Abstract
The cystic fibrosis transmembrane conductance regulator (CFTR) is a chloride ion channel that also serves as a receptor for entry of Pseudomonas aeruginosa and Salmonella enterica serovar Typhi into epithelial cells. To evaluate heterogeneity in CFTR protein expression in cultured cells and the effect of heterogeneity on internalization of different P. aeruginosa and serovar Typhi strains, we used two-color flow cytometry and confocal laser microscopy to study bacterial uptake by Madin-Darby canine kidney (MDCK) type I epithelial cells stably expressing a green fluorescent protein (GFP)-CFTR fusion construct (MDCK–GFP-CFTR cells). We found a strong correlation between cell size and GFP-CFTR protein expression, with 60 to 70% of cells expressing low levels of GFP-CFTR protein, 20 to 30% expressing intermediate levels, and <10% expressing high levels. The cells were sorted into low-, intermediate-, or high-level producers of CFTR protein; in vitro growth of each sorted population yielded the same distribution of CFTR protein expression as that in the original population. Cells expressing either low or high levels of CFTR protein internalized bacteria poorly; maximal bacterial uptake occurred in the cells expressing intermediate levels of CFTR protein. Treatment of MDCK cells with sodium butyrate markedly enhanced the production of CFTR protein without increasing cell size; butyrate treatment also increased the proportion of cells with internalized bacteria. However, there were fewer bacteria per butyrate-treated cell and, for P. aeruginosa, there was an overall decrease in the total level of bacterial uptake. The most highly ingested bacterial strains were internalized by fewer total MDCK–GFP-CFTR cells, indicating preferential bacterial uptake by a minority of epithelial cells within a given culture. Confocal fluorescence microscopy showed that P. aeruginosa and serovar Typhi induced cytoplasmic accumulation of CFTR protein close to the plasma membrane where the bacteria were adherent. These results show that within a population of MDCK–GFP-CFTR cells, there are cells with markedly different abilities to ingest bacteria via CFTR, the majority of the P. aeruginosa and serovar Typhi cells are ingested by the one-fourth to one-third of the cells that exhibit an intermediate size and level of CFTR protein expression, and overexpression of the CFTR receptor does not increase total bacterial uptake but rather allows more epithelial cells to ingest fewer total bacteria.
Although the cystic fibrosis transmembrane conductance regulator (CFTR) has been principally characterized as a chloride ion channel, it also functions as an epithelial cell receptor for internalization of Pseudomonas aeruginosa and Salmonella enterica serovar Typhi. P. aeruginosa is the predominant cause of chronic respiratory infections leading to pulmonary failure and death in patients with CF (16, 18). We have proposed that epithelial cell uptake of P. aeruginosa is critical for normal bacterial clearance from the lung epithelium, promoting microbial elimination by both desquamation of internalized bacterial organisms and initiation of innate antibacterial host inflammatory responses (17). Most mutations in the CFTR gene that lead to the severe phenotype, characterized by chronic mucoid P. aeruginosa pulmonary infection, result in little or no CFTR membrane protein (e.g., the ΔF508 CFTR allele). Among common bacterial respiratory pathogens, CFTR protein has been shown to be a receptor only for P. aeruginosa (18). Lack of CFTR protein-mediated uptake of P. aeruginosa may account for the strong association between severe CF and infection with this pathogen.
CFTR protein-mediated gastrointestinal epithelial cell internalization of serovar Typhi results in submucosal translocation of this pathogen (15). Uptake and translocation of serovar Typhi is markedly reduced in heterozygote murine tissues expressing one wild-type and one ΔF508 Cftr allele and is completely absent in transgenic mice homozygous for the ΔF508 Cftr allele (15). These findings are the basis for the hypothesis that increased resistance to typhoid fever provides the heterozygote advantage for maintaining the ΔF508 CFTR allele at levels of 4 to 5% in some populations of European ancestry.
Previous experiments analyzing bacterium-CFTR protein interactions have used gentamicin exclusion assays, in which internalized bacteria are enumerated differentially from extracellular bacteria by the use of gentamicin to kill the extracellular bacteria 3 to 4 h after inoculation onto a population of 105 to 106 epithelial cells. While this is a highly useful assay to measure overall bacterial uptake, it does not allow analysis of nonuniform responses of individual epithelial cells to the bacterial inoculum. Thus, important aspects of the dynamics of CFTR protein responses to bacterial infection among cells ingesting bacteria would be missed in such assays, as would an understanding of the variance within an epithelial cell population in the capacity of individual cells to ingest bacteria. Most epithelial cells express only low levels of CFTR protein, but accumulation and turnover of this protein through the plasma membrane are markedly enhanced by infection (16). Madin-Darby canine kidney (MDCK) type I epithelial cells have been studied by others for their interactions with P. aeruginosa (2, 5, 8), but these studies have focused on the effects of cell polarity and susceptibility of MDCK cells to bacterial binding and cytotoxicity. Therefore, to clarify further the manner by which CFTR protein responds to, interacts with, and mediates translocation of P. aeruginosa and serovar Typhi from the cell surface into the cell, we analyzed bacterial uptake by two-color flow cytometry and confocal fluorescence microscopy with stained bacteria and MDCK cells stably transfected with a green fluorescent protein (GFP)-CFTR expression vector in which GFP is ligated to the N terminus of wild-type CFTR protein.
MATERIALS AND METHODS
Bacterial strains.
The P. aeruginosa strains used were PAO1, a serogroup O5 laboratory strain; PAC557, a strain that produces high levels of the bacterial ligand for CFTR protein, comprising the complete lipopolysaccharide (LPS) core but no O side chains; strains 324 and 149, clinical isolates from infected patients with CF obtained early in the course of infection (LPS smooth, nonmucoid strains); strain 6294, a serogroup O6 isolate from a corneal infection; and P. aeruginosa PAO1 algC::tet, a transposon mutant lacking a complete LPS core (9). Serovar Typhi strains were Ty2 and 1068, the former obtained from the American Type Culture Collection (Manassas, Va.) and the latter provided by Elizabeth Hohmann (Massachusetts General Hospital, Boston, Mass.). Strain 1068 is a clinical isolate from an infected patient whose case has been described previously (13).
Cell culture.
MDCK type I cells were obtained from the American Type Culture Collection (CCL-34) and grown in minimal essential medium with Earle's salts containing 10% fetal bovine serum, 50 U of penicillin/ml, 50 μg of streptomycin/ml, and 2 mM l-glutamine. MDCK cells stably expressing GFP-CFTR fusion protein (11) were provided by Bruce A. Stanton (Dartmouth Medical School, Hanover, N.H.) and grown in the same medium also containing 150 μg of G418/ml. The cells were grown in 5% CO2-balanced air at 37°C.
Epithelial cell ingestion assay.
Cells were released from monolayers growing in tissue-culture flasks by treatment with trypsin-EDTA (0.5% trypsin and 0.53 mM EDTA in Hanks' balanced salt solution), washed, counted, seeded into 96-well tissue culture plates (105 cells/well) in the Earle's salts medium described above, and incubated overnight at 37°C in 5% CO2. In some experiments, CFTR protein levels were increased by adding 5 mM (final concentration) sodium butyrate to the cells during the overnight incubation. The butyrate was washed out 2 h before the ingestion assays were started. Fresh, overnight bacterial cultures prepared in tryptic soy broth (serovar Typhi) or on a tryptic soy agar plate (P. aeruginosa) were used to formulate an inoculum containing 5 × 107 CFU/ml. Bacterial cells were recovered from the broth by centrifugation or from the plates by suspension in Earle's salts medium and diluted to contain 1 × 107 to 5 × 107 CFU/ml; 100 μl of this suspension was added to the wells containing MDCK cells. The bacteria were allowed to interact with the epithelial cells for 3 to 4 h at 37°C, after which the nonadherent bacteria were removed by washing. To kill the extracellular organisms, the cell cultures were treated with 300 μg of gentamicin/ml for 45 min. The cells were then washed to remove the antibiotic and lysed with 100 μl of 0.5% Triton X-100 to release the intracellular organisms. The lysates were diluted and plated to quantify intracellular bacteria. Other wells were used to determine the total bacterial growth, obtained by lysing the cells after 3 h of invasion without removing the nonadherent bacteria, and the total killing, where gentamicin was added after the cells were lysed to insure that sufficient antibiotic was provided to kill all extracellular bacterial cells. Duplicate counts of bacteria in a minimum of three replicates were obtained per experimental condition.
Inhibition of bacterial uptake mediated by binding to CFTR protein was determined by the use of ingestion inhibition assays, as described previously (15, 18). In brief, a peptide corresponding to amino acids 103 to 117 of CFTR protein, predicted to be in the first extracellular domain, was synthesized and 1 to 25 nM was added to bacterial cultures 30 min before the addition of the cultures to epithelial cells for uptake assays. The control was a scrambled version of the peptide, as described previously (16), composed of the same amino acids but in random order. Duplicate counts of bacteria in a minimum of three replicates were obtained per experimental condition.
Flow cytometric analysis of epithelial cell ingestion.
MDCK cells stably expressing GFP-CFTR fusion protein were trypsinized and seeded into six-well tissue culture plates (5 × 105 cells/well). As described above, for some experiments, sodium butyrate was added to cultures overnight to increase CFTR protein expression. The butyrate was washed out 2 h before the addition of bacterial cells for ingestion experiments. Bacteria from overnight tryptic soy agar plates were suspended to 109 CFU/ml in 10 ml of PBS (0.1 M phosphate buffer, 0.15 M sodium chloride, pH 7.2), and 0.1 ml of a 3 mM suspension of 4′,6)-diamidino-2-phenylindole (DAPI; Molecular Probes, Eugene, Oreg.) in water was added to give a final concentration of 30 μM DAPI. This bacterial suspension was mixed and incubated at room temperature for 45 min in the dark. Unbound DAPI was washed away by repeated centrifugation and resuspension of bacterial cells in PBS (eight washes minimum), and the stained bacterial cells were finally resuspended in 10 ml of Earle's salts medium and kept in the dark. The epithelial-cell cultures were exposed to 1 ml of a 1:20 dilution of this bacterial suspension, containing 5 × 107 CFU of bacteria/ml, for 0.25, 0.5, 1, 2, 3, 4, 5, 6, 7, or 8 h at 37°C. After the nonadherent bacteria were removed by washing, extracellular organisms were killed by treatment of the cultures with 400 μg of gentamicin/ml for 45 min. The cells were then washed, trypsinized, and fixed in 1% paraformaldehyde in PBS. This process eliminated all extracellular bacterial fluorescence. Bacterial and GFP fluorescence was measured by flow cytometry (FACSVantage; Becton Dickinson Immunocytometry Systems, San Jose, Calif.) with the 488-nm excitation laser line; emission data were collected by using a standard fluorescein isothiocyanate filter set (530 ± 15 nm). DAPI fluorescence was measured by using the UV lines of an argon laser; emission was collected with a 450 ± 25-nm bandpass filter set. To analyze bacterial uptake by the MDCK–GFP-CFTR cells, the level on the y axis detecting DAPI fluorescence was set at the beginning of each experiment (time zero) such that, given the initial distribution of DAPI fluorescence, ≤0.6% of the epithelial cells would be counted as having internalized bacterial cells. At this set point, ingestion of about two to three bacterial cells per epithelial cell would increase the fluorescence intensity of a single MDCK–GFP-CFTR cell sufficiently to place it in the category of having ingested bacterial cells. In addition, the fluorescence-activated cell sorter (FACs) machine was adjusted to measure outcomes only in living cells. The FACScan software package (Becton Dickinson) was used for data analysis.
Confocal microscopy.
MDCK cells stably expressing GFP-CFTR fusion protein were trypsinized and seeded into 35-mm-diameter experimental chambers (5 × 104 cells/chamber). Live cells were observed on the microscope stage at 37°C. For bacterial translocation assays, DAPI-stained bacteria were allowed to invade the epithelial cells for 15 min to 3 h at 37°C prior to observation. Images were acquired with a Zeiss Axiovert microscope equipped with a 63× C-Apochromat/1.2 NA water immersion objective that was interfaced with an MRC-1024/2P multiphoton instrument (Bio-Rad, Hercules, Calif.). GFP was excited with the 488-nm laser line of a krypton-argon laser, and the emission was collected with a 530 ± 30-nm bandpass filter. DAPI was excited with the instrument in multiphoton mode with a femtosecond-pulsed Ti-sapphire laser tuned to 735 nm, and the emission was collected with a 460-nm-long pass filter.
Statistical analysis.
To determine whether histograms of fluorescence intensity (x axis) against the count of cells with that intensity (y axis) differed between two populations of cells analyzed by flow cytometry, the Kolmogorov-Smirnov (K-S) two-sample test was used. Indices of similarities for two curves were determined by D/s(n), where D is the K-S statistic reflecting the greatest difference between two curves and s(n) equals the square root of (n1 + n2), where n1 and n2 are the number of events in each data set. Larger values of D/s(n) indicate greater levels of dissimilarity between two curves (i.e., D/s(n) is 0 when two curves are identical). These calculations were part of the FACScan software. Differences in cellular uptake of bacterial cells were analyzed by either unpaired t tests or analysis of variance along with the Fisher predicted least significant difference statistic for pairwise differences, using the Statview Software on a Macintosh computer.
RESULTS
Comparison of bacterial uptake by MDCK and MDCK–GFP-CFTR cells.
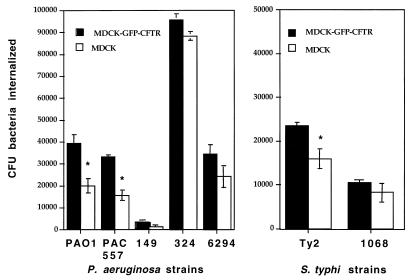
To determine whether the stable transfection of MDCK cells with DNA encoding GFP-CFTR protein markedly affected bacterial uptake, we compared the MDCK and MDCK–GFP-CFTR cell lines for the ability to ingest a variety of P. aeruginosa and serovar Typhi strains (Fig. 1). The final total amount of bacterial cells in the culture wells during the interaction increased comparably (∼10-fold) in both cell lines, indicating that GFP-CFTR gene transfection had no effect on bacterial survival or growth. When cells were lysed before the addition of gentamicin, >99.9999% of the bacteria were always killed, regardless of cell line, indicating that sufficient amounts of antibiotic had been added to the assay to kill all of the bacterial cells if none of them had been ingested by the MDCK cells. For all of the bacterial strains, the MDCK–GFP-CFTR cell line ingested more bacteria than the nontransfected parental line, but this difference was statistically significant for only three of the seven strains tested (Fig. 1). The efficiencies with which different bacterial strains were ingested by the two MDCK cell lines were identical, indicating that the cell lines were similar in their interactions with P. aeruginosa and serovar Typhi. For further analysis we chose P. aeruginosa 324 (high ingestion) and PAO1 (intermediate ingestion) and the serovar Typhi Ty2 strain (low ingestion), which are bacterial strains we have studied extensively for their interactions with CFTR protein (15, 16).
FIG. 1.
Internalization of different bacterial strains by MDCK type I cells and MDCK cells stably expressing GFP-CFTR fusion protein. The bars represent the means of six to nine replicates, the error bars represent standard errors, and asterisks indicate points significantly different from others in the same group by an unpaired t test (P < 0.001).
Studies of the inhibition of bacterial uptake were used to analyze the role of CFTR protein in MDCK cell ingestion of these bacterial strains. A synthetic peptide containing amino acids 103 to 117 of the first extracellular CFTR protein domain was used as the cognate inhibitor. This peptide has been shown to interact directly with both P. aeruginosa (16) and serovar Typhi (15), and a BLAST search (1) conducted in December 1999 showed that this peptide is found only in CFTR protein molecules of mammalian and amphibian origin. Addition of ≥10 nM cognate peptide into bacterial ingestion assays with either MDCK cells or MDCK–GFP-CFTR cells inhibited bacterial uptake from 54 to 98%, whereas use of a scrambled version of the same peptide containing the amino acids in a random order was without significant effect (<10% inhibition of ingestion) (P < 0.001; analysis of variance and Fisher predicted least significant difference). This ability of a specific CFTR peptide to inhibit bacterial ingestion by MDCK cells indicates a role for this molecule in bacterial uptake by the cells.
Analysis of GFP-CFTR protein expression by transfected cells.
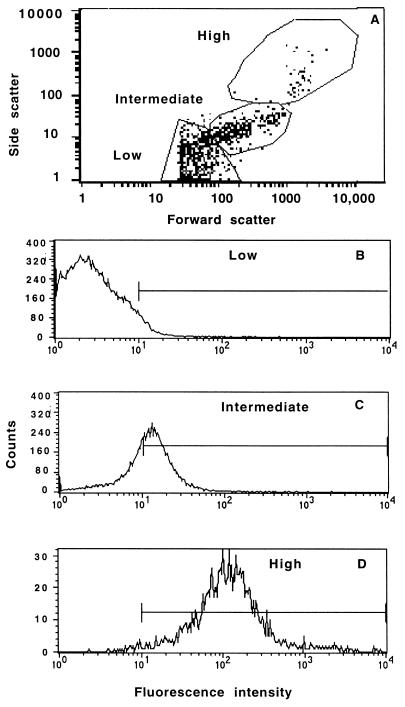
When uninfected MDCK–GFP-CFTR cells were analyzed by flow cytometry, they were found to have a nonhomogeneous distribution in the baseline levels of forward and side scatter, parameters that indicate cellular size and granule content, respectively. A representative analysis is shown in Fig. 2A. Essentially all of the cells in the population were viable, as indicated by a forward scatter of ≥25 U. When the cells were divided into three populations representing those with low, intermediate, or high levels of forward and side scatter, there was a strong correlation between these levels and the level of GFP-CFTR protein fluorescence (Fig. 2). This correlation suggested that the cytoplasmic concentration of GFP-CFTR protein (i.e., the amount of GFP-CFTR protein per unit cell volume) was fairly uniform among the three cell populations. In six separate experiments, measurement of the GFP-CFTR protein fluorescence intensity in the three cell populations showed that 60 to 70% of the cells had a low mean fluorescence intensity (2 to 5 U [Fig. 2B]), 20 to 30% had an intermediate mean intensity (15 to 25 U [Fig. 2C]), and 5 to 10% had a very high mean intensity (∼100 U [Fig. 2D]). Control cultures with parental MDCK cells showed background levels of fluorescence, with a distribution comparable to that of the MDCK–GFP-CFTR cells with the lowest mean fluorescence intensity (data not shown).
FIG. 2.
Heterogeneity of GFP-CFTR protein expression in cultures of MDCK–GFP-CFTR cells. (A) Distribution of GFP-CFTR protein in 96,839 cells based on forward and side scatter, which indicate cell size and granularity, respectively. The cells were divided into the indicated regions for analysis of bacterial uptake by these different subpopulations of MDCK–GFP-CFTR cells, all of which were routinely found within a culture. One percent of total dots are shown. (B to D) Fluorescence intensity indicative of the level of the GFP-CFTR protein from the cell populations in panel A that had a low (B), intermediate (C), or high (D) level of forward scatter, indicative of cell size. The bars indicate the proportion of cells in each population with an intensity of >10 U.
To confirm that the variation in cell size and fluorescence intensity was not due to use of a nonclonal cell line, we sorted the cells into different populations based on their relative fluorescences, cultured them again in vitro, and reanalyzed the distribution of GFP-CFTR protein after the cultures reached confluence. All sorted populations regenerated essentially the original fluorescence distribution intensity (Fig. 2), indicating that the nonhomogeneous baseline levels of GFP-CFTR protein expression were reproducible, stable upon subculture, and not due to cultures containing subpopulations of transfected MDCK cells with varying levels of GFP-CFTR protein expression. Indeed, in all of our flow cytometry experiments conducted over an 8-month period, the same relative baseline distribution of GFP-CFTR protein expression was always seen at the start of each experiment.
Analysis of bacterial uptake by MDCK–GFP-CFTR cells.
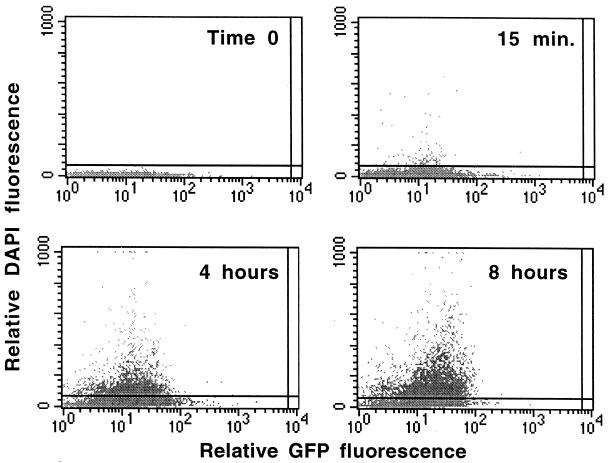
Analysis of the kinetics of bacterial uptake, the proportion of cells ingesting P. aeruginosa and serovar Typhi, and the relationship between the levels of expression of CFTR protein and bacterial ingestion revealed that the bacterial-epithelial cell interactions were rapid but nonhomogeneous. In all experiments, DAPI-stained, internalized bacteria were detected as early as 15 min after addition to cell monolayers (Fig. 3). The mean fluorescence intensity of the DAPI signal within the MDCK–GFP-CFTR cells generally increased steadily up to 4 h following infection, after which there were differences that varied with the individual bacterial strains. As one example, there was a further 20 to 25% increase in internalization of bacteria over an additional 4 h of incubation with P. aeruginosa PAO1 (Fig. 3).
FIG. 3.
Ingestion of DAPI-labelled P. aeruginosa PAO1 by MDCK–GFP-CFTR cells. The horizontal line was set at a level, at time zero, above which <0.6% of bacterial cells would be counted as internalized by epithelial cells; this level was equal to 51 U. The grey dots represent individual epithelial cells. One percent of all cells are shown.
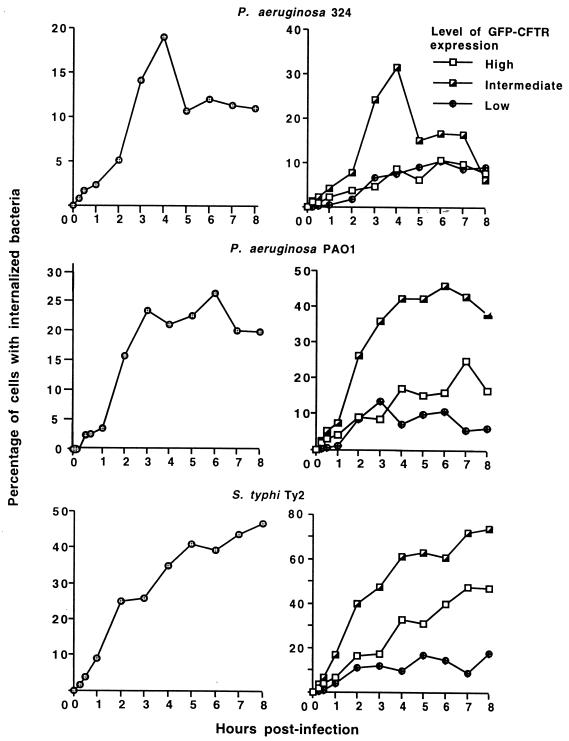
For all three bacterial strains, only a fraction of the epithelial cells in the culture ingested bacteria (Fig. 4). For P. aeruginosa 324, which had the highest total bacterial cell uptake by the MDCK–GFP-CFTR cells (Fig. 1), just under 20% of the epithelial cells internalized bacteria by 4 h after infection, and from 5 to 8 h postinfection, only about 10% of the epithelial cells contained internalized bacteria. For P. aeruginosa PAO1, the percentage of cells with internalized bacteria stabilized at 20 to 25% of the total population from 3 to 8 h after infection, indicating either that the internalization capacity of the MDCK–GFP-CFTR cells was saturated or that a stable system was created in which exit of bacteria from cells by exocytosis or lysis was approximately counterbalanced by ingestion of bacteria by other epithelial cells in the culture. The total uptake of the serovar Typhi strain Ty2 was slightly less than that of the two P. aeruginosa strains (Fig. 1), but the proportion of MDCK–GFP-CFTR cells with internalized serovar Typhi progressively increased over the entire time course, reaching just under 50% of cells with ingested bacteria by 8 h after infection (Fig. 4). Thus, for P. aeruginosa 324, fewer MDCK–GFP-CFTR cells ingested more bacteria, while for serovar Typhi, more epithelial cells ingested fewer bacteria; the proportion of MDCK–GFP-CFTR cells ingesting P. aeruginosa PAO1 was intermediate between the proportions of cells ingesting the other two bacterial strains.
FIG. 4.
Kinetics of ingestion of three different bacterial strains by MDCK–GFP-CFTR cells. The left-hand panels show percent ingestion by all of the cells in the population. The right-hand panels show percent ingestion by cells expressing low (mean <5 fluorescence units throughout the assay), intermediate (mean, 10 to 15 fluorescence units throughout the assay), or high (>85 fluorescence units throughout the assay) levels of GFP-CFTR protein. The points indicate means. Each point represents >3,000 events. Standard error bars fall within the symbol for each point.
Quantitative confirmation of these differences was achieved by analyzing the mean fluorescence intensity per MDCK–GFP-CFTR cell in the region of DAPI fluorescence above the time zero set point of fluorescence intensity (where ≤0.6% of the cells had fluorescence intensities above the set point; see Materials and Methods and Fig. 3, Time 0). MDCK–GFP-CFTR cells ingesting P. aeruginosa 324 had the highest mean fluorescence intensity per cell (8.27 × 10−3 U), cells ingesting P. aeruginosa PAO1 had an intermediate value (7.58 × 10−3 U), and cells ingesting serovar Typhi Ty2 had the lowest value (4.82 × 10−4 U). Statistical analysis of the curves obtained by plotting total internalized bacterial fluorescence over time showed significant differences in uptake among the three bacterial strains at a P value of <0.001 by the K-S test. The K-S statistic, D/s(n), which indicates the degree of statistical similarity between curves, was largest when comparing the two strains most disparate in total ingestion, P. aeruginosa 324 and serovar Typhi Ty2 (D/s(n) = 62.36), intermediate when comparing strain Ty2 and P. aeruginosa PAO1 (D/s(n) = 56.63), and lowest when comparing the two strains most similar in total uptake levels and kinetics, P. aeruginosa PAO1 and 324 (D/s(n) = 12.06).
We also analyzed the ingestion by MDCK–GFP-CFTR cells of P. aeruginosa PAO1 algC::tet, a transposon mutant lacking the complete LPS core ligand that mediates bacterial binding to CFTR protein (18) due to insertion of a tetracycline resistance gene into the algC gene needed for complete LPS core synthesis (9). As expected, the uptake of P. aeruginosa PAO1 algC::tet was only about 10% that of wild-type strain PAO1, and fewer than 8% of MDCK–GFP-CFTR cells contained measurable internalized mutant bacteria expressing only the incomplete LPS core oligosaccharide.
Bacterial-cell uptake by subpopulations of the MDCK–GFP-CFTR cells expressing low, intermediate, or high levels of CFTR protein were next analyzed for the ability to ingest P. aeruginosa and serovar Typhi. For all three bacterial strains, the populations of cells expressing intermediate levels of CFTR protein were best able to ingest bacteria (Fig. 4, right-hand panels) (P < 0.001 compared to cells expressing low or high levels of CFTR protein; K-S test). The proportion of cells with intermediate levels of GFP-CFTR protein that ingested bacteria ranged from 30% for P. aeruginosa 324 4 h after infection to 70% for serovar Typhi Ty2 7 to 8 h after infection. As might be expected, the MDCK–GFP-CFTR cells expressing low levels of CFTR protein were the least able to ingest either P. aeruginosa or serovar Typhi; typically, <10% of these cells contained detectable intracellular bacteria. It was of interest that, among the cells with the highest expression of CFTR protein, ingestion of bacteria was also fairly low. The proportion of these cells showing ingested P. aeruginosa was either the same or only marginally greater than that shown by the cells with the lowest level of CFTR protein expression. For serovar Typhi, the cells expressing the highest level of CFTR protein had significantly more internalized bacteria than those expressing low levels of CFTR protein (P < 0.001; K-S test), but the former cells were still less efficient at bacterial uptake than the cells with intermediate levels of CFTR protein. Thus, the cells with intermediate levels of GFP-CFTR protein expression accounted for most of the bacterial ingestion measured in the entire epithelial cell population.
The proportions of cells expressing high, intermediate, and low levels of GFP-CFTR protein were unchanged from those shown in Fig. 1 over an 8-h period of incubation with bacterial strains (data not shown). This was also true of control cultures incubated for 8 h in the absence of bacteria. These data do not mean, however, that there could not have been increased or decreased CFTR protein production among individual cells; rather, the relative proportions of the cells in the entire population producing low, intermediate, and high levels of GFP-CFTR protein remained constant throughout the experiment.
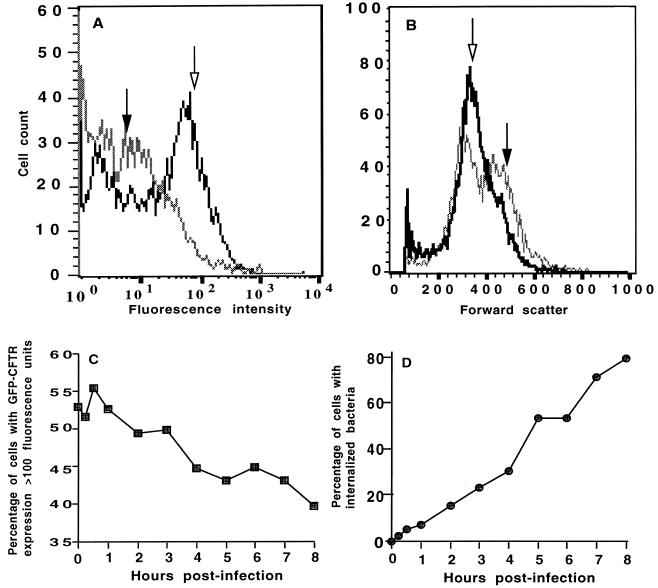
Because the fraction of cells expressing high levels of GFP-CFTR protein were also the largest cells in the culture, we could not determine whether the suboptimal uptake of bacteria by these cells was due to increased cell size or to overproduction of GFP-CFTR protein. To formally evaluate these possibilities, MDCK–GFP-CFTR cells were incubated with 5 mM sodium butyrate overnight to enhance CFTR protein expression, and the butyrate was removed 2 h before the ingestion assays. The CFTR gene promoter, like many eukaryotic promoters, is activated by exposure to butyrate (4, 14). We found that, in MDCK–GFP-CFTR cell populations grown overnight with 5 mM butyrate, over 50% of the cells expressed high levels (mean, ∼100 fluorescence units) of CFTR protein; these data should be compared with ≤10% of cells expressing high levels of GFP-CFTR protein when butyrate was not used (Fig. 5A). Butyrate treatment did not affect cell size, however, as the butyrate-treated and untreated cells had a comparable distribution of forward scatter in each population (Fig. 5B). The proportion of butyrate-treated cells with high-level CFTR protein expression decreased to 40% over the 8-h period of infection with bacteria (Fig. 5C), likely due to the removal of butyrate from the cultures. The proportion of butyrate-treated MDCK–GFP-CFTR cells that ingested P. aeruginosa PAO1 was nearly 80% at the conclusion of this experiment (Fig. 5D), compared with only 20% of epithelial cells containing P. aeruginosa PAO1 (Fig. 4) when cells were grown without butyrate. Comparable results were obtained with P. aeruginosa 324 and serovar Typhi Ty2, with a six- and twofold respective increase in the proportion of butyrate-treated MDCK–GFP-CFTR cells that showed internalized bacteria (data not shown). Increasing CFTR protein expression with sodium butyrate was also associated with a 20 to 75% decrease in overall ingestion of P. aeruginosa and, while there was no decrease in uptake of serovar Typhi, there was also no increase in uptake in spite of the increased levels of GFP-CFTR protein. Thus, by increasing CFTR protein expression, the proportion of epithelial cells in the culture that were able to ingest bacterial cells was increased by up to sixfold but the total bacterial uptake by the MDCK–GFP-CFTR cells was reduced, indicating fewer bacteria per epithelial cell.
FIG. 5.
Effect of treatment with 5 mM sodium butyrate on distribution of GFP-CFTR protein, cell size, and kinetics of ingestion of P. aeruginosa PAO1 by MDCK–GFP-CFTR cells. (A) Distribution of GFP-CFTR protein in cells grown without butyrate (solid arrow) or with 5 mM butyrate (open arrow). (B) Distribution of cell size in cells grown without butyrate (solid arrow) or with 5 mM butyrate (open arrow). (C) Changes in percentage of butyrate-treated cells with high levels of GFP-CFTR protein during the course of infection. (D) Percentage of butyrate-treated MDCK–GFP-CFTR cells with internalized P. aeruginosa. Each point represents >10,000 events, and the error bars are within each symbol.
Analysis of bacterial uptake by confocal microscopy.
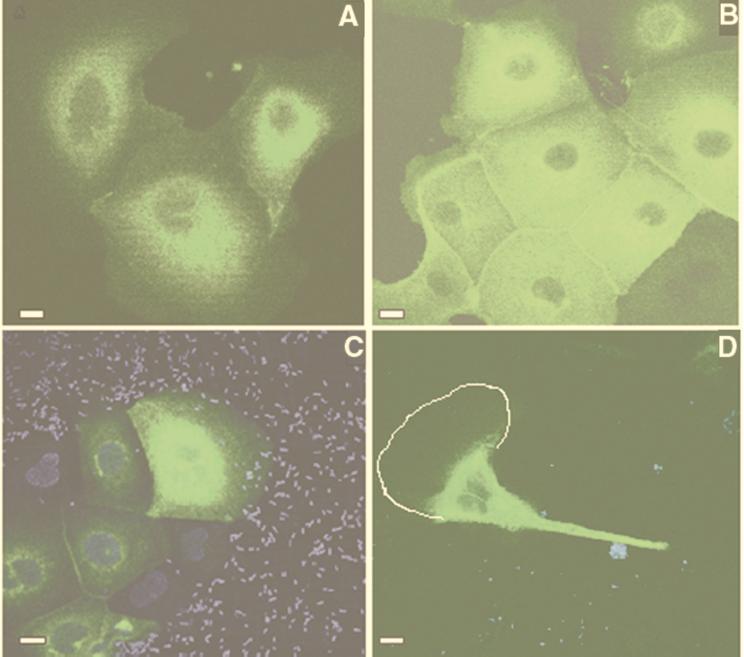
To obtain visual confirmation of these findings, confocal microscopy was used to observe MDCK–GFP-CFTR cells ingesting bacteria. Confirming the flow cytometry data, heterogeneity in the expression of GFP-CFTR protein in the MDCK cells was observed, such that only 15 to 30% of the cells appeared to express GFP at the start of an assay. In those cells with observable fluorescence, GFP fluoresced throughout the cell with a perinuclear concentration seen in sections taken through the middle of the cell (Fig. 6A). In butyrate-treated MDCK–GFP-CFTR cultures, large clusters of cells with an increased expression of GFP fluorescence were routinely seen (Fig. 6B), consistent with the increase in GFP-CFTR protein levels induced by butyrate treatment of the cells. In many sections, starting as early as 15 min after infection, colocalization of CFTR protein and DAPI-stained P. aeruginosa was observed (not shown). Images of cells prior to washing away unbound bacteria showed a homogeneous distribution of bacterial cells against the nonuniform epithelial cell expression of GFP-CFTR protein (Fig. 6C). One notable observation was cells in which the cytoplasmic CFTR protein could be seen to localize in the part of the cell where clusters of P. aeruginosa were in close proximity to the plasma membrane (Fig. 6D). Sections of confocal images taken through a single cell showed GFP-CFTR protein accumulating on the same side of the cell as a cluster of P. aeruginosa organisms (Fig. 7). The P. aeruginosa PAO1 LPS mutant strain lacking the ligand for CFTR protein showed essentially no bacterial interaction with GFP-CFTR protein or entry of bacteria into MDCK–GFP-CFTR cells (data not shown).
FIG. 6.
Visualization of CFTR protein distribution in MDCK–GFP-CFTR cells interacting with P. aeruginosa. (A) Uninfected MDCK–GFP-CFTR cells. (B) Uninfected MDCK–GFP-CFTR cells treated overnight with 5 mM butyrate to increase GFP-CFTR protein expression. In panels A and B, sections were taken through the middle of the cell, showing the distribution of GFP-CFTR protein. (C) MDCK–GFP-CFTR cell incubated for 45 min with DAPI-stained P. aeruginosa 324 prior to removal of bacterial cells by washing. (D) Translocation of GFP-CFTR protein toward sites where large clusters of DAPI (blue)-stained P. aeruginosa PAO1 bacteria were found on or near the cell membrane. The white line outlines the plasma membrane as visualized in the image captured from the microscope; this outline was too faint for adequate reproduction in the printed image. Bars = 1 μm.
FIG. 7.
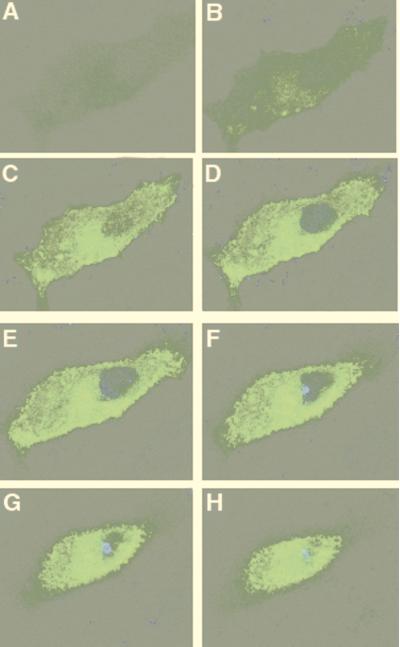
Accumulation of GFP-CFTR protein on the apical portion of an MDCK–GFP-CFTR cell where a cluster of DAPI-stained P. aeruginosa cells had adhered. The confocal images are 1-μm-thick sections through the cell, starting at the basal side (A) and finishing at the apical side (H). Magnification, ×6,300.
DISCUSSION
Bacterial interactions with host cells, particularly ingestion by epithelial and phagocytic cells, have been a mainstay of research in microbial pathogenesis. However, most of the protocols used in measuring these interactions rely either on cultures with large numbers of bacterial and eukaryotic cells contained in a single tube or vessel or on interactions among a few cells, usually analyzed by microscope-based procedures. We used flow cytometry to analyze the interaction of two different bacterial pathogens, P. aeruginosa and serovar Typhi, with large numbers of individual epithelial cells expressing a labelled receptor for internalization of these pathogens. Flow cytometry allows easy analysis of 10,000 or more individual eukaryotic and prokaryotic cells. We found substantial heterogeneity among the cells in regard to internalization of bacteria. We determined that populations of MDCK–GFP-CFTR cells in culture wells had a nonhomogeneous expression of GFP-CFTR protein, which correlated with cell size. However, this overall distribution of GFP-CFTR protein expression was fairly stable upon multiple subcultures of the cells and even upon sorting of cells with differing levels of GFP-CFTR protein expression and regrowth of the sorted populations in vitro. Those MDCK–GFP-CFTR cells expressing an intermediate level of CFTR protein were best able to ingest P. aeruginosa and serovar Typhi, while cells expressing little to no CFTR protein, as well as the minority of cells with very high-level expression of CFTR protein, ingested bacteria poorly. Furthermore, an unexpected finding was that, for those bacterial strains that were better internalized by the MDCK–GFP-CFTR cells, ingestion of the larger amount of bacteria was due not to an increase in the proportion of epithelial cells internalizing bacteria but rather to an increase in the quantity of bacterial cells entering into the subpopulation of epithelial cells with the best capacity to ingest bacteria. Use of a mutant strain of P. aeruginosa lacking the bacterial ligand for CFTR protein demonstrated the specificity of the measured interactions because the mutant strain entered MDCK–GFP-CFTR cells poorly and, by confocal microscopy, was not seen binding to GFP-CFTR protein or entering cells.
The addition of the gene encoding the GFP-CFTR molecule did not markedly alter the abilities of MDCK–GFP-CFTR cells to ingest bacteria. Although MDCK–GFP-CFTR cells ingested three of seven strains better than the parental MDCK cells, increased ingestion was not consistent across bacterial strains, indicating a reasonable conservation of function in spite of the genetic change to the MDCK–GFP-CFTR cells. More importantly, the transfected cells ingested the seven bacterial pathogens in a pattern identical to that of the parental MDCK cell line. Moreover, any increased or altered level of CFTR protein expression in the transfected cells from that of the parental cells did not compromise the cells' abilities to ingest bacteria, because the transfected cells performed comparably to or slightly better than the parental cells in bacterial-ingestion assays. Thus, it is reasonable to conclude that the MDCK–GFP-CFTR cells were phenotypically comparable to the parental cells and therefore useful in measuring the interaction of CFTR protein with P. aeruginosa and serovar Typhi.
The fact that cells with low levels of CFTR protein ingested P. aeruginosa and serovar Typhi poorly is consistent with the finding that this protein is a major epithelial cell receptor for ingestion of these pathogens. Consistent with previous results (21), there does not appear to be any significant activation of transcription of the CFTR gene and increased protein production due to addition of P. aeruginosa to cell cultures. The majority of the bacterial ingestion occurred in the 20 to 30% of cells with some measurable expression of CFTR protein. There was a small proportion of large cells that expressed high total levels of CFTR protein but ingested bacteria poorly; it is likely, however, that the higher total amount of CFTR in this population reflected merely the greater cell volume. The basis for the suboptimal uptake of bacteria by these large cells was not determined, but as they represented <10% of the total cells, it is unlikely that their suboptimal ingestion of bacteria had a major impact on the overall biology of the system. In addition, increased size, granularity, and total CFTR protein expression in this population were not fixed properties, because these cells could be sorted and regrown into populations with the same distributions of size, granularity, and GFP-CFTR protein expression as those found in the initial culture. One potential explanation for the poor uptake of bacteria by these larger cells is that they are physiologically quite distinct from the rest of the population because they have entered the cell cycle leading to division, a state preceded by an increase in cell size.
Studies in which sodium butyrate was used to produce a high-level expression of CFTR protein without affecting cell size showed that cellular ingestion of P. aeruginosa and serovar Typhi was compromised under these conditions. Butyrate treatment increased the expression of GFP-CFTR protein and the overall proportion of epithelial cells internalizing some bacteria while reducing the total amount of P. aeruginosa internalized by 20 to 75% and having no positive effect on the uptake of serovar Typhi. Thus, butyrate treatment led to more cells showing internalized P. aeruginosa but fewer internalized bacteria per epithelial cell, indicating that increasing the cellular receptor for P. aeruginosa actually decreased the ability of individual cells to ingest this organism. While butyrate treatment of epithelial cells did not decrease serovar Typhi uptake, it also did not increase uptake, indicating that other cellular processes may have been affected by the butyrate treatment such that the increased levels of the bacterial receptor for ingestion were counterbalanced by changes reducing the epithelial cells' ability to ingest bacteria. Consistent with our finding that increased CFTR expression actually decreased its biologic activity is the recent finding of Moyer et al. (12), who showed that butyrate increases apical-membrane expression of CFTR in MDCK–GFP-CFTR cells by 25-fold but decreases Cl− ion transport. Thus, overexpression of CFTR in cells adversely affects both ion transport properties and bacterial-ingestion activity, indicating that properly regulated levels of CFTR protein are required for its biological activities to be optimal.
Nonquantitative confocal-microscopy experiments confirmed some of the observations made by using flow cytometry with the MDCK–GFP-CFTR cells. Bacteria were preferentially seen in cells expressing green fluorescence, and in some cells, translocation of cytoplasmic CFTR protein toward sites of bacterial accumulation on the membrane was observed. The latter finding could indicate a cytoskeletal reorganization within cells that had adherent P. aeruginosa or serovar Typhi, with movement of CFTR protein toward the area of bacterial binding. While several studies of CFTR protein trafficking have measured a rapid membrane turnover rate in cultured cells (19, 20), we know of no studies such as the one described here measuring CFTR protein trafficking in the presence of bacterial pathogens. In addition, while membrane CFTR protein levels are generally thought to be low in uninfected tissues (6, 7, 10), our confocal-microscopy experiments with MDCK–GFP-CFTR cells showed extensive cytoplasmic stores of CFTR protein in a good proportion of the cells, consistent with the observations of intracellular stores of CFTR protein (3) and a possible role for intracellular CFTR protein in normal cellular functions.
The use of MDCK–GFP-CFTR cells to study the interaction of CFTR protein with bacterial pathogens known to use this molecule as a receptor for cellular entry revealed a fair amount of heterogeneity within small populations of cells growing in tissue culture wells and dishes. We do not know whether the heterogeneity we observed in both CFTR protein expression and cellular uptake of bacteria reflects the situation in vivo, although it is likely that, within a host tissue, there is considerable heterogeneity among epithelial cells in both physiological state and ability to respond to stimuli such as bacterial pathogens. Nonetheless, the ability to quantify large numbers of epithelial-bacterial interactions and to analyze the relationship between receptor expression and bacterial uptake will be important tools for evaluating and testing of hypotheses in further experiments. Extension of the findings with cultured cells reported here to intact tissues and organs should provide greater insight into microbial pathogenesis and host-microbe interactions.
ACKNOWLEDGMENTS
We thank Bruce Stanton of Dartmouth Medical School, Hanover, N.H. for provision of MDCK–GFP-CFTR cells and for reviewing the manuscript.
This work was supported by an Interdisciplinary Seed Grant from Brigham and Women's Hospital and by NIH grants AI 22806, HL 58398, HL 32854, and HL 15157.
The images were obtained at the Brigham and Women's Hospital Confocal Microscopy Core Facility.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apodaca G, Bomsel M, Lindstedt R, Engel J, Frank D, Mostov K E, Wiener-Kronish J. Characterization of Pseudomonas aeruginosa-induced MDCK cell injury: glycosylation-defective host cells are resistant to bacterial killing. Infect Immun. 1995;63:1541–1551. doi: 10.1128/iai.63.4.1541-1551.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradbury N A. Intracellular CFTR: localization and function. Physiol Rev. 1999;79:S175–S191. doi: 10.1152/physrev.1999.79.1.S175. [DOI] [PubMed] [Google Scholar]
- 4.Cheng S H, Fang S L, Zabner J, Marshall J, Piraino S, Schiavi S C, Jefferson D M, Welsh M J, Smith A E. Functional activation of the cystic fibrosis trafficking mutant delta F508-CFTR by overexpression. Am J Physiol. 1995;268:L615–L624. doi: 10.1152/ajplung.1995.268.4.L615. [DOI] [PubMed] [Google Scholar]
- 5.Comolli J C, Waite L L, Mostov K E, Engel J N. Pili binding to asialo-GM1 on epithelial cells can mediate cytotoxicity or bacterial internalization by Pseudomonas aeruginosa. Infect Immun. 1999;67:3207–3214. doi: 10.1128/iai.67.7.3207-3214.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denning G M, Ostedgaard L S, Cheng S H, Smith A E, Welsh M J. Localization of cystic fibrosis transmembrane conductance regulator in chloride secretory epithelia. J Clin Investig. 1992;89:339–349. doi: 10.1172/JCI115582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denning G M, Ostedgaard L S, Welsh M J. Abnormal localization of cystic fibrosis transmembrane conductance regulator in primary cultures of cystic fibrosis airway epithelia. J Cell Biol. 1992;118:551–559. doi: 10.1083/jcb.118.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleiszig S M, Evans D J, Do N, Vallas V, Shin S, Mostov K E. Epithelial cell polarity affects susceptibility to Pseudomonas aeruginosa invasion and cytotoxicity. Infect Immun. 1997;65:2861–2867. doi: 10.1128/iai.65.7.2861-2867.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldberg J B, Hatano K, Pier G B. Synthesis of lipopolysaccharide O side chains by Pseudomonas aeruginosa PAO1 requires the enzyme phosphomannomutase. J Bacteriol. 1993;175:1605–1611. doi: 10.1128/jb.175.6.1605-1611.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marshall J, Fang S, Ostedgaard L S, O'Riordan C R, Ferrara D, Amara J F, Hoppe IV H, Scheule R K, Welsh M J, Smith A E. Stoichiometry of recombinant cystic fibrosis transmembrane conductance regulator in epithelial cells and its functional reconstitution into cells in vitro. J Biol Chem. 1994;269:2987–2995. [PubMed] [Google Scholar]
- 11.Moyer B D, Loffing J, Schwiebert E M, Loffing-Cueni D, Halpin P A, Karlson K H, Ismailov I I, Guggino W B, Langford G M, Stanton B A. Membrane trafficking of the cystic fibrosis gene product, cystic fibrosis transmembrane conductance regulator, tagged with green fluorescent protein in Madin-Darby canine kidney cells. J Biol Chem. 1998;273:21759–21768. doi: 10.1074/jbc.273.34.21759. [DOI] [PubMed] [Google Scholar]
- 12.Moyer B D, Loffing-Cueni D, Loffing J, Reynolds D, Stanton B A. Butyrate increases apical membrane CFTR but reduces chloride secretion in MDCK cells. Am J Physiol. 1999;277:F271–F276. doi: 10.1152/ajprenal.1999.277.2.F271. [DOI] [PubMed] [Google Scholar]
- 13.New England Journal of Medicine. Case Reports of the Massachusetts General Hospital. Weekly clinicopathological exercises. Case 8-1999. A 28-year-old man with gram-negative sepsis of uncertain cause. N Engl J Med. 1999;340:869–876. doi: 10.1056/NEJM199903183401108. [DOI] [PubMed] [Google Scholar]
- 14.Olsen J C, Sechelski J. Use of sodium butyrate to enhance production of retroviral vectors expressing CFTR cDNA. Hum Gene Ther. 1995;6:1195–1202. doi: 10.1089/hum.1995.6.9-1195. [DOI] [PubMed] [Google Scholar]
- 15.Pier G B, Grout M, Zaidi T, Meluleni G, Mueschenborn S S, Banting G, Ratcliff R, Evans M J, Colledge W H. Salmonella typhi uses CFTR to enter intestinal epithelial cells. Nature. 1998;392:79–82. doi: 10.1038/30006. [DOI] [PubMed] [Google Scholar]
- 16.Pier G B, Grout M, Zaidi T S. Cystic fibrosis transmembrane conductance regulator is an epithelial cell receptor for clearance of Pseudomonas aeruginosa from the lung. Proc Natl Acad Sci USA. 1997;94:12088–12093. doi: 10.1073/pnas.94.22.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pier G B, Grout M, Zaidi T S, Goldberg J B. How mutant CFTR may contribute to Pseudomonas aeruginosa infection in cystic fibrosis. Am J Respir Crit Care Med. 1996;154:S175–S182. doi: 10.1164/ajrccm/154.4_Pt_2.S175. [DOI] [PubMed] [Google Scholar]
- 18.Pier G B, Grout M, Zaidi T S, Olsen J C, Johnson L G, Yankaskas J R, Goldberg J B. Role of mutant CFTR in hypersusceptibility of cystic fibrosis patients to lung infections. Science. 1996;271:64–67. doi: 10.1126/science.271.5245.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prince L S, Peter K, Hatton S R, Zaliauskiene L, Cotlin L F, Clancy J P, Marchase R B, Collawn J F. Efficient endocytosis of the cystic fibrosis transmembrane conductance regulator requires a tyrosine-based signal. J Biol Chem. 1999;274:3602–3609. doi: 10.1074/jbc.274.6.3602. [DOI] [PubMed] [Google Scholar]
- 20.Prince L S, Workman R B, Jr, Marchase R B. Rapid endocytosis of the cystic fibrosis transmembrane conductance regulator chloride channel. Proc Natl Acad Sci USA. 1994;91:5192–5196. doi: 10.1073/pnas.91.11.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaidi T S, Lyczak J, Preston M, Pier G B. Cystic fibrosis transmembrane conductance regulator-mediated corneal epithelial cell ingestion of Pseudomonas aeruginosa is a key component in the pathogenesis of experimental murine keratitis. Infect Immun. 1999;67:1481–1492. doi: 10.1128/iai.67.3.1481-1492.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]