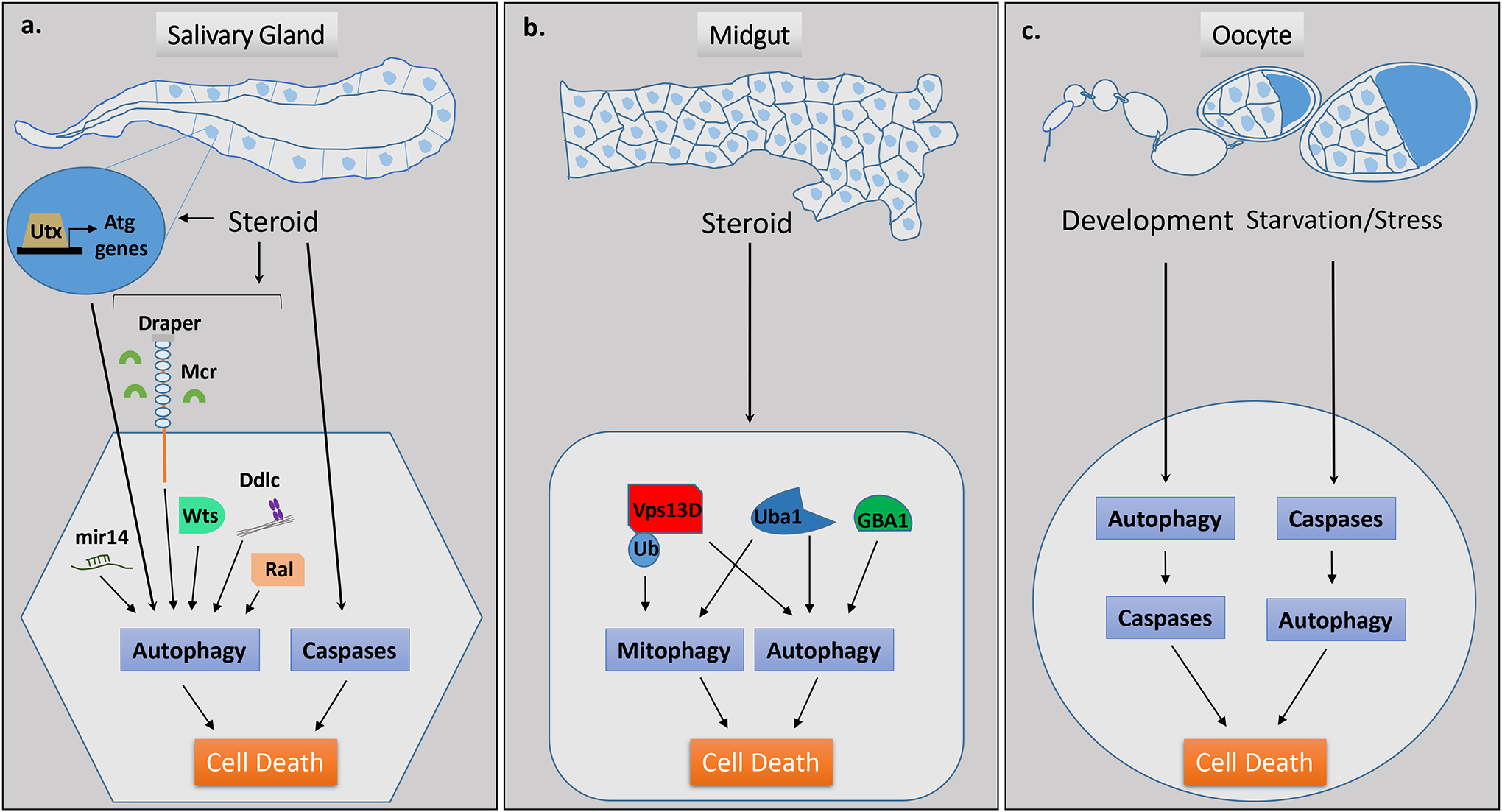
a, Drosophila developmental salivary gland cell death occurs in response to steroid signaling and is dependent upon both autophagy and caspases. Several context-specific regulators of autophagic cell death have been identified from screens performed during developmental salivary gland cell death, including Utx, mir-14, Draper/Mcr, Wts, Ddlc, and Ral. b, Drosophila developmental midgut regression and mitochondrial clearance through mitophagy occurs in response to steroid signaling, is dependent upon autophagy but not caspases, and involves factors including Vps13D, Uba1 and GBA1. c, Autophagy contributes to developmental cell death in the Drosophila oocyte through degradation of an anti-apoptotic factor and subsequent activation of caspases resulting in apoptosis. During starvation and/or stress, autophagic cell death occurs in the oocyte and this is dependent upon caspases.
Drosophila larval midgut degradation also occurs in response to the steroid ecdysone (Fig. 1). Although increased caspase levels occur during midgut degradation, caspase inhibition does not prevent midgut cell size reduction or degradation50. However, autophagy inhibition prevents cell size reduction and degradation, and mitochondrial clearance51. Intriguingly, neither autophagy nor mitophagy in this context requires the conserved E1 enzyme encoded by Atg7, the E2 enzyme encoded by Atg3, or Atg8a lipidation, although these genes are required for salivary gland degradation and starvation-induced autophagy in the fly fatbody51. These findings highlight the cell type- and context-specific differences in the utilization of autophagic machinery.
The Drosophila ovary provides valuable insight into the complex mechanisms underlying cell death (Fig. 1). Autophagy occurs during Drosophila oogenesis to promote the degradation of ovarian support nurse cells and subsequent oocyte maturation52. Autophagic cell death per se does not occur in this context; rather autophagy facilitates oogenesis by degrading the anti-apoptotic factor dBruce to allow for caspase-dependent degradation and nurse cell removal. Autophagy also degrades anti-apoptotic factorsin mammalian cells53. During Drosophila germarium and mid-stage oogenesis, stress or nutrient deprivation can induce autophagic cell death, and in this context autophagic cell death is regulated by the Death caspase-1 (Dcp-1) and the anti-apoptotic factor dBruce54. These studies highlight the complex relationship between autophagy and caspases during Drosophila developmental PCD. Autophagy acts in parallel with caspases during salivary gland cell death, independent of apoptosis during midgut cell death and, depending on the context, autophagy functions upstream or downstream of caspases during oogenesis. In addition to its role in developmental PCD, autophagy also drives adult epithelial cell death in Drosophila during tissue aging55.
Context-specific regulators of autophagy during cell death
Whereas the core autophagy genes are conserved in most situations, studies in Drosophila salivary gland, midgut, and fatbody have uncovered autophagy regulatory genes with tissue and/or context specificity56.
Several screens identified regulators of developmental PCD in Drosophila salivary glands47, 48, 57–60. Proteomic analyses revealed the tumor suppressor Warts is required for cell growth arrest, regulation of caspases, and autophagy during salivary gland cell death61 (Fig. 1). This function is independent of other components of the classic Warts pathway, but dependent upon class I PI3K signaling. An RNAi screen to identify Jumonji HDM family members involved in Drosophila PCD identified the histone demethylase Utx as required for transcriptional regulation of apoptosis and autophagy genes during salivary gland degradation62, 63(Fig. 1). A forward genetic screen uncovered dynein light chain and Ral, a member of the Ras superfamily of small GTPases, as regulators of autophagy in dying salivary glands59, 64(Fig. 1). In contrast to studies in mammalian cell culture, Ral is dispensable for starvation-induced autophagy in the Drosophila fatbody, highlighting the importance of in vivo studies64. Gene expression profiling identified microRNAs as being required for salivary gland degradation60 (Fig. 1). miR-14 regulates autophagy by targeting ip3-kinase 2 which regulates inositol 1,4,5-triphosphate signaling and calcium release from the ER during salivary gland degradation60. Genome wide DNA microarray analyses of dying salivary glands showed enrichment of several engulfment factors displaying no detectable changes in expression upon starvation-induced autophagy48. Subsequently, the phagocytic receptor Draper, the orthologue of human Megf10, was shown to be present on the surface of cells and act in a cell autonomous manner upstream of the autophagy pathway to initiate degradation and clearance of salivary glands65(Fig. 1). The complement ortholog Mcr also plays an essential role in salivary gland cell death and embryonic wound healing by influencing autophagy in neighboring cells66. In both contexts, the activity of Mcr is dependent upon the immune receptor Draper, suggesting a relationship between autophagy and inflammatory mechanisms66.
Intriguingly, several of the aforementioned genes are dispensable for autophagy-dependent midgut regression, which occurs in a caspase- and Atg3/Atg7-independent manner, and for starvation-induced autophagy in the Drosophila fat body, where autophagy promotes cell survival not cell death60, 64, 66, 67.
Autophagy regulators required for PCD during midgut degradation, and not during salivary gland degradation or starvation-induced autophagy, have also been identified. An RNAi screen to identify E1 activating enzymes that function in place of Atg7 during PCD in the midgut uncovered Uba1, the E1 enzyme used in ubiquitylation, as a regulator of cell size reduction, autophagy, and mitophagy in the midgut51(Fig. 1). The ubiquitin-binding domain encoding gene VPS13D is essential for autophagy, mitochondrial clearance and mitochondrial size in the midgut68(Fig. 1). Notably, human cells lacking VPS13D have similar mitochondrial defects to those in the Drosophila midgut and two recent studies, one performed in Drosophila and one in humans, associated mutations in VPS13D with movement disorders69, 70. Another study linking the ubiquitin proteosome system to autophagic degradation showed loss of the receptor protein tyrosine phosphatase, PTP52F, delayed midgut degradation and this was due to dephosphorylation of Ter94, a regulator of the ubiquitin proteosome system71. A Drosophila RNAi screen for genes involved in resveratrol-triggered autophagic cell death of lung carcinoma cells identified Glucocerebrosidase 1 (GBA1), a lysosomal enzyme glucocerebrosidase, as a positive regulator of autophagic cell death during fly midgut regression72(Fig. 1). Homozygous mutations in GBA1 result in the lysosomal storage disorder Gaucher Disease, and mutations in GBA1 pose a significant risk for Parkinsons Disease, both of which show signs of defective autophagy72. These studies underscore the usefulness of Drosophila as a physiologically relevant system to identify novel cell death regulators and aid potential therapeutic developments.
Autophagy in mammalian cell death
Few examples of autophagic cell death during mammalian development exist. In the context of impaired apoptosis, autophagy contributes to developmental regression of mouse embryonic interdigital webs73. However, it is unclear if autophagy is the primary mechanism or if it facilitates an alternative form of cell death. Atg9a acts in an autophagy independent manner to drive necrosis during developmental bone formation in mice74. Although evidence for a role of autophagy in mammalian development is sparse, numerous studies reveal context-specific roles for autophagy and/or autophagy genes during mammalian cell death.
In apoptosis resistant bax−/−;bak−/− Mouse Embryonic Fibroblasts (MEFs), chemotherapeutic agents induce cell death accompanied by numerous autophagic structures and necrosis like features, and cell death can be blocked by inhibiting autophagy75. In apoptosis competent 293T cells, a short mitochondrial isoform of the tumor suppressor p19Arf depolarizes mitochondria inducing caspase-independent cell death accompanied by increased autophagic vesicles76. Atg5 or Beclin-1 knockdown partially rescues cell death suggesting that autophagy acts in parallel with other factors to control cell death76. Apoptosis-independent autophagic cell death also occurs in adult rat hippocampal neural stem cells in response to insulin withdrawal. The extent of cell death correlates with levels of autophagy, and knockdown of Atg7 reduces cell death77. In neonatal rat hippocampal neurons, hypoxic/ischemic brain injury induces autophagy- and caspase-dependent neuronal cell death78. Atg7 deletion inhibits autophagy, partially rescues tissue loss, and partially inhibits both caspase-dependent and caspase-independent cell death78.
In Cnot3 depleted mouse hearts mRNA deadenylation is disrupted and Atg7 mRNA levels increase resulting in autophagic vacuole formation, cardiomyocyte death and lethal heart failure79. Atg7 interacts with and regulates p53 activity to induce expression of cell death promoting genes. Genetic ablation of Atg7 increases survival and cardiac function79. A similar regulatory mechanism occurs during Drosophila oogenesis, in which Orb and the CCR4 deadenylase repress translation of Atg1280. In orb mutant ovaries, increased Atg12 mRNA results in increased cell death, and decreasing mRNA levels of either Atg12 or other autophagy genes inhibits cell death80. These data suggest post-transcriptional control of autophagy genes is an important regulatory mechanism utilized in developing and differentiated tissues, and increased autophagy gene levels are partly responsible for cell death.
A form of autophagy-dependent cell death named “autosis” was recently discovered81. Autosis is induced in vitro by starvation or with the autophagy inducing peptide tat-beclin, and in vivo in neurons of mice exposed to cerebral hypoxic ischemia81 (Fig. 2a). Autosis is independent of apoptosis and necroptosis, dependent upon Na+/K+ATPase, and results in the disappearance of the ER and perinuclear focal swelling. Genetic impairment of core autophagy genes inhibits autotic cell death while blocking of autophagosome/lysosome fusion with bafilomycin A does not, suggesting only the early autophagic machinery is necessary for autosis to occur81. A similar mechanism of cell death requiring Na+/K+-ATPase dependent autophagy has also been shown to preferentially kill HIV-infected macrophages and prevent viral rebound82. Further studies will reveal if autosis is a common or unusual context-specific form of cell death.
Figure 2. Autophagy is at the interface of various forms of cell death.
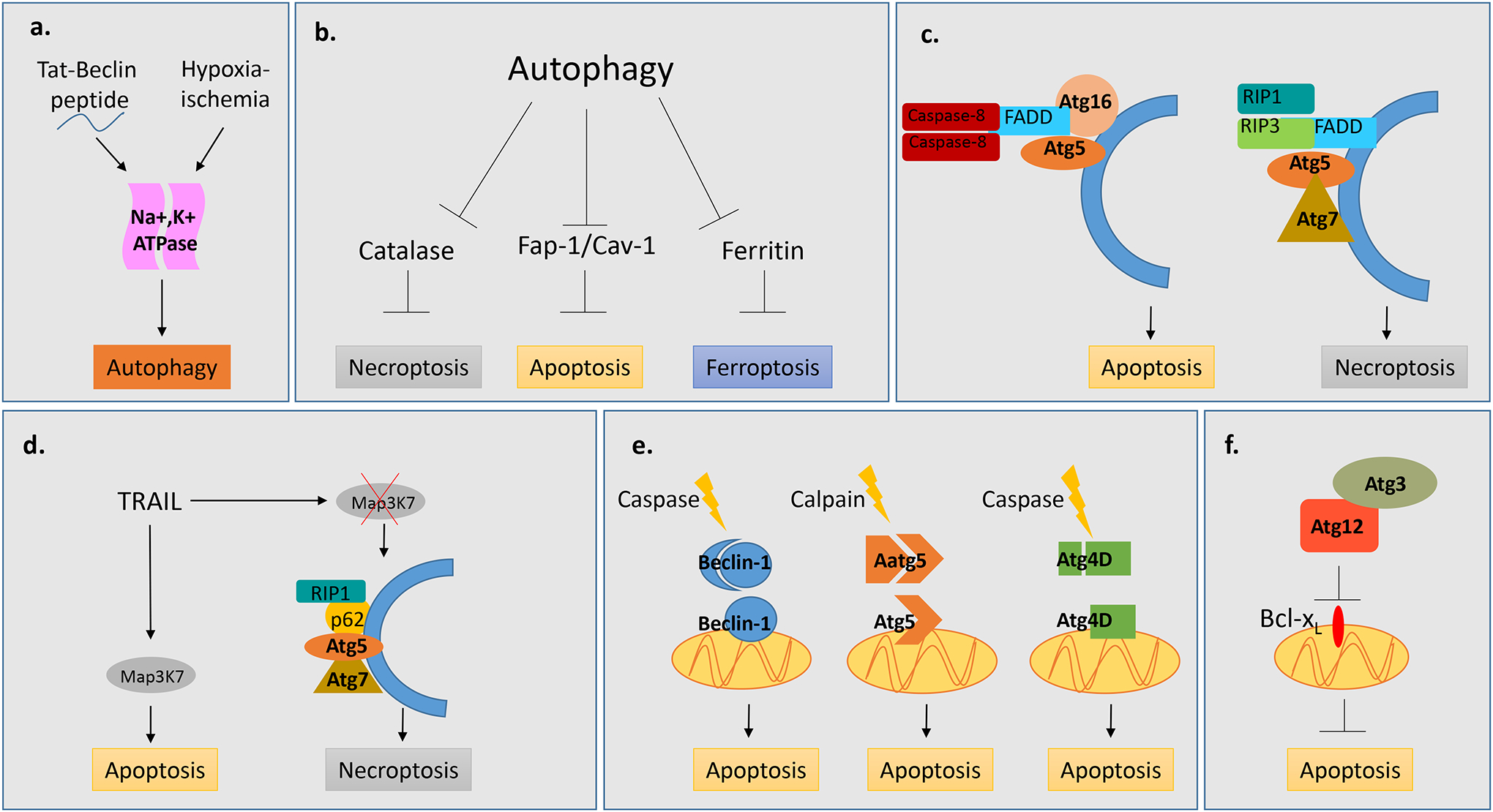
a, Tat-Beclin, hypoxia and ischemia can drive autophagic cell death which is dependent upon the Na+,K+ ATPase. b, Autophagy facilitates cell death by degrading anti-apoptotic (Fap-1/Cav-1), anti-ferroptotic (Ferritin) and anti-necroptotic (Catalase) factors. c, Components of the autophagic machinery serve as a scaffold for apoptotic and necroptotic machinery. Atg5 and Atg16 serve as a scaffold for FADD and Caspase-8 to promote apoptosis. Atg5 and Atg7 serve as a scaffold for necrosome components, RIP1, RIP3 and FADD to promote necroptosis. d, In response to TRAIL, autophagy acts as a switch between the apoptotic and necroptotic machinery by serving as a scaffold for the necrosome component RIP1 and p62 in the absence of Map3K7.. e, Cleavage of autophagy factors, Atg5, Beclin-1 and Atg4D results in products which localize to mitochondria and drive apoptotic cell death. f, Non-canonical conjugation of Atg12 with Atg3 drives intrinsic apoptosis through downstream repression of Bcl-xl.
Autophagic cell death occurs in several cancer cell lines. Human ovarian epithelial cancer cells overexpressing oncogenic H-ras undergo caspase-independent cell death exhibiting features of autophagy83. Cell death results from increased NOXA levels resulting in Beclin-1 dissociation from Mcl-1 and activation of autophagy. Knockdown of core autophagy genes or overexpression of Beclin-1 inhibitory factors attenuate H-Ras-induced cell death83. Expression of oncogenic H-ras or K-ras in three different normal fibroblast cell lines induced caspase-independent cell death accompanied by accumulation of autophagosomes and upregulation of Atg5, all of which are attenuated by inhibition of autophagy84, 85.
In transformed HEK293, U87 and HeLa cells, hydrogen peroxide or 2-Mercaptoethanol treatment induced ROS, autophagy and caspases86. Inhibition of ROS or autophagy by chemical and genetic means blocked cell death, but inhibition of caspases did not. Furthermore, blocking autophagy did not inhibit ROS formation, placing ROS upstream of autophagy induction86. Phycocyanin treatment inhibits tumorigenic potential in pancreatic cancer cells by increasing both autophagic and apoptotic cell death87. Increased autophagy levels are dependent upon Akt/mTOR/p70S6K pathway inhibition and NF-κB pathway stimulation. Beclin-1 knockdown partially prevented cell death and inhibited LC3 puncta formation whereas inhibition of both autophagy and apoptosis further inhibited cell death87.
Depending on the state of the cells, the same stimuli can elicit different death mechanisms. In apoptotic competent breast cancer, glioma, and embryonic cell lines under hypoxic stress, etoposide induces autophagic rather than apoptotic cell death88. Genetic and chemical inhibition of autophagy prevented cell death whereas inhibition of apoptosis did not. By contrast, under normoxic conditions, etoposide induced apoptosis88.
Interplay between autophagy, apoptosis and necrosis
Recent studies have revealed a complex and multifaceted relationship between autophagy, apoptosis and necrosis. Above we discussed examples demonstrating a role for autophagy in cell death. In these contexts, the presence of autophagy markers, combined with loss of autophagy accompanied by an increase in cell survival has been characterized as autophagic cell death. However, recent studies reveal that autophagy may facilitate other forms of cell death rather than acting as the primary form of death, and this may be the case with some of the previously mentioned examples. For instance, autophagy initiates several other forms of cell death by selective degradation of molecules involved in those processes (Fig. 2b). Inhibition of apoptosis in L929 cells triggers autophagic degradation of Catalase resulting in increased ROS and cell death89, 90. Similarly, in erastin treated MEFs, autophagic degradation of ferritin results in increased levels of labile iron and ROS leading to ferroptosis, a PCD characterized by iron accumulation and lipid peroxidation91.
Autophagy also promotes apoptosis through the degradation of anti-apoptotic and cell survival factors. In rat hippocampal astrocytes, autophagic degradation of Caveolin-1 contributes to palmitic acid induced inflammation and apoptosis92. Autophagic degradation of Fap-1, an inhibitor of Fas-mediated apoptosis, sensitizes Type I cells to Fas induced apoptosis53. This mechanism depends upon levels of autophagy occurring in the cell and, in a population of proliferating cells, varying levels of autophagy within individual cells results in different cell fates in response to Fas53. These studies highlight the importance of uncovering not only the means of cell death but also the autophagic substrate. Notably, autophagic degradation of certain substrates also promotes cell survival93, 94.
In addition to degradative functions, autophagy proteins promote cell death by providing a scaffold for cell death complexes and signaling molecules (Fig. 2c). In MEFs treated with sphingosine kinase inhibitor (SK-I), which promotes cell death through the suppression of sphingosine 1-phosphate, caspase-dependent cell death occurs only in the presence of functional autophagy95. SK-I induces the translocation of caspase-8 homocomplex and Fas-Associated protein with Death Domain (FADD) to Atg5 and Atg16L-positive autophagosomal membranes which provide a scaffold for the efficient formation of an intracellular Death-Inducing Signaling Complex (iDISC)95. Autophagosomes also provide a scaffold for the necroptosis machinery. In rhabdomyosarcoma cells, Obatoclax, an antagonist of Bcl-2 family proteins, activates autophagy and necroptotic cell death96. This necroptosis is dependent upon the recruitment of the necrosome, a complex comprised of FADD, RIPK1 and RIPK3, to the autophagosomal membrane96.
Autophagy also determines the means of cell death by serving as a switch between apoptosis and necroptosis97 (Fig. 2d). TNF-Related Apoptosis-Induced Ligand (TRAIL)-induced cell death generally occurs through apoptosis, and loss of autophagy sensitizes cells to TRAIL through upregulation of PUMA93. However, in mouse prostate cells mutant for the tumor suppressor Map3k7, cell death occurs via necroptosis rather than apoptosis and this switch in cell death mechanism is facilitated by autophagy98. Interestingly, in Map3k7−/− cells knockdown of genes involved in the early steps of autophagy, Beclin1, Atg5 and Atg7, prevents necroptotic cell death and results in cell survival. However, inhibition of later autophagic events, such as autophagosome fusion with the lysosome, results in increased TRAIL-induced cell death indicating a protective role for autophagic flux98. In this situation, autophagosomes were found to serve as a scaffold for the necroptosis machinery-mediated by p62 recruitment of RIPK1 to the autophagosome. When necrosome formation is inhibited, cell death can occur through apoptosis. Thus, the cells are apoptosis competent but in the absence of Map3k7 utilize an alternative cell death mechanism98. This study reveals that autophagy can act in a pro-survival and pro-death manner within the same cell. The absence of autophagic flux enhances cell death indicating a protective role for autophagy. However, autophagy also facilitates cell death by providing a scaffold for the cell death machinery. These data reveal the autophagosome may serve broader functions than just removal of cellular trash.
Individual autophagy genes also play roles in cell death independent of the intrinsic functions of autophagy in autophagosome formation and degradation (Fig. 2e). Protease cleavage of core autophagy genes represents a complex regulatory interplay between autophagy and apoptosis. Calpain-mediated cleavage of Atg5 results in a pro-apoptotic protein that associates with anti-apoptotic Bcl-xl to stimulate cyctochrome c release, caspase activation and intrinsic apoptosis99. Similarly, Beclin 1 contains two caspase cleavage sites and upon cleavage the C-terminal fragment translocates to the mitochondria and induces apoptosis through the release of pro-apoptotic factors100, 101. Caspase cleavage of Atg4 results in a cytotoxic protein that is transiently recruited to mitochondria102. However, in contrast to cleavage of Beclin1 and Atg5 which abrogates their autophagic function, Atg4 cleavage results in increased autophagy which occurs independently of its cytotoxicity.
Autophagy-independent conjugation systems between components of the core autophagy machinery also promote cell death. Atg12 and Atg3 are essential components of the autophagy ubiquitin-like conjugation machinery required for autophagosome formation18. However, conjugation of Atg12 to Atg3 does not occur during starvation-induced autophagy and is not necessary for lipidation of Atg8103. Conjugation between Atg3 and Atg12 functions during mitochondrial mediated apoptosis103 (Fig. 2f). Cells expressing a mutant form of Atg3 that is unable to conjugate to Atg12 exhibit increased mitochondrial mass and fragmentation, increased levels of anti-apoptotic Bcl2 family member proteins and increased resistance to death mediated by mitochondrial pathways. Interestingly, conjugation of Atg12 to Atg3 is not required for death receptor mediated apoptosis103.
Autophagy function in engulfment and inflammation
Efficient removal of dead, damaged or old cells is critical to tissue homeostasis and immune regulation in mammals, and persistence of dead or damaged cells can drive auto-inflammatory responses and autoimmune disease104, 105. Among the many roles autophagy genes play in cell death they also facilitate cell corpse engulfment and degradation, immunogenic cell death and LC3 associated phagocytosis (LAP)105.
Autophagy can function in the dying cell as well as the phagocytic cell to ensure proper cell corpse engulfment and degradation (Fig. 3a). In chick embryo retinas and mammalian embryoid bodies, autophagy contributes to apoptotic corpse engulfment by exposing energy-dependent engulfment signals on the surface of dying cells106, 107. Similarly, autophagy facilitates the immunogenic release of ATP from tumor cells allowing for the recruitment of dendritic cells and T lymphocytes, and subsequent engulfment and immunogenic cell death of tumor cells108, 109. Autophagy genes also act within macrophages and other engulfing cells to drive cell death in a cell non-autonomous manner. Beclin-1 is required in macrophages and fibroblasts to facilitate actin dynamics and membrane phospholipid synthesis during the engulfment of apoptotic cells, and Beclin-1 knockdown results in decreased apoptotic cell internalization110. C. elegans studies demonstrate that depending on the context, autophagy genes are required in engulfing cells for engulfment and/or degradation of cell corpses110–113. Autophagy also acts within adult Drosophila enterocytes to drive the engulfment of dying stem cells114. Interestingly, the phagocytic receptor Draper, required for autophagy during salivary gland clearance, also acts upstream of autophagy in engulfing enterocytes.114.
Figure 3. Autophagy, cell recognition and cell engulfment.
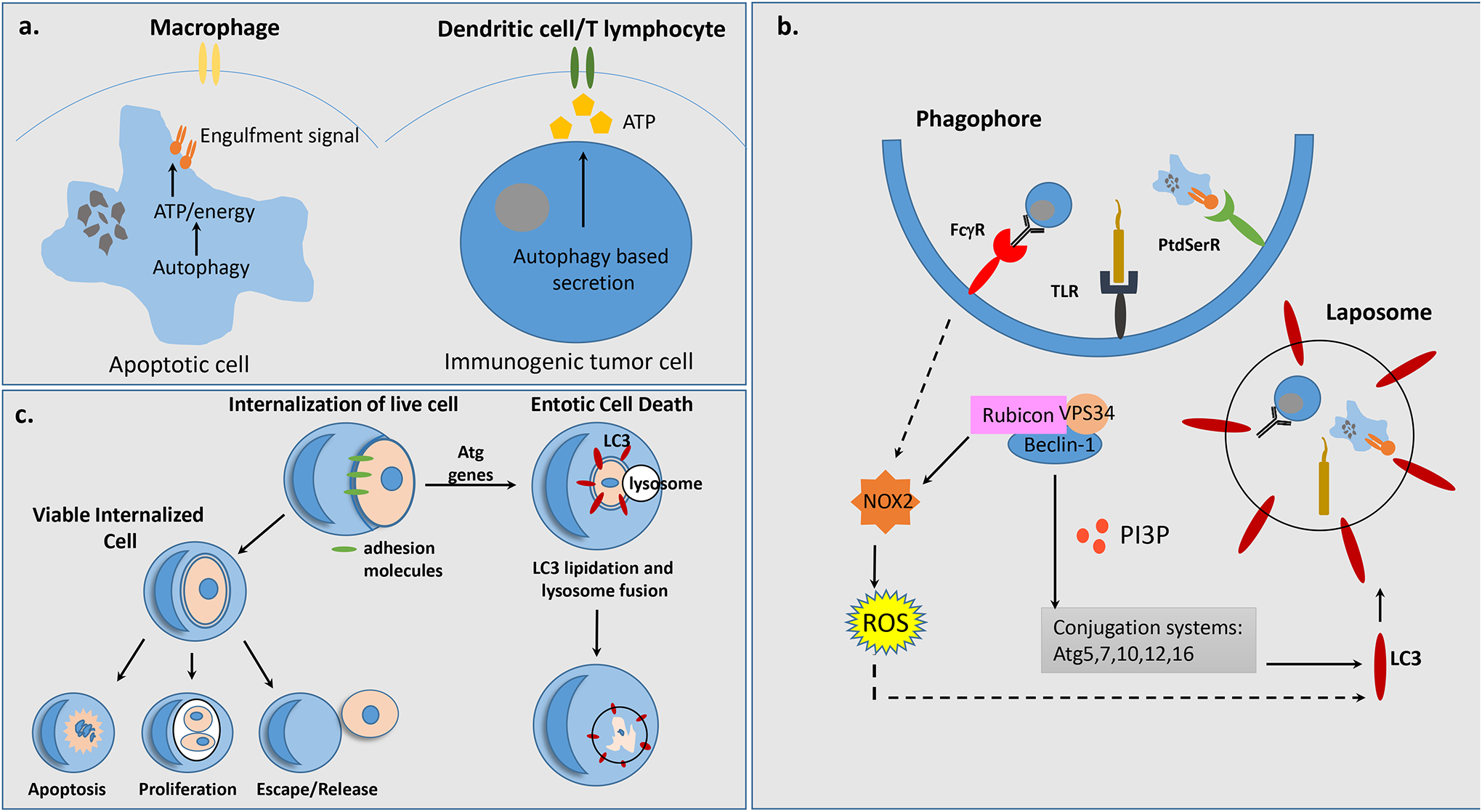
a, Autophagy regulates apoptotic cell clearance and immunogenic cell death by enabling the exposure of energy-dependent engulfment signals and release of ATP from dying cells and live tumor cells respectively, which are recognized and engulfed either by macrophages or dendritic cells/T lymphocytes. b, Autophagy facilitates cell death by acting within engulfing cells to promote LC3 Associated Phagocytosis (LAP) in response to FcγR, TLR or PtdSerR signalingLocalization of PI(3)P, results in sustained NOX2 and increased ROS as well as recruitment of downstream autophagy factors, Atg5, Atg7, Atg10, Atg12 and Atg16, allowing for LC3 lipidation of a single membrane vesicle, termed the laposome. c, Entosis is a sub-type of LAP which utilizes adhesion machinery to internalize live neighboring cells. The internalized cells often undergo entotic cell death through LAP involving LC3 lipidation and autophagosome-lysosome fusion. However, it is possible for internalized cells to undergo apoptotic cell death, proliferate within the host cell, or escape the host cell.
LC3 associated phagocytosis (LAP) is a form of engulfment dependent upon non-canonical autophagy (Fig. 3b), initially identified by autophagy factor recruitment to the phagosome during Toll Like Receptor signaling in murine macrophages115 116. LAP is distinct from autophagy because it involves a single membrane vesicle, not a double membrane autophagosome, occurs in nutrient rich environments in the presence of activated mTORC1 kinase, and does not require the early autophagy factors Ulk1/2, Fip200, Atg13 and Atg101115, 117 116, 118. Instead, recruitment of Rubicon to the phagosome leads to activation of the Class III PI3K complex comprised of Beclin-1, UVRAG and VPS34119. This complex facilitates sustained localization of PI(3)P necessary for the recruitment of downstream autophagy conjugation factors and stabilization of NOX2 required for ROS production119. This allows for LC3 lipidation and recruitment to phagosomes enabling lysosome fusion and efficient corpse degradation105, 118. LAP occurs in a variety of different contexts including phagocytosis of dead or dying cells, Fc-gamma receptor mediated phagocytosis of live tumor cells, removal of invading pathogens, antigen presentation, modulation of cytokine release, and entosis34, 117, 120–125. In conjunction with these various roles, LAP is required to regulate the appropriate immune response and aberrant inflammation and pathogenicity occur in the absence of LAP105.
In addition to removal of apoptotic cells, LAP also aids in the engulfment of non-apoptotic, live cells through a process termed entosis126 (Fig. 3c). During entosis, cells utilize adhesion machinery to ingest and kill neighboring cells to scavenge for nutrients and support their own proliferation127, 128. The ingested cells actively partake in their own ingestion and are typically killed by the host cell in a LAP-dependent manner126. However, the ingested cell may also be released or it can result in death of the engulfing cell. Entosis actsas a tumor suppressive mechanism by inducing cell death and inhibiting transformation, but may also promote tumor activity by disrupting cell division of live engulfed cells resulting in ploidy changes. Similar to autophagy, entosis is regulated by nutrient sensing pathways and is induced by glucose withdrawal through AMPK, a positive regulator of autophagy129. TM9SF4, a nine transmembrane segment protein required for phagocytosis in amoeba and Drosophila macrophages and engulfment activity of malignant human cancer cells, regulates autophagy under nutrient deprivation130–134. TM9SF4 localizes to lysosomes and initiates autophagy through mTORC1 inhibition130. These data raise the possibility that autophagy, entosis and other forms of cannabalism are co-regulated. Determining the relationship between autophagy, phagocytic processes, and immune responses could inform therapeutic treatments for cancers and autoimmune diseases.
Concluding remarks
The modulation of autophagy not only influences the health of a cell but also how a cell dies. However, outstanding questions remain, including whether autophagic cell survival and autophagic cell death utilize the same molecular machinery, and what is being degraded in each instance. It is unclear what external stimuli or cues promote autophagic cell death, and whether there is a threshold level of autophagy which, once passed, becomes detrimental to the cell. It will be important to elucidate how autophagy influences other forms of cell death and vice versa. Future studies directed towards answering these critical questions could offer therapeutic potential for the treatment of neurodegenerative diseases, cancer, and inflammatory disorders. Considering the highly contextual nature of autophagy and the autophagic machinery, it is imperative these studies be performed in physiologically relevant contexts.
References
- 1.Green DR & Llambi F Cell Death Signaling. Cold Spring Harb Perspect Biol 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kerr JF, Wyllie AH & Currie AR Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer 26, 239–257 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galluzzi L et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ 25, 486–541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi Y Mechanisms of caspase activation and inhibition during apoptosis. Mol Cell 9, 459–470 (2002). [DOI] [PubMed] [Google Scholar]
- 5.De Duve C & Wattiaux R Functions of lysosomes. Annu Rev Physiol 28, 435–492 (1966). [DOI] [PubMed] [Google Scholar]
- 6.Proskuryakov SY, Konoplyannikov AG & Gabai VL Necrosis: a specific form of programmed cell death? Exp Cell Res 283, 1–16 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Denton D, Nicolson S & Kumar S Cell death by autophagy: facts and apparent artefacts. Cell Death Differ 19, 87–95 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizushima N & Komatsu M Autophagy: renovation of cells and tissues. Cell 147, 728–741 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Thumm M et al. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett 349, 275–280 (1994). [DOI] [PubMed] [Google Scholar]
- 10.Tsukada M & Ohsumi Y Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 333, 169–174 (1993). [DOI] [PubMed] [Google Scholar]
- 11.Russell RC et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 15, 741–750 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizushima N et al. A protein conjugation system essential for autophagy. Nature 395, 395–398 (1998). [DOI] [PubMed] [Google Scholar]
- 13.Mizushima N, Noda T & Ohsumi Y Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J 18, 3888–3896 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirisako T et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol 147, 435–446 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang WP, Scott SV, Kim J & Klionsky DJ The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J Biol Chem 275, 5845–5851 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Lang T et al. Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO J 17, 3597–3607 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ichimura Y et al. A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Ohsumi Y Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2, 211–216 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Glick D, Barth S & Macleod KF Autophagy: cellular and molecular mechanisms. J Pathol 221, 3–12 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu L, Chen Y & Tooze SA Autophagy pathway: Cellular and molecular mechanisms. Autophagy 14, 207–215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inoki K, Zhu T & Guan KL TSC2 mediates cellular energy response to control cell growth and survival. Cell 115, 577–590 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Kroemer G & Levine B Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol 9, 1004–1010 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boya P et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol 25, 1025–1040 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka Y et al. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature 406, 902–906 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Zhu H et al. The fusion of autophagosome with lysosome is impaired in L-arginine-induced acute pancreatitis. Int J Clin Exp Pathol 8, 11164–11170 (2015). [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzalez-Polo RA et al. The apoptosis/autophagy paradox: autophagic vacuolization before apoptotic death. J Cell Sci 118, 3091–3102 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Schweichel JU & Merker HJ The morphology of various types of cell death in prenatal tissues. Teratology 7, 253–266 (1973). [DOI] [PubMed] [Google Scholar]
- 28.Fulda S & Kogel D Cell death by autophagy: emerging molecular mechanisms and implications for cancer therapy. Oncogene 34, 5105–5113 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Xu T, Nicolson S, Denton D & Kumar S Distinct requirements of Autophagy-related genes in programmed cell death. Cell Death Differ 22, 1792–1802 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das G, Shravage BV & Baehrecke EH Regulation and function of autophagy during cell survival and cell death. Cold Spring Harb Perspect Biol 4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neufeld TP & Baehrecke EH Eating on the fly: function and regulation of autophagy during cell growth, survival and death in Drosophila. Autophagy 4, 557–562 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H & Baehrecke EH Eaten alive: novel insights into autophagy from multicellular model systems. Trends Cell Biol 25, 376–387 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anding AL & Baehrecke EH Autophagy in Cell Life and Cell Death. Curr Top Dev Biol 114, 67–91 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Zhao Z et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe 4, 458–469 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller BC et al. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy 4, 309–314 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Subramani S & Malhotra V Non-autophagic roles of autophagy-related proteins. EMBO Rep 14, 143–151 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cadwell K & Debnath J Beyond self-eating: The control of nonautophagic functions and signaling pathways by autophagy-related proteins. J Cell Biol 217, 813–822 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Codogno P, Mehrpour M & Proikas-Cezanne T Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat Rev Mol Cell Biol 13, 7–12 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Scarlatti F, Maffei R, Beau I, Ghidoni R & Codogno P Non-canonical autophagy: an exception or an underestimated form of autophagy? Autophagy 4, 1083–1085 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Fuchs Y & Steller H Programmed cell death in animal development and disease. Cell 147, 742–758 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nezis IP, Vaccaro MI, Devenish RJ & Juhasz G Autophagy in development, cell differentiation, and homeodynamics: from molecular mechanisms to diseases and pathophysiology. Biomed Res Int 2014, 349623 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang C, Baehrecke EH & Thummel CS Steroid regulated programmed cell death during Drosophila metamorphosis. Development 124, 4673–4683 (1997). [DOI] [PubMed] [Google Scholar]
- 43.Martin DN & Baehrecke EH Caspases function in autophagic programmed cell death in Drosophila. Development 131, 275–284 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Lee CY, Simon CR, Woodard CT & Baehrecke EH Genetic mechanism for the stage- and tissue-specific regulation of steroid triggered programmed cell death in Drosophila. Dev Biol 252, 138–148 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Lee CY, Cooksey BA & Baehrecke EH Steroid regulation of midgut cell death during Drosophila development. Dev Biol 250, 101–111 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Berry DL & Baehrecke EH Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell 131, 1137–1148 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gorski SM et al. A SAGE approach to discovery of genes involved in autophagic cell death. Curr Biol 13, 358–363 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Lee CY et al. Genome-wide analyses of steroid- and radiation-triggered programmed cell death in Drosophila. Curr Biol 13, 350–357 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Lee CY & Baehrecke EH Steroid regulation of autophagic programmed cell death during development. Development 128, 1443–1455 (2001). [DOI] [PubMed] [Google Scholar]
- 50.Denton D et al. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr Biol 19, 1741–1746 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang TK et al. Uba1 functions in Atg7- and Atg3-independent autophagy. Nat Cell Biol 15, 1067–1078 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nezis IP et al. Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis. J Cell Biol 190, 523–531 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gump JM et al. Autophagy variation within a cell population determines cell fate through selective degradation of Fap-1. Nat Cell Biol 16, 47–54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hou YC, Chittaranjan S, Barbosa SG, McCall K & Gorski SM Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J Cell Biol 182, 1127–1139 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scherfer C, Han VC, Wang Y, Anderson AE & Galko MJ Autophagy drives epidermal deterioration in a Drosophila model of tissue aging. Aging (Albany NY) 5, 276–287 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McPhee CK & Baehrecke EH Autophagy in Drosophila melanogaster. Biochim Biophys Acta 1793, 1452–1460 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin DN et al. Proteomic analysis of steroid-triggered autophagic programmed cell death during Drosophila development. Cell Death Differ 14, 916–923 (2007). [DOI] [PubMed] [Google Scholar]
- 58.McPhee CK et al. Identification of factors that function in Drosophila salivary gland cell death during development using proteomics. Cell Death Differ 20, 218–225 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Batlevi Y et al. Dynein light chain 1 is required for autophagy, protein clearance, and cell death in Drosophila. Proc Natl Acad Sci U S A 107, 742–747 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nelson C, Ambros V & Baehrecke EH miR-14 regulates autophagy during developmental cell death by targeting ip3-kinase 2. Mol Cell 56, 376–388 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dutta S & Baehrecke EH Warts is required for PI3K-regulated growth arrest, autophagy, and autophagic cell death in Drosophila. Curr Biol 18, 1466–1475 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Denton D et al. UTX coordinates steroid hormone-mediated autophagy and cell death. Nat Commun 4, 2916 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ress C, Holtmann M, Maas U, Sofsky J & Dorn A 20-Hydroxyecdysone-induced differentiation and apoptosis in the Drosophila cell line, l(2)mbn. Tissue Cell 32, 464–477 (2000). [DOI] [PubMed] [Google Scholar]
- 64.Tracy K, Velentzas PD & Baehrecke EH Ral GTPase and the exocyst regulate autophagy in a tissue-specific manner. EMBO Rep 17, 110–121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McPhee CK, Logan MA, Freeman MR & Baehrecke EH Activation of autophagy during cell death requires the engulfment receptor Draper. Nature 465, 1093–1096 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin L et al. Complement-Related Regulates Autophagy in Neighboring Cells. Cell 170, 158–171 e158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McPhee CK & Baehrecke EH The engulfment receptor Draper is required for autophagy during cell death. Autophagy 6, 1192–1193 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anding AL et al. Vps13D Encodes a Ubiquitin-Binding Protein that Is Required for the Regulation of Mitochondrial Size and Clearance. Curr Biol 28, 287–295 e286 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seong E et al. Mutations in VPS13D lead to a new recessive ataxia with spasticity and mitochondrial defects. Ann Neurol (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gauthier J et al. Recessive mutations in >VPS13D cause childhood onset movement disorders. Ann Neurol (2018). [DOI] [PubMed] [Google Scholar]
- 71.Santhanam A et al. Ecdysone-induced receptor tyrosine phosphatase PTP52F regulates Drosophila midgut histolysis by enhancement of autophagy and apoptosis. Mol Cell Biol 34, 1594–1606 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dasari SK et al. Death by over-eating: The Gaucher disease associated gene GBA1, identified in a screen for mediators of autophagic cell death, is necessary for developmental cell death in Drosophila midgut. Cell Cycle 16, 2003–2010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arakawa S et al. Role of Atg5-dependent cell death in the embryonic development of Bax/Bak double-knockout mice. Cell Death Differ 24, 1598–1608 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Imagawa Y, Saitoh T & Tsujimoto Y Vital staining for cell death identifies Atg9a-dependent necrosis in developmental bone formation in mouse. Nat Commun 7, 13391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shimizu S et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol 6, 1221–1228 (2004). [DOI] [PubMed] [Google Scholar]
- 76.Reef S et al. A short mitochondrial form of p19ARF induces autophagy and caspase-independent cell death. Mol Cell 22, 463–475 (2006). [DOI] [PubMed] [Google Scholar]
- 77.Yu SW et al. Autophagic death of adult hippocampal neural stem cells following insulin withdrawal. Stem Cells 26, 2602–2610 (2008). [DOI] [PubMed] [Google Scholar]
- 78.Xie C et al. Neuroprotection by selective neuronal deletion of Atg7 in neonatal brain injury. Autophagy 12, 410–423 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yamaguchi T et al. The CCR4-NOT deadenylase complex controls Atg7-dependent cell death and heart function. Sci Signal 11 (2018). [DOI] [PubMed] [Google Scholar]
- 80.Rojas-Rios P et al. Translational Control of Autophagy by Orb in the Drosophila Germline. Dev Cell 35, 622–631 (2015). [DOI] [PubMed] [Google Scholar]
- 81.Liu Y et al. Autosis is a Na+,K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc Natl Acad Sci U S A 110, 20364–20371 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang G, Luk BT, Hamidy M, Zhang L & Spector SA Induction of a Na(+)/K(+)-ATPase-Dependent Form of Autophagy Triggers Preferential Cell Death of Human Immunodeficiency Virus Type-1-Infected Macrophages. Autophagy (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Elgendy M, Sheridan C, Brumatti G & Martin SJ Oncogenic Ras-induced expression of Noxa and Beclin-1 promotes autophagic cell death and limits clonogenic survival. Mol Cell 42, 23–35 (2011). [DOI] [PubMed] [Google Scholar]
- 84.Byun JY et al. The Rac1/MKK7/JNK pathway signals upregulation of Atg5 and subsequent autophagic cell death in response to oncogenic Ras. Carcinogenesis 30, 1880–1888 (2009). [DOI] [PubMed] [Google Scholar]
- 85.Byun JY et al. Oncogenic Ras signals through activation of both phosphoinositide 3-kinase and Rac1 to induce c-Jun NH2-terminal kinase-mediated, caspase-independent cell death. Mol Cancer Res 7, 1534–1542 (2009). [DOI] [PubMed] [Google Scholar]
- 86.Chen Y, McMillan-Ward E, Kong J, Israels SJ & Gibson SB Oxidative stress induces autophagic cell death independent of apoptosis in transformed and cancer cells. Cell Death Differ 15, 171–182 (2008). [DOI] [PubMed] [Google Scholar]
- 87.Liao G et al. Phycocyanin Inhibits Tumorigenic Potential of Pancreatic Cancer Cells: Role of Apoptosis and Autophagy. Sci Rep 6, 34564 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Azad MB et al. Hypoxia induces autophagic cell death in apoptosis-competent cells through a mechanism involving BNIP3. Autophagy 4, 195–204 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yu L et al. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science 304, 1500–1502 (2004). [DOI] [PubMed] [Google Scholar]
- 90.Yu L et al. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci U S A 103, 4952–4957 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hou W et al. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy 12, 1425–1428 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen Z et al. The autophagic degradation of Cav-1 contributes to PA-induced apoptosis and inflammation of astrocytes. Cell Death Dis 9, 771 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thorburn J et al. Autophagy controls the kinetics and extent of mitochondrial apoptosis by regulating PUMA levels. Cell Rep 7, 45–52 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jiang K et al. Autophagic degradation of FOXO3a represses the expression of PUMA to block cell apoptosis in cisplatin-resistant osteosarcoma cells. Am J Cancer Res 7, 1407–1422 (2017). [PMC free article] [PubMed] [Google Scholar]
- 95.Young MM et al. Autophagosomal membrane serves as platform for intracellular death-inducing signaling complex (iDISC)-mediated caspase-8 activation and apoptosis. J Biol Chem 287, 12455–12468 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Basit F, Cristofanon S & Fulda S Obatoclax (GX15–070) triggers necroptosis by promoting the assembly of the necrosome on autophagosomal membranes. Cell Death Differ 20, 1161–1173 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sakamaki JI & Ryan KM Autophagy Determines the Path on the TRAIL to Death. Dev Cell 37, 291–293 (2016). [DOI] [PubMed] [Google Scholar]
- 98.Goodall ML et al. The Autophagy Machinery Controls Cell Death Switching between Apoptosis and Necroptosis. Dev Cell 37, 337–349 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yousefi S et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol 8, 1124–1132 (2006). [DOI] [PubMed] [Google Scholar]
- 100.Zhu Y et al. Beclin 1 cleavage by caspase-3 inactivates autophagy and promotes apoptosis. Protein Cell 1, 468–477 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wirawan E et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis 1, e18 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Betin VM & Lane JD Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J Cell Sci 122, 2554–2566 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Radoshevich L et al. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell 142, 590–600 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Szondy Z, Sarang Z, Kiss B, Garabuczi E & Koroskenyi K Anti-inflammatory Mechanisms Triggered by Apoptotic Cells during Their Clearance. Front Immunol 8, 909 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Green DR, Oguin TH & Martinez J The clearance of dying cells: table for two. Cell Death Differ 23, 915–926 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Qu X et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell 128, 931–946 (2007). [DOI] [PubMed] [Google Scholar]
- 107.Mellen MA, de la Rosa EJ & Boya P The autophagic machinery is necessary for removal of cell corpses from the developing retinal neuroepithelium. Cell Death Differ 15, 1279–1290 (2008). [DOI] [PubMed] [Google Scholar]
- 108.Michaud M et al. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science 334, 1573–1577 (2011). [DOI] [PubMed] [Google Scholar]
- 109.Ko A et al. Autophagy inhibition radiosensitizes in vitro, yet reduces radioresponses in vivo due to deficient immunogenic signalling. Cell Death Differ 21, 92–99 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Konishi A, Arakawa S, Yue Z & Shimizu S Involvement of Beclin 1 in engulfment of apoptotic cells. J Biol Chem 287, 13919–13929 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huang S, Jia K, Wang Y, Zhou Z & Levine B Autophagy genes function in apoptotic cell corpse clearance during C. elegans embryonic development. Autophagy 9, 138–149 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ruck A et al. The Atg6/Vps30/Beclin 1 ortholog BEC-1 mediates endocytic retrograde transport in addition to autophagy in C. elegans. Autophagy 7, 386–400 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li W et al. Autophagy genes function sequentially to promote apoptotic cell corpse degradation in the engulfing cell. J Cell Biol 197, 27–35 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Singh SR et al. The lipolysis pathway sustains normal and transformed stem cells in adult Drosophila. Nature 538, 109–113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sanjuan MA, Milasta S & Green DR Toll-like receptor signaling in the lysosomal pathways. Immunol Rev 227, 203–220 (2009). [DOI] [PubMed] [Google Scholar]
- 116.Sanjuan MA et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450, 1253–1257 (2007). [DOI] [PubMed] [Google Scholar]
- 117.Martinez J et al. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci U S A 108, 17396–17401 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim SE & Overholtzer M Autophagy proteins regulate cell engulfment mechanisms that participate in cancer. Semin Cancer Biol 23, 329–336 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Martinez J et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat Cell Biol 17, 893–906 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 120.Huang J et al. Activation of antibacterial autophagy by NADPH oxidases. Proc Natl Acad Sci U S A 106, 6226–6231 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ma J, Becker C, Lowell CA & Underhill DM Dectin-1-triggered recruitment of light chain 3 protein to phagosomes facilitates major histocompatibility complex class II presentation of fungal-derived antigens. J Biol Chem 287, 34149–34156 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Romao S et al. Autophagy proteins stabilize pathogen-containing phagosomes for prolonged MHC II antigen processing. J Cell Biol 203, 757–766 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Florey O, Kim SE, Sandoval CP, Haynes CM & Overholtzer M Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat Cell Biol 13, 1335–1343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gutierrez MG et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119, 753–766 (2004). [DOI] [PubMed] [Google Scholar]
- 125.Nakagawa I et al. Autophagy defends cells against invading group A Streptococcus. Science 306, 1037–1040 (2004). [DOI] [PubMed] [Google Scholar]
- 126.Overholtzer M et al. A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell 131, 966–979 (2007). [DOI] [PubMed] [Google Scholar]
- 127.Sun Q, Cibas ES, Huang H, Hodgson L & Overholtzer M Induction of entosis by epithelial cadherin expression. Cell Res 24, 1288–1298 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang M et al. Impaired formation of homotypic cell-in-cell structures in human tumor cells lacking alpha-catenin expression. Sci Rep 5, 12223 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hamann JC et al. Entosis Is Induced by Glucose Starvation. Cell Rep 20, 201–210 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sun L et al. TM9SF4 is a novel factor promoting autophagic flux under amino acid starvation. Cell Death Differ 25, 368–379 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bergeret E et al. TM9SF4 is required for Drosophila cellular immunity via cell adhesion and phagocytosis. J Cell Sci 121, 3325–3334 (2008). [DOI] [PubMed] [Google Scholar]
- 132.Cornillon S et al. Phg1p is a nine-transmembrane protein superfamily member involved in dictyostelium adhesion and phagocytosis. J Biol Chem 275, 34287–34292 (2000). [DOI] [PubMed] [Google Scholar]
- 133.Lozupone F et al. TM9SF4 is a novel V-ATPase-interacting protein that modulates tumor pH alterations associated with drug resistance and invasiveness of colon cancer cells. Oncogene 34, 5163–5174 (2015). [DOI] [PubMed] [Google Scholar]
- 134.Lozupone F et al. The human homologue of Dictyostelium discoideum phg1A is expressed by human metastatic melanoma cells. EMBO Rep 10, 1348–1354 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]


