Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) enters cells using angiotensin-converting enzyme 2 (ACE2) and neuropilin-1 (NRP-1) as the primary receptor and entry co-factor, respectively. Cell entry is the first and major step in initiation of the viral life cycle, representing an ideal target for antiviral interventions. In this study, we used a recombinant replication-deficient vesicular stomatitis virus-based pseudovirus bearing the spike protein of SARS-CoV-2 (SARS2-S) to screen a US Food and Drug Administration-approved drug library and identify inhibitors of SARS-CoV-2 cell entry. The screen identified 24 compounds as primary hits, and the largest therapeutic target group formed by these primary hits was composed of seven dopamine receptor D2 (DRD2) antagonists. Cell-based and biochemical assays revealed that the DRD2 antagonists inhibited both fusion activity and the binding of SARS2-S to NRP-1, but not its binding to ACE2. On the basis of structural similarity to the seven identified DRD2 antagonists, which included six phenothiazines, we examined the anti-SARS-CoV-2 activity of an additional 15 phenothiazines and found that all the tested phenothiazines shared an ability to inhibit SARS2-S-mediated cell entry. One of the phenothiazines, alimemazine, which had the lowest 50% effective concentration of the tested phenothiazines, exhibited a clear inhibitory effect on SARS2-S–NRP-1 binding and SARS-CoV-2 multiplication in cultured cells but not in a mouse infection model. Our findings provide a basis for the development of novel anti-SARS-CoV-2 therapeutics that interfere with SARS2-S binding to NRP-1.
Keywords: SARS-CoV-2, COVID-19, Phenothiazine, Neuropilin-1
1. Introduction
The rapid global spread of coronavirus disease 2019 (COVID-19) caused by infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) resulted in an unprecedented pandemic with more than 639 million cases and more than 6 million deaths to date (https://www.who.int/emergencies/diseases/novel-coronavirus-2019). Several COVID-19 vaccines, chiefly mRNA-based vaccines, have been used worldwide and have significantly contributed to the prevention of SARS-CoV-2 infection and COVID-19 progression to severe disease (National Center for, 2020). However, the continuous emergence of SARS-CoV-2 variants with accumulated mutations that attenuate vaccine efficacy and the incompatibility of vaccine strategies for immunocompromised patients underscore the need for safe, effective, and readily available antivirals against SARS-CoV-2. Remdesivir, a nucleoside analog prodrug, originally developed as an Ebola virus disease treatment, is the first US Food and Drug Administration (FDA)-approved anti-SARS-CoV-2 drug (Beigel et al., 2020; Rubin et al., 2020). Remdesivir, which needs to be administrated intravenously, may be used for COVID-19 patients who require oxygen supplementation or have a pre-existing disease that could increase their risk of serious illness (Schooley et al., 2021). Baricitinib, a Janus kinase (JAK) inhibitor, is an oral medication initially approved to treat rheumatoid arthritis (Al-Salama and Scott, 2018). Recently, baricitinib was approved by the FDA for the treatment of COVID-19 (Rubin, 2022). However, use of baricitinib is limited to hospitalized adults with COVID-19 requiring oxygen supplementation, non-invasive or invasive mechanical ventilation, or extracorporeal membrane oxygenation (ECMO). These limitations further emphasize the importance of expanding the number of potential anti-SARS-CoV-2 drug candidates. Given that the COVID-19 pandemic is an ongoing crisis, the repositioning of already existing, clinically used drugs with known safety profiles is the most practical approach for the rapid development of COVID-19 treatments (Riva et al., 2020).
The SARS-CoV-2 life cycle can be divided into several steps: cell entry, genome translation, subgenomic transcription, genome replication, and progeny virion formation. Of those distinct steps of the virus life cycle, cell entry is the first, and this critical step in the initiation of the viral life cycle represents an ideal target for antiviral interventions. The spike protein of SARS-CoV-2 (SARS2-S) is responsible for both receptor recognition and membrane fusion. SARS2-S is proteolytically processed in-cis by the host protease furin at the S1/S2 boundary, generating the mature virion surface glycoprotein complex composed of noncovalently associated S1 and S2. Subsequently, S2 further undergoes priming at the S2′ site in-trans by host serin proteases, which include TMPRSS2 at the plasma membrane and cathepsins in the endosomes, to expose the fusion peptide of S2; this process is required for membrane fusion (Hoffmann et al., 2020a). S1 binds to the cell surface angiotensin-converting enzyme 2 (ACE2) (Yu et al., 2022), which was identified as the primary receptor for SARS-CoV as well (Li et al., 2003). In contrast with the spike protein of SARS-CoV, SARS2-S can be cleaved by furin, generating an Arg-Arg-Ala-Arg (RRAR) sequence at the C-terminus end, and this sequence matches an [R/K]XX [R/K] motif, called the C-end Rule (CendR). As with other proteins that conform to the CendR, the furin-cleaved S1 can bind to neuropilin-1 (NRP-1), and the S1–NRP-1 interaction facilitates ACE2-dependent cell entry (Cantuti-Castelvetri et al., 2020; Daly et al., 2020).
In the present study, we have used a recombinant replication-deficient vesicular stomatitis virus (VSV)-based pseudovirus bearing SARS2-S to screen a US FDA-approved drug library and identify novel drug candidates that specifically inhibit SARS2-S-mediated cell entry.
2. Materials and methods
2.1. ELISA-based SARS2-S–NRP-1 binding assay
The effects of the test compounds on SARS2-S binding to NRP-1 were analyzed with a RayBio COVID-19 Spike-NRP1 binding assay kit (RayBiotech, Peachtree Corners, GA, USA). Compounds were separately added to the recombinant NRP-1 (rNRP-1)-coated microplate in the presence of recombinant SARS2-S S1 domain (rS1), and the microplate was incubated at room temperature for 2.5 h. The microplate was then washed four times to remove unbound rS1, and rNRP-1-bound rS1 was reacted with a mouse anti-S1 IgG detection antibody, followed by an HRP-conjugated anti-mouse secondary IgG. TMB (3,3′, 5,5′-tetramethyl benzidine) substrate was used to detect HRP activity, which was halted by the addition of the Stop Solution (0.2 M sulfuric acid). The absorbance at 450 nm was measured using a multi-mode microplate reader (SpectraMax iD5, Molecular Devices, San Jose, CA, USA).
2.2. Statistical analysis
GraphPad Prism 9 (GraphPad, San Diego, CA, USA) was used for all the statistical analyses. Statistical significance was analyzed by one-way ANOVA, and statistically significant differences were determined by a Dunnett's multiple comparisons test unless otherwise indicated.
The detailed materials and methods are described in SI Materials and Methods.
3. Results
3.1. Screening of a US FDA-approved library to identify inhibitors of SARS2-S-mediated cell entry
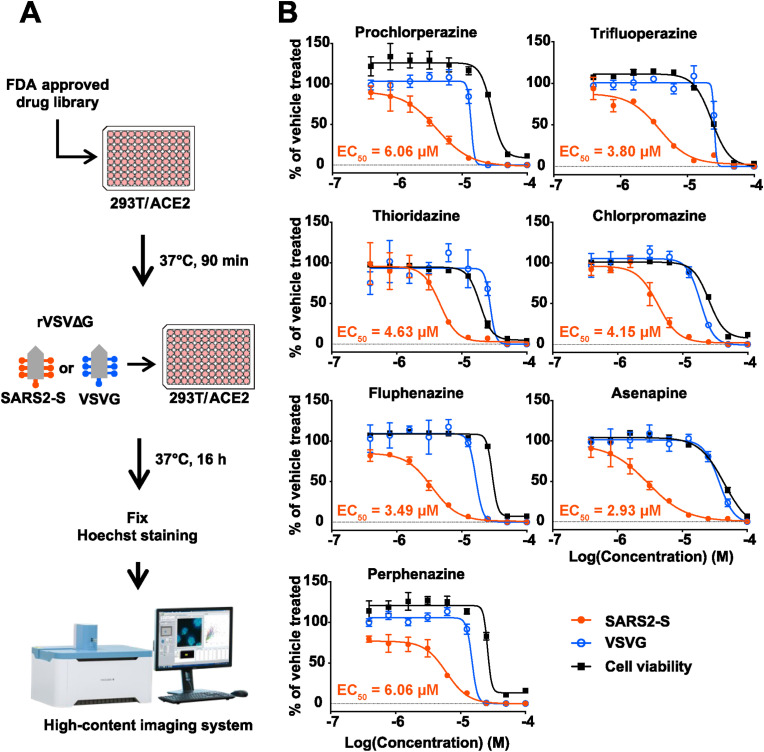
We conducted VSV pseudotype-based screening using a US FDA-approved drug library containing 1,061 compounds to identify novel inhibitors of SARS2-S-mediated cell entry. We used a VSV pseudotype encoding green fluorescent protein (GFP) instead of the VSV glycoprotein (VSVG) and bearing a SARS2-S (rVSVΔG-GFP/SARS2-S) from the original strain of SARS-CoV-2 (Wu et al., 2020) containing a D614G substitution. The D614G substitution contributes to increased viral transmissibility and has been retained in all SARS-CoV-2 variants of concern (VOCs) (Korber et al., 2020). We also used a VSV pseudotype bearing VSVG (rVSVΔG-GFP/VSVG), which allowed us to identify the compounds that had non-specific or VSV replication-specific inhibitory effects. VSV pseudotypes were used to inoculate HEK293T cells constitutively expressing human ACE2 (HEK293T/ACE2) that had been treated with each compound at 10 μM. At 16 h post-infection (hpi), the GPF-positive (VSV pseudotype-infected) cell and nuclei numbers were determined by high-content imaging analysis (Fig. 1 A). The number of nuclei was determined as an initial assessment of the cytotoxicity of the test compounds. Primary hits were selected on the basis of a strong reduction in the rVSVΔG-GFP/SARS2-S-infected cell number (<25%), but limited impacts on rVSVΔG-GFP/VSVG-infected cell (>75%) number and number of nuclei (>75%) compared with vehicle treatment (Table 1 and Appendix B, Dataset S1).
Fig. 1.
Screening of a US FDA-approved drug library to identify inhibitors of SARS-CoV-2 spike protein (SARS2-S)-mediated cell entry. (A) Screening flow chart. (B) Dose-dependent inhibitory effects of dopamine receptor D2 (DRD2) antagonists on pseudovirus infection. HEK293T/ACE2 cells that had been treated with two-fold serial dilutions of each of the DRD2 antagonists or with vehicle (0.1% DMSO) for 90 min were inoculated with vesicular stomatitis virus (VSV)-based pseudovirus bearing SARS2-S (rVSVΔG-GFP/SARS2-S, SARS2-S) or the VSV glycoprotein (rVSVΔG-GFP/VSVG, VSVG). The DRD2 antagonists and DMSO were present throughout the experimental period. At 16 h post-inoculation, the cells were fixed, and their nuclei were stained with Hoechst 33342. The GFP-positive (virally infected) cells were quantified with a high-content imaging system. The mean value of vehicle-treated, VSV-based pseudovirus-inoculated cells was set to 100%. The cell viability of HEK293T/ACE2 cells treated with two-fold serial dilutions of each of the DRD2 antagonists or with vehicle (0.1% DMSO) for 17.5 h was determined with CellTiter 96 AQueous One Solution Reagent. The mean value of vehicle-treated, uninfected cells was set to 100%. The presented data are the mean ± SD of the results of three replicates. EC50 values for the rVSVΔG-GFP/SARS2-S-inoculated cells are indicated in each graph.
Table 1.
Inhibitory potencya of 24 primary hits against pseudovirus infection identified in the US FDA-approved drug library screening and their pharmacological target molecules.
| Compound | Target | rVSVΔG-GFP/SARS2-S |
rVSVΔG-GFP/VSVG |
||
|---|---|---|---|---|---|
| GFP (%) | Hoechst (%) | GFP (%) | Hoechst (%) | ||
| Bazedoxifene acetate | NR3A (ESR) | 12.26 | 88.96 | 115.01 | 102.51 |
| Toremifene | NR3A (ESR) IGF1R (CD221) |
21.77 | 101.27 | 142.24 | 90.44 |
| Tamoxifen citrate | NR3A (ESR) | 16.27 | 99.67 | 105.72 | 97.14 |
| Toremifene citrate | NR3A (ESR) IGF1R (CD221) |
8.32 | 99.98 | 109.35 | 98.74 |
| Raloxifene hydrochloride | NR3A (ESR) | 9.66 | 101.70 | 97.55 | 103.53 |
| Vandetanib (ZD6474) | VEGFR2 EGFR RET |
20.29 | 104.11 | 107.38 | 99.04 |
| AZD-9291 mesylate | EGFR | 5.36 | 98.67 | 112.06 | 98.67 |
| Prochlorperazine Maleate | DRD2 | 16.56 | 97.05 | 131.30 | 93.26 |
| Thioridazine hydrochloride | DRD2 | 15.80 | 98.59 | 118.64 | 95.38 |
| Fluphenazine hydrochloride | DRD2 | 19.31 | 103.35 | 106.20 | 100.59 |
| Perphenazine | DRD2 | 17.90 | 104.54 | 113.49 | 86.64 |
| Trifluoperazine dihydrochloride | DRD2 | 8.63 | 107.56 | 129.52 | 97.29 |
| Chlorpromazine hydrochloride | HTR2 HRH1 ADRA2 DRD2 CHRM |
24.88 | 98.41 | 95.70 | 97.98 |
| Asenapine Maleate | HTR1A HTR2A HTR2C HTR6 HTR7 DRD2 |
14.81 | 98.20 | 150.43 | 95.98 |
| Sertraline hydrochloride | SLC6A4 (HTT) | 15.85 | 101.27 | 104.62 | 97.33 |
| Trimipramine maleate | SLC6A2 (NAT1) SLC6A4 (HTT) |
23.16 | 103.55 | 90.44 | 99.90 |
| Nortriptyline hydrochloride | SLC6A2 (NAT1) SLC6A4 (HTT) |
17.44 | 81.23 | 91.71 | 76.77 |
| Amoxapine | SLC6A4 (HTT) SLC6A2 (NAT1) |
23.68 | 102.06 | 116.48 | 99.31 |
| Paroxetine hydrochloride | SLC6A4 (HTT) | 21.00 | 102.73 | 112.39 | 98.03 |
| Clemastine fumarate | HRH1 | 12.46 | 106.94 | 104.53 | 101.47 |
| Dronedarone hydrochloride | CACNA1-L KCND3 KCNH2 KCNQ1 KCNJ3 KCNJ5 KCNJ11 ADRA1 ADRB1 |
10.87 | 99.72 | 138.68 | 101.34 |
| Azithromycin dihydrate | 50 S ribosomal subunit | 18.36 | 100.22 | 101.10 | 101.30 |
| Cinacalcet hydrochloride | CASR | 23.68 | 97.33 | 131.09 | 95.94 |
| Imatinib Mesylate (STI571) | BCR-ABL FIP1L1-PDGFRA KIT (CD117) |
20.79 | 96.53 | 103.15 | 94.49 |
The mean value of vehicle-treated cells was set to 100%.
The classification of the 24 primary hits by their target molecules uncovered dopamine receptor D2 (DRD2) as the most common therapeutic target of the primary hits. One of the DRD2 antagonists, asenapine (maleate salt), was also identified as one of four top hits in another US FDA-approved drug library screening (hydrochloride salt version in a previous study) (Xiong et al., 2020), supporting the robustness of our screening system. These seven DRD2 antagonists were selected for further study, and experiments to validate their specificity and assess the correlation between the efficacy and cytotoxicity were conducted with freshly prepared compounds. All seven DRD2 antagonists exhibited dose-dependent inhibitory effects on rVSVΔG-GFP/SARS2-S with 50% effective concentrations (EC50s) ranging from 2.93 to 6.06 and selective indexes (SIs) [SI = 50% cytotoxic concentration (CC50)/EC50] ranging from 4.08 to 14.50 at concentrations that did not cause a significant reduction in rVSVΔG-GFP/VSVG infectivity (Fig. 1B and Table S1).
3.2. The DRD2 antagonists inhibit SARS2-S-mediated membrane fusion without affecting SARS2-S–ACE2 binding
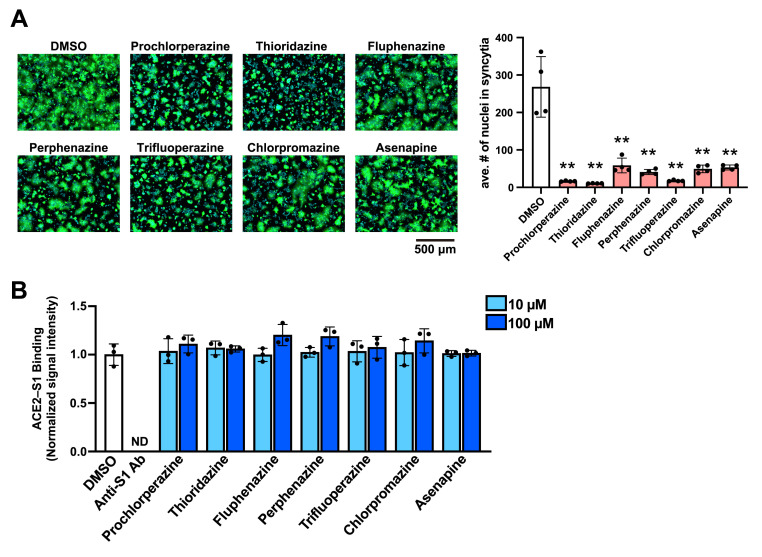
SARS-CoV-2 enters the cell via the cell surface pathway or the endocytic pathway depending on the location where the S2′ priming takes place (Hoffmann et al., 2020a). In either mode of cell entry, membrane fusion is a final and critical process, by which genome RNA is released from the viral particle into the cell cytoplasm for the initiation of genome RNA translation. To determine the effect of the seven DRD2 antagonists on SARS2-S-mediated membrane fusion, we examined whether treatment with the DRD2 antagonists prevented SARS2-S-mediated cell fusion following the co-culture of SARS2-S-expressing HEK293T and HEK293T/ACE2 cells (Ou et al., 2020). HEK293T transfectants expressing SARS2-S and ZsGreen were added onto HEK293T/ACE2 cells treated with the DRD2 antagonists at a concentration of 10 μM. ZsGreen was expressed to fluorescently visualize the area of cytoplasm. After a 4 h co-culture, the magnitude of membrane fusion was evaluated by the average number of nuclei in each syncytium. Consistent with the VSV-pseudotype infection assay results, all of the DRD2 antagonists strongly inhibited SARS2-S-mediated membrane fusion (Fig. 2 A). One of the DRD2 antagonists, chlorpromazine, has been reported to inhibit clathrin-mediated endocytosis (Daniel et al., 2015), and could have affected the cell surface expression of host cell proteins including ACE2 during pretreatment. To address this issue, we examined the effects of the DRD2 antagonists on the cell surface expression of ACE2 by detecting the ACE2 levels in biotinylated cell surface proteins (Fig. S1A) and by flow cytometry using an antibody that recognizes the extracellular domain of ACE2 (Fig. S1B). We confirmed that treatment with any of the DRD2 antagonists did not affect the cell surface expression of ACE2. In agreement with these results, we confirmed the inhibitory effect of the DRD2 antagonists in a fusion assay without pretreatment (Fig. S1C).
Fig. 2.
The dopamine receptor D2 (DRD2) antagonists inhibit the SARS-CoV-2 spike protein (SARS2-S)-mediated membrane fusion but not SARS2-S binding to ACE2. (A) Effect of the DRD2 antagonists on SARS2-S-mediated membrane fusion. HEK293T cells transfected with plasmids expressing SARS2-S and ZsGreen (ZsG) were detached with trypsin and overlaid on HEK293T/ACE2 cells that had been pretreated with 0.1% DMSO or one of the DRD2 antagonists (10 μM) for 90 min. DMSO and the DRD2 antagonists were present throughout the experimental period. After a 4-h incubation, the cells were fixed, and their nuclei were stained with Hoechst 33342. Fluorescent images of the cells were captured (left), and the average number (ave. #) of nuclei in each syncytium was determined with a high-content imaging system (right). The presented data are the mean ± SD of the results of four independent experiments. Statistical significance was determined by comparing the average nuclei numbers of the DRD2 antagonist-treated samples with that of the DMSO-treated samples. **p < 0.01. (B) Inhibitory effect of the DRD2 antagonists on the binding of SARS2-S to ACE2. An ELISA-based SARS2-S–ACE2 binding assay was performed in the presence of 0.1% DMSO, the neutralizing antibody anti-SARS2-S S1 domain (anti-S1 Ab, 5 μg/mL), or one of the DRD2 antagonists (10 μM or 100 μM). The mean chemiluminescent signal intensity of DMSO-treated samples was set to 1. The presented data are the mean ± SD of the results of three replicates. ND, not detected.
Several of the previously reported inhibitors of SARS-CoV-2 cell entry were suggested to exert their inhibitory effect by disturbing the interaction between SARS2-S and the primary cell entry receptor, ACE2 (Fu et al., 2021; Taha et al., 2022; Wang et al., 2021). We then examined whether the DRD2 antagonists interrupted the SARS2-S–ACE2 interaction by performing an enzyme-linked immunosorbent assay (ELISA)-based SARS2-S–ACE2 binding assay. Recombinant SARS2-S receptor-binding domain (rRBD)-coated microplate was incubated with recombinant ACE2 fused with a His tag (rACE2-His) in the presence of a DRD2 antagonist, an anti-S1 neutralization antibody (Anti-S1 Ab), or the treatment vehicle (DMSO). After the microplate was extensively washed, the rRBD-bound rACE2-His was detected using HRP-conjugated anti-His antibody. As expected, anti-S1 Ab strongly blocked rRBD binding to rACE2-His (Fig. 2B). However, the DRD2 antagonists did not affect the interaction between rRBD and rACE2-His at concentrations as high as 100 μM, indicating that the DRD2 antagonists disrupt the SARS2-S binding to a cell host protein other than ACE2.
3.3. The DRD2 antagonists block SARS2-S–NRP-1 interaction
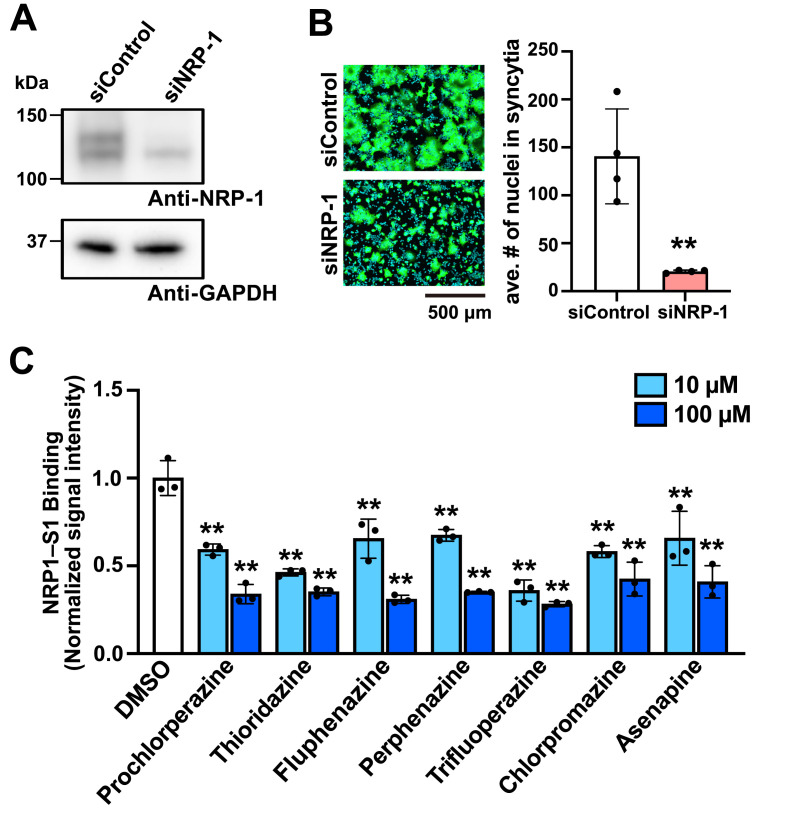
SARS-CoV-2 uses NRP-1 to facilitate ACE2-mediated cell entry. Although there are conflicting results regarding NRP-1 expression in HEK293T cells (Cantuti-Castelvetri et al., 2020; Daly et al., 2020), the inability of the DRD2 antagonists to block SARS2-S binding to ACE2 led us to explore the possibility that the SARS2-S–NRP-1 interaction was disrupted by the DRD2 antagonists. NRP-1 was readily detected in HEK293T/ACE2 cell lysates by western blotting, and its expression was significantly reduced by NRP-1-specific small interfering RNA (siRNA) treatment (Fig. 3 A). In the fusion assay, the formation of syncytia was suppressed when SARS2-S- and ZsGreen-expressing HEK293T cells and HEK293T/ACE2 cells were treated with siRNA against NRP-1 as compared with that when these cells were treated with non-specific siRNA (siControl) (Fig. 3B), indicating that NRP-1 plays an important role in SARS2-S-mediated cell entry in our experimental system. We then asked whether the DRD2 antagonists could inhibit the binding of SARS2-S to NRP-1. For this, we employed an ELISA-based SARS2-S–NRP-1 binding assay, where a rNRP-1-coated microplate was incubated with rS1 in the presence of the DRD2 antagonists and rNRP-1-binding rS1 was detected using anti-S1 Ab. The DRD2 antagonists inhibited the interaction between SARS2-S and NRP-1 at the concentrations as low as 10 μM (Fig. 3C). EG00229 is a small compound that is reported to bind to the b1 domain of NRP-1 preventing SARS2-S–NRP-1 binding (Daly et al., 2020), and can be a benchmark to evaluate the inhibitory potency of the DRD2 compounds on SARS2-S–NRP-1 binding. In line with the previous observation that EG00229 inhibited SARS-CoV-2 infection at 100 μM (Daly et al., 2020), EG00229 required a higher concentration (1 mM) to inhibit SARS2-S–NRP-1 binding (Fig. S2), compared with the DRD2 antagonists, suggesting a correlation between inhibitory effects of the compounds on SARS2-S–NRP-1 binding and SARS2-mediated cell entry.
Fig. 3.
The dopamine receptor D2 (DRD2) antagonists block binding of the spike protein of SARS-CoV-2 (SARS2-S) to neuropilin-1 (NRP-1). (A and B) Role of NRP-1 in SARS2-S-mediated membrane fusion with HEK293T/ACE2 cells. HEK293T/ACE2 cells were transfected with siRNA against NRP-1 (siNRP-1) or with non-targeting siRNA (siControl). At 48 h post-transfection, total cell lysate was prepared, and the protein levels of NRP-1 and GAPDH in the cell lysates were examined by western blotting (A). siNRP-1-treated HEK293T cells transfected with plasmids expressing SARS2-S and ZsGreen (ZsG) were detached with trypsin and overlaid on siNRP-1-treated HEK293T/ACE2 cells. For a control, siControl was used instead of siNRP-1. After a 4-h incubation, the cells were fixed, and their nuclei were stained with Hoechst 33342. Fluorescent images of the cells were captured (B, left), and the average number (ave. #) of nuclei in each syncytium was determined with a high-content imaging system (B, right). The presented data are the mean ± SD of the results of four independent experiments. Statistical significance was determined by a Student's t-test. **p < 0.01. (C) Effects of the DRD2 antagonists on the binding of SARS2-S to NRP-1. An ELISA-based SARS2-S-NRP-1 binding assay was performed in the presence of 0.1% DMSO or one of the DRD2 antagonists (10 μM or 100 μM). The mean absorbance value of the DMSO-treated samples was set to 1. The presented data are the mean ± SD of the results of three replicates. Statistical significance was determined by comparing the absorbance values of the DRD2 antagonist-treated samples with the value for the DMSO-treated samples. **p < 0.01.
Taken together, these results indicate that the tested DRD2 antagonists inhibit SARS2-S-mediated cell entry by blocking the SARS2-S–NRP-1 interaction.
3.4. Alimemazine inhibits SARS-CoV-2 infection
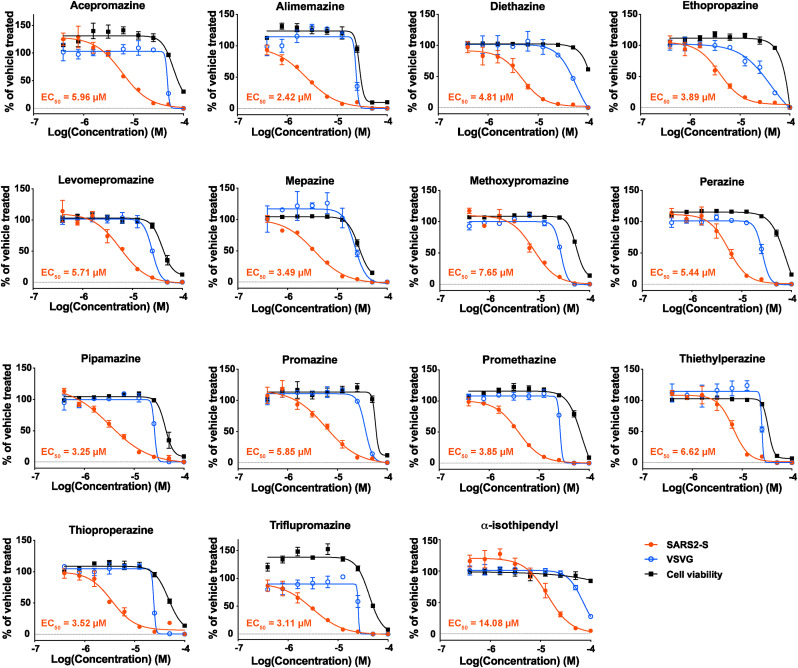
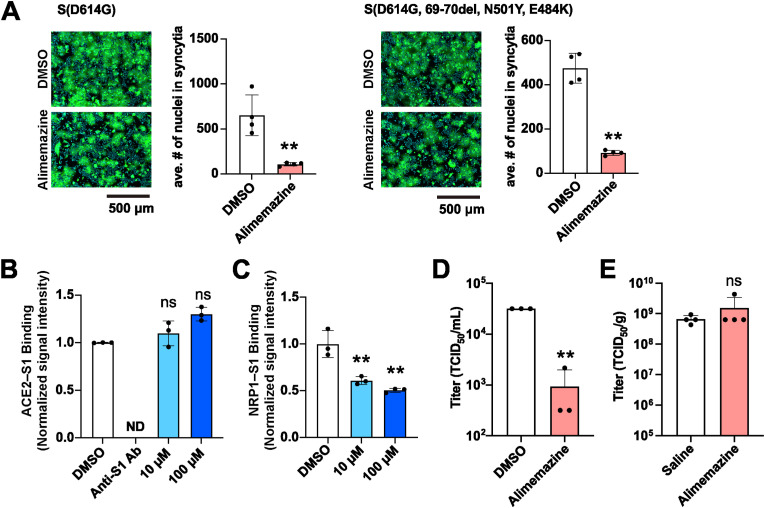
Phenothiazines are a group of compounds that include several antipsychotic drugs already used in clinics. Notably, six out of the seven DRD2 antagonists identified by our screen are phenothiazine derivatives, indicating that phenothiazines may share the ability to inhibit SARS2-S-mediated cell entry. To investigate this possibility, the EC50s and CC50s for an additional 15 commercially available phenothiazines were determined. As expected, all of these additional phenothiazines specifically inhibited SARS2-S-mediated cell entry with variable potencies (EC50 = 2.42–14.08 μM) but did not affect VSVG-mediated cell entry (Fig. 4 and Table S2). Among the compounds we tested, alimemazine (tartrate salt) exhibited the lowest EC50 (2.42 μM) and had a high SI (12.2); thus, we selected it for use in downstream validation assays. Using the fusion assay, we confirmed that alimemazine treatment reduced the formation of syncytia induced by the expression of SARS2-S as well as by a mutant SARS2-S containing a deletion in aa positions 69 to 70 (Δ69–70) and the substitutions of N501Y and E484K, the characteristic mutations found in SARS-CoV-2 VOCs, suggesting that alimemazine could have an inhibitory effect on cell entry by a broad range of SARS-CoV-2 VOCs (Fig. 5 A). In addition, alimemazine inhibited the binding activity of SARS2-S to NRP-1 but not to ACE2 (Fig. 5B and C), suggesting that alimemazine inhibits SARS2-S-mediated cell entry through a mechanism similar to that used by the DRD2 antagonists. The serine protease TMPRSS2 plays a critical role in SARS-CoV-2 infection in vivo (Iwata-Yoshikawa et al., 2022a). We then assessed the inhibitory effect of alimemazine on VSV-based pseudovirus cell entry in Vero E6 cells expressing TMPRSS2 (VeroE6/TMPRSS2). Alimemazine significantly inhibited the SARS2-S-mediated, but not VSVG-mediated, cell entry in VeroE6/TMPRSS2 cells (Fig. S3A) at the concentration (10 μM) that did not affect the VeroE6/TMPRSS2 cell viability (Fig. S3B).
Fig. 4.
Dose-dependent inhibitory effects of phenothiazines on pseudovirus infection. HEK293T/ACE2 cells that had been treated with two-fold serial dilutions of a phenothiazine or with vehicle (0.1% DMSO) for 90 min were inoculated with vesicular stomatitis virus (VSV)-based pseudovirus bearing the spike protein of SARS-CoV-2 (rVSVΔG-GFP/SARS2-S, SARS2-S) or the VSV glycoprotein (rVSVΔG-GFP/VSVG, VSVG). The tested phenothiazine or DMSO was present throughout the experimental period. At 16 h post-inoculation, the cells were fixed, and their nuclei were stained with Hoechst 33342. The number of GFP-positive (virally infected) cells was determined with a high-content imaging system. The mean value of vehicle-treated, VSV-based pseudovirus-inoculated cells was set to 100%. The cell viability of HEK293T/ACE2 cells treated with two-fold serial dilutions of each of the phenothiazines or with vehicle (0.1% DMSO) for 17.5 h was determined with CellTiter 96 AQueous One Solution Reagent. The mean value of the vehicle-treated, uninfected cells was set to 100%. The presented data are the mean ± SD of the results of three replicates. EC50 values for the rVSVΔG-GFP/SARS2-S-inoculated cells are indicated in each graph.
Fig. 5.
Antiviral effect of alimemazine on SARS-CoV-2 multiplication. (A) Effect of alimemazine on SARS2-S-mediated membrane fusion. HEK293T cells transfected with a plasmid expressing SARS2-S (left) or SARS2-S containing 69–70 deletion and the N501Y and E484K substitutions in addition to the D614G substitution (right) together with a plasmid expressing ZsGreen (ZsG) were overlaid on HEK293T/ACE2 cells that had been pretreated with 0.1% DMSO or 10 μM alimemazine for 90 min. DMSO and alimemazine were present throughout the experimental period. After a 4-h incubation, fluorescent images of the cells were captured (left), and the average number (ave. #) of nuclei in each syncytium was determined with a high-content imaging system (right). The presented data are the mean ± SD of the results of four independent experiments. Statistical significance was determined by a Student's t-test. **p < 0.01. (B and C) Effect of alimemazine on SARS2-S binding to ACE2 (B) or to NRP-1 (C). ELISA-based SARS2-S–ACE2 and SARS2-S–NRP-1 binding assays were performed in the presence of DMSO (0.1%), the neutralizing antibody anti-SARS2-S S1 domain (anti-S1 Ab; 5 μg/mL, used only in B), or alimemazine (10 μM or 100 μM). The mean chemiluminescence (B) and absorbance (C) values of DMSO-treated samples were each set to 1. The presented data are the mean ± SD of the results of three replicates. Statistical significance was determined by comparing the chemiluminescence (B) and absorbance (C) values of the experimentally treated samples with the values for the DMSO-treated samples. ns, p > 0.05, not significant; **p < 0.01; ND, not detected. (D) Effect of alimemazine on SARS-CoV-2 multiplication in cultured cells. HEK293T/ACE2 cells that had been pretreated with 0.1% DMSO or 10 μM alimemazine for 90 min were inoculated (MOI = 0.1) with SARS-CoV-2 TY7-501 strain. After 24 h, the cells in the tissue culture supernatant were collected, and the virus titers were determined by a TCID50 assay. The presented data are the mean and SD of the results of three independent experiments. Statistical significance was determined by a Student's t-test. **p < 0.01. (E) Effect of alimemazine on SARS-CoV-2 multiplication in mice. Eight-week-old BALB/c mice were treated with alimemazine (10 mg/kg) or saline by oral gavage 1 h prior to and 24 h after intranasal inoculation with SARS-CoV-2 TY7-501 strain (2 × 104 TCID50 per mouse). At 2 days post-viral inoculation, lung tissues were harvested, and the tissue viral titers were determined by a TCID50 assay. The presented data are the mean and SD of the results from four mice per group. Statistical significance was determined by a Student's t-test.
Next, we assessed the effect of alimemazine on SARS-CoV-2 multiplication in cultured cells. HEK293T/ACE2 or VeroE6/TMPRSS2 cells were inoculated with a gamma SARS-CoV-2 variant, strain TY7-501, in the presence of alimemazine. Viral titers in the tissue culture supernatants were determined at 24 hpi. Consistent with the inhibitory effect of alimemazine on rVSVΔG-GFP/SARS2-S infectivity, alimemazine significantly reduced the production of infectious SARS-CoV-2 progeny (Fig. 5D and Fig. S3C). This clear inhibitory effect of alimemazine on SARS-CoV-2 multiplication in HEK293T/ACE2 and VeroE6/TMPRSS2 cells motivated us to investigate the efficacy of alimemazine in vivo. For this, we employed a mouse infection model of SARS-CoV-2 gamma variants to gain an assessment of the in vivo efficacy of alimemazine. SARS-CoV-2 gamma variants contain an N501Y mutation in the spike protein, which significantly increases the affinity to mouse ACE2 (Imai et al., 2021; Li et al., 2021; Winkler et al., 2022). Accordingly, intranasal inoculation of a SARS-CoV-2 gamma variant to a wildtype (WT) mouse results in a productive infection in the lungs, albeit without developing systemic manifestations, such as bodyweight loss (Imai et al., 2021). To investigate the antiviral effect of alimemazine in vivo, WT BALB/c mice were intranasally inoculated with SARS-CoV-2 strain TY7-501, and the lung viral titers were determined at 2 days post-inoculation. Treatment with two doses of 10 mg/kg alimemazine (at 1 h prior to and 24 h after virus inoculation, respectively) did not decrease the lung viral titer (Fig. 5E).
4. Discussion
Substantial drug repurposing efforts have resulted in the identification of several candidate drugs that inhibit SARS-CoV-2 cell entry. The mechanisms of action of those identified cell entry inhibitors involve disruption of the SARS2-S–ACE2 interaction (Fu et al., 2021; Taha et al., 2022; Wang et al., 2021) and reduction of the host protease activity (Chen et al., 2021; Hoffmann et al., 2020b; Yu et al., 2022) but are otherwise not well defined. In the present study, we showed that phenothiazines inhibit SARS2-S-mediated cell entry and block SARS2-S–NRP-1 binding. This novel mechanism of action may permit the use of phenothiazines not only as monotherapy options but also as drugs to use in combination with other antivirals that have distinct mechanisms of action to ideally produce synergistic antiviral effects. One advantage of combination therapy lies in its use of lower doses of each drug, thus reducing the risk of severe side effects and emergence of drug-resistant viral variants. Additionally, phenothiazines can be used as a molecular probe to facilitate the investigation of NRP-1-dependent cell entry by SARS-CoV-2.
The phenothiazine moiety has been observed to exhibit versatile biological properties, which led to the synthesis of its derivatives. Many such drugs are now used in clinics for the treatment of a variety of diseases, including psychotic diseases, targeting several neurotransmitter receptors and transporters, such as dopamine and histamine receptors (Jaszczyszyn et al., 2012; Pluta et al., 2011). Furthermore, several phenothiazines have been reported to have antiviral potential against hepatitis B virus, influenza virus, measles virus, human immunodeficiency virus, John Cunningham virus, Japanese encephalitis virus, hepatitis C virus, mouse hepatitis virus, chikungunya virus, SARS-CoV, Middle East respiratory syndrome coronavirus (MERS-CoV), Zika virus, dengue virus, Ebola virus, and Epstein-Barr virus (EBV) (Anderson et al., 2019; Chu et al., 2006; Nugent and Shanley, 1984; Otreba et al., 2020; Varga et al., 2017). On the basis of the diverse biological activities of phenothiazines, their anti-SARS-CoV-2 activities have been explored in other work (Lu et al., 2021). Phenothiazines were previously shown to associate with the membrane fraction of ACE2-expressing HEK293T cells by cell membrane chromatography (CMC) (Lu et al., 2021). In contrast to our results, trifluoperazine, chlorpromazine, and thioridazine reportedly bound to purified ACE2 with dissociation constant (KD) values of 33.3, 13.8, and 7.88 μM, respectively, indicating that these phenothiazines exert an antiviral effect through their binding to ACE2. However, the relatively high KD values compared with EC50 values determined in this study suggest that the contribution of the association of phenothiazines with ACE2 to the inhibition of SARS-CoV-2 cell entry may be limited. Moreover, the membrane fraction used in the CMC may have contained NRP-1 and, therefore, the strong retention of phenothiazines in the column could have been due to their association with NRP-1 rather than with ACE2.
HEK293T cells expressing SARS2-S were reported to fuse to ACE2-expessing HEK293T cells without trypsin treatment or the presence of TMPRSS2, but those expressing the SARS-CoV spike protein (SARS1-S) were unable to fuse (Ou et al., 2020). While the relevance of trypsin-uncleaved SARS2-S-mediated membrane fusion in SARS-CoV-2 cell entry remains to be determined, insertion of the multibasic furin cleavage site in SARS2-S is responsible for the marked phenotypic difference between SARS2-S and SARS1-S in the fusion assay (Hornich et al., 2021; Xia et al., 2020). SARS2-S-mediated cell entry requires proteolytic cleavage of SARS2-S by cellular proteases including cathepsins and TMPRSS2 (Hoffmann et al., 2020a). To mimic SARS2-S processing by cellular proteases, we treated SARS2-S-expressing cells with trypsin in the fusion assay. Previous reports showed that trypsin treatment did not significantly promote SARS2-S-mediated membrane fusion in some cell lines expressing TMPRSS2, indicating that trypsin treatment can be a functional surrogate of SARS2-S cleavage by TMPRSS2 (Koch et al., 2021; Ou et al., 2020). In our fusion assay, SARS2-S-expressing HEK293T cells were treated with trypsin and co-cultured with HEK293T/ACE2 for 4 h prior to cell fixation to evaluate the levels of membrane fusion. It should be noted that SARS2-S newly expressed on the cell surface during the 4-h co-culture was not exposed to trypsin, and trypsin-uncleaved SARS2-S may have partially contributed to membrane fusion in the fusion assay. As such, the substantial reduction in syncytia formation observed following treatment with the DRD2 antagonists, siNRP-1, or alimemazine indicates that the reduction was due to the inhibition of membrane fusion mediated by trypsin-cleaved and trypsin-uncleaved SARS2-S.
Several cationic amphiphilic drugs (CADs) have been reported to exhibit antiviral activity against diverse enveloped viruses, including Ebola and Marburg (MARV) viruses, Lassa virus, hepatitis C virus, Japanese encephalitis virus, SARS-CoV, MERS-CoV, and EBV (Klintworth et al., 2015; Nemerow and Cooper, 1984; Salata et al., 2017). A comprehensive physicochemical analysis of CADs in MARV cell entry revealed that the antiviral activity of CADs is associated with their ability to induce cellular phospholipidosis (Gunesch et al., 2020). In line with this study, Tummino et al. demonstrated a correlation, in the same concentration range, between the drug-induced phospholipidosis and anti-SARS-CoV-2 effects of CADs (Tummino et al., 2021). Phospholipidosis may disrupt lipid homeostasis, causing the disruption of double membrane vesicles (DMVs) and therefore suppressing the propagation of viruses whose life cycle is dependent on DMVs. The cationic amphiphilic nature of phenothiazines raises the possibility that phenothiazines exert their anti-SARS-CoV-2 activity via the induction of phospholipidosis. However, we found that phenothiazines blocked the interaction between purified recombinant SARS2-S and NRP-1, strongly suggesting that they have specific target-based antiviral activity. The diverse structural features of anti-SARS-CoV-2 phenothiazines will facilitate structure-activity relationship (SAR) approaches to the development of novel antivirals with reduced risks of undesirable pharmacological actions, including phospholipidosis.
The availability of small animal models that mimic the natural course of infection is a critical factor in promoting the rapid development of antivirals. A mouse infection model in particular would be ideal because vast resources for physiological, biochemical, immunological, and genetic analyses in mice already exist. However, mice are refractory to infection with the original strain of SARS-CoV-2, which can be explained mainly by the low binding affinity of SARS2-S to mouse ACE2 (mACE2) (Zhou et al., 2020). To address this issue, genetically engineered mice in which human ACE2 (hACE2) is systemically expressed (hACE2-transgenic mice) or is expressed in place of mACE2 (hACE2-knock-in mice) have been developed (Bao et al., 2020; Jiang et al., 2020; Winkler et al., 2022). In addition, mouse-adapted strains of SARS-CoV-2 have been generated by the serial passage of SARS-CoV-2 in mice (Gu et al., 2020; Huang et al., 2021; Iwata-Yoshikawa et al., 2022b; Leist et al., 2020). Genetic and biochemical analyses of the mouse-adapted SARS-CoV-2 strains have identified several mutations in the spike protein that contribute to an enhanced binding of SARS2-S to mACE2 and thereby promote productive SARS-CoV-2 infection in mice. Those identified mutations include the N501Y substitution that is also present in SARS-CoV-2 lineages B.1.1.7, B.1.351 (alpha variants), and P.1 (gamma variant). In this study, we used a lineage P.1 TY7-501 gamma variant of SARS-CoV-2, which has the N501Y mutation, to examine the in vivo efficacy of alimemazine. However, the clear in vitro antiviral effect of alimemazine did not translate in this mouse infection model. Interestingly, three amino acid differences, between human NRP-1 (hNRP-1) and NRP-2 (hNRP-2), located in the L1 loop of b1 domain, dictate the binding capacity for VEGF-A, which shares the same binding site as SARS2-S (Parker et al., 2012a, 2012b). Likewise, mouse NRP-1 (mNRP-1) contains in its L1 loop an amino acid that is different from the one at the corresponding position of hNRP-1. This indicates that SARS2-S binds to mNRP-1 in a manner distinct from that of SARS2-S binding to hNRP-1. Therefore, possible explanations for the lack of in vivo efficacy of alimemazine are that alimemazine cannot interfere with the SARS2-S–mNRP-1 interaction or that SARS-CoV-2 enters the cells via a mNRP-1-independent manner in the mouse infection model. The development of a novel mouse model that can reproduce human NRP-1-mediated cell entry by SARS-CoV-2 will significantly facilitate research on phenothiazine-based, and other novel, antivirals against SARS-CoV-2 that disrupt the SARS2-S–NRP-1 interaction.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Yoshiharu Matsuura for providing us with a pseudotyped VSV system. We also thank Katie Oakley, PhD, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript. This research was supported in part by the Japan Agency for Medical Research and Development (AMED) (JP20nk0101602 and JP20nf0101623) (M.I.).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2022.105481.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Al-Salama Z.T., Scott L.J. Baricitinib: a review in rheumatoid arthritis. Drugs. 2018;78:761–772. doi: 10.1007/s40265-018-0908-4. [DOI] [PubMed] [Google Scholar]
- Anderson A.G., Gaffy C.B., Weseli J.R., Gorres K.L. 2019. Inhibition of Epstein-Barr Virus Lytic Reactivation by the Atypical Antipsychotic Drug Clozapine. Viruses 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L.N., Deng W., Huang B.Y., Gao H., Liu J.N., Ren L.L., Wei Q., Yu P., Xu Y.F., Qi F.F., Qu Y.J., Li F.D., Lv Q., Wang W.L., Xue J., Gong S.R., Liu M.Y., Wang G.P., Wang S.Y., Song Z.Q., Zhao L.N., Liu P.P., Zhao L., Ye F., Wang H.J., Zhou W.M., Zhu N., Zhen W., Yu H.S., Zhang X.J., Guo L., Chen L., Wang C.H., Wang Y., Wang X.M., Xiao Y., Sun Q.M., Liu H.Q., Zhu F.L., Ma C.X., Yan L.M., Yang M.L., Han J., Xu W.B., Tan W.J., Peng X.Z., Jin Q., Wu G.Z., Qin C. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M.D., Ruiz-Palacios G.M., Benfield T., Fatkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C., Members A.-S.G. Remdesivir for the treatment of covid-19 - final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M., Smura T., Levanov L., Szirovicza L., Tobi A., Kallio-Kokko H., Osterlund P., Joensuu M., Meunier F.A., Butcher S.J., Winkler M.S., Mollenhauer B., Helenius A., Gokce O., Teesalu T., Hepojoki J., Vapalahti O., Stadelmann C., Balistreri G., Simons M. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Lear T.B., Evankovich J.W., Larsen M.B., Lin B., Alfaras I., Kennerdell J.R., Salminen L., Camarco D.P., Lockwood K.C., Tuncer F., Liu J., Myerburg M.M., McDyer J.F., Liu Y., Finkel T., Chen B.B. A high-throughput screen for TMPRSS2 expression identifies FDA-approved compounds that can limit SARS-CoV-2 entry. Nat. Commun. 2021;12:3907. doi: 10.1038/s41467-021-24156-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu V.C., McElroy L.J., Ferguson A.D., Bauman B.E., Whittaker G.R. Avian infectious bronchitis virus enters cells via the endocytic pathway. Adv. Exp. Med. Biol. 2006;581:309–312. doi: 10.1007/978-0-387-33012-9_54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly J.L., Simonetti B., Klein K., Chen K.E., Williamson M.K., Anton-Plagaro C., Shoemark D.K., Simon-Gracia L., Bauer M., Hollandi R., Greber U.F., Horvath P., Sessions R.B., Helenius A., Hiscox J.A., Teesalu T., Matthews D.A., Davidson A.D., Collins B.M., Cullen P.J., Yamauchi Y. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J.A., Chau N., Abdel-Hamid M.K., Hu L., von Kleist L., Whiting A., Krishnan S., Maamary P., Joseph S.R., Simpson F., Haucke V., McCluskey A., Robinson P.J. Phenothiazine-derived antipsychotic drugs inhibit dynamin and clathrin-mediated endocytosis. Traffic. 2015;16:635–654. doi: 10.1111/tra.12272. [DOI] [PubMed] [Google Scholar]
- Fu W., Chen Y., Wang K., Hettinghouse A., Hu W., Wang J.Q., Lei Z.N., Chen Z.S., Stapleford K.A., Liu C.J. Repurposing FDA-approved drugs for SARS-CoV-2 through an ELISA-based screening for the inhibition of RBD/ACE2 interaction. Protein Cell. 2021;12:586–591. doi: 10.1007/s13238-020-00803-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.Q., Wang Y., Teng Y., Zhao Z., Cui Y., Li Y., Li X.F., Li J., Zhang N.N., Yang X., Chen S., Guo Y., Zhao G., Wang X., Luo D.Y., Wang H., Yang X., Li Y., Han G., He Y., Zhou X., Geng S., Sheng X., Jiang S., Sun S., Qin C.F., Zhou Y. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunesch A.P., Zapatero-Belinchon F.J., Pinkert L., Steinmann E., Manns M.P., Schneider G., Pietschmann T., Bronstrup M., von Hahn T. Filovirus antiviral activity of cationic amphiphilic drugs is associated with lipophilicity and ability to induce phospholipidosis. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00143-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280 e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Schroeder S., Kleine-Weber H., Muller M.A., Drosten C., Pohlmann S. Nafamostat mesylate blocks activation of SARS-CoV-2: new treatment option for COVID-19. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00754-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornich B.F., Grosskopf A.K., Schlagowski S., Tenbusch M., Kleine-Weber H., Neipel F., Stahl-Hennig C., Hahn A.S. SARS-CoV-2 and SARS-CoV spike-mediated cell-cell fusion differ in their requirements for receptor expression and proteolytic activation. J. Virol. 2021;95 doi: 10.1128/JVI.00002-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K., Zhang Y., Hui X., Zhao Y., Gong W., Wang T., Zhang S., Yang Y., Deng F., Zhang Q., Chen X., Yang Y., Sun X., Chen H., Tao Y.J., Zou Z., Jin M. Q493K and Q498H substitutions in Spike promote adaptation of SARS-CoV-2 in mice. EBioMedicine. 2021;67 doi: 10.1016/j.ebiom.2021.103381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Halfmann P.J., Yamayoshi S., Iwatsuki-Horimoto K., Chiba S., Watanabe T., Nakajima N., Ito M., Kuroda M., Kiso M., Maemura T., Takahashi K., Loeber S., Hatta M., Koga M., Nagai H., Yamamoto S., Saito M., Adachi E., Akasaka O., Nakamura M., Nakachi I., Ogura T., Baba R., Fujita K., Ochi J., Mitamura K., Kato H., Nakajima H., Yagi K., Hattori S.I., Maeda K., Suzuki T., Miyazato Y., Valdez R., Gherasim C., Furusawa Y., Okuda M., Ujie M., Lopes T.J.S., Yasuhara A., Ueki H., Sakai-Tagawa Y., Eisfeld A.J., Baczenas J.J., Baker D.A., O'Connor S.L., O'Connor D.H., Fukushi S., Fujimoto T., Kuroda Y., Gordon A., Maeda K., Ohmagari N., Sugaya N., Yotsuyanagi H., Mitsuya H., Suzuki T., Kawaoka Y. Characterization of a new SARS-CoV-2 variant that emerged in Brazil. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2106535118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Yoshikawa N., Kakizaki M., Shiwa-Sudo N., Okura T., Tahara M., Fukushi S., Maeda K., Kawase M., Asanuma H., Tomita Y., Takayama I., Matsuyama S., Shirato K., Suzuki T., Nagata N., Takeda M. Essential role of TMPRSS2 in SARS-CoV-2 infection in murine airways. Nat. Commun. 2022;13:6100. doi: 10.1038/s41467-022-33911-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Yoshikawa N., Shiwa N., Sekizuka T., Sano K., Ainai A., Hemmi T., Kataoka M., Kuroda M., Hasegawa H., Suzuki T., Nagata N. A lethal mouse model for evaluating vaccine-associated enhanced respiratory disease during SARS-CoV-2 infection. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abh3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaszczyszyn A., Gąsiorowski K., Świątek P., Malinka W., Cieślik-Boczula K., Petrus J., Czarnik-Matusewicz B. Chemical structure of phenothiazines and their biological activity. Pharmacol. Rep. 2012;64:16–23. doi: 10.1016/s1734-1140(12)70726-0. [DOI] [PubMed] [Google Scholar]
- Jiang R.D., Liu M.Q., Chen Y., Shan C., Zhou Y.W., Shen X.R., Li Q., Zhang L., Zhu Y., Si H.R., Wang Q., Min J., Wang X., Zhang W., Li B., Zhang H.J., Baric R.S., Zhou P., Yang X.L., Shi Z.L. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell. 2020;182:50–58 e58. doi: 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klintworth A., Nolden T., Westhaus S., Rohrmann K., David S., Manns M.P., Finke S., Ciesek S., von Hahn T. Cationic amphiphilic drugs enhance entry of lentiviral particles pseudotyped with rabies virus glycoprotein into non-neuronal cells. Antivir. Res. 2015;124:122–131. doi: 10.1016/j.antiviral.2015.10.017. [DOI] [PubMed] [Google Scholar]
- Koch J., Uckeley Z.M., Doldan P., Stanifer M., Boulant S., Lozach P.Y. TMPRSS2 expression dictates the entry route used by SARS-CoV-2 to infect host cells. EMBO J. 2021;40 doi: 10.15252/embj.2021107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Sheffield C.-G.G., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827 e819. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist S.R., Dinnon K.H., 3rd, Schafer A., Tse L.V., Okuda K., Hou Y.J., West A., Edwards C.E., Sanders W., Fritch E.J., Gully K.L., Scobey T., Brown A.J., Sheahan T.P., Moorman N.J., Boucher R.C., Gralinski L.E., Montgomery S.A., Baric R.S. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell. 2020;183:1070–1085 e1012. doi: 10.1016/j.cell.2020.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Nie J., Wu J., Zhang L., Ding R., Wang H., Zhang Y., Li T., Liu S., Zhang M., Zhao C., Liu H., Nie L., Qin H., Wang M., Lu Q., Li X., Liu J., Liang H., Shi Y., Shen Y., Xie L., Zhang L., Qu X., Xu W., Huang W., Wang Y. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell. 2021;184:2362–2371 e2369. doi: 10.1016/j.cell.2021.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Hou Y., Ge S., Wang X., Wang J., Hu T., Lv Y., He H., Wang C. Screened antipsychotic drugs inhibit SARS-CoV-2 binding with ACE2 in vitro. Life Sci. 2021;266 doi: 10.1016/j.lfs.2020.118889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Center for Immunization and Respiratory Diseases (NCIRD), Division of Viral Diseases Science brief: COVID-19 vaccines and vaccination, CDC COVID-19 science briefs. Centers for Disease Control and Prevention (US) 2020 [PubMed] [Google Scholar]
- Nemerow G.R., Cooper N.R. Infection of B lymphocytes by a human herpesvirus, Epstein-Barr virus, is blocked by calmodulin antagonists. Proc. Natl. Acad. Sci. U. S. A. 1984;81:4955–4959. doi: 10.1073/pnas.81.15.4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent K.M., Shanley J.D. Verapamil inhibits influenza A virus replication. Arch. Virol. 1984;81:163–170. doi: 10.1007/BF01309305. [DOI] [PubMed] [Google Scholar]
- Otreba M., Kosmider L., Rzepecka-Stojko A. Antiviral activity of chlorpromazine, fluphenazine, perphenazine, prochlorperazine, and thioridazine towards RNA-viruses. A review. Eur. J. Pharmacol. 2020;887 doi: 10.1016/j.ejphar.2020.173553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M.W., Xu P., Guo H.F., Vander Kooi C.W. Mechanism of selective VEGF-A binding by neuropilin-1 reveals a basis for specific ligand inhibition. PLoS One. 2012;7 doi: 10.1371/journal.pone.0049177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M.W., Xu P., Li X., Vander Kooi C.W. Structural basis for selective vascular endothelial growth factor-A (VEGF-A) binding to neuropilin-1. J. Biol. Chem. 2012;287:11082–11089. doi: 10.1074/jbc.M111.331140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta K., Morak-Mlodawska B., Jelen M. Recent progress in biological activities of synthesized phenothiazines. Eur. J. Med. Chem. 2011;46:3179–3189. doi: 10.1016/j.ejmech.2011.05.013. [DOI] [PubMed] [Google Scholar]
- Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Pache L., Burgstaller-Muehlbacher S., De Jesus P.D., Teriete P., Hull M.V., Chang M.W., Chan J.F., Cao J., Poon V.K., Herbert K.M., Cheng K., Nguyen T.H., Rubanov A., Pu Y., Nguyen C., Choi A., Rathnasinghe R., Schotsaert M., Miorin L., Dejosez M., Zwaka T.P., Sit K.Y., Martinez-Sobrido L., Liu W.C., White K.M., Chapman M.E., Lendy E.K., Glynne R.J., Albrecht R., Ruppin E., Mesecar A.D., Johnson J.R., Benner C., Sun R., Schultz P.G., Su A.I., Garcia-Sastre A., Chatterjee A.K., Yuen K.Y., Chanda S.K. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin R. Baricitinib is first approved COVID-19 immunomodulatory treatment. JAMA. 2022;327:2281. doi: 10.1001/jama.2022.9846. [DOI] [PubMed] [Google Scholar]
- Rubin D., Chan-Tack K., Farley J., Sherwat A. FDA approval of remdesivir - a step in the right direction. N. Engl. J. Med. 2020;383:2598–2600. doi: 10.1056/NEJMp2032369. [DOI] [PubMed] [Google Scholar]
- Salata C., Calistri A., Parolin C., Baritussio A., Palu G. Antiviral activity of cationic amphiphilic drugs. Expert Rev. Anti Infect. Ther. 2017;15:483–492. doi: 10.1080/14787210.2017.1305888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooley R.T., Carlin A.F., Beadle J.R., Valiaeva N., Zhang X.Q., Clark A.E., McMillan R.E., Leibel S.L., McVicar R.N., Xie J., Garretson A.F., Smith V.I., Murphy J., Hostetler K.Y. Rethinking remdesivir: synthesis, antiviral activity, and pharmacokinetics of oral lipid prodrugs. Antimicrob. Agents Chemother. 2021;65 doi: 10.1128/AAC.01155-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha Z., Arulanandam R., Maznyi G., Godbout E., Carter-Timofte M.E., Kurmasheva N., Reinert L.S., Chen A., Crupi M.J.F., Boulton S., Laroche G., Phan A., Rezaei R., Alluqmani N., Jirovec A., Acal A., Brown E.E.F., Singaravelu R., Petryk J., Idorn M., Potts K.G., Todesco H., John C., Mahoney D.J., Ilkow C.S., Giguere P., Alain T., Cote M., Paludan S.R., Olagnier D., Bell J.C., Azad T., Diallo J.S. Identification of FDA-approved bifonazole as a SARS-CoV-2 blocking agent following a bioreporter drug screen. Mol. Ther. 2022 doi: 10.1016/j.ymthe.2022.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummino T.A., Rezelj V.V., Fischer B., Fischer A., O'Meara M.J., Monel B., Vallet T., White K.M., Zhang Z., Alon A., Schadt H., O'Donnell H.R., Lyu J., Rosales R., McGovern B.L., Rathnasinghe R., Jangra S., Schotsaert M., Galarneau J.R., Krogan N.J., Urban L., Shokat K.M., Kruse A.C., Garcia-Sastre A., Schwartz O., Moretti F., Vignuzzi M., Pognan F., Shoichet B.K. Drug-induced phospholipidosis confounds drug repurposing for SARS-CoV-2. Science. 2021;373:541–547. doi: 10.1126/science.abi4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga B., Csonka A., Csonka A., Molnar J., Amaral L., Spengler G. Possible biological and clinical applications of phenothiazines. Anticancer Res. 2017;37:5983–5993. doi: 10.21873/anticanres.12045. [DOI] [PubMed] [Google Scholar]
- Wang G., Yang M.L., Duan Z.L., Liu F.L., Jin L., Long C.B., Zhang M., Tang X.P., Xu L., Li Y.C., Kamau P.M., Yang L., Liu H.Q., Xu J.W., Chen J.K., Zheng Y.T., Peng X.Z., Lai R. Dalbavancin binds ACE2 to block its interaction with SARS-CoV-2 spike protein and is effective in inhibiting SARS-CoV-2 infection in animal models. Cell Res. 2021;31:17–24. doi: 10.1038/s41422-020-00450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler E.S., Chen R.E., Alam F., Yildiz S., Case J.B., Uccellini M.B., Holtzman M.J., Garcia-Sastre A., Schotsaert M., Diamond M.S. SARS-CoV-2 causes lung infection without severe disease in human ACE2 knock-in mice. J. Virol. 2022;96 doi: 10.1128/JVI.01511-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Lan Q., Su S., Wang X., Xu W., Liu Z., Zhu Y., Wang Q., Lu L., Jiang S. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal Transduct. Targeted Ther. 2020;5:92. doi: 10.1038/s41392-020-0184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H.L., Cao J.L., Shen C.G., Ma J., Qiao X.Y., Shi T.S., Ge S.X., Ye H.M., Zhang J., Yuan Q., Zhang T.Y., Xia N.S. Several FDA-approved drugs effectively inhibit SARS-CoV-2 infection in vitro. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.609592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Zheng X., Zhou B., Li J., Chen M., Deng R., Wong G., Lavillette D., Meng G. SARS-CoV-2 spike engagement of ACE2 primes S2' site cleavage and fusion initiation. Proc. Natl. Acad. Sci. U. S. A. 2022;119 doi: 10.1073/pnas.2111199119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.