Summary
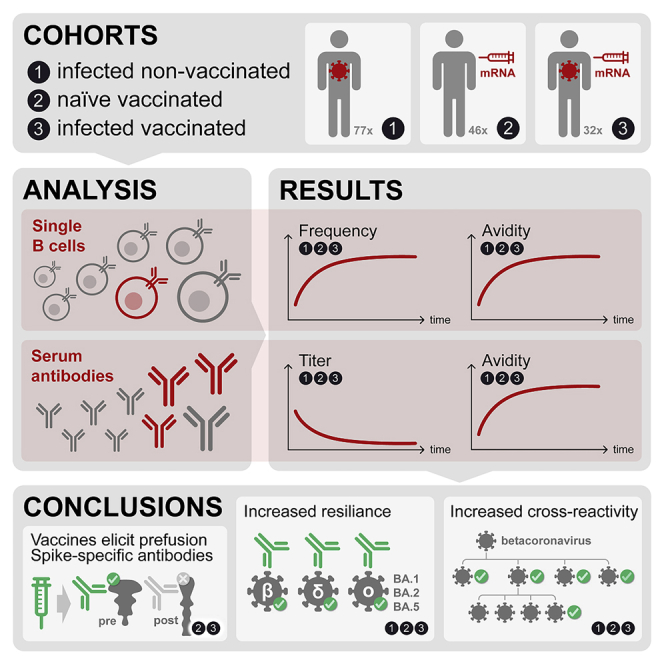
Memory B cells (MBCs) generate rapid antibody responses upon secondary encounter with a pathogen. Here, we investigated the kinetics, avidity, and cross-reactivity of serum antibodies and MBCs in 155 SARS-CoV-2 infected and vaccinated individuals over a 16-month time frame. SARS-CoV-2-specific MBCs and serum antibodies reached steady-state titers with comparable kinetics in infected and vaccinated individuals. Whereas MBCs of infected individuals targeted both prefusion and postfusion Spike (S), most vaccine-elicited MBCs were specific for prefusion S, consistent with the use of prefusion-stabilized S in mRNA vaccines. Furthermore, a large fraction of MBCs recognizing postfusion S cross-reacted with human betacoronaviruses. The avidity of MBC-derived and serum antibodies increased over time resulting in enhanced resilience to viral escape by SARS-CoV-2 variants, including Omicron BA.1 and BA.2 sublineages, albeit only partially for BA.4 and BA.5 sublineages. Overall, the maturation of high-affinity and broadly reactive MBCs provides the basis for effective recall responses to future SARS-CoV-2 variants.
Subject areas: Immunology, Virology
Graphical abstract
Highlights
-
•
SARS-CoV-2 infection and vaccines induce specific memory B cells that reach steady-state levels
-
•
mRNA COVID-19 vaccines elicit prefusion Spike-specific memory B cell responses
-
•
Postfusion Spike-specific memory B cells cross-react with human betacoronaviruses
-
•
Antibody avidity rises over time raising resilience to escape by variants of concern
Immunology; Virology
Introduction
Since its appearance in 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has rapidly spread worldwide resulting in more than 500 million infections and 6.2 million deaths. The virus has evolved into variants of concern (VOC), including the currently circulating Omicron (B.1.1.529) sublineages, which have infected many convalescent and vaccinated individuals.1,2 These VOC have accrued several mutations, in particular in the Spike (S), resulting in the reduction or complete loss of neutralizing activity of polyclonal serum and monoclonal antibodies of individuals who were infected or vaccinated with the prototypic SARS-CoV-2 S.3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19 Besides serum antibodies, memory B cells (MBCs) induced by infection or vaccination play a major role in humoral immunity through recall responses to a second encounter with the same or a related pathogen. Although several studies reported a progressive decline of serum antibody titers over time in convalescent and vaccinated individuals, SARS-CoV-2-specific MBCs have been shown to increase or remain stable in number and to produce neutralizing antibodies.14,20,21,22,23,24,25,26
In this study, we performed single-MBC repertoire analysis from longitudinal samples of convalescent or healthy individuals receiving up to three doses of the Pfizer/BioNtech BNT162b2 mRNA vaccine. We found that, while SARS-CoV-2-specific serum antibodies waned, MBCs increased progressively in frequency and avidity, reaching steady-state levels that remained stable up to 16 months after infection or after the third vaccine dose. Vaccination-induced MBCs mainly targeted the prefusion SARS-CoV-2 S conformation, while infection-induced MBCs recognized also the postfusion conformation and cross-reacted with the S of human betacoronaviruses HCoV-HKU1 and HCoV-OC43. We show that the increased avidity of MBC-derived antibodies provides a mechanism of resilience against emerging variants, including Omicron BA.1 and BA.2 sublineages.
Results
Progressive maturation of memory B cells and serum antibodies following SARS-CoV-2 infection
Blood samples were collected from 64 individuals diagnosed with COVID-19 between March and November 2020 and from 13 individuals diagnosed between December 2020 and January 2021 after an outbreak of the SARS-CoV-2 Alpha variant (Table S1). Peripheral blood mononuclear cells (PBMCs) were isolated for antigen-specific memory B cell repertoire analysis (AMBRA)27 (Figure 1A). PBMCs were stimulated in multiple cultures with IL-2 and the Toll-like receptor 7/8-agonist R848 to promote the selective proliferation and differentiation of MBCs into antibody-secreting cells. The culture supernatants were collected on day 10 and screened in parallel using multiple ELISA assays to detect antibodies of different specificities and to determine the frequencies of antigen-specific MBCs expressed as a fraction of total IgG+ MBCs (Figures S1A-S1C).
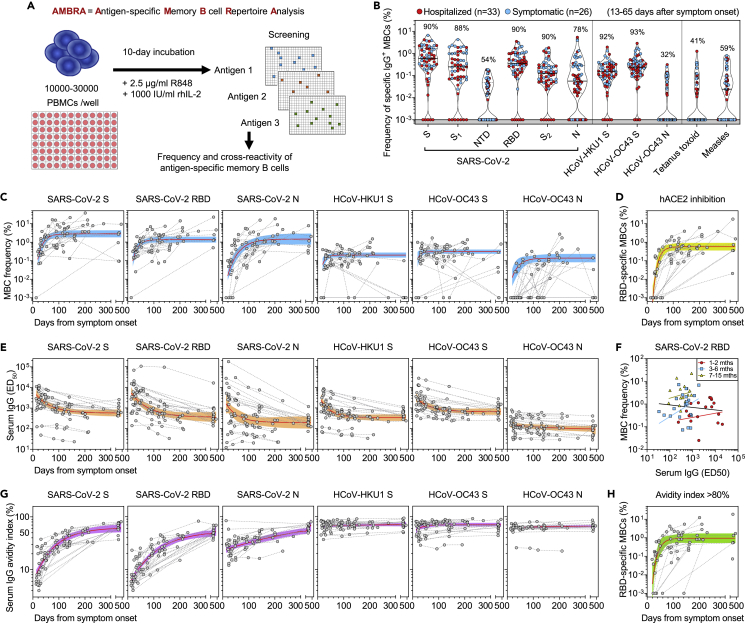
Figure 1.
Early response, RBD immunodominance, kinetics, and affinity maturation of memory B cells primed by Wuhan-Hu-1 SARS-CoV-2
(A) Scheme of the AMBRA method used in this study. PBMC, peripheral blood mononuclear cells. R848, agonist of Toll-like receptors 7 and 8. rhIL2, recombinant human interleukin-2.
(B) Frequency of SARS-CoV-2-specific MBCs isolated between 13 and 65 days after symptom onset from n = 59 donors (33 hospitalized, red, and 26 symptomatic, blue) after the analysis of 5664 MBC cultures. Shown is the reactivity to antigens of SARS-CoV-2 and other betacoronaviruses (HCoV-HKU1 and HCoV-OC43): Spike (S), S1 domain, N-terminal domain (NTD), receptor-binding domain (RBD), S2 domain, Nucleoprotein (N). Reactivities to Tetanus toxoid and to Measles virus (lysate) are included as controls. Median and quartiles are shown as plain and dotted lines, respectively. Percentages of donors with detectable specific MBCs are indicated above each set of data.
(C) Frequency of MBCs specific for SARS-CoV-2 S, RBD and N, HCoV-HKU1 S, HCoV-OC43 S and N from n = 23 donors followed-up up to 469 days after symptom onset. Frequencies were obtained from the analysis of 6336 MBC cultures (66 samples, minimum 2 samples per donor). Black dotted lines connect samples from the same donor. A one-phase association kinetics model (red line) was calculated from all the non-null values of each sample. The area within 95% confidence bands is shown in blue.
(D) Frequency of SARS-CoV-2 RBD-specific MBC-producing antibodies showing the inhibition of RBD binding to ACE2 from n = 23 donors. A one-phase association kinetics model (red line) was calculated from all the non-null values of each sample and the area within 95% confidence bands is shown in yellow.
(E) Serum IgG ED50 titers to SARS-CoV-2 S, RBD and N, HCoV-HKU1 S, HCoV-OC43 S and N of samples collected from 29 donors analyzed up to 469 days after symptom onset. A one-phase decay kinetics model (red line) was calculated from all the non-null values of each sample and the area within 95% confidence bands is shown in orange.
(F) Correlation analysis between frequency of SARS-CoV-2 RBD-specific MBCs and serum RBD-specific IgG titers of n = 56 samples from n = 18 donors collected at different time points. All samples (black line): Spearman r = −0.102 (95% confidence interval −0.363 to 0.173; non-significant p = 0.45). Samples at 1-2 months (n = 18, red line): Spearman r = 0.112 (95% confidence interval −0.387 to 0.561; non-significant p = 0.66). Samples at 3-6 months (n = 23, blue line): Spearman r = 0.214 (95% confidence interval −0.229 to 0.584; non-significant p = 0.33). Samples at 7-15 months (n = 15, yellow line): Spearman r = 0.221 (95% confidence interval −0.343 to 0.668; non-significant p = 0.43).
(G) Serum IgG avidity indexes to SARS-CoV-2 S, RBD and N, HCoV-HKU1 S, HCoV-OC43 S and N of samples collected from 29 donors analyzed up to 469 days after symptom onset. A one-phase association kinetics model (red line) was calculated from all the non-null values of each sample and the area within 95% confidence bands is shown in violet.
(H) Frequency of SARS-CoV-2 RBD-specific B cells with an avidity index greater than 80%. A one-phase association kinetics model (red line) was calculated from all the non-null values of each sample and the area within 95% confidence bands is shown in green. See also Figure S1 and Table S1.
Repertoire analysis of MBCs collected at early time points after infection with SARS-CoV-2 Wuhan-Hu-1 (13-65 days after symptom onset) showed that 90% of the donors had detectable MBCs specific for the prefusion-stabilized SARS-CoV-2 S ectodomain trimer28 (Figures 1B and S1D). The magnitudes of MBC responses were heterogeneous with frequencies ranging between 0 and 6.6% for S-specific MBCs across individuals. In most cases receptor-binding domain (RBD)-specific MBCs dominated the response to S, whereas N-terminal domain (NTD)- and S2-specific MBCs were present at lower frequencies, concurring with the fact that most mAbs cloned from the memory B cells of previously infected subjects target the RBD.13,29 Overall, S-specific MBCs were present at higher frequencies than N-specific MBCs (median: 0.59 vs 0.06%, respectively). A similar magnitude and reactivity of the MBC response were observed in the individuals infected with the SARS-CoV-2 Alpha variant, with a higher frequency of MBCs specific for RBD carrying the N501Y mutation as compared to the Wuhan-Hu-1 RBD (Figure S1E).
By analyzing longitudinal samples collected up to 16 months after infection, we found that S-, RBD-, NTD-, S2- and nucleoprotein (N)-specific MBCs progressively rose in frequency in the first 6-8 months, reaching up to 20% of total IgG MBCs in some cases, followed by a plateau.24 Conversely, the frequency of MBCs specific for the S and N proteins of common cold coronaviruses remained largely constant over time (Figures 1C and S1F). The expansion of RBD-specific MBCs was accompanied by an increase of MBCs producing antibodies that blocked RBD binding to human ACE2, a correlate of neutralization29 (Figures 1D and S1G).
Analysis of longitudinal serum samples showed that IgG antibodies to SARS-CoV-2 S, RBD, and N progressively decreased and reached a plateau, which paralleled the rise of MBCs, consistent with previous reports21,30,31 (Figure 1E). Similarly, IgG antibodies recognizing HCoV-HKU1 and HCoV-OC43 S rapidly dropped early after SARS-CoV-2 infection to levels maintained until the end of the observation period (Figure 1E). No correlation was observed between serum IgG S antibody levels and MBC frequencies in the subjects tested with both assays over the 500-day period analyzed (Figure 1F).
The binding avidity of serum- and MBC-derived specific antibodies was expressed as an avidity index by measuring antibody binding in presence of a chaotropic agent (Figure S1H). The avidity of serum IgG antibodies to SARS-CoV-2 S, RBD, and N increased over time reaching levels comparable to those observed for serum IgG antibodies to HKU1 and OC43 antigens (Figure 1G). Similarly, the frequency of high-avidity MBC-derived RBD-specific antibodies increased over time reaching a plateau after approximately 3-to-4 months after infection (Figure 1H).
The rapid decline of serum antibodies is consistent with an initial generation of many short-lived plasma cells from either naive B cells or cross-reactive MBCs. However, the clonal analysis of serial samples reveals a rapid expansion of S- and RBD-specific MBCs followed by a progressive maturation consistent with a germinal center reaction leading to the generation of plasma cells and MBCs with increased affinity.
Three mRNA-vaccine doses induce high-avidity memory B cells and serum antibodies
The frequency and fine specificity of MBCs were analyzed after the first, second, and third administration of the Pfizer/BioNtech BNT162b2 mRNA vaccine in two cohorts of healthy individuals. Vaccine recipients were either naive (n = 46) or immune (n = 32) to SARS-CoV-2 due to infection occurring 53-389 days before the first dose. In most naive individuals, the first vaccine dose induced SARS-CoV-2 S-specific MBCs at frequencies comparable to those found in samples collected from convalescent individuals at similar timepoints post-antigen exposure (Figure 2A). The second dose elicited a robust response in about half of the donors with a 5-fold increase in the median MBC frequency (Figure 2A). The third dose induced a further 2-fold increase of the median MBC frequency resulting in a robust response in all the naive donors analyzed (Figure 2A). As expected, vaccination of naive donors did not elicit N-specific MBCs, the few exceptions likely reflecting cross-reactivity with other betacoronaviruses (Figure S2A). Remarkably, the first vaccine dose induced very high MBC S-specific frequencies in previously infected donors, exceeding by ∼40-fold those found in naive vaccinated or convalescent individuals. Additional vaccine doses did not result in further MBC increase and did not alter the frequency of MBCs specific for the S of the common cold coronaviruses HCoV-HKU1 and HCoV-OC43 (Figures 2B and S2B).
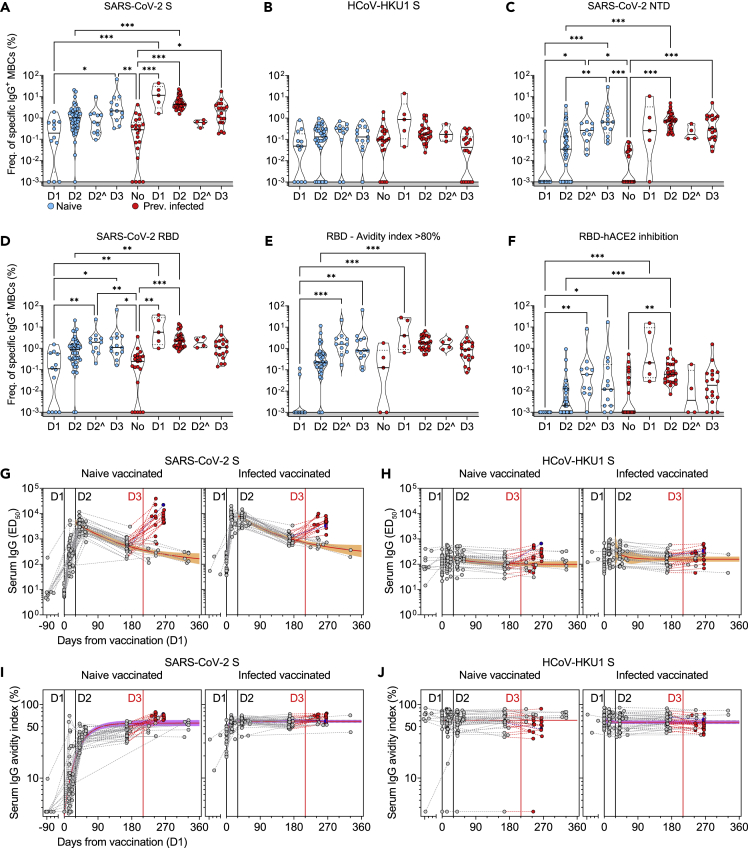
Figure 2.
Characterization of vaccine-induced MBC- and serum-derived antibody response in naive and SARS-CoV-2 immune donors
(A-D) Frequency of MBCs specific for SARS-CoV-2 S (A), HCoV-HKU1 S (B), SARS-CoV-2 NTD (C) and RBD (D) of n = 12, 45, and 13 naive donors and 5, 31, and 18 previously infected donors 10-35 days after the first (D1), the second (D2) and the third dose (D3) of Pfizer/BioNtech BNT162b2 mRNA vaccine, respectively. Shown are also 11 naive and 4 immune donors whose MBCs were isolated 125-293 days after the second dose (D2ˆ). Median frequencies are compared withing donor groups and between respective vaccine doses as well as to a group of n = 21 convalescent donors at 18-30 days after symptom onset. Significant differences are indicated as ∗∗∗ (p <0.001); ∗∗ (p < 0.002), ∗ (p < 0.033).
(E) Frequency of SARS-CoV-2 RBD-specific MBCs with an avidity index greater than 80%.
(F) Frequency of SARS-CoV-2 RBD-specific MBCs inhibiting binding of RBD to ACE2.
(G and H) Serum IgG ED50 titers to SARS-CoV-2 S (G) and HCoV-HKU1 S (H) of samples collected from n = 47 naive (left) and 32 immune donors (right) 10-35 days after the first (D1), the second (D2) and the third dose (D3) of Pfizer/BioNtech BNT162b2 mRNA vaccine, respectively. A one-phase decay kinetics model (red line) was calculated from all the non-null values of each sample and the area within 95% confidence bands is shown in orange. 37 samples collected from individuals who received a third dose (red) or had a second SARS-CoV-2 infection (blue) were excluded from the decay analysis.
(I and J) Serum IgG avidity indexes to SARS-CoV-2 S (I) and HCoV-HKU1 S (J) of the same samples shown in panels G-H. A one-phase association kinetics model (red line) was calculated from all the non-null values of each sample and the area within 95% confidence bands is shown in violet. See also Figure S2 and Table S1.
After the administration of two vaccine doses, the response in both naive and infected donors was dominated by RBD-specific MBCs, while MBCs specific for the NTD or the S2 subunit were present at low to undetectable levels (Figures 2C, 2D, and S2C). Interestingly, NTD-specific, but not S2-, MBCs increased over time in naive donors (Figures 2C and S2C). High-avidity RBD-specific and ACE2-blocking antibodies were detected only after the second dose in naive individuals, whereas these antibodies were detected after one dose in all the previously infected donors and were not further boosted upon subsequent immunizations (Figures 2E and 2F).
When we analyzed longitudinal samples of serum antibodies against SARS-CoV-2 S and RBD, we observed that a single immunization induced highly heterogeneous antibody levels in naive donors that were further increased in all samples after the second and the third immunization (Figures 2G and S2D). In infected donors, the titers of serum antibodies had reached the maximal level after the first immunization, with no further increase after the second or the third immunization. The serum titers of S-specific antibodies declined similarly over time in naive and infected donors with a half-life of 4 months. As expected, no overall variation in N-specific antibody titers was observed (Figure S2E). While the avidity of SARS-CoV-2 S- and RBD-specific serum antibodies in naive donors rapidly increased after vaccination, the avidity in infected donors was found to be high before vaccination with no further increase over time (Figures 2I, S2F, and S2G). In both naive and infected donors, vaccination did not increase the presence or avidity of antibodies specific for the S of the human betacoronaviruses HCoV-HKU1 and HCoV-OC43, concurring with recent data14 (Figures 2H and 2J, S2H, and S2I).
Collectively, these findings indicate that, while in infected donors a single dose of an mRNA vaccine is sufficient to boost a high-avidity antibody response to SARS-CoV-2 S, in naive donors such response is elicited only upon three rounds of immunization.
The antibody response in naive vaccinated individuals is skewed toward prefusion SARS-CoV-2 S
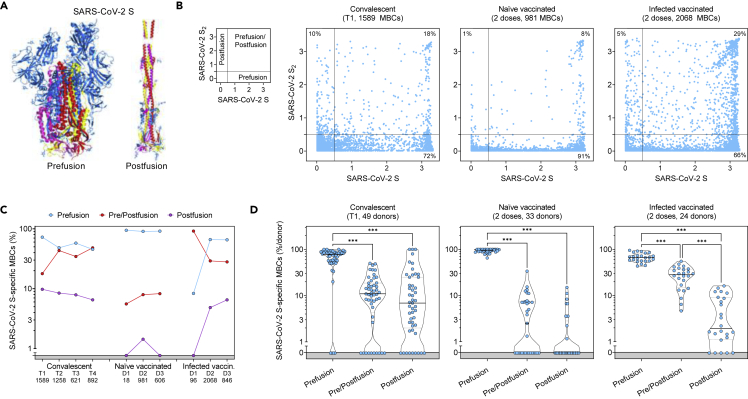
The Pfizer/BioNtech BNT162b2 mRNA vaccine was designed to express the full-length SARS-CoV-2 S stabilized in its prefusion conformation through the 2P mutations32,33 and recent data suggest that vaccination induces high titers of prefusion S-specific plasma antibodies.34 To assess whether vaccination also induced an MBC response preferentially targeting the prefusion conformation of S as compared to that elicited by natural infection, we analyzed the MBC-derived antibodies for their binding to either the prefusion-stabilized SARS-CoV-2 S, postfusion S2, or both in cohorts of convalescent donors and in naive or infected vaccinated individuals (Figure 3A). We found a strong correlation between antibodies binding to postfusion S2 and a structurally validated postfusion SARS-CoV-2 S2,34 thus supporting the use of S2 as a proxy for the postfusion conformation of S (Figure S3A). Across all individuals, most MBCs induced by natural infection and/or vaccination were specific for the prefusion conformation (Figures 3B-3D and S3B). However, while convalescent and infected vaccinated individuals had 10 and 5% of their MBCs specific for S2, respectively, only 1% of MBCs from naive vaccinated donors were postfusion S2-specific. A fraction of MBCs recognized S epitopes shared between prefusion and postfusion conformations. Collectively, these data show that mRNA vaccines primarily induce MBC responses skewed to the prefusion conformation of SARS-CoV-2 S, as recently reported.35
Figure 3.
Comparison of the prefusion and postfusion S-specific MBC responses after vaccination or natural infection
(A) Structural representation of SARS-CoV-2 S in its prefusion and postfusion conformation (adapted from PBD 7tat and 7e9t). The three S2 domains that are maintained in both conformations are colored red, yellow, and pink.
(B) MBC cross-reactivity between SARS-CoV-2 S (prefusion S) and S2 (postfusion S). Shown are average OD values as measured by ELISA with blank subtracted from n = 2 replicates of 1589, 981, and 2068 MBC cultures analyzed from 49 convalescent, 33 naive and 24 infected vaccinated donors. Cumulative fraction of MBCs reactive to either prefusion or postfusion S or both is indicated as a percentage in the respective quadrant. The small panel on the left describes the distribution of prefusion and/or postfusion S-specific MBCs in the different quadrants.
(C) Cumulative fraction of S-specific MBCs reactive to prefusion and/or postfusion S at different timepoints after natural infection (T1, T2, T3 and T4) or vaccine doses (D1, D2, D3).
(D) Individual fractions of S-specific MBCs reactive to prefusion and/or postfusion S in 49 convalescent, 33 naive, and 24 infected vaccinated donors. Significant differences are indicated as ∗∗∗ (p-value < 0.001); ∗∗ (p < 0.002), ∗ (p < 0.033). See also Figure S3.
Infection- and vaccine-induced memory B cells show variable degrees of cross-reactivity against other betacoronaviruses
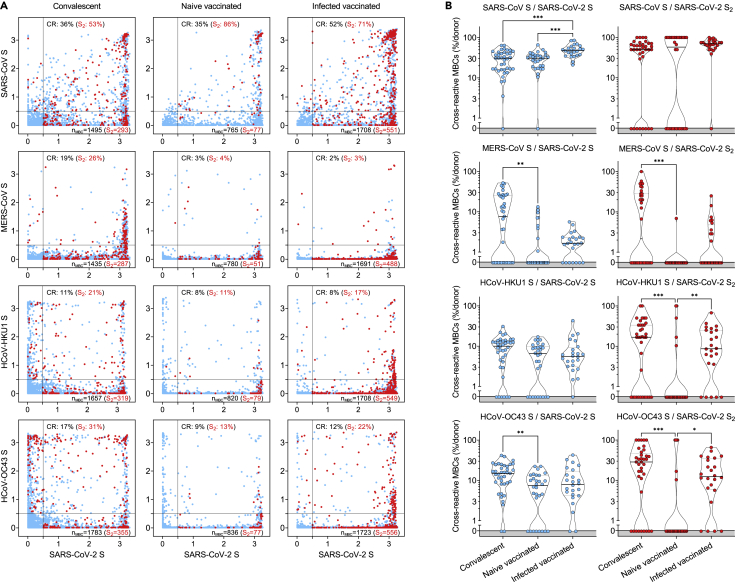
Unlike serological analyses, the AMBRA method is suitable to dissect the cross-reactivity of individual MBC-derived antibodies, at the single clone level, against a panel of human and animal betacoronaviruses.36 Analysis of MBCs from SARS-CoV-2-convalescent and from naive or infected donors receiving two vaccine doses revealed a high frequency (35-52%) of SARS-CoV-2 S-specific MBCs that cross-reacted with SARS-CoV S, consistent with the high level of S sequence similarity with SARS-CoV-2 (Figures 4A and 4B). Conversely, we observed a low frequency (2-19%) of SARS-CoV-2 S-specific MBCs that cross-reacted with the more divergent S of MERS-CoV, HCoV-HKU1 and HCoV-OC43 betacoronaviruses. A similar trend was observed at late time points in convalescent donors as well as after the third vaccine dose in vaccinated individuals (Figure S4). Deconvolution of SARS-CoV-2 S-reactivity revealed a higher frequency of cross-reactive antibodies among the subset of S2-specific MBCs (Figures 4A and 4B).
Figure 4.
Cross-reactivity to betacoronaviruses of MBCs primed by SARS-CoV-2 infection and/or vaccination
(A) Cumulative MBC cross-reactivity between SARS-CoV-2 S and the four betacoronaviruses SARS-CoV, MERS-CoV, HCoV-HKU1, and HCoV-OC43. Shown are average OD values as measured by ELISA with blank subtracted from n = 2 replicates of 3744, 2880, and 2304 MBC cultures analyzed from 39 convalescent, 30 naive, and 24 infected donors after two vaccine doses. S2-specific MBCs are shown in red. Numbers of S- and S2-specific MBCs are indicated in the bottom-right quadrants of each panel. Cumulative fractions of S- and S2-cross-reactive (CR) MBCs are indicated as a percentage in the top-right quadrant.
(B) Individual fractions of SARS-CoV-2 S- (left) and S2-specific MBCs (right) that cross-react with the four betacoronaviruses in 39 convalescent, 30 naive and 24 infected vaccinated donors. Significant differences are indicated as ∗∗∗ (p-value < 0.001); ∗∗ (p < 0.002), ∗ (p < 0.033). See also Figure S4.
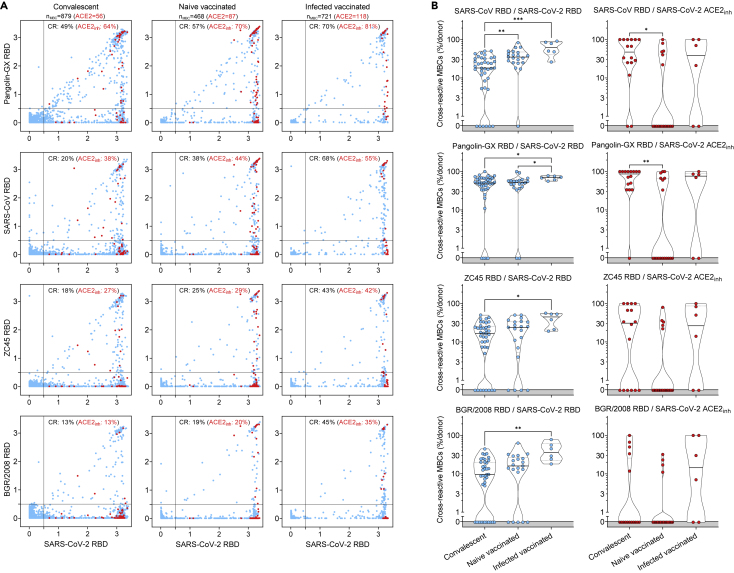
The same analysis was performed on RBDs of sarbecoviruses representative of clades 1a (SARS-CoV), 1b (Pangolin Guangxi), 2 (bat ZC45), and 3 (bat BM48-31/BGR/2008) comparing the reactivity of MBCs from SARS-CoV-2-convalescent and from naive or infected donors receiving two vaccine doses. We found that the frequencies of antibodies cross-reactive to heterologous sarbecovirus RBDs, including those inhibiting binding of RBD to human ACE2, were progressively lower as a function of the decreasing sequence identity with SARS-CoV-2 (Figure 5A, 5B, S5A, and S5B). Infected vaccinees had the highest frequency of cross-reactive MBCs against diverse RBDs, suggesting that increased avidity resulting from multiple and diverse antigenic stimulations may have also contributed to broadening the reactivity toward heterologous sarbecoviruses.
Figure 5.
Cross-reactivity to sarbecoviruses of MBCs primed by SARS-CoV-2 infection and/or vaccination
(A) Cumulative MBC cross-reactivity between SARS-CoV-2 RBD and four sarbecoviruses representative of clades 1a (SARS-CoV), 1b (Pangolin Guangxi), 2 (ZC45) and 3 (BM48-31/BGR/2008). Shown are average OD values as measured by ELISA with blank subtracted from n = 2 replicates of 3744, 2016 and 576 MBC cultures analyzed from 39 convalescent donors at two different timepoints and from 21 naive and 6 infected donors after receiving two vaccine doses. RBD-specific MBCs showing the inhibition of binding to ACE2 are shown in red. Cumulative fractions of total and ACE2-inhibiting RBD-cross-reactive (CR) MBCs are indicated as a percentage in the top-right quadrant.
(B) Individual fractions of SARS-CoV-2 RBD-specific MBCs (left) and those showing the inhibition of binding to ACE2 (right) that cross-react with the four representative sarbecoviruses in 39 convalescent, 21 naive and 6 infected vaccinated donors. Significant differences are indicated as ∗∗∗ (p-value < 0.001); ∗∗ (p < 0.002), ∗ (p < 0.033). See also Figure S5.
Affinity maturation of receptor-binding domain-specific memory B cells leads to resilience to viral escape by SARS-CoV-2 variants of concern
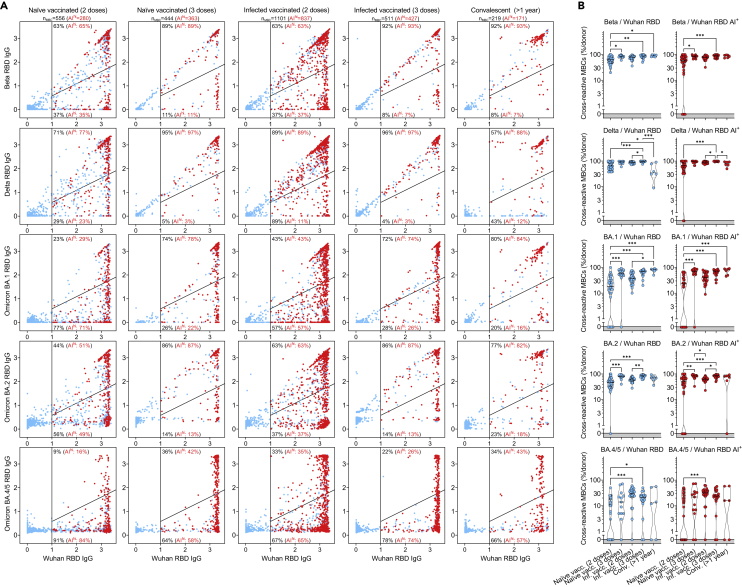
In view of the continuing emergence of SARS-CoV-2 VOC, we analyzed at a clonal level the extent to which MBCs elicited by infection with Wuhan-Hu-1-related viruses in early 2020 or by mRNA vaccines could cross-react with RBDs of progressively diverging VOC. We, therefore, tested MBC-derived antibodies for their ability to retain binding (here measured as less than 2-fold loss compared to Wuhan-Hu-1) to RBD of different VOC, including Beta (B.1.351), Delta (B.1.617.2) and Omicron (B.1.1.529) BA.1, BA.2, BA.3 and BA.4/BA.5 sublineages (Figures 6A, 6B, and S6A). In naive or infected individuals receiving two doses of the Pfizer/BioNtech BNT162b2 mRNA vaccine, mutations in the VOC RBDs were poorly tolerated with the greatest loss of binding observed against Omicron sublineages. Importantly, the third vaccine dose substantially increased the resilience to VOC escape from binding antibodies in both naive and infected individuals to a degree similar to that observed in convalescent individuals more than one year after infection, in line with the analysis of neutralizing antibody responses.14,15,17,37,38,39,40 Interestingly, in individuals given three vaccine doses, the higher fraction of MBCs cross-reactive with VOC RBDs was characterized by high-avidity and ACE2-blocking activity (Figures 6A, 6B, S6A, and S6B). However, in all cohorts analyzed, the binding of MBC-derived antibodies was substantially reduced when tested against BA.4/BA.5 RBDs (Figure 6A).
Figure 6.
Resilience to viral escape of VOC by high-avidity MBCs primed by SARS-CoV-2 infection and/or vaccination
(A) Cumulative MBC cross-reactivity between RBD from Wuhan-Hu-1 SARS-CoV-2 and Beta, Delta, Omicron BA.1, BA.2, and BA.4/5 VOC. Shown are average OD values as measured by ELISA with blank subtracted from n = 2 replicates of 2976, 1248, 2304, 1728, and 576 MBC cultures analyzed from 31 to 13 naive donors, 24 and 18 infected donors after receiving two and three vaccine doses and from 6 convalescent donors at 376-469 days from symptom onset. RBD-specific MBCs showing high avidity index (AI>80%) are shown in red. Numbers of total and high-avidity RBD-specific MBCs are indicated in the top-left quadrants. Cumulative fractions of total and high-avidity RBD-specific MBCs maintaining or losing binding to the VOC RBD are indicated as a percentage in the top-right and bottom-right quadrants.
(B) Individual fractions of total (left) and high-avidity (right) SARS-CoV-2 RBD-cross-reactive MBCs that maintain binding with the RBDs of different VOC in 31 and 13 naive donors, 24 and 18 infected vaccinated donors after receiving two and three vaccine doses and in 6 convalescent donors at 376-469 days from symptom onset. Significant differences are indicated as ∗∗∗ (p-value < 0.001); ∗∗ (p < 0.002), ∗ (p < 0.033). See also Figure S6.
Taken together, these findings indicate that long-lasting affinity maturation upon infection by Wuhan-Hu-1 SARS-CoV-2 and multiple vaccinations can drive the development of MBCs with greater resilience to viral escape.
Discussion
Kinetics of serum- and MBC-derived antibodies to SARS-CoV-2 after infection and vaccination have been extensively described.14,20,21,22,23,24,25,26 In this study, we provide an in-depth characterization of the maturation of the memory B cell response to SARS-CoV-2, supporting evidence of how the MBC repertoire is shaped to broaden recall responses to other betacoronaviruses and future variants of concern. Compared to flow-cytometry-based methods,22,23,24,25,41 the antigen-specific memory B cell repertoire analysis (AMBRA) has the advantage of analyzing very large numbers of MBCs at the single-cell level, thus allowing unbiased direct comparisons of multiple specificities and functional properties of MBC-derived antibodies.
Documenting the reciprocal kinetics of serum antibodies and MBCs to SARS-CoV-2 antigens illustrates a fundamental aspect of the antibody response. While serum antibodies produced by the first wave of short-lived plasma cells decline over time, MBCs increase in numbers reaching up to 20% of total IgG MBCs in a few months after SARS-CoV-2 infection before their frequencies stabilize. Importantly, this time-dependent increase of MBCs is accompanied by affinity maturation and breadth expansion. As a consequence, while serum antibodies decline, the immune system builds up the capacity to mount a very potent secondary memory response. Accordingly, high numbers of MBCs and breadth against VOC are characteristic of donors who had hybrid immunity due to infection followed by vaccination.42,43 Conversely, naive donors require a longer time and multiple immunizations to develop an MBC response of magnitude and breadth, which are comparable to those of infected individuals.38,39 The increased avidity resulting from multiple antigenic stimulations may therefore contribute to broaden the reactivity toward heterologous sarbecoviruses as well as to generate resilience to new VOC,38,44,45 including Omicron sublineages. This is consistent with the notion that cross-reactive MBCs are primarily induced by repeated antigenic stimulations leading to epitope spread and affinity maturation, which are fundamental for an effective and long-lasting recall response to future SARS-CoV-2 variants. However, the less pronounced resilience observed against the recently emerging BA.4 and BA.5 variants suggests that immune escape, even from high-avidity antibodies, may be a major driver for the evolution of Omicron sublineages.
Limitations of the study
The AMBRA method requires in vitro restimulation of memory B cells, which has a cloning efficiency of approximately 30%,27 thus potentially introducing some biases in the responding B cells subsets. The low volume of B cell supernatants obtained after stimulation also limits the number of antigens and antibody properties that can be analyzed from the same sample. Binding analysis does not necessarily reflect affinity or neutralization. The ACE2 inhibition assay used as a proxy for neutralization might not have captured all the neutralizing antibodies targeting epitopes on the SARS-CoV-2 Spike that are not involved in the interaction with the ACE2 receptor.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD3 | BioLegend | OKT3; Cat#317328; RRID: AB_2562907 |
| CD4 | BioLegend | RM4-5; Cat#100526; RRID: AB_312727 |
| CD19 | BioLegend | SJ25C1; Cat#363024; RRID: AB_2564253 |
| CD16 | BioLegend | 3G8; Cat#302044; RRID: AB_2563802 |
| CD14 | BioLegend | M5E2; Cat#301838; RRID: AB_2562909 |
| IgG | BD Biociences | G18-145; Cat#555786; RRID: AB_396121 |
| Goat F(ab')2 Anti-Mouse IgG(H + L), Human ads-AP | Bioconcept | Cat#1032-04; RRID: AB_2794320 |
| Biological samples | ||
| Donors’ PBMCs | This study | |
| Donors’ sera | This study | |
| Chemicals, peptides, and recombinant proteins | ||
| R848 (Resiquimod) | InvivoGen | Cat#tlrl-r848-5 |
| Recombinant human IL-2 | ImmunoTools | Cat#11340027 |
| RBD, mouse Fc Tag | Sino Biological | Cat#40592-V05H |
| pNPP | Sigma-Aldrich | Cat#71768-25G |
| SARS-CoV-2 S2 | The Native Antigen Company | Cat#REC31807-500 |
| SARS-CoV-2 S1 | The Native Antigen Company | Cat#40150-V08B1 |
| SARS-CoV-2 N | The Native Antigen Company | Cat#REC31812 |
| HCoV-OC43 N | The Native Antigen Company | Cat#REC31857 |
| SARS-CoV-2 NTD | McCallum et al.13 | |
| SARS-CoV-2 S | Walls et al.28,47 | |
| SARS-CoV S | Walls et al.48 | |
| HCoV-OC43 S | Pinto et al.49 | |
| MERS-CoV S | Walls et al.48 | |
| HCoV-HKU1 S | Pinto et al.49 | |
| SARS-CoV-2 RBD | Piccoli et al.29 | |
| BM48-31/BGR/2008 RBD | This study | |
| Pangolin GX RBD | This study | |
| ZC45 RBD | This study | |
| SARS-CoV RBD | This study | |
| Beta RBD | This study | |
| Delta RBD | This study | |
| Omicron BA.1 RBD | This study | |
| Omicron BA.2 RBD | This study | |
| Omicron BA.3 RBD | This study | |
| Omicron BA.4/5 RBD | This study | |
| Human ACE2 | Piccoli et al.29 | |
| Blocker™ Casein in PBS | Thermo Fisher Scientific | Cat#37528 |
| Tween 20 | Sigma Aldrich | Cat#93773-1KG |
| Goat Anti-Human IgG-AP | Bioconcept | Cat#2040-04; RRID: AB_2795643 |
| Sodium thiocyanate | Sigma-Aldrich | Cat#251410-2.5KG |
| Zombie Aqua Fixable Viability Kit | BioLegend | Cat#423101 |
| Ficoll-Paque PLUS (6 x500ml) | VWR International | Cat#17-1440-03 |
| RPMI-1640 W/O L-Glutamine (10x500ml) | Life Technologies Europe BV | Cat#31870074 |
| HyClone Fetal Bovine Serum, US Origin 500ml, | VWR International | Cat#SH30070.03 |
| DPBS w/o Ca and Mg (500ml), | Chemie Brunschwig | Cat#P04-36500 Pan Biotech |
| MEM NEAA Solution 100x, 100ml, | Bioconcept | Cat#5-13K00-H |
| Stable Glutamine solution (L-Ala/L-Gln)100x, 100ml | Bioconcept | Cat#5-10K50-H |
| Penicillin-Streptomycin | Bioconcept | Cat#4-01F00-H |
| Kanamycin (5,000ug/ml), 100ml | Bioconcept | Cat#4-08F00-H |
| Transferrin (Holo) from human serum | LuBioscience | Cat#0905-100 |
| 2-Mercaptoethanol 50MM | Bioconcept | Cat#5-69F00-E |
| Sodium Pyruvate (100mM, 100 ml) | Bioconcep | Cat#5-60F00-H |
| Experimental models: Cell lines | ||
| FreeStyleTM 293-F Cells | ThermoFisher Scientific | Cat# R79007; RRID: CVCL_D603 |
| Expi293FTM Cells | ThermoFisher Scientific | Cat# A14527; RRID: CVCL_D615 |
| ExpiCHO-STM | ThermoFisher Scientific | Cat# A29127; RRID: CVCL_5J31 |
| Recombinant DNA | ||
| SARS-CoV-2 NTD pCMV plasmid | McCallum et al.13 | |
| SARS-CoV-2 S phCMV1 plasmid | Walls et al. and Starr et al.28,47 | |
| SARS-CoV S phCMV1 plasmid | Walls et al.48 | |
| HCoV-OC43 S phCMV1 plasmid | Pinto et al.49 | |
| MERS-CoV S phCMV1 plasmid | Walls et al.48 | |
| HCoV-HKU1 S phCMV1 plasmid | Pinto et al.49 | |
| SARS-CoV-2 RBD phCMV1 plasmid | Piccoli et al.29 | |
| BM48-31/BGR/2008 RBD phCMV1 plasmid | This study | |
| Pangolin GX RBD phCMV1 plasmid | This study | |
| ZC45 RBD phCMV1 plasmid | This study | |
| SARS-CoV RBD phCMV1 plasmid | This study | |
| Beta RBD phCMV1 plasmid | This study | |
| Delta RBD phCMV1 plasmid | This study | |
| Omicron BA.1 RBD phCMV1 plasmid | This study | |
| Omicron BA.2 RBD phCMV1 plasmid | This study | |
| Omicron BA.3 RBD phCMV1 plasmid | This study | |
| Omicron BA.4/5 RBD phCMV1 plasmid | This study | |
| Human ACE2 phCMV1 plasmid | Piccoli et al.29 | |
| Software and algorithms | ||
| Flowjo (v10.7.1) | FlowJo | https://www.flowjo.com |
| GraphPad Prism (v9.3.1) | GraphPad | https://www.graphpad.com |
| Everest (v3.0) | Bio-Rad | https://www.bio-rad.com |
| Microsoft Excel for Microsoft 365 MSO (Version 2204 Build 16.0.15128.20240) | Microsoft | https://www.microsoft.com |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Luca Piccoli (lpiccoli@vir.bio).
Materials availability
Materials generated in this study will be made available on request and may require a material transfer agreement.
Experimental model and subject details
Cell lines
Cell lines used in this study were obtained from ThermoFisher Scientific (FreeStyleTM 293-F Cells, Expi293FTM Cells and ExpiCHO-STM).
Study participants and ethics statement
Samples were obtained from 3 cohorts of Wuhan-Hu-1 SARS-CoV-2-infected individuals, 1 cohort of Alpha SARS-CoV-2-infected individuals and 2 cohorts of individuals vaccinated with Pfizer/BioNtech BNT162b2 mRNA COVID-19 vaccine under study protocols approved by the local Institutional Review Boards (Canton Ticino Ethics Committee, Switzerland and the Ethical committee of Luigi Sacco Hospital, Milan, Italy). COVID-19 was diagnosed by PCR with primers specific for the detection of Wuhan-Hu-1 or Alpha SARS-CoV-2 in nasal swaps. All donors provided written informed consent for the use of blood and blood components (such as PBMCs, sera or plasma) and were recruited at hospitals or as outpatients. Based on their availability, participants were enrolled and allocated to either single blood draws or longitudinal follow-up.
Method details
Isolation of peripheral blood mononuclear cells (PBMCs), plasma and sera
PBMCs and plasma were isolated from blood draw performed using tubes or syringes pre-filled with heparin or sodium EDTA, followed by Ficoll-Paque PLUS (6x500 ml) (VWR International, 17-1440-03) density gradient centrifugation. Sera were obtained from blood collected using tubes containing clot activator, followed by centrifugation. PBMCs, plasma and sera were stored in liquid nitrogen and -80°C freezers until use, respectively.
Immunophenotyping
PBMCs were thawed and washed twice with RPMI-1640 W/O L-Glutamine (10x500 ml) (Life Technologies Europe BV, 31870074) 10% HyClone Fetal Bovine Serum, US Origin 500ml (VWR International, SH30070.03), and incubated in the same medium for 2 h at 37°C. Live PBMCs were counted post thawing and seeded at 1 million into round-bottom 96-well plates (Corning, 3799). PBMCs were stained with LIFE/DEAD marker (Zombie Aqua Fixable Viability Kit, BioLegend 423101) in Dulbecco's phosphate-buffered saline (DPBS) w/o Ca and Mg (500ml), (Chemie Brunschwig, P04-36500 Pan Biotech) for 30’ at RT, washed in MACS buffer (PBS 2% HyClone, 2 mM EDTA), and stained with antibodies to CD3, CD4, CD19, CD16, CD14, (BioLegend), IgG (BD Bioscience) (key resources table) for 30’ at 4°C. Cells were then washed and resuspended in MACS buffer for data acquisition at ZE5 cytometer (Bio-Rad). Data were analysed with FlowJo software.
Memory B cell culture
Replicate cultures of total PBMCs were set at different cell densities (10,000-30,000 cells/culture) in 96 U-bottom plates (Corning, 3799). Cells were cultured at 37°C in RPMI 10% Hyclone, 1% Stable Glutamine, 1% Sodium Pyruvate, 1% MEM NEAA, 1% Pen-Strep, 1% Kanamycin, 30 μg/ml Transferrin Holo, 50 μM 2-Mercaptoethanol (50 mM), and stimulated with 2.5 μg/mL R848 (Invivogen, tlrl-r848-5) and 1,000 U/mL human recombinant IL-2 (ImmunoTools, 11340027). Supernatants were harvested after 10 days.
Plasmid design
The SARS-CoV-2, SARS-CoV, MERS-CoV, HCoV-HKU1 and HCoV-OC43 prefusion S and the SARS-CoV-2 postfusion S ectodomains were synthetized by Genscript or GeneArt and cloned in the phCMV1 vector, as previously described.28,34,48,49,47,46,50 The Wuhan-Hu-1 SARS-CoV-2 RBD plasmid, which encodes for the S residues 328–531, was synthetized by Genscript and cloned in the phCMV1 vector, as previously described.29 Plasmids encoding for RBDs of different sarbecoviruses were synthetized by Genscript and cloned in the phCMV1 vector. Plasmids encoding for the RBD of SARS-CoV-2 Beta, Delta, Omicron BA.1, BA.2, BA3 and BA.4/5 variants were generated by overlap PCR.6 The SARS-CoV-2 NTD plasmid, which encodes for the S residues 14–307, was synthetized by GeneArt and cloned in the pCMV vector, as previously described.13
Recombinant glycoprotein production
All SARS-CoV-2 S ectodomains were produced in 500-mL cultures of FreeStyle™ 293-F cells (ThermoFisher Scientific) grown in suspension using FreeStyle 293 expression medium (ThermoFisher Scientific) at 37°C in a humidified 8% CO2 incubator rotating at 130 r.p.m. Cells grown to a density of 2.5 million cells per mL were transfected using PEI (9 μg/mL) or 293fectin and respective plasmids, and cultivated for 3-4 days. The supernatant was harvested and, for some productions, cells were resuspended for another three days, yielding two harvests. SARS-CoV-2 S ectodomain was purified from clarified supernatants using a 5-mL C-tag affinity matrix column (Thermo-Fischer). SARS-CoV, MERS-CoV and HCoV-HKU1 S ectodomains were purified using a cobalt affinity column (Cytiva, HiTrap TALON crude). HCoV-OC43 S ectodomain was purified using a 1-ml StrepTrap column (GE Healthcare). All purified proteins were then concentrated using a 100 kDa centrifugal filter (Amicon Ultra 0.5 mL centrifugal filters, MilliporeSigma). Concentrated SARS-CoV-2 S was further purified by a sizing step, using a Superose 6 Increase 10/300 GL column (Cytiva) with 50 mM Tris pH 8, 200 mM NaCl as a running buffer. Peak fractions corresponding to homogeneous spike trimer were pooled. All the proteins were flash frozen in liquid nitrogen and stored for further usage at −80°C.
Postfusion SARS-CoV-2 S, all RBDs and the NTD were produced in Expi293F™ Cells (ThermoFisher Scientific) grown in suspension using Expi293™ Expression Medium (ThermoFisher Scientific) at 37°C in a humidified 8% CO2 incubator rotating at 130 r.p.m. Cells grown to a density of 3 million per mL were transfected using the respective plasmids with the ExpiFectamine™ 293 Transfection Kit (ThermoFisher Scientific) and cultivated for 5 days. SARS-CoV-2 S (used to prepare postfusion S) was purified using a nickel HisTrap HP affinity column (Cytiva) and then incubated with 1:1 w/w S2X58-Fab47 and 10 ug/mL trypsin for 1 h at 37°C before size exclusion on a Superose 6 Increase column (Cytiva). Supernatants containing RBDs were harvested five days after transfection, equilibrated with 0.1 M Tris-HCl, 0.15 M NaCl, 10 mM EDTA, pH 8.0 and supplemented with a biotin blocking solution (IBA Lifesciences). RBDs were purified by affinity chromatography on a Strep-Trap HP 5 ml column followed by elution with 50 mM biotin and buffer exchange into PBS. The NTD domain was purified from clarified supernatants using 2 ml of cobalt resin (Takara Bio TALON), washing with 50 column volumes of 20 mM HEPES-HCl pH 8.0 and 150 mM NaCl and eluted with 600 mM imidazole. Purified protein was concentrated using a 30 kDa centrifugal filter (Amicon Ultra 0.5 mL centrifugal filters, MilliporeSigma), the imidazole was washed away by consecutive dilutions in the centrifugal filter unit with 20 mM HEPES-HCl pH 8.0 and 150 mM NaCl, and finally concentrated to 20 mg/ml and flash frozen.
Recombinant human ACE2 was expressed in Expi293FTM or ExpiCHO-STM cells transiently transfected with a plasmid encoding for ACE2 residues 19-615, as previously described.29 Supernatant was collected 6-8 days after transfection, supplemented with buffer to a final concentration of 80 mM Tris-HCl pH 8.0, 100 mM NaCl, and then incubated with BioLock (IBA GmbH) solution. ACE2 was purified using StrepTrap High Performance columns (Cytiva) followed by isolation of the monomeric ACE2 by size exclusion chromatography using a Superdex 200 Increase 10/300 GL column (Cytiva) pre-equilibrated in PBS or 20 mM Tris-HCl pH 7.5, 150 mM NaCl.
Enzyme-linked immunosorbent assay (ELISA)
Spectraplate-384 with high protein binding treatment (custom made from Perkin Elmer) were coated overnight at 4°C with 1 μg/ml of SARS-CoV-2 S (produced in house), SARS-CoV-2 S2 (The Native Antigen Company, REC31807-500), S1 (The Native Antigen Company, 40150-V08B1), NTD (produced in house), N (The Native Antigen Company, REC31812), SARS-CoV S (produced in house), MERS-CoV S (produced in house), HCoV-HKU1 S (produced in house), HCoV-OC43 S (produced in house) and HCoV-OC43 N (The Native Antigen Company, REC31857), 5 μg/ml of RBD from SARS-CoV-2 (produced in house), SARS-CoV (produced in house), PangolinGX (produced in house), ZC45 (produced in house) and BM48-31/BGR/2008 (produced in house). For VOC comparison, plates were coated with 1 μg/ml of SARS-CoV-2 RBD (produced in house), Beta RBD (produced in house), Delta RBD (produced in house), Omicron BA.1, BA.2, BA.3 and BA.4/5 RBD (produced in house) in PBS pH 7.2 or PBS alone as control. Plates were subsequently blocked with Blocker Casein (1%) in PBS (Thermo Fisher Scientific, 37528) supplemented with 0.05% Tween 20 (Sigma Aldrich, 93773-1KG). The coated plates were incubated with diluted B cell supernatant for 1 h at RT. Plates were washed with PBS containing 0.05 % Tween20 (PBS-T), and binding was revealed using secondary goat anti-human IgG-AP (Southern Biotech, 2040-04). After washing, pNPP substrate (Sigma-Aldrich, 71768-25G) was added and plates were read at 405 nm after 1 h or 30’. For chaotropic ELISA, after coating with 5 μg/ml of SARS-CoV-2 RBD, blocking and incubation with B-cell supernatants, plates were washed and incubated with 1 M solution of sodium thiocyanate (NaSCN) (Sigma-Aldrich, 251410-2.5KG) for 1 h.
Blockade of RBD binding to human ACE2
Plasma or memory B cell culture supernatants were diluted in PBS and mixed with SARS-CoV-2 RBD mouse Fc-tagged antigen (Sino Biological, 40592-V05H, final concentration 20 ng/ml) and incubated for 30 min at 37°C. The mix was added for 30 min to ELISA 384-well plates (NUNC, P6366-1CS) pre-coated overnight at 4°C with 4 μg/ml human ACE2 (produced in house) in PBS. Plates were washed with PBS containing 0.05 % Tween20 (PBS-T), and RBD binding was revealed using secondary goat anti-mouse IgG-AP (Southern Biotech, 1032-04). After washing, pNPP substrate (Sigma-Aldrich, 71768-25G) was added and plates were read at 405 nm after 1 h.
Quantification and statistical analysis
Data management and statistical analysis were carried out by in-house software based on PostgreSQL and Scala.51 Positive cultures of antigen-specific MBCs were identified from those showing OD values >0.5 by ELISA. This cut-off was determined from 3 times the average OD of pre-pandemic controls. The percentage of inhibition of RBD binding to human ACE2 was calculated as follow: (1−(OD sample−OD neg ctr)/(OD pos ctr−OD neg ctr)]) × 100. The frequency of B cells precursors specific for a given antigen was calculated assuming a Poisson distribution with the following equation: % of antigen-specific MBCs = -100∗(LN(number of negative wells/number of total seeded wells))/number of IgG+ MBCs per well. Other statistical and data analyses were performed using GraphPad Prism (v9.3.1) and Microsoft Excel for Microsoft 365 MSO (Version 2204 Build 16.0.15128.20240). Nonparametric Kruskal-Wallis test was used to analyze statistical differences between groups analyzed. Correction for multiple comparison was performed with Dunn’s test. Statistical significance was defined as p <0.05. ED50 values were determined by non-linear regression analysis (log(agonist) versus response - Variable slope (four parameters)). Avidity Index was calculated as the ratio (%) between the ED50 in presence and the ED50 in absence of NaSCN. Variation of frequencies and serum titers or avidity over time was determined by one-phase association or decay kinetics models from all the non-null values of each sample.
Acknowledgments
We thank the personnel from the EOC Institute of Laboratory Medicine (EOLAB) for taking blood samples and Ms. Sarita Prosperi for logistics support. F.S. is supported by the Helmut Horten Foundation. O.G. is supported by the Swiss Kidney Foundation. This project has been funded in part with federal funds from the NIAID/NIH under Contract No. DP1AI158186 and HHSN272201700059C to D.V., a Pew Biomedical Scholars award (D.V.), an Investigators in the Pathogenesis of Infectious Disease award from the Burroughs Wellcome Fund (D.V.), Fast Grants (D.V.), and the Natural Sciences and Engineering Research Council of Canada (M.M.). D.V. is an Investigator of the Howard Hughes Medical Institute.
Author contributions
Conceptualization, R.M., J.B., C.S.-F., D.C. and L.Pi.; methodology, R.M., J.B., C.S.-F., F.M., A.C., J.S.L. and L.Pi.; formal analysis, R.M., J.B., C.S.-F., I.B., D.V., A.T., D.C., and L.Pi.; investigation, R.M., J.B., C.S.-F., and F.M.; resources - donors’ recruitment and sample collection, L.Pe., T.T., V.L., M.T., A.R., M.B., A.F.-P., C.G., P.F., A.C., and O.G.; resources - protein expression and purification, K.C., N.S., G.L., C.S., E.C., A.C.W., M.M., M.A.T., J.E.B., E.A.D.J., J.R.D., and N.C.; data curation, I.B. and L.Pi.; writing - original draft, C.H.-D., A.A., H.W.V., F.S., D.V., A.L., D.C., and L.Pi.; writing - review & editing, D.V., A.L., D.C., and L.Pi.; visualization, D.V., A.L., D.C., and L.Pi.; supervision, C.G., O.G., A.T., H.W.V., F.S., D.V., A.L., D.C., and L.Pi.
Declaration of interests
R.M., J.B., C.S.-F., I.B., F.M., K.C., N.S., G.L., C.S., E.C., E.A.D.J., J.R.D., N.C., C.H.-D., A.T., A.A., H.W.V., A.L., D.C., and L.Pi. are or were employees of Vir Biotechnology Inc. and may hold shares in Vir Biotechnology Inc. C.G. is an external scientific consultant to Humabs BioMed SA. The other authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: January 20, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105726.
Supplemental information
Data and code availability
Data and code generated in this study will be made available on request and may require a material transfer agreement.
References
- 1.Viana R., Moyo S., Amoako D.G., Tegally H., Scheepers C., Althaus C.L., Anyaneji U.J., Bester P.A., Boni M.F., Chand M., et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022;603:679–686. doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tegally H., Moir M., Everatt J., Giovanetti M., Scheepers C., Wilkinson E., Subramoney K., Makatini Z., Moyo S., Amoako D.G., et al. Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat. Med. 2022;28:1785–1790. doi: 10.1038/s41591-022-01911-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 4.Supasa P., Zhou D., Dejnirattisai W., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Nutalai R., Tuekprakhon A., et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell. 2021;184:2201–2211.e7. doi: 10.1016/j.cell.2021.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–2361.e6. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collier D.A., De Marco A., Ferreira I.A.T.M., Meng B., Datir R.P., Walls A.C., Kemp S.A., Bassi J., Pinto D., Silacci-Fregni C., et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593:136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cele S., Gazy I., Jackson L., Hwa S.H., Tegally H., Lustig G., Giandhari J., Pillay S., Wilkinson E., Naidoo Y., et al. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature. 2021;593:142–146. doi: 10.1038/s41586-021-03471-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Beltran W.F., Lam E.C., St Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383.e9. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen X., Tang H., McDanal C., Wagh K., Fischer W., Theiler J., Yoon H., Li D., Haynes B.F., Sanders K.O., et al. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe. 2021;29:529–539.e3. doi: 10.1016/j.chom.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rees-Spear C., Muir L., Griffith S.A., Heaney J., Aldon Y., Snitselaar J.L., Thomas P., Graham C., Seow J., Lee N., et al. The effect of spike mutations on SARS-CoV-2 neutralization. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCallum M., De Marco A., Lempp F.A., Tortorici M.A., Pinto D., Walls A.C., Beltramello M., Chen A., Liu Z., Zatta F., et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347.e16. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walls A.C., Sprouse K.R., Bowen J.E., Joshi A., Franko N., Navarro M.J., Stewart C., Cameroni E., McCallum M., Goecker E.A., et al. SARS-CoV-2 breakthrough infections elicit potent, broad, and durable neutralizing antibody responses. Cell. 2022;185:872–880.e3. doi: 10.1016/j.cell.2022.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowen J.E., Addetia A., Dang H.V., Stewart C., Brown J.T., Sharkey W.K., Sprouse K.R., Walls A.C., Mazzitelli I.G., Logue J.K., et al. Omicron spike function and neutralizing activity elicited by a comprehensive panel of vaccines. Science. 2022;377:890–894. doi: 10.1126/science.abq0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Q., Guo Y., Iketani S., Nair M.S., Li Z., Mohri H., Wang M., Yu J., Bowen A.D., Chang J.Y., et al. Antibody Evasion by SARS-CoV-2 Omicron Subvariants BA.2.12.1, BA.4, & BA.5. Nature. 2022;608:603–608. doi: 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., di Iulio J., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng B., Abdullahi A., Ferreira I.A.T.M., Goonawardane N., Saito A., Kimura I., Yamasoba D., Gerber P.P., Fatihi S., Rathore S., et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature. 2022;603:706–714. doi: 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCallum M., Czudnochowski N., Rosen L.E., Zepeda S.K., Bowen J.E., Walls A.C., Hauser K., Joshi A., Stewart C., Dillen J.R., et al. Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Science. 2022;375:864–868. doi: 10.1126/science.abn8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Röltgen K., Powell A.E., Wirz O.F., Stevens B.A., Hogan C.A., Najeeb J., Hunter M., Wang H., Sahoo M.K., Huang C., et al. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci. Immunol. 2020;5:eabe0240. doi: 10.1126/sciimmunol.abe0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T.Y., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodda L.B., Netland J., Shehata L., Pruner K.B., Morawski P.A., Thouvenel C.D., Takehara K.K., Eggenberger J., Hemann E.A., Waterman H.R., et al. Functional SARS-CoV-2-specific immune memory persists after mild COVID-19. Cell. 2021;184:169–183.e17. doi: 10.1016/j.cell.2020.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sokal A., Chappert P., Barba-Spaeth G., Roeser A., Fourati S., Azzaoui I., Vandenberghe A., Fernandez I., Meola A., Bouvier-Alias M., et al. Maturation and persistence of the anti-SARS-CoV-2 memory B cell response. Cell. 2021;184:1201–1213.e14. doi: 10.1016/j.cell.2021.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371:eabf4063. doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goel R.R., Apostolidis S.A., Painter M.M., Mathew D., Pattekar A., Kuthuru O., Gouma S., Hicks P., Meng W., Rosenfeld A.M., et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naive and recovered individuals following mRNA vaccination. Sci. Immunol. 2021;6:eabi6950. doi: 10.1126/sciimmunol.abi6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goel R.R., Painter M.M., Apostolidis S.A., Mathew D., Meng W., Rosenfeld A.M., Lundgreen K.A., Reynaldi A., Khoury D.S., Pattekar A., et al. mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science. 2021;374:abm0829. doi: 10.1126/science.abm0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinna D., Corti D., Jarrossay D., Sallusto F., Lanzavecchia A. Clonal dissection of the human memory B-cell repertoire following infection and vaccination. Eur. J. Immunol. 2009;39:1260–1270. doi: 10.1002/eji.200839129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Achiron A., Gurevich M., Falb R., Dreyer-Alster S., Sonis P., Mandel M. SARS-COV-2 antibody dynamics and B-cell memory response over-time in COVID-19 convalescent subjects. Clin. Microbiol. Infect. 2021;27:1349.e1–1349.e6. doi: 10.1016/j.cmi.2021.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 32.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vogel A.B., Kanevsky I., Che Y., Swanson K.A., Muik A., Vormehr M., Kranz L.M., Walzer K.C., Hein S., Güler A., et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature. 2021;592:283–289. doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- 34.Bowen J.E., Park Y.J., Stewart C., Brown J.T., Sharkey W.K., Walls A.C., Joshi A., Sprouse K.R., McCallum M., Tortorici M.A., et al. SARS-CoV-2 spike conformation determines plasma neutralizing activity elicited by a wide panel of human vaccines. Sci. Immunol. 2022:eadf1421. doi: 10.1126/sciimmunol.adf1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaku C.I., Bergeron A.J., Ahlm C., Normark J., Sakharkar M., Forsell M.N.E., Walker L.M. Recall of preexisting cross-reactive B cell memory after Omicron BA.1 breakthrough infection. Sci. Immunol. 2022;7:eabq3511. doi: 10.1126/sciimmunol.abq3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. USA. 2020;117:9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park Y.J., Pinto D., Walls A.C., Liu Z., De Marco A., Benigni F., Zatta F., Silacci-Fregni C., Bassi J., Sprouse K.R., et al. Imprinted antibody responses against SARS-CoV-2 Omicron sublineages. Science. 2022;378:619–627. doi: 10.1126/science.adc9127. [DOI] [PubMed] [Google Scholar]
- 38.Goel R.R., Painter M.M., Lundgreen K.A., Apostolidis S.A., Baxter A.E., Giles J.R., Mathew D., Pattekar A., Reynaldi A., Khoury D.S., et al. Efficient recall of Omicron-reactive B cell memory after a third dose of SARS-CoV-2 mRNA vaccine. Cell. 2022;185:1875–1887.e8. doi: 10.1016/j.cell.2022.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muecksch F., Wang Z., Cho A., Gaebler C., Ben Tanfous T., DaSilva J., Bednarski E., Ramos V., Zong S., Johnson B., et al. Increased memory B cell potency and breadth after a SARS-CoV-2 mRNA boost. Nature. 2022;607:128–134. doi: 10.1038/s41586-022-04778-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y., Chen L., Yin S., Tao Y., Zhu L., Tong X., Mao M., Li M., Wan Y., Ni J., et al. The Third dose of CoronVac vaccination induces broad and potent adaptive immune responses that recognize SARS-CoV-2 Delta and Omicron variants. Emerg. Microb. Infect. 2022;11:1524–1536. doi: 10.1080/22221751.2022.2081614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z., Muecksch F., Schaefer-Babajew D., Finkin S., Viant C., Gaebler C., Hoffmann H.H., Barnes C.O., Cipolla M., Ramos V., et al. Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature. 2021;595:426–431. doi: 10.1038/s41586-021-03696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crotty S. Hybrid immunity. Science. 2021;372:1392–1393. doi: 10.1126/science.abj2258. [DOI] [Google Scholar]
- 43.Rodda L.B., Morawski P.A., Pruner K.B., Fahning M.L., Howard C.A., Franko N., Logue J., Eggenberger J., Stokes C., Golez I., et al. Imprinted SARS-CoV-2-specific memory lymphocytes define hybrid immunity. Cell. 2022;185:1588–1601.e14. doi: 10.1016/j.cell.2022.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sokal A., Barba-Spaeth G., Fernández I., Broketa M., Azzaoui I., de La Selle A., Vandenberghe A., Fourati S., Roeser A., Meola A., et al. mRNA vaccination of naive and COVID-19-recovered individuals elicits potent memory B cells that recognize SARS-CoV-2 variants. Immunity. 2021;54:2893–2907.e5. doi: 10.1016/j.immuni.2021.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stamatatos L., Czartoski J., Wan Y.H., Homad L.J., Rubin V., Glantz H., Neradilek M., Seydoux E., Jennewein M.F., MacCamy A.J., et al. mRNA vaccination boosts cross-variant neutralizing antibodies elicited by SARS-CoV-2 infection. Science. 2021;372:1413–1418. doi: 10.1126/science.abg9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W., et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Starr T.N., Czudnochowski N., Liu Z., Zatta F., Park Y.J., Addetia A., Pinto D., Beltramello M., Hernandez P., Greaney A.J., et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature. 2021;597:97–102. doi: 10.1038/s41586-021-03807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walls A.C., Xiong X., Park Y.J., Tortorici M.A., Snijder J., Quispe J., Cameroni E., Gopal R., Dai M., Lanzavecchia A., et al. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176:1026–1039.e15. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pinto D., Sauer M.M., Czudnochowski N., Low J.S., Tortorici M.A., Housley M.P., Noack J., Walls A.C., Bowen J.E., Guarino B., et al. Broad betacoronavirus neutralization by a stem helix-specific human antibody. Science. 2021;373:1109–1116. doi: 10.1126/science.abj3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lempp F.A., Soriaga L.B., Montiel-Ruiz M., Benigni F., Noack J., Park Y.J., Bianchi S., Walls A.C., Bowen J.E., Zhou J., et al. Lectins enhance SARS-CoV-2 infection and influence neutralizing antibodies. Nature. 2021;598:342–347. doi: 10.1038/s41586-021-03925-1. [DOI] [PubMed] [Google Scholar]
- 51.Odersky M., Altherr P., Cremet V., Emir B., Maneth S., Micheloud S., Mihaylov N., Schinz M., Stenman E., Zenger M. Ecole Polytéchnique Fédérale de Lausanne; 2004. An Overview Of The Scala Programming Language. IC/2004/64. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and code generated in this study will be made available on request and may require a material transfer agreement.