Abstract
Neoantigens derived from mutant proteins in tumour cells could elicit potent personalized anti‐tumour immunity. Nevertheless, the layout of vaccine vehicle and synthesis of neoantigen are pivotal for stimulating robust response. The power of synthetic biology enables genetic programming bacteria to produce therapeutic agents under contol of the gene circuits. Herein, we genetically engineered bacteria to synthesize fusion neoantigens, and prepared bacteria derived vesicles (BDVs) presenting the neoantigens (BDVs‐Neo) as personalized therapeutic vaccine to drive systemic antitumour response. BDVs‐Neo and granulocyte‐macrophage colony‐stimulating factor (GM‐CSF) were inoculated subcutaneously within hydrogel (Gel), whereas sustaining release of BDVs‐Lipopolysaccharide (LPS) and GM‐CSF recruited the dendritic cells (DCs). Virtually, Gel‐BDVs‐Neo combined with the programmed cell death protein 1 (PD‐1) antibody intensively enhanced proliferation and activation of tumour‐infiltrated T cells, as well as memory T cell clone expansion. Consequently, BDVs‐Neo combining with checkpoint blockade therapy effectively prevented tumour relapse and metastasis.
Keywords: bacteria derived vesicles (BDVs), cancer vaccines, checkpoint blockade, immunotherapy, neoantigen
1. Introduction
Cancer vaccines are expected to exert the protective or therapeutic efficacy as that of pathogenic microorganisms (Melero et al., 2014; Palena et al., 2006; Sahin & Tureci, 2018). However, the dearth of potent anti‐tumour response of the tumour‐associated antigens (TAAs), derived from self‐antigens that are abnormally expressed in tumour cells, leads to the termination of most TAAs‐vaccines‐associated clinical trials in the past decades (Blass & Ott, 2021; Hollingsworth & Jansen, 2019; Kreiter et al., 2015; Saxena et al., 2021; Schumacher & Schreiber, 2015). Virtually, the antitumour immune responses from TAAs are basically unsatisfactory in the clinical, which is probably ascribed to the fact that TAA‐specific T cells suffer central and/or peripheral tolerance (Blass & Ott, 2021; Hollingsworth & Jansen, 2019). Recently, the mutant antigens, referred as neoantigens, have been identified from individual tumour tissue by exome sequencing (Kreiter et al., 2015). Neoantigen is an abnormal protein exclusively produced by cancer cells through ‘non‐synonymous mutation’ (Kreiter et al., 2015; Yang et al., 2019). Hence, T cells enable eradicate cancer cells presenting neoantigens, which could drive potent anti‐tumour response (Hu et al., 2021; Keskin et al., 2019; Schumacher & Schreiber, 2015; Yang et al., 2019). Neoantigen‐based vaccines have been shown to specifically induce tumour antigen‐specific CD4+ and CD8+ T cell responses, which could improve patient survival combining with immune checkpoint inhibition (ICI) therapy (Saxena et al., 2021). Intriguingly, neoantigen‐specific T cells can avoid ‘off‐target’ damage to normal tissues, since neoantigens circumvent T cell central tolerance to self‐epitopes (Blass & Ott, 2021). Currently, neoantigen vaccines developed with poly‐neo‐epitope RNA or peptide sequestered in liposome enable boost tumour‐specific immune responses (Kreiter et al., 2015; Sahin et al., 2020). However, spontaneous immune recognition of neo‐epitope is inefficient if no proper immunologic adjuvant is utilized. Hence, it is critical to devise an adjuvant vehicle for the unmet demand of neoantigen vaccines.
Lipid‐based nanoparticles (NPs) such as liposomes, exosomes, tumour cell membrane vesicles and bacterial outer membrane vesicles (OMVs), have been intensively explored as cancer antigen vehicles to elicit systemic antitumour immunity (Cheng et al., 2021; Valentine et al., 2016; Ye et al., 2019). Intriguingly, OMVs released from Gram‐negative bacteria contain foreign coymponents and thus could potentiate immune response (Carvalho et al., 2019; Kuehn & Kesty, 2005; Lee et al., 2008; Rosenthal et al., 2014; Zanella et al., 2021). As a major component of OMVs, lipopolysaccharide (LPS) is a potent activator of immune cells, such as monocytes/macrophages (Kulp & Kuehn, 2010; Sartorio et al., 2021). However, this inflammatory activation can also result in high vaccine reactogenicity. Exception of LPS, OMVs also contain pathogen‐associated antigens and lipoproteins, which could modulate and activate innate immunity by the Toll‐like receptor 2 (TLR2) signal pathway (Bachmann & Jennings, 2010; Kovacs‐Simon et al., 2011; Mancini et al., 2020). Based on the characteristics of OMVs, it has inspired researchers to explore their application in tumour immunity (Kim et al., 2017; Huang et al., 2020; Li, Zhou et al., 2020; Li, Zhao et al., 2020; Peng et al., 2020). Recently, in order to enhance the immunization efficacy of the antigens, amounts of studies have focused on integrating antigens into OMVs. For example, Salmonella OMVs which presenting an antigen of Mycobacterium tuberculosis named Ag85B were constructed by Luirink et al., the study resulting in the the engineered OMVs could activate the bone marrow‐derived dendritic cells (BMDCs), and the antigen are subsequently presented to the navie T cells (Daleke‐Schermerhorn et al., 2014). E. coli OMVs displaying Omp22 could protect mice from Acinetobacter baumannii‐induced lethal challenge and trigger a robust immune response after immunization (Huang et al., 2016). Moreover, OMVs carrying Cytolysin A fusion proteins have been investigated for their potential as anti‐tumour agents (Murase, 2022). In addition, the design of hybrid biomimetic nanovesicles has been explored by fusion of eukaryotic and prokaryotic cell derived membrane vesicles (Chen, Huang et al., 2020; Park et al., 2021). Alternatively, OMVs have been developed as a novel type of drug delivery vehicles for enhanced enhancing cancer immunotherapy (Chen, Bai et al., 2020; Gujrati et al., 2014; Huang et al., 2020; Patel et al., 2019).
Synthetic biology, a promising discipline, aims to harness living cells including bacteria to produce chemicals, materials and biological therapeutic agents under the control of gene circuits (Chowdhury et al., 2019; Cubillos‐Ruiz et al., 2021). Currently, synthetic gene circuits are devised to program bacteria to generate therapeutic agents such as antibodies, immunologic factors, and even L‐arginine in tumour microenvironment to eradicate cancer (Canale et al., 2021; Gurbatri et al., 2020; McNerney et al., 2021). The exploitation of natural or bioengineered bacteria membrane derived vesicle as therapeutic bacteria vaccines has shown considerable prospects. Herein, we consulted the nucleotide sequence of mutations identified by exome sequencing (Kreiter et al., 2015) and utilized the non‐pathogenic Escherichia coli (E. coli) BL21(DE3)plysS strain to engineer bacteria derived vesicles (BDVs) displaying the neoantigen peptide. BDVs‐Neo and granulocyte‐macrophage colony‐stimulating factor (GM‐CSF) were loaded into a thermosensitive hydrogel as antigen delivery vehicle, then was accompanied with PD‐1 antibody to improve the anticancer efficacy and long‐term memory immunity (Scheme 1). PD‐1 antibody protected CD8+ T cells from exhaustion by blockade of PD‐1/PD‐L1 axis, that facilitated cytotoxic T cells (CTLs) to recognize and eliminate the surgical residual tumour cells. Collectively, the unique features of BDVs contribute to boosting ICI therapy and reactivating the T cell antitumour immunity (Chen & Han, 2015; Gong et al., 2018; Wei et al., 2021; Zou et al., 2016).
SCHEME 1.
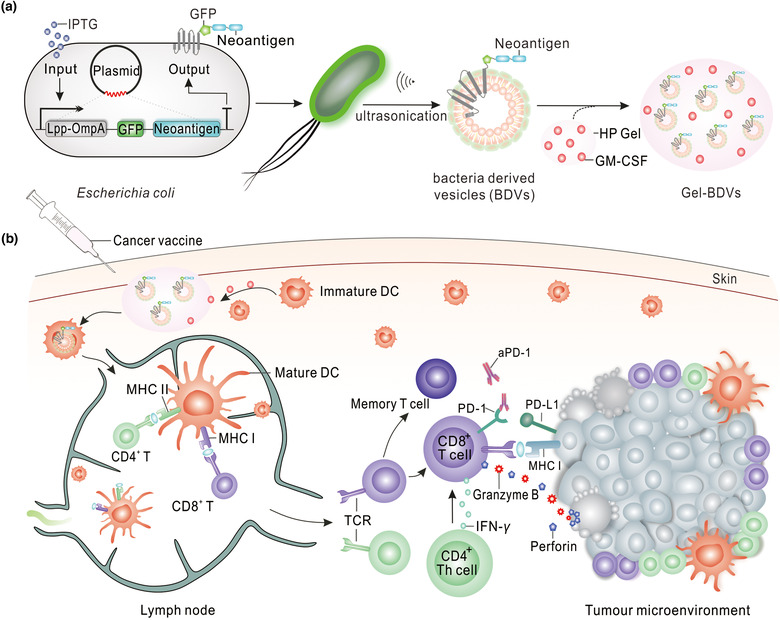
Schematic of preparation of Gel‐GFP‐Mutation‐M33‐M47 BDVs combined with PD‐1 antibody for immunotherapy of melanoma. (a) Preparation process of Gel‐BDVs. (b) Effector T cell were actived by Gel‐GFP‐Mutation‐M33‐M47 BDVs to against melanoma
2. MATERIALS AND METHODS
2.1. Plasmid construction
The nucleotide sequences of neoantigens were based on the next‐generation sequencing (Kreiter et al., 2015). DNA fragments encoding for the Lpp‐OmpA‐Green Fluorescent Proteins (GFP)‐M33‐M47 (neoantigen) protein were synthesized commercially by Yingmao, Inc. (Chongqing, China). Using gene recombinant techniques, wild type epitopes (M33‐Valine; M47‐Alanine) and the mutated epitopes (M33‐Asparticacid; M47‐Glycine) gene segments were linked into pET21a vector between NdeI and Xhol I sites, respectively (Table S1). The recombinant plasmids were transformed into competent E. coli BL21(DE3)plysS cells (TransGen Biotech).
2.2. Expression of GFP‐neoantigen and the preparation of BDVs
The recombinant cells were grown in LB medium until the OD600 value reached 0.6, and then the expression of GFP‐neoantigen was induced by addition of 1 mM isopropyl β‐d‐1‐thiogalactopyranoside (IPTG) at 16°C for 6 h. After induction, recombinant cells stably expressing GFP‐neoantigen were collected by centrifugation at 5000 × g for 10 min. The cells were resuspended in cold phosphate‐buffered saline (PBS) and washed three times. Then we suspended the cells with cold HM lysis buffer (0.25 M sucrose, 1 mM Ethylene Diamine Tetraacetic Acid (EDTA), 20 mM HEPES‐NaOH, pH7.4, and 1х protease inhibitor cocktail (Beyotime, 100х)). Afterwards, the mixed solution was transferred to the ultrasonic cell disruptor in ice bath for ultrasonic decomposition (power: 30%, time: 2 min). The entire solution was centrifuged at 1000 × g for 5 min in order to eliminate cells. Next, the supernatant was centrifuged at 3000 × g for 5 min. Carefully aspirate the supernatant to a new tube and then centrifuged at 15,000 × g for 0.5 h to collected the sediment. The sediment was resuspended in cold HM buffer, passing through 0.8 μm and 0.22 μm filters at least 10 times in turn to get the BDVs. The BCA protein assay kit (Beyotime) was used to quantitatively calculate the total protein concentration of BDVs. At last, The BDVs were stored in HM buffer at −80°C. Fluorescence images were taken on the confocal microscope (Zeiss) to verify the expression of GFP. The preparation of thermosensitive HP hydrogel (Hyaluronic Acid, 2 wt%, Macklin; Pluronic F‐127, 25 wt%, Sigma Aldrich) was performed using a cold method as described by Ye et al (Ye et al., 2019). In brief, HA and Pluronic F‐127 were mixed in cold distilled water and then stored in 4°C refrigerator until the polymer had completely dissolved. Murine GM‐CSF (20 ng/ml, PeproTech) and BDVs (25 μg) were then added in the 150 μl HP hydrogel, and the mixture was stored at 4°C before use. In 37°C incubator, thermosensitive HP hydrogel could rapidly form the hydrogel and act as an embedding carrier for long‐term drug release.
2.3. Separation of outer and inner membrane
Two liters of recombinant cells were harvested by centrifugation at 4°C. The cells were washed with 10 mM Tris‐(hydroxymethyl)‐aminomethane‐hydrochloride (Tris‐HCL buffer, pH 8.0) and resuspended in 20 ml of 20% (wt/vol) sucrose (50 μg/ml Deoxyribonuclease I). Add 2 ml of 1.5 mg/ml lysozyme (final concentration of 150 μg/ml) and 200 μl 100х protease inhibitor cocktail to the resuspension. Afterwards, the mixed solution was transferred to the ultrasonic cell disruptor in ice bath for ultrasonic decomposition (power: 30%, time: 2 min). Cell debris was then removed by centrifugation at 2,000 x g for 10 min, and the supernatant was decanted to a new centrifuge tube. Eight milliliters were then layered onto a sucrose step gradient containing 8 ml of 70% (wt/vol) sucrose and 10 ml of 50% (wt/vol) sucrose in Tris‐HCL buffer, and then the tubes were centrifuged at 184,500 × g in a Beckman Type 70i (70 Ti) rotor for 18 h. The two bands were observed and then remove the upper‐brown inner membrane (IM) layer using the pipette by suction from above. Repeat the pipetting procedure for the outer membrane (OM) fraction. The individual samples were diluted in distilled water and collected by centrifuged at 184,500 × g in a Beckman 70 Ti rotor. This procedure is a modification of the method described by Hancock (Hancock & Nikaido, 1978) and Cian (Cian et al., 2020).
2.4. Morphology and characterization
The recombinant E. coli cells were washed with PBS and fixed with 4% paraformaldehyde for 15 min. Then cells were stained with 4′,6‐diamidino‐2‐phenylindole (DAPI, Beyotime) for 15 min at room temperature and washed with PBS buffer. Fluorescence of GFP and DAPI were observed by confocal microscope (Zeiss) with 40× objective. The morphology of E. coli cells and BDVs, negatively stained with 1% uranyl acetate, were observed using transmission electron microscopy (TEM; Shiyanjia Lab (www.shiyanjia.com) for the TEM image). In addition, the size distribution of BDVs were measured by the dynamic light scattering (DLS, Nano‐ZS, model MPT2;). The zeta potential of the BDVs were measured by Malvern Zetasizer (Malvern Instruments).
2.5. Western blot
The recombinant protein of E. coli cells, BDVs, OM and IM were boiled with 1x loading buffer and separated on a 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS‐PAGE) gel. Then the protein was transferred onto the polyvinylidene fluoride (PVDF) membrane, which was blocked with 5% skimmed milk (tris buffer solution tween (TBST, Tween 20, 0.1%) buffer) for 2 h at room temperature. The membrane was hybridized with EGFP (Abmart) or ompF (Thermo Fisher PA5‐121442) antibodies (1:1000) overnight at 4°C. Then the membrane was washed three times for about 15 min in TBST buffer and incubated with secondary antibody for 1 h at room temperature. After three washes for 5 min each with TBST, the imaging was performed using enhanced chemiluminescence detection (ECL, FDbio‐Pico).
2.6. In vitro BMDCs isolation and uptake
BMDCs were isolated and collected from the bone marrow which was flushed from the tibias and femurs of female C57BL/6 mice (6‐week‐old). The harvested cells were resuspended in red blood cell (RBC) Lysis Buffer (Sigma‐Aldrich) for 10 min to eliminate RBCs. The remaining cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 supplemented with 10% fetal bovine serum (FBS), 1% penicillin‐streptomycin, 10 ng/ml interleukin 4 (IL‐4, PeproTech), and 20 ng/ml GM‐CSF in 6‐well plates at a concentration of 1 × 106 cells. All of the floating cells were removed and replaced with fresh medium after 24 h. The nonadherent cells under the plate were recognized as bone marrow‐derived dendritic cells. Engineered BDVs (25 μg) presenting GFP were added into the plate to verify the phagocytosis by BMDCs. The cells were incubated with BDVs for 24 h, subsequently stained with wheat germ agglutinin Alexa Fluor 647 conjugate (WGA AF647, Invitrogen, 1833658) and DAPI and analyzed by confocal microscope (Zeiss, LSN880).
To study the activation of DCs, the cells were divided into four groups: PBS (G1), blank‐BDVs (G2), Normal‐M33‐M47 BDVs (G3), Mutation‐M33‐M47 BDVs (G4). The BMDCs (1 × 106 cells/ml) were incubated with the different formulations (with same mass of BDVs, 25 μg) for 12 h and then collected, washed three times with PBS. The cells were divided equally into three parts and stained with anti‐CD11c‐Allophycocyanin (APC), anti‐CD40‐Phycoerythrin (PE), anti‐MHC II‐PE, anti‐CD80‐fluorescein isothiocyanate (FITC) and anti‐CD86‐PE antibodies (Biolegend) according to the manufacturer's protocols. The stained cells were sorted using Beckman CytoFlex flow cytometer and analyzed with FlowJo software.
2.7. In vivo activation
Six‐week‐old female mice were randomly separated into five groups (n = 5) and subjected to different treatments: Gel (150 μl, #1), Gel‐GM‐CSF (150 μl, 20 ng/ml GM‐CSF, #2), Gel‐blank BDVs + GM‐CSF (150 μl, 25 μg BDVs and 20 ng/ml GM‐CSF, #3), Gel‐Normal‐M33‐M47 BDVs + GM‐CSF (150 μl, 25 μg BDVs, 20 ng/ml GM‐CSF, #4), Gel‐Mutation‐M33‐M47 BDVs + GM‐CSF (150 μl, 25 μg BDVs and 20 ng/ml GM‐CSF, #5). Different types of Gel‐formulas were injected on the left side of the back via subcutaneous injection. To assess the recruitment and maturation of dendritic cells, the skin of the injection area and inguinal lymph nodes were respectively harvested after treatment for 72 h. The skin of mice was harvested and snap frozen in optimal cutting temperature (OCT) compound. The immunofluorescent assay of skin was carried out according to the description below.
On the other hand, the inguinal lymph nodes were collected from each group, and monocellular suspensions were prepared to assess the maturation of dendritic cell. Cells were stained with anti‐CD11c‐APC, anti‐CD40‐PE, anti‐CD80‐FITC and anti‐CD86‐PE antibodies (Biolegend). All samples were sorted using Beckman CytoFlex flow cytometer and analyzed with FlowJo software.
2.8. In vivo tumour model
Animal experiments were performed under an approved protocol by the Administrative Committee of Animal Research in the Sun Yat‐Sen University (SYSU‐IACUC‐2021‐000874). In order to study the anti‐tumour therapeutic efficacy of BDVs in vivo, 6‐week‐old female C57BL/6 mice in SPF grade were purchased from Guangdong Medical Laboratory Animal Center (Guangdong, China) and kept in SPF level Laboratory Animal Center of Sun Yat‐sen University. Mice were cared and performed according to the instructions and approval of the Institutional Animal Care and Use Committee of Sun Yat‐sen University. Mice were challenged with 1 × 106 B16F10‐luc cells on the right side of the back on day 0. After 7 days, approximately 1 mm3 in volume of the tumour was remained to simulate the presence of the residual microtumour in the clinical operation. The wound was stitched up skillfully with three surgical suture nails. Metastasis mouse model were established by intravenous injection of 5 × 105 B16F10‐luc via the tail vein. The mice were separated randomly into six groups (n = 7 or 10): Gel‐PBS (150 μl, 20 ng/ml GM‐CSF, #1), Gel‐blank BDVs (150 μl, 25 μg BDVs and 20 ng/ml GM‐CSF, #2), Gel‐Normal‐M33‐M47 BDVs (150 μl, 25 μg BDVs, 20 ng/ml GM‐CSF, #3), Gel‐Mutation‐M33‐M47 BDVs (150 μl, 25 μg BDVs and 20 ng/ml GM‐CSF, #4), aPD‐1 (anti‐PD‐1 antibody, GoInVivo Purified anti‐mouse CD279 (PD‐1), Biolegend, 114115, 2.5 mg/kg, #5) and Gel‐Mutation‐M33‐M47 BDVs + aPD‐1 (150 μl, 25 μg BDVs and 20 ng/ml GM‐CSF, 2.5 mg/kg, #6). Gel‐formulations were injected on the left side of the back via subcutaneous injection, and aPD‐1 was injected via the tail‐vein. Mice were anesthetized with isoflurane and bioluminescence images of melanomas were captured by IVIS fluorescence imaging system based on XenoLight D‐Luciferin‐K+ Salt Bioluminescent Substrate. The mice weight and tumour size were measured every other day. The tumour size was measured by digital caliper, and the tumour volume (mm3) was calculated as follows: width2 × length /2. When the tumour volume of tumour‐bearing mouse exceeds 1500 mm3, the suffering mice should be humanely euthanized and the tumours and organs were collected. The numbers of lung metastatic nodules in different groups were counted carefully. The organs (heart, liver, spleen, kidney and lung) of surgical mice and lung tissue of lung tumours mice were stained with hematoxylin‐eosin staining (H&E).
2.9. Tissue immunofluorescent assay
The analysis of tissue immunofluorescent was performed following the instructions. First of all, skin and tumours were collected and frozen in optimal cutting temperature (OCT) compound (SAKURA). The proper thickness section (8 μm) was obtained through rotary freezing microtome. Secondly, the slices from different groups were washed with PBS buffer in order to clean off the embedding medium. Then 4% paraformaldehyde was used to fix the slices, whereafter, 3% bovine serum albumin (BSA, containing 0.2% Triton X‐100) was applied to block the slices. Thirdly, the slices of skin were then incubated with CD11c primary antibody (1:200 in 1.5% BSA, Abcam, ab254183) overnight at 4°C. Whereas, the slices of tumour were incubated with CD4 (1:100 in 1.5% BSA, Abcam, ab183685) and CD8 primary antibody (1:100 in 1.5% BSA, Abcam, ab217344) under the same conditions. After washing 3 times with PBS buffer, the secondary antibodies (1:500 in 1.5% BSA, ThermoFisher) were incubated with skin or tumour specimens for about 2 h in the darkness. Finally, DAPI staining was used to locate the position of cell nucleus and ensure the depth of shooting field. Fluorescence images were observed on the confocal microscope (Zeiss, LSN880).
2.10. Statistical analysis
The data values were presented as the mean ± s.d. or the mean ± s.e.m.. Comparisons between the experimental groups were performed using the one‐way analysis of variance (ANOVA) followed by the Tukey post‐hoc tests. Comparisons of survival curves were performed using the Long‐Rank (Mantel‐Cox) test. All statistical analyses were performed using GraphPad Prism 8 software (*P < 0.05, **P < 0.01, and ***P < 0.001). Unless otherwise stated, all experiments were performed in biological replicates.
3. RESULTS
3.1. Preparation and characterization of Mutation‐M33‐M47 BDVs
OMVs are naturally released from the outer membrane of Gram‐negative bacteria (Carvalho et al., 2019; Kuehn & Kesty, 2005; Lee et al., 2008; Rosenthal et al., 2014; Zanella et al., 2021). To synthesize fusion neoantigens and display the neoantigens on the outer membrane of the bacteria, we subcloned pET‐21a vector containing two neoantigen nucleotide sequences of M33 and M47 that had been identified from B16F10 cells (Kreiter et al., 2015). The Lpp‐OmpA‐based display system behind the IPTG promoter were employed to make the fusion proteins localize on the outer membrane, while GFP was fused on the N‐terminal of the neoantigen as tracking tag (Scheme 1). Then the plasmid was transformed into competent BL21(DE3)plysS cells. The transfected BL21(DE3)plysS cells were then induced by IPTG for 16 h, and were observed by confocal microscope. It showed that the Lpp‐OmpA‐GFP‐M33‐M47 fusion protein was successfully expressed and localized on the outer membrane in engineered E. coli (Figure 1a, b). The morphology of GFP‐Mutation‐M33‐M47 E. coli was characterized by TEM (Figure 1c). The BDVs were prepared by ultrasonication and hypothermal differential centrifugation and then were filtered through 0.8 μm and 0.22 μm pore‐sized hydrophilic membrane. Hereafter, we characterized the morphology of the negatively stained GFP‐Mutation‐M33‐M47 E. coli BDVs by TEM and confirmed that the BDVs akin to OMVs (Figure 1d). We further validated that GFP‐Mutation‐M33‐M47 fusion protein had been presented on BDVs by the green fluorescence signals of GFP (Figure 1e). In addition, the size distribution of BDVs were measured by the dynamic light scattering (DLS), which indicated that the average diameter of BDVs was ∼120 nm (Figure 1f). The zeta potential of different formulation of BDVs was around −12 mV using Malvern Zetasizer Nano ZS ZETA (Figure 1g). The characterizations of BDVs were same as the cell membrane vesicles and natural OMVs (Li et al., 2021; Park et al., 2021; Xue et al., 2022). And on top of that, we also successfully extracted the inner and outer membrane of E. Coli (Figure S1). Analysis of the OM and IM proteins by SDS‐polyacrylamide gel electrophoresis showed a little bit different (Figure S2). Ultimately, western blot analysis showed that the Lpp‐OmpA‐GFP‐M33‐M47 fusion protein and outer membrane protein F (ompF) were efficiently expressed in E. coli BL21(DE3)plysS cells, BDVs and OM, respectively (Figure 1h, i).
FIGURE 1.
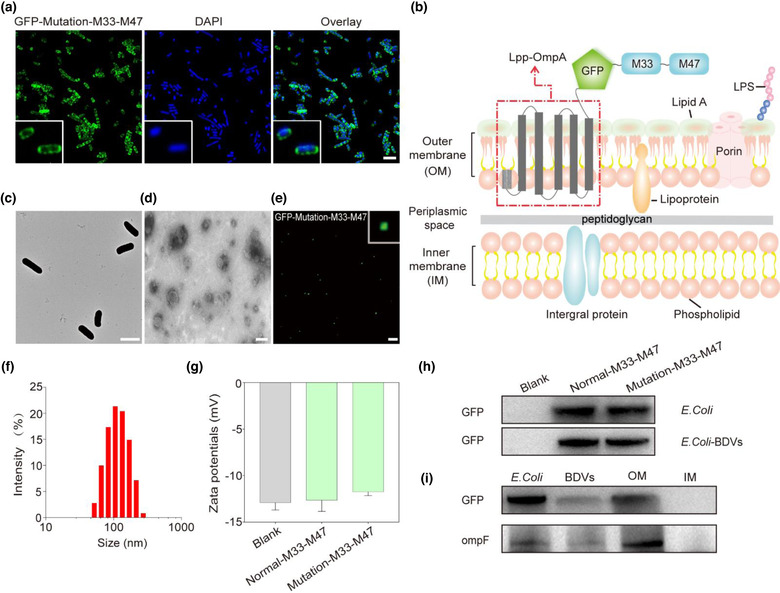
Characterizations of GFP‐Mutation‐M33‐M47 E. coli BDVs. (a) Establishment of E. coli stably expressing GFP (Scale bar: 2 μm). (b) Schematic diagram illustration the presentation of neoantigens using Lpp‐OmpA system on E. coli outer membrane. (c) The TEM images shown the size and morphology of GFP‐Mutation‐M33‐M47 E. coli (Scale bar: 2 μm). (d) The TEM images shown the size and morphology of GFP‐Mutation‐M33‐M47 E. coli BDVs (Scale bar: 100 nm). (e) The confocal image of GFP‐Mutation‐M33‐M47 E. coli BDVs (Scale bar: 5 μm). (f) The size distribution of GFP‐Mutation‐M33‐M47 E. coli BDVs measured by dynamic light scattering (DLS). (g) The zeta potential of the E. coli BDVs. Blank (BL21(DE3)plysS): non‐heat shock transformation control, worked as a negative control. (h) Western blot analysis indicated the expression of GFP on E. coli and BDVs. (i) Western blot analysis indicated the expression of GFP and ompF on E. coli, BDVs, OM and IM
3.2. In vitro and in vivo immune response of mutation‐M33‐M47 BDVs
Dendritic cells (DCs) enable internalizing tumour antigen and priming T cells to generate tumour antigen‐specific T cells (Banchereau & Palucka, 2005). As the well‐equipped professional antigen‐presenting cells, DCs play crucial roles in the adaptive immune response. BMDCs from female C57BL/6 mice were isolated to evaluate the endocytosis of the BDVs by antigen‐presenting cells. To test whether the BDVs could be internalized by BMDCs in vitro, GFP‐Mutation‐M33‐M47 BDVs were incubated with BMDCs at 37°C for 24 h. As shown in Figure 2a and Figure S3, BMDCs could efficiently internalize BDVs. Furthermore, the expression of costimulatory markers CD11c, CD40, MHC‐II, CD80 and CD86 on BMDCs were employed to assess whether the BDVs could promote the maturation of BMDCs by flow cytometry. The cells received different treatments were collected after co‐incubation for 24 h. Compared with PBS (G1), the cells received blank BDVs (G2), Normal‐M33‐M47 BDVs (G3), or Mutation‐M33‐M47 BDVs (G4) all showed a higher proportion of mature DCs, demonstrating that BDVs could effectively promote the maturation of DCs (Figure 2b, c and Figures S4, S5).
FIGURE 2.
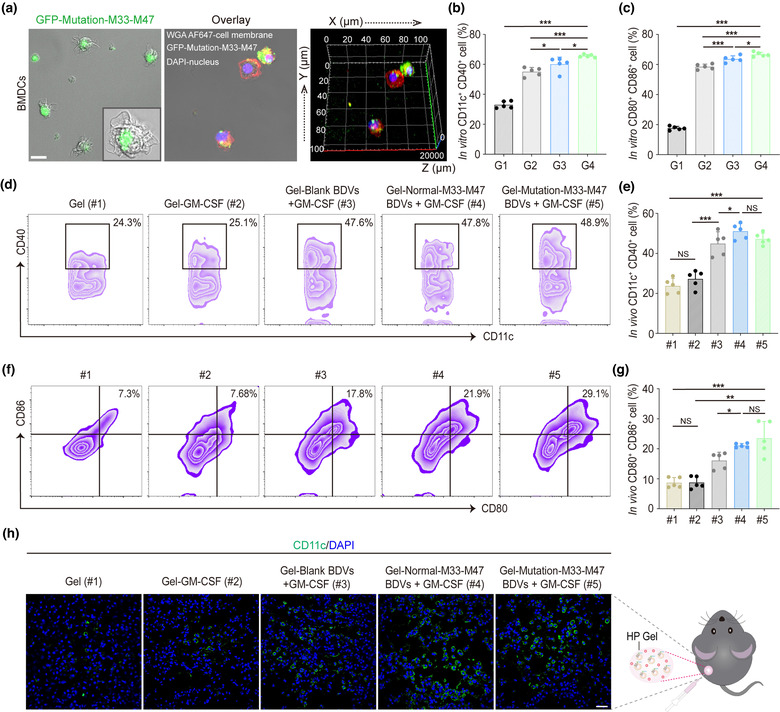
Dendritic cell activity and maturation in vitro and in vivo. (a) Confocal 3D image of BMDCs uptake of GFP‐Mutation‐M33‐M47 E. coli BDVs in vitro (Scale bar: 10 μm). Green: BDVs. Red: cell membrane. Blue: cell nucleus. (b, c) Quantitative analysis of CD11c+ CD40+ cells (b) and mature DCs (c) from different treatment groups in vitro (gated on CD11c+ cells, n = 5). Error bar, mean ± s.d.. PBS (G1), Blank BDVs (G2), Normal‐M33‐M47 BDVs (G3), Mutation‐M33‐M47 BDVs (G4). (d, e) Representative flow cytometry data (d) and statistical data (e) to show CD11c+ CD40+ cells induced by different formulations of BDVs in vivo (in lymph nodes) on day 3 post‐ injection (gated on CD11c+ cells, n = 5). Cells were stained with anti‐CD11c‐APC, anti‐CD40‐PE antibodies (Biolegend). (f, g) Representative flow cytometry data (f) and statistical data (g) to show DC maturation induced by different formulations of BDVs in vivo (in lymph nodes) on day 3 post‐injection (gated on CD11c+ cells, n = 5). Cells were stained with anti‐CD11c‐APC, anti‐CD80‐FITC, anti‐CD86‐PE antibodies (Biolegend). Error bar, mean ± s.d.. (h) Immunofluorescence analysis of CD11c+ DCs in the skin after the treatments (Scale bar: 50 μm). Gel (#1), Gel‐GM‐CSF (#2), Gel‐Blank BDVs + GM‐CSF (#3), Gel‐Normal‐M33‐M47 BDVs + GM‐CSF (#4), Mutation‐M33‐M47 BDVs + GM‐CSF (#5). mean ± s.d., n = 5. NS: no significant, *P < 0.05, **P < 0.01, ***P < 0.001. One‐way ANOVA with Tukey post‐hoc tests (b, c, e, g)
In order to assess the immune stimulation capacity of BDVs vaccines, the mice were subcutaneously inoculated with Gel (#1), Gel‐GM‐CSF (#2), Gel‐blank‐BDVs + GM‐CSF (#3), Gel‐Normal‐M33‐M47 BDVs + GM‐CSF (#4), or Gel‐Mutation‐M33‐M47 BDVs + GM‐CSF (#5), respectively. After inoculation, the activation extent of the immune system was investigated with skin and inguinal lymph nodes. Flow cytometry analysis data showed that neither Gel (#1) nor Gel‐GM‐CSF (#2) could induce the maturation and activation of DCs effectively. On the contrary, BDVs could significantly induce the maturation and activation of DCs. The activation markers of DCs including CD11c+ CD40+ and CD80+ CD86+ were intensively increased in the mature DCs in lymph nodes, indicating BDVs could promote the maturation of DCs efficiently in vivo (Figure 2d–g). Moreover, the mice inoculated with Gel‐Mutation‐M33‐M47 BDVs plus GM‐CSF significantly enhanced the maturation of DCs compared to those receiving Gel‐Normal‐M33‐M47 BDVs plus GM‐CSF. These data indicated that BDVs with neoantigens could promote the antigen recognition and maturation of DCs, while GM‐CSF facilitated the recruitment and stimulation of the clone expansion of DCs. Next, to assess the recruitment of DCs, the skin of the injected area in experimental group was harvested and analyzed by immunofluorescence. Similar to the results of FACS analysis, the density of CD11c+ cells with immunofluorescence staining in skin was obviously higher in the Gel‐BDVs treatment groups than in the Gel‐PBS group (Figure 2h). Taken together, these data indicated that injection of BDVs with GM‐CSF could trigger a robust immune response both in vitro and in vivo.
3.3. In vivo therapeutic efficacy of mutation‐M33‐M47 BDVs combined with PD‐1 blockade for anti‐tumour relapse
Neoantigens were identified by exome sequencing of the individual surgical tumour tissue (Kreiter et al., 2015). Thus, the tumour vaccines leveraging neoantigen could be potent for protecting individual from tumour relapes and tumour metastasis (Blass & Ott, 2021). To evaluate whether BDVs‐neoantigen vaccine could prevent tumour relapse after the surgery, B16F10‐tumour xenograft mouse model was employed in the present experiments. Majority of the tumour (∼90%) was surgically removed on day 8, and the mice were randomly assigned into 6 different treatment groups: Gel‐PBS (#1), Gel‐blank‐BDVs (#2), Gel‐Normal‐M33‐M47 BDVs (#3), Gel‐Mutation‐M33‐M47 BDVs (#4), aPD‐1 (2.5 mg/kg, #5), Gel‐Mutation‐M33‐M47 BDVs + aPD‐1 (2.5 mg/kg, #6) (Figure 3a). Tumour growth were monitored by bioluminescence imaging and tumour volume (Figure 3b, c). It was worth noting that mice treated with Gel‐Mutation‐M33‐M47 BDVs combined aPD‐1 (#6) showed the best inhibition of tumour growth compared with the mice received other treatments. Meanwhile, mice treated with Gel‐Mutation‐M33‐M47 BDVs (#4) and aPD‐1 (#5) also exhibited better ability of antitumour response than Gel‐PBS (#1), Gel‐blank‐BDVs (#2) and Gel‐Normal‐M33‐M47 BDVs (#3), indicating that the neoanitgens and PD‐1 antibody both contributed to therapeutic effect in repressing tumour growth. According to Figure 3d–f, mice treated with Gel‐Mutation‐M33‐M47 BDVs combined aPD‐1 (#6) not only exhibited the strongest anti‐tumour relapse efficacy, but also benifited higher survival rate. In contrast, mice in other groups all died within 36 days post‐challenge (Figure 3f). Additionally, the mice had no obvious weight changes throughout the treatment (Figure S6). Meanwhile, the major organs of the mice, including heart, liver, spleen, kidney and lung were harvested and performed the H&E staining, and no abnormal phenomenon were observed, indicating that the BDVs formulations had no toxic to mice (Figure S7). These data indicated that BDVs‐neoantigen vaccines combined with PD‐1 antibody intensively inreased the number and activity of CD8+ CTLs, and thus improved the efficacy of cancer immunotherapy.
FIGURE 3.
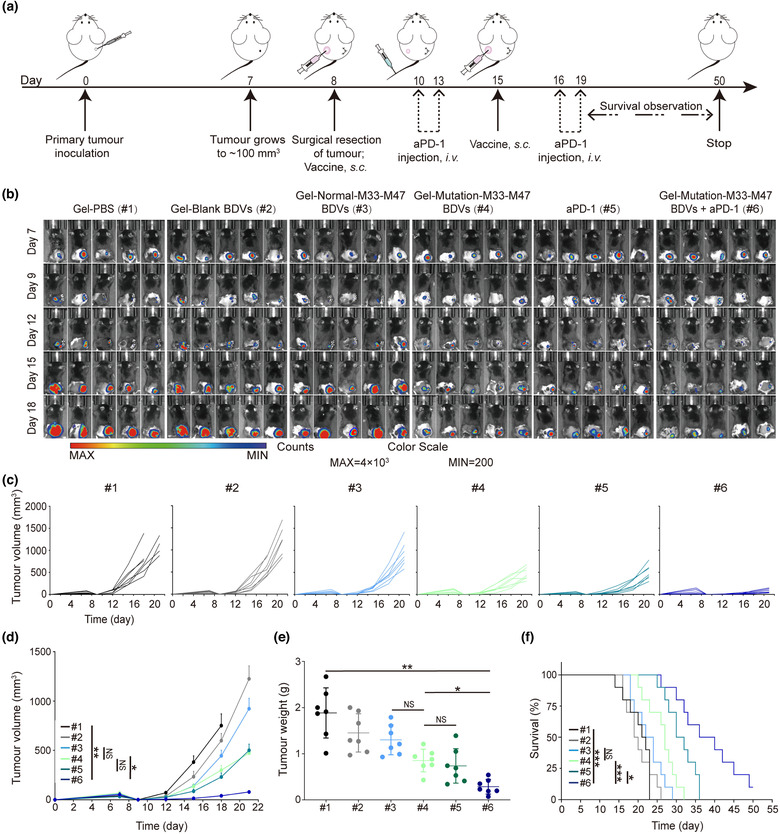
Antitumour recurrence effect of BDVs‐triggered cancer immunotherapy in the surgical bed of B16F10‐luc model. (a) Schematic illustration of BDVs‐triggered cancer vaccine in an incomplete‐surgery B16F10‐luc tumour model. (b) In vivo bioluminescence images of the B16F10‐luc tumour‐bearing mice (n = 5). (c) Single tumour volume of each mouse in different groups. (d) Tumour growth curves in B16F10‐luc model (n = 7), Error bar, mean ± s.e.m.. (e) Tumour weights in different treatment groups (n = 7), Error bar, mean ± s.d.. (f) Survival curves in different groups (n = 10) (#1) Gel‐PBS, (#2) Gel‐Blank BDVs, (#3) Gel‐Normal‐M33‐M47 BDVs, (#4) Gel‐Mutation‐M33‐M47 BDVs, (#5) aPD‐1 (anti‐PD‐1 antibody), (#6) Gel‐Mutation‐M33‐M47 BDVs + aPD‐1. NS: no significant, *P < 0.05, **P < 0.01, ***P < 0.001. One‐way ANOVA with Tukey post‐hoc tests (d, e) or the Long‐Rank (Mantel‐Cox) test (f)
To further explore the mechanism and synergistic therapeutic effect of the Gel‐Mutation‐M33‐M47 BDVs in combination with aPD‐1, immunofluorescence and flow cytometry were used to evaluate the situation of tumour‐infiltrating lymphocytes (TILs) in the relapsed tumours. T lymphocyte is the main effector cell of anti‐tumour immunity (Farhood et al., 2019; McLane et al., 2019), thus the relapsed tumour infiltrated CD4+ and CD8+ T cells infiltrating into the relapsed tumours were analysed by flow cytometry. Notably, the number of CD4+ and CD8+ T cells were significantly increased in mice that were treated with the Gel‐Mutation‐M33‐M47 BDVs combined with aPD‐1 (Figure 4a, b and Figure S8). Moreover, the proliferation of CD8+ T cells in the relapsed tumours of mice treated with Gel‐Mutation‐M33‐M47 BDVs combined with aPD‐1 (#6) were significantly increased (Figure 4c, d). Gel‐Mutation‐M33‐M47 BDVs combined with aPD‐1 treatment could activate the native CD8+ T cells in vivo. CD69 is one of the markers of T lymphocytes activation, therefore the number of active CD8+ CD69+ T cells were significantly increased in the FACS detection (Figure 4e, f). In addition, to further assess the mechanism of the anti‐tumour effect of Gel‐BDV formulations, we also examined the central memory CD8+ T (TCM) cells. Virtually, the population of CD44+ CD62L+ TCM cells showed a intensive increase in the Gel‐Mutation‐M33‐M47 BDVs with aPD‐1‐treated mice compared with the PBS control (Figure 4g, h), indicating BDVs combination therapy could generate a strong immune memory effect to protect mice from tumour recurrence. Importantly, we found the proliferation of CD4+ T cells in relapsed tumours of mice treated with Gel‐Mutation‐M33‐M47 BDVs with aPD‐1 was also significantly increased (Figure S9a, b). Nonetheless, the number of regulatory T cells Tregs (CD4+ CD25+ Foxp3+ T cells) was decreased (Figure S9c, d).
FIGURE 4.
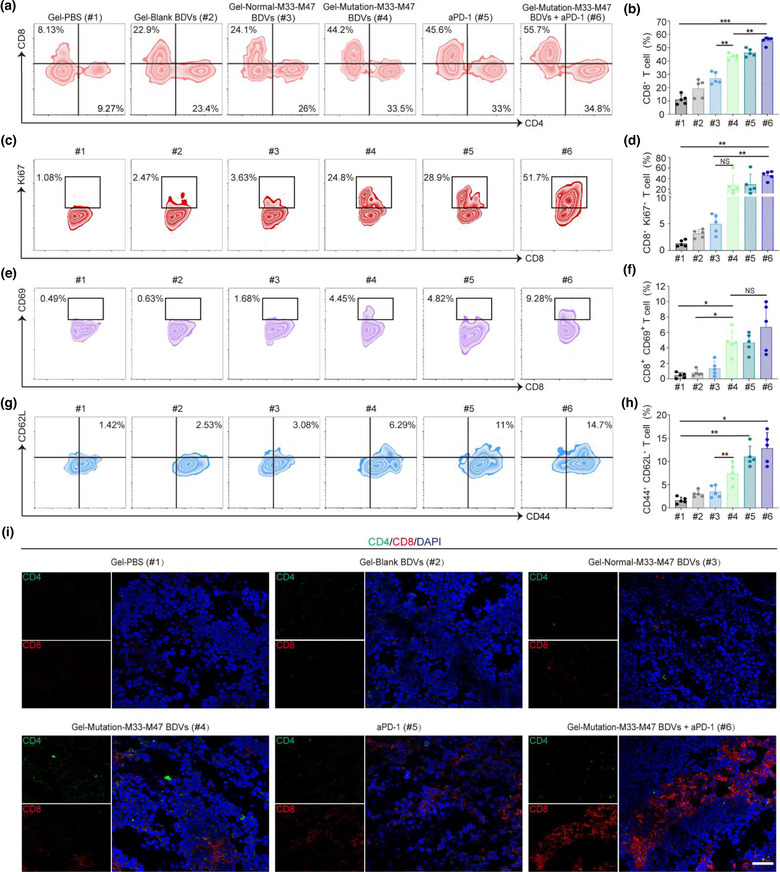
BDV‐neoantigen vaccine induced a robust antitumour immune response. (a, b) Representative flow cytometry plots (a) and ratios (b) of different groups of T cells in the residual tumours from different groups (gated on CD3+ T cells, n = 5). Cells were stained with anti‐CD3‐FITC, anti‐CD4‐Brilliant Violet 421, anti‐CD8a‐APC/Fire750 antibodies (Biolegend). Error bar, mean ± s.d.. (c, d) Representative flow cytometry plots (c) and ratios (d) of different groups of CD8+ Ki67+ T cells in tumours (gated on CD3+ CD8+ T cells, n = 5). Cells were stained with anti‐CD3‐FITC, anti‐CD8‐APC/Fire750, anti‐Ki67‐Alexa Fluor® 647 antibodies (Biolegend). Error bar, mean ± s.d.. (e, f) Representative flow cytometry plots (e) and ratios (f) of different groups of CD8+ CD69+ T cells in tumours (Gated on CD3+ CD8+ T cells, n = 5). Cells were stained with anti‐CD3‐FITC, anti‐CD8‐APC/Fire750, anti‐CD69‐APC antibodies (Biolegend). Error bar, mean ± s.d.. (g, h) Representative flow cytometry plots (g) and ratios (h) of different groups of CD44+ CD62L+ T cells infiltrating in tumours (Gated on CD3+ CD8+ T cells, n = 5). Cells were stained with anti‐CD3‐FITC, anti‐CD8‐APC/Fire750, anti‐CD44‐PE, anti‐CD62L‐Alexa Fluor® 700 antibodies (Biolegend). Error bar, mean ± s.d.. (i) The immunofluorescence images of CD4+ CD8+ T cells infiltrating in tumours (Scale bar: 50 μm). (#1) Gel‐PBS, (#2) Gel‐Blank BDVs, (#3) Gel‐Normal‐M33‐M47 BDVs, (#4) Gel‐Mutation‐M33‐M47 BDVs, (#5) aPD‐1, (#6) Gel‐Mutation‐M33‐M47 BDVs + aPD‐1. Cells were stained with anti‐CD11c‐APC, anti‐CD40‐PE, anti‐CD80‐FITC and anti‐CD86‐PE antibodies (Biolegend). All samples were sorted using Beckman CytoFlex flow cytometer and analyzed with FlowJo software. NS: no significant, *P < 0.05, **P < 0.01, ***P < 0.001. One‐way ANOVA with Tukey post‐hoc tests (b, d, f, h)
Cytotoxic proteins such as granzyme B (Gzm B) and perforin can directly kill tumour cells by activating caspase enzymes mediated apoptosis (Russell & Ley, 2002; Voskoboinik et al., 2015). Thus, we detected the expressions of GzmB in the T cells after the treatment. Flow cytometry data showed that there was a intensive increase of Gzm B+ CD8+ T cells and perforin+ CD8+ T cells in the mice treated with Gel‐Mutation‐M33‐M47 BDVs combined with aPD‐1 (Figure S10a–d). Eventually, immunostaining of the CD4+ and CD8+ T cells in the tumour tissue section showed similar results compared with FACS analysis, and the number of CD8+ T cells significatly significantly increased in the margin of the tumour section (Figure 4i). Collectively, these results indicated that the Gel‐Mutation‐M33‐M47 BDVs combined with aPD‐1 blockade could trigger anti‐tumour immune response and effectively prevent tumour relapse.
3.4. In vivo therapeutic efficacy of engineered BDVs for metastatic tumours
Subsequently, we investigated whether the BDVs‐neoantigen vaccine could prevent tumour metastasis. We established a metastatic mice mouse model by i.v. injection of B16F10‐luc cells (∼1 × 106) to mimic the metastasis of cancer cells. The mice were randomly devided into six experimental groups and received the same treatment as described above (Figure 5a). The growth and spread of tumours were monitored by bioluminescence of B16F10‐luc cells. As shown in Figure 5b, mice treated with PBS showed obvious cancer metastasis and all of them died within 16 days. On the contrary, Gel‐Mutation‐M33‐M47 BDVs combined with aPD‐1 showed the relatively weak bioluminescence signal and exhibited much better anti‐tumour ability. Notably, the typical lung metastatic tumour images showed that mice received Gel‐Mutation‐M33‐M47 BDVs combined with aPD‐1 treatment had few metastatic tumour nodules and achieved promising anti‐tumour effect in the lung metastatic model (Figure 5c). Hereafter, all lungs harvested from the mice received treatments were fixed with 4% paraformaldehyde, and numbers of metastatic nodules were counted at the end time. The results revealed that the mice treated with Gel‐Mutation‐M33‐M47 BDVs, aPD‐1 or Gel‐Mutation‐M33‐M47 BDVs combined with aPD‐1 had few metastatic nodules compared to those received other formulations treatment (Figure 5d). Moreover, the mice treated with Gel‐Mutation‐M33‐M47 BDVs combined with aPD‐1 also had a higher proportion (∼70%) of survical rate after tumour injection (Figure 5e). Virtually, H&E staining results showed that mice with Gel‐Mutation‐M33‐M47 BDVs, aPD‐1 and Gel‐Mutation‐M33‐M47 BDVs combined with aPD‐1 treatments had less lung metastases, indicating both Gel‐Mutation‐M33‐M47 BDVs and aPD‐1 had conspicuously better effect in delaying the development and progress of lung metastasis (Figure 5f).
FIGURE 5.
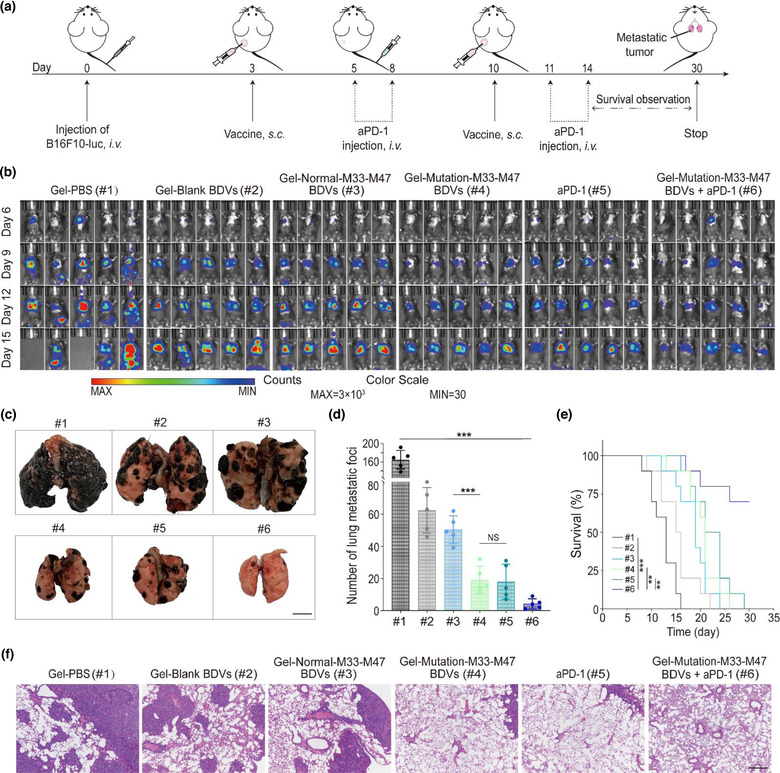
In vivo suppression of antimetastatic tumour effect of BDV‐neoantigen vaccine. (a) Schematic illustration of the therapy of BDV‐neoantigen vaccine in B16F10‐luc model of metastasis. (b) In vivo bioluminescence images of the B16F10‐luc lung metastasis in different groups (n = 5). (c) Representative images of the lung metastatic nodules (Scale bar: 5 mm). (d) Numbers of lung metastatic nodules in different groups (n = 5), Error bar, mean ± s.d.. (e) Survival curves of different groups (n = 10). Day 0 is the day of tumour cells injection via tail vein. (f) H&E‐stained lung slices (Scale bar: 100 μm). (#1) Gel‐PBS, (#2) Gel‐Blank BDVs, (#3) Gel‐Normal‐M33‐M47 BDVs, (#4) Gel‐Mutation‐M33‐M47 BDVs, (#5) aPD‐1, (#6) Gel‐Mutation‐M33‐M47 BDVs + aPD‐1. NS: no significant, *P < 0.05, **P < 0.01, ***P < 0.001. One‐way ANOVA with Tukey post‐hoc tests (d) or the Long‐Rank (Mantel‐Cox) test (e)
4. DISSCUSSION
In summary, we genetically programed bacteria to synthesize fusion neoantigens, and prepared BDVs displaying neoantigen (BDVs‐Neo) as personalized therapeutic vaccine to elicit systemic anti‐tumour immunity. BDVs‐Neo and GM‐CSF loaded within thermosensitive hydrogel could recruit and stimulate the maturature of DCs within subcutaneous. Notably, BDVs also exhibited spherical structures the same as exosome and showed similar diameter as natural OMVs (Chen, Bai et al., 2020; Li et al., 2022). Moreover, BDVs‐Neo not only could be easily internalized into BMDCs in vitro, but also stimulated DC maturation efficiently in vivo. BDVs‐Neo vaccine formulation could improve the anticancer efficacy and long‐term memory immunity through combination treatment with PD‐1 antibody. CTLs could recognize and eliminate residual tumour cells through cytotoxic effects and the secretion of cytokines, such as IFN‐γ. Flow cytometry and immunofluorescence analysis of tumour tissue showed that BDVs‐Neo vaccine could improve CD8+ T cell activity. And beyond that, BDVs‐Neo vaccine also increased the number of central memory T cells, which protected the mice from tumour relapse and metasitasis. BDVs‐Neo vaccine formulation improved the anti‐cancer efficacy and long‐term memory immunity through combination treatment. Collectively, BDVs‐Neo is a promising personalized cancer vaccine formulation that could induce robust anti‐tumour response.
AUTHOR CONTRIBUTIONS
X.Z. and X.L. contributed conception and design of the study. F.M., L.L., Z.Z., Z. L. and J.Z. performed the experiments. F.M., X.S., T.X. and C.X. interpreted the data. Z. L. assisted with some parts of experiments and data analysis. F.M. wrote the first draft of the manuscript. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
CONFLICT OF INTERESTS
The authors declare that no competing interests exist.
Supporting information
SUPPORTING INFORMATION
ACKNOWLEDGEMENTS
This work was supported by grants from Shenzhen Science and Technology Program (Grant No. RCYX20200714114643121), the National Natural Science Foundation of China (31971268,32201084), Science, Technology & Innovation Commission of Shenzhen Municipality (JCYJ20200109142610136), Basic Research Program of Shenzhen (JCYJ20180507181654186), the Natural Science Foundation of Guangdong Province (Nos 2019A1515010790,2019A1515010855, 1515010802, 2022A1515012289), Guangdong Basic and Applied Basic Research Foundation (2020A1515110166), Health system scientific research project of Shenzhen Guangming District Science and innovation Bureau (2020R01073), Special fund for economic development of Shenzhen Guangming District (2021R01128),University of Chinese Academy of Sciences‐Shenzhen Hospital Research Funding (HRF‐2020004) and the Fundamental Research Funds for the Central Universities (19lgzd45), Doctoral personnel scientific research start‐up Fund project of Guangdong Medical University (GDMUB2022037).
Meng, F. , Li, L. , Zhang, Z. , Lin, Z. , Zhang, J. , Song, X. , Xue, T. , Xing, C. , Liang, X. , & Zhang, X. (2022). Biosynthetic neoantigen displayed on bacteria derived vesicles elicit systemic antitumour immunity. Journal of Extracellular Vesicles, 11, e12289. 10.1002/jev2.12289
Fanqiang Meng, Liyan Li and Zhirang Zhang contributed equally to this work.
Contributor Information
Xin Liang, Email: liangxingibh@gdmu.edu.cn.
Xudong Zhang, Email: zhangxd56@mail.sysu.edu.cn.
REFERENCES
- Bachmann, M. F. , & Jennings, G. T. (2010). Vaccine delivery: A matter of size, geometry, kinetics and molecular patterns. Nature Reviews Immunology, 10, 787–796. [DOI] [PubMed] [Google Scholar]
- Banchereau, J. , & Palucka, A. K. (2005). Dendritic cells as therapeutic vaccines against cancer. Nature Reviews Immunology, 5, 296–306. [DOI] [PubMed] [Google Scholar]
- Blass, E. , & Ott, P. A. (2021). Advances in the development of personalized neoantigen‐based therapeutic cancer vaccines. Nature Reviews Clinical Oncology, 18, 215–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cian, M. B. , Giordano, N. P. , Mettlach, J. A. , Minor, K. E. , & Dalebroux, Z. D. (2020). Separation of the cell envelope for gram‐negative bacteria into inner and outer membrane fractions with technical adjustments for Acinetobacter baumannii . Jove‐Journal of Visualized Experiments, 158, e60517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canale, F. P. , Basso, C. , Antonini, G. , Perotti, M. , Li, N. , Sokolovska, A. , Neumann, J. , James, M. J. , Geiger, S. , & Jin, W. J. (2021). Metabolic modulation of tumours with engineered bacteria for immunotherapy. Nature, 598, 662–666. [DOI] [PubMed] [Google Scholar]
- Carvalho, A. L. , Fonseca, S. , Miquel‐Clopés, A. , Cross, K. , Kok, K. S. , Wegmann, U. , Gil‐Cordoso, K. , Bentley, E. G. , Al Katy, S. H. M. , & Coombes, J. L. (2019). Bioengineering commensal bacteria‐derived outer membrane vesicles for delivery of biologics to the gastrointestinal and respiratory tract. Journal of Extracell Vesicles, 8, 1632100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. P. , & Han, X. (2015). Anti‐PD‐1/PD‐L1 therapy of human cancer: Past, present, and future. J Clin Invest, 125, 3384–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Q. , Bai, H. Z. , Wu, W. T. , Huang, G. J. , Li, Y. , Wu, M. , Tang, G. P. , & Ping, Y. (2020). Bioengineering bacterial vesicle‐coated polymeric nanomedicine for enhanced cancer immunotherapy and metastasis prevention. Nano Letters, 20, 11–21. [DOI] [PubMed] [Google Scholar]
- Chen, Q. , Huang, G. J. , Wu, W. T. , Wang, J. W. , Hu, J. W. , Mao, J. M. , Chu, P. K. , Bai, H. Z. , & Tang, G. P. (2020). A hybrid eukaryotic‐prokaryotic nanoplatform with photothermal modality for enhanced antitumour vaccination. Advanced Materials, 32, e1908185. [DOI] [PubMed] [Google Scholar]
- Cheng, K. M. , Zhao, R. F. , Li, Y. , Qi, Y. Q. , Wang, Y. Z. , Zhang, Y. L. , Qin, H. , Qin, Y. T. , Chen, L. , & Li, C. (2021). Bioengineered bacteria‐derived outer membrane vesicles as a versatile antigen display platform for tumour vaccination via Plug‐and‐Display technology. Nature Communications, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury, S. , Castro, S. , Coker, C. , Hinchliffe, T. E. , Arpaia, N. , & Danino, T. (2019). Programmable bacteria induce durable tumour regression and systemic antitumour immunity. Nature Medicne, 25, 1057–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos‐Ruiz, A. , Guo, T. X. , Sokolovska, A. , Miller, P. F. , Collins, J. J. , Lu, T. K. , & Lora, J. M. (2021). Engineering living therapeutics with synthetic biology. Nature Reviews Drug Discovery, 20, 941–960. [DOI] [PubMed] [Google Scholar]
- Daleke‐Schermerhorn, M. H. , Felix, T. , Soprova, Z. , ten Hagen‐Jongman, C. M. , Vikstrom, D. , Majlessi, L. , Beskers, J. , Follmann, F. , de Punder, K. , & van der Wel, N. N. (2014). Decoration of outer membrane vesicles with multiple antigens by using an autotransporter approach. Applied and Environmental Microbiology, 80, 5854–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhood, B. , Najafi, M. , & Mortezaee, K. (2019). CD8+ cytotoxic T lymphocytes in cancer immunotherapy: A review. Journal of Cellular Physiology, 234, 8509–8521. [DOI] [PubMed] [Google Scholar]
- Gong, J. , Chehrazi‐Raffle, A. , Reddi, S. , & Salgia, R. (2018). Development of PD‐1 & PD‐L1 inhibitors as a form of cancer immunotherapy: A comprehensive review of registration trials and future considerations. Journal for ImmunoTherapy of Cancer, 6, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujrati, V. , Kim, S. , Kim, S. H. , Min, J. J. , Choy, H. E. , Kim, S. C. , & Jon, S. (2014). Bioengineered bacterial outer membrane vesicles as cell‐specific drug‐delivery vehicles for cancer therapy. ACS Nano, 8, 1525–1537. [DOI] [PubMed] [Google Scholar]
- Gurbatri, C. R. , Lia, I. , Vincent, R. , Coker, C. , Castro, S. , Treuting, P. M. , Hinchliffe, T. E. , Arpaia, N. , & Danino, T. (2020). Engineered probiotics for local tumour delivery of checkpoint blockade nanobodies. Science Translational Medicine, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock, R. E. , & Nikaido, H. (1978). Outer membranes of gram‐negative bacteria. xix. Isolation from pseudomonas aeruginosa pao1 and use in reconstitution and definition of the permeability barrier. Journal of Bacteriology, 136, 381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth, R. E. , & Jansen, K. (2019). Turning the corner on therapeutic cancer vaccines. Npj Vaccines, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Z. T. , Leet, D. E. , Allesoe, R. L. , Oliveira, G. , Li, S. Q. , Luoma, A. M. , Liu, J. Y. , Forman, J. , Huang, T. , & Iorgulescu, J. B. (2021). Personal neoantigen vaccines induce persistent memory T cell responses and epitope spreading in patients with melanoma. Nature Medicne, 27, 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W. W. , Wang, S. J. , Yao, Y. F. , Xia, Y. , Yang, X. , Li, K. , Sun, P. Y. , Liu, C. B. , Sun, W. J. , & Bai, H. M. (2016). Employing Escherichia coli‐derived outer membrane vesicles as an antigen delivery platform elicits protective immunity against Acinetobacter baumannii infection. Scientific Reports Uk, 6, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W. W. , Zhang, Q. S. , Li, W. R. , Yuan, M. C. , Zhou, J. X. , Hua, L. Q. , Chen, Y. J. , Ye, C. , & Ma, Y. B. (2020). Development of novel nanoantibiotics using an outer membrane vesicle‐based drug efflux mechanism. Journal of Controlled Release, 317, 1–22. [DOI] [PubMed] [Google Scholar]
- Keskin, D. B. , Anandappa, A. J. , Sun, J. , Tirosh, I. , Mathewson, N. D. , Li, S. Q. , Oliveira, G. , Giobbie‐Hurder, A. , Felt, K. , & Gjini, E. (2019). Neoantigen vaccine generates intratumoural T cell responses in phase Ib glioblastoma trial. Nature, 565, 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, O. Y. , Park, H. T. , Dinh, N. T. H. , Choi, S. J. , Lee, J. , Kim, J. H. , Lee, S. W. , & Gho, Y. S. (2017). Bacterial outer membrane vesicles suppress tumour by interferon‐gamma‐ mediated antitumour response. Nature Communications, 8, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs‐Simon, A. , Titball, R. W. , & Michell, S. L. (2011). Lipoproteins of Bacterial Pathogens. Infection and Immunity, 79, 548–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiter, S. , Vormehr, M. , van de Roemer, N. , Diken, M. , Lower, M. , Diekmann, J. , Boegel, S. , Schrors, B. , Vascotto, F. , & Castle, J. C. (2015). Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature, 520, 692–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn, M. J. , & Kesty, N. C. (2005). Bacterial outer membrane vesicles and the host‐pathogen interaction. Gene Development, 19, 2645–2655. [DOI] [PubMed] [Google Scholar]
- Kulp, A. , & Kuehn, M. J. (2010). Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annual Review of Microbiology, 64, 163–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, E. Y. , Choi, D. S. , Kim, K. P. , & Gho, Y. S. (2008). Proteomics in gram‐negative bacterial outer membrane vesicles. Mass Spectrometry Reviews, 27, 535–555. [DOI] [PubMed] [Google Scholar]
- Li, B. Q. , Fang, T. L. , Li, Y. , Xue, T. Y. , Zhang, Z. R. , Li, L. Y. , Meng, F. Q. , Wang, J. Q. , Hou, L. L. , Liang, X. , & Zhang, X. D. (2022). Engineered T cell extracellular vesicles displaying PD‐1 boost anti‐tumour immunity. Nano Today, 46, 101606. [Google Scholar]
- Li, L. Y. , Miao, Q. W. , Meng, F. Q. , Li, B. Q. , Xue, T. Y. , Fang, T. L. , Zhang, Z. R. , Zhang, J. X. , Ye, X. Y. , & Kang, Y. (2021). Genetic engineering cellular vesicles expressing CD64 as checkpoint antibody carrier for cancer immunotherapy. Theranostics, 11, 6033–6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. , Zhou, H. , Yang, C. , Wu, Y. , Zhou, X. C. , Liu, H. , & Wang, Y. C. (2020). Bacterial outer membrane vesicles as a platform for biomedical applications: An update. Journal of Controlled Release, 323, 253–268. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Zhao, R. F. , Cheng, K. M. , Zhang, K. Y. , Wang, Y. Z. , Zhang, Y. L. , Li, Y. J. , Liu, G. N. , Xu, J. C. , & Xu, J. Q. (2020). Bacterial outer membrane vesicles presenting programmed death 1 for improved cancer immunotherapy via immune activation and checkpoint inhibition. ACS Nano, 14, 16698–16711. [DOI] [PubMed] [Google Scholar]
- Mancini, F. , Rossi, O. , Necchi, F. , & Micoli, F. (2020). OMV vaccines and the role of TLR agonists in immune response. International Journal of Molecular Sciences, 21, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLane, L. M. , Abdel‐Hakeem, M. S. , & Wherry, E. J. (2019). CD8 T cell exhaustion during chronic viral infection and cancer. Annual Review of Immunology, 37, 457–495. [DOI] [PubMed] [Google Scholar]
- McNerney, M. P. , Doiron, K. E. , Ng, T. L. , Chang, T. M. Z. , & Silver, P. A. (2021). Theranostic cells: Emerging clinical applications of synthetic biology. Nature Reviews Genetics, 22, 730–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melero, I. , Gaudemack, G. , Gerritsen, W. , Huber, C. , Parmiani, G. , Scholl, S. , Thatcher, N. , Wagstaff, J. , Zielinski, C. , & Faulkner, I. (2014). Therapeutic vaccines for cancer: An overview of clinical trials. Nature Reviews Clinical Oncology, 11, 509–524. [DOI] [PubMed] [Google Scholar]
- Murase, K. (2022). Cytolysin A (ClyA): A bacterial virulence factor with potential applications in nanopore technology, vaccine development, and tumour therapy. Toxins, 14, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palena, C. , Abrams, S. I. , Schlom, J. , & Hodge, J. W. (2006). Cancer vaccines: Preclinical studies and novel strategies. Advances In Cancer Research, 95, 115–145. [DOI] [PubMed] [Google Scholar]
- Park, K. S. , Svennerholm, K. , Crescitelli, R. , Lasser, C. , Gribonika, I. , & Lotvall, J. (2021). Synthetic bacterial vesicles combined with tumour extracellular vesicles as cancer immunotherapy. Journal of Extracell Vesicles, 10, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, R. B. , Ye, M. Z. , Carlson, P. M. , Jaquish, A. , Zangl, L. , Ma, B. , Wang, Y. Y. , Arthur, I. , Xie, R. S. , & Brown, R. J. (2019). Development of an in situ cancer vaccine via combinational radiation and bacterial‐membrane‐coated nanoparticles. Advanced Materials, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, L. H. , Wang, M. Z. , Chu, Y. , Zhang, L. , Niu, J. , Shao, H. T. , Yuan, T. J. , Jiang, Z. H. , Gao, J. Q. , & Ning, X. H. (2020). Engineering bacterial outer membrane vesicles as transdermal nanoplatforms for photo‐TRAIL‐programmed therapy against melanoma. Science Advances, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal, J. A. , Chen, L. X. , Baker, J. L. , Putnam, D. , & DeLisa, M. P. (2014). Pathogen‐like particles: Biomimetic vaccine carriers engineered at the nanoscale. Current Opinion in Biotechnology, 28, 51–58. [DOI] [PubMed] [Google Scholar]
- Russell, J. H. , & Ley, T. J. (2002). Lymphocyte‐mediated cytotoxicity. Annual Review of Immunology, 20, 323–370. [DOI] [PubMed] [Google Scholar]
- Sahin, U. , Oehm, P. , Derhovanessian, E. , Jabulowsky, R. A. , Vormehr, M. , Gold, M. , Maurus, D. , Schwarck‐Kokarakis, D. , Kuhn, A. N. , & Omokoko, T. (2020). An RNA vaccine drives immunity in checkpoint‐inhibitor‐treated melanoma. Nature, 585, 107–112. [DOI] [PubMed] [Google Scholar]
- Sahin, U. , & Tureci, O. (2018). Personalized vaccines for cancer immunotherapy. Science, 359, 1355–1360. [DOI] [PubMed] [Google Scholar]
- Sartorio, M. G. , Pardue, E. J. , Feldman, M. F. , & Haurat, M. F. (2021). Bacterial outer membrane vesicles: from discovery to applications. Annual Review of Microbiology, 75, 609–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena, M. , van der Burg, S. H. , Melief, C. J. M. , & Bhardwaj, N. (2021). Therapeutic cancer vaccines. Nat Rev Cancer, 21, 360–378. [DOI] [PubMed] [Google Scholar]
- Schumacher, T. N. , & Schreiber, R. D. (2015). Neoantigens in cancer immunotherapy. Science, 348, 69–74. [DOI] [PubMed] [Google Scholar]
- Valentine, J. L. , Chen, L. X. , Perregaux, E. C. , Weyant, K. B. , Rosenthal, J. A. , Heiss, C. , Azadi, P. , Fisher, A. C. , Putnam, D. , & Moe, G. R. (2016). Immunization with outer membrane vesicles displaying designer glycotopes yields class‐switched, glycan‐specific antibodies. Cell Chemical Biology, 23, 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskoboinik, I. , Whisstock, J. C. , & Trapani, J. A. (2015). Perforin and granzymes: function, dysfunction and human pathology. Nature Reviews Immunology, 15, 388–400. [DOI] [PubMed] [Google Scholar]
- Wei, Z. H. , Zhang, X. Q. , Yong, T. Y. , Bie, N. N. , Zhan, G. T. , Li, X. , Liang, Q. L. , Li, J. Y. , Yu, J. J. , & Huang, G. (2021). Boosting anti‐PD‐1 therapy with metformin‐loaded macrophage‐derived microparticles. Nature Communications, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, T. , Zhang, Z. , Fang, T. , Li, B. , Li, Y. , Li, L. , Jiang, Y. , Duan, F. , Meng, F. , & Liang, X. (2022). Cellular vesicles expressing PD‐1‐blocking scFv reinvigorate T cell immunity against cancer. Nano Research, . [Google Scholar]
- Yang, W. , Lee, K. W. , Srivastava, R. M. , Kuo, F. S. , Krishna, C. , Chowell, D. , Makarov, V. , Hoen, D. , Dalin, M. G. , & Wexler, L. (2019). Immunogenic neoantigens derived from gene fusions stimulate T cell responses. Nature Medicne, 25, 767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, X. Y. , Liang, X. , Chen, Q. , Miao, Q. W. , Chen, X. L. , Zhang, X. D. , & Mei, L. (2019). Surgical tumour‐derived personalized photothermal vaccine formulation for cancer immunotherapy. ACS Nano, 13, 2956–2968. [DOI] [PubMed] [Google Scholar]
- Zanella, I. , Konig, E. , Tomasi, M. , Gagliardi, A. , Frattini, L. , Fantappie, L. , Irene, C. , Zerbini, F. , Caproni, E. , & Isaac, S. J. (2021). Proteome‐minimized outer membrane vesicles from Escherichia coli as a generalized vaccine platform. Journal of Extracell Vesicles, 10, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, W. P. , Wolchok, J. D. , & Chen, L. P. (2016). PD‐L1 (B7‐H1) and PD‐1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Science Translational Medicine, 8, . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPORTING INFORMATION


