Abstract
Matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI-TOF MS) is a broadly used technique for identification and typing of microorganisms. However, its application to filamentous fungi has been delayed. The objective of this study was to establish a data library for rapid identification of the genus Aspergillus sect. Nigri using MALDI-TOF MS. With respect to sample preparation, we compared the utility of using mature mycelia, including conidial structures, to accumulate a wider range of proteins versus the conventional method relying on young hyphae. Mass spectral datasets obtained for 61 strains of 17 species were subjected to cluster analysis and compared with a phylogenetic tree based on calmodulin gene sequences. Specific and frequent mass spectral peaks corresponding to each phylogenetic group were selected (superspectra for the SARAMIS system). Fifteen superspectra representing nine species were ultimately created. The percentage of correct identification for 217 spectra was improved from 36.41% to 86.64% using the revised library. Additionally, 2.76% of the spectra were assigned to candidates that comprised several related species, including the correct species.
Keywords: Eurotiales, food-borne fungi, laboratory techniques, profiling, protein fingerprint
1. Introduction
The use of proteome analysis based on matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) for the rapid identification of clinically important fungi is continually advancing. This technique has been developed on the detection of ribosomal protein, the most abundant cellular protein of bacteria, from target organisms cultivated under various growth conditions (Ryzhov & Fenselau, 2001), with subsequent comparison of the mass spectral peaks against an existing database (fingerprinting). In many cases, however, spectral databases and methodological information are inadequate, especially for fungi used in food and industrial manufacturing. Unlike bacteria and yeasts, the mass spectra (MS) of filamentous fungi are significantly affected by culture conditions, because the cellular abundance of various proteins and secondary metabolites in these fungi seem to be higher than that of ribosomal and house-keeping proteins. On the basis of manufacturers’ guidelines and previous experiments, the use of primary hyphae incubated for 2 to 3 d—prior to the production of such impurities—is thus recommended (Rahi, Prakash, & Shouche, 2016). Nevertheless, stable mass spectrometry detection has not been achieved by technicians and laboratories, and pretreatment protocols need improvement. Furthermore, the number of constructed libraries, all based on various studies of fungi is insufficient yet (De Respinis et al., 2010; Rodrigues, Santos, Venâncio, & Lima, 2011; Normand et al., 2017).
Black aspergilli, Aspergillus sect. Nigri (Gams, Christensen, Onions, Pitt, & Samson, 1986 [1985]), comprise a group of fungi of major importance to humans. The type species of Aspergillus sect. Nigri, A. niger Tiegh., is ubiquitous in soil and air. Sometimes acting as a plant pathogen, A. niger is also responsible for food spoilage, as it produces mycotoxins such as ochratoxin A. This species as well as A. tubingensis Mosseray ocasionally causes lung disease in humans and animals (Schuster, Dunn-Coleman, Frisvad, & Van Dijck, 2002; Gautier et al., 2016; Mirhendi, Zarei, Motamedi, & Nouripour-Sisakht, 2016). Conversely, A. niger is used for the biotechnological fermentation of citric acid, gluconic acid, and hydrolytic enzymes (Schuster et al., 2002; Varga et al., 2007). Several species are common in fermented food especially in East Asia, such as A. luchuensis Inui, A. acidus Kozak., and A. kawachii Kitahara & Yoshida (Hong et al., 2013). The taxonomy of this group has been revised several times (Raper & Fennell, 1965; Varga et al., 2011; Samson et al., 2014), and 29 species are recognized in the latest study (Houbraken et al., 2020).
Superspectra used by the SARAMIS (Anagnos-Tec, Potsdam, Germany) and Vitek MS (bioMérieux, Marcy l’Étoile, France) systems were developed as a library of biomarker protein mass patterns (Stephan, Ziegler, Pfluger, Vogel, & Lehner, 2010; Lima-Neto et al., 2014). Inter- and intra-species specific mass peaks as superspectra can be selected and weighted according to their specificity (Kallow, Erhard, Dieckmann, & Sauermann, 2004). Then the superspectra are registered into the software database to improve future fingerprinting accuracy (Emonet, Shah, Cherkaoui, & Schrenzel, 2010). The inclusion of additional mass peaks from total proteins would increase the number of species identified, only a few investigators have exploited superspectra and the above platforms for the identification of fungal species (Erhard, Hipler, Burmester, Brakhage, & Wöstemeyer, 2008; De Respinis et al., 2013, 2014; Nenoff et al., 2013; Américo et al., 2019). We have previously established an in-house superspectral library for the rapid identification of A. flavus Link and A. oryzae (Ahlb.) Cohn. For species-level discrimination, the best culture conditions for sample preparation have been obtained from 10-d-old mycelia on an agar plate (data available at https://www.nite.go.jp/nbrc/industry/maldi/maldi.html, in Japanese). In the present study, we aimed to establish a superspectral library for more accurate identification of closely related species of Aspergillus sect. Nigri.
2. Materials and methods
2.1. Fungal strains
In this study, we analyzed 61 strains of 17 species of Aspergillus sect. Nigri preserved at the Biological Resource Center, National Institute of Technology and Evaluation (NBRC), Chiba, Japan (Supplementary Table S1). Strains stored in a freezer at -80 °C were pre-incubated on potato dextrose agar (PDA; Nissui Pharmaceutical, Tokyo, Japan) plates at 25±2 °C for more than 5 d. Conidia were scraped from the pre-incubation plates with an L-shaped needle and transferred by three-point inoculation to a new agar plate (9 cm diam, 20 mL, 25±2 °C, dark conditions) for extraction for MALDI-TOF MS. Ribosomal RNA 5.8S and internal transcribed spacer 1 and 2 (ITS), calmodulin, and β-tubulin gene sequences were used for confirmation strains and phylogenetic analyses per modern classifications (Samson et al., 2014; Houbraken et al., 2020). DNA extraction and sequencing and sequence assembly were carried out as described by Yamazaki and Kawasaki (2014). The primer sets used were the same as described in Samson et al. (2014). The phylogenetic analysis was conducted using MEGA 7 software (Kumar, Stecher, & Tamura, 2016).
2.2. MALDI-TOF MS
To determine conditions yielding the best profile spectra, a preliminary MALDI-TOF MS analysis was performed using representative strains cultured on different substrate media (PDA plates or Sabouraud’s dextrose broth [SDB; BD, JK, USA]) for various incubation times (2, 5, 7, 10, or 14 d). Conidial formation was usually observed after 3 to 5 d, but varied by a few days depending on minor environmental variation.
Sample preparation was a modified version of the protocols of De Respinis et al. (2014). A 3-cm2 area of growing mycelium, starting from the center to the edge of the colony, was collected with sterilized wet swabs, with care taken not to remove agar. The collected mycelium was suspended in 250 μL of distilled water in an acid-resistant plastic tube (Eppendorf, Hamburg, Germany). Ethanol (99.5%, 1 mL) was then added to give a final concentration of 80% to remove oily metabolites. The solution was centrifuged (13,000 g for 2 min) and the supernatant was discarded. Fresh 70% aqueous solution of formic acid (50 μL) was added using acid-resistant microtips (Sorenson BioScience, Salt Lake City, UT, USA), and the mixture was vortexed. An equal volume of acetonitrile (50 μL; Fujifilm Wako Pure Chemical, Osaka, Japan) was added, followed by vortexing and centrifugation (approximately 15,000 g for 2 min). The supernatant (1 μL) was spotted onto wells of a single-use FlexiMass-DS target plate (Shimadzu Scientific Instrument, Kyoto, Japan) and allowed to evaporate for approximately 5 to 10 min. Next, 1 μL of a matrix solution of α-cyano-4-hydroxycinnamic acid (CHCA; Shimadzu GLC, Tokyo, Japan) was added to each sample. The solvent was allowed to evaporate at room temperature in a draft chamber (approximately 5 to 10 min), with crystal formation controlled visually. Cells of Escherichia coli NBRC 3301 (K-12 derived strain) were used as a standard for calibration.
Mass spectra analyses were performed using an AXIMA Performance mass spectrometer (Shimadzu, Kyoto, Japan) in positive linear mode with a mass-to-change ratio (m/z) ranging from 3,000 to 20,000 and equipped with a 50-Hz nitrogen laser. Laser settings were as follows: power, 53; profiles, 100 per sample; and 5 shots accumulated per profile. Averaged profile spectra fulfilling quality criteria were collected from 20 laser shot cycles. For each strain, at least 16 averaged profile spectra (4 sample spots × 2 repeat MS measurements × 2 independent consecutive examinations) were stored and used in subsequent analyses. All MS were processed via baseline collection, peak filtering, and smoothing using MALDI-TOF MS Launchpad 2.9.3 software (Kratos Analytical, Manchester, UK).
2.3. Construction of a superspectral library
Mass spectra of individual samples were imported into the SARAMIS Premium software package (v.1.1.0) and subjected to cluster analysis. The similarities among mass peak lists were calculated by the software without considering intensity. The similarity was converted into a distance, and the pair of two data having the smallest distance was merged into a minimum common peak list. This calculation sequence was repeated until the pair became one (single-linkage method). A dendrogram was constructed based on the respective distances. We selected MS exhibiting more than 70% similarity within a given strain and cultural periods. All molecular ion peaks of a given strain under the same medium and incubation periods were aggregated, with the error range of each peak calibrated with a mass tolerance limit of ±800 ppm. Supermass was calculated from the aggregated MS of each strain. A cluster dendrogram of individual supermasses were generated using the SARAMIS software for comparison with a phylogenetic tree based on calmodulin (Supplementary Fig. S1). For each species, at least three strains having ≥ 70.0% supermass similarity to one another were chosen for subsequent construction of primary superspectra. To generate each superspectrum, we identified consensus peaks without considering intensity, i.e., peaks common to multiple strains but unique to a species—typically 35 to 49 per species. We then adopted weighting factors (scores) based on the number of peaks overlapping with the most closely related species, as defined by the manufacturer, in the SARAMIS database.
2.4. Superspectral library validation
Primary superspectra of the 17 studied species of Aspergillus sect. Nigri were constructed and integrated into the database. As an initial verification, we generated and examined a cluster dendrogram of all superspectra of Aspergillus sect. Nigri generated in this study and those in the commercial database. Next, we re-checked the identity of the 61 well-identified strains of Aspergillus sect. Nigri on the basis of their raw MS data using the newly constructed superspectra. For this validation, we randomly selected two raw MS datasets from each culture subjected to different incubation periods (5 and 10 d). For identification with the SARAMIS software, a “confidence level” was calculated based on the sum of weight values of matching peaks, converted to a percentage of up to 99.9%, then displayed with the species name (as described in the SARAMIS user manual). If a validation did not result in a high-confidence (> 90.0%) identification, the combination of strains or the peak selection criterion (frequency) was changed, the superspectrum was reconstructed, and the validation was repeated.
3. Results
3.1. Influence of incubation conditions
Less than 70 MS ions (data counts) were obtained from mycelia of twelve strains incubated for 3 d on SDB (Table 1), and no species could be identified when these data were compared against the already installed SARAMIS dataset. Compared with our previous unpublished study, in which high-quality MS data (data counts > 80) were obtained from mycelia of Aspergillus sect. Flavi incubated on SDB (data not shown), few data counts were obtained from sect. Nigri. Additionally, the number of ions composing spectra of Aspergillus sect. Nigri cultured on PDA for 10 d was generally lower (average 94.58) than the number (mostly > 150) obtained from sect. Flavi under the same culturing medium and incubation periods. In Aspergillus sect. Nigri, except for one strain of A. carbonarius NBRC 4030, the number of ions was statistically significantly higher when PDA was used compared with SDB.
Table 1. Difference of the average of the numbers of peaks (datacount) between liquid broth (SDB, 3 d) and solid medium (PDA, 10 d).
|
Species |
NBRC No. |
SDB 3 d (n=8) |
PDA 10 d (n=16 a/ n=20) |
P value |
|
Aspergillus niger |
105649 |
49.88 |
55.31 a |
0.33958 |
|
6428 |
47.38 |
139.25 |
1.243E-11 |
|
|
A. niger var. niger f. hennebergii |
4043 |
47.75 |
68.45 |
0.00154 |
|
A. brasiliensis |
6341 |
40.50 |
84.60 |
0.00596 |
|
9455 |
31.13 |
67.10 a |
0.00203 |
|
|
A. luchuensis |
4281 T |
50.88 |
105.60 |
0.00447 |
|
4314 |
42.13 |
115.80 a |
0.00215 |
|
|
A. tubingensis |
4407 |
53.50 |
149.70 |
5.297E-6 |
|
A. carbonarius |
4030 |
66.13 |
74.70 |
0.40288 b |
|
4038 |
40.25 |
100.15 |
4.749E-6 |
|
|
A. aculeatus (clade VI) |
5330 |
55.25 |
75.90 |
0.00425 |
|
A. aculeatus (clade VII) |
31348 |
68.13 |
98.45 |
2.836E-8 |
|
Mean |
49.41 |
94.58 |
0.00034 |
a The population parameter was 16.
b It was indicated no significant difference between the datacount of mass spectra (P > 0.05).
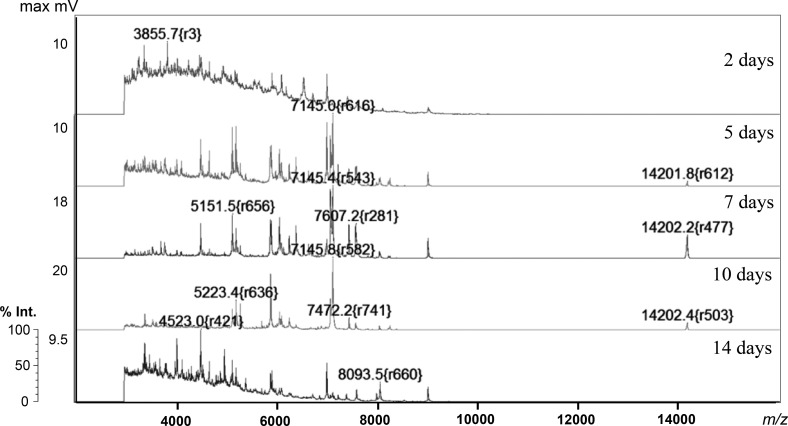
Mass spectra patterns of most fungi are dependent on the incubation period. The average number and stability index of ions obtained from 8 or 12 replicates is shown in Table 2. Stability index, which is the degree of similarity of data replicates cultured under the same conditions, was classified into categories of 100–90.0%, 89.9–80.0%, 79.9–70.0%, and < 69.9%. Representative MALDI-TOF MS spectra are shown in Fig. 1. At the early stage of growth (2 d), no strains produced conidia, and only a few ions (average 67.54) between m/z 3,000 and 4,000 were generally detected (Table 2). Following the start of conidial production (after 5 d), the number of ions increased, and the waveform shifted up to m/z 14,000. Two replicates were analyzed from 5-d and 10-d incubation conditions, and the results were affected by the absence or presence of conidia. In addition, the similarity of the two datasets obtained from the same period was sometimes not > 70.0%. As indicated by the stability index values in Table 2, the MS from the 2-d samples, but not the 10-d ones, were relatively stable. Stability was restored on 14 d as a result of sudden decrease spore production. Similar to the situation shown in Table 2, a higher number of molecules was generally detected after 5 d compared to 2 d (p = 0.002579, one-way analysis of variance), and the larger the number, the better for satisfactory characterization of species and strains. The choice of samples to be used was thus a tradeoff between quantity (more ions) and quality (higher stability). Strains resulting in low MS stability under the same situation and/or with poor conidial production were therefore excluded from subsequent examinations. A dataset obtained from 5-d and 10-d cultures of 42 strains was finally selected for supermass calculations.
Table 2. Influence of incubation period (2, 5, 7, 10 and 14 d) on the numbers of peaks (datacount) and their stability.
|
Species |
NBRC No. |
Average of datacount, stability index a |
|||||||||
|
2 d (n=8) |
5 d (n=12) |
7 d (n=8) |
10 d (n=12) |
14 d (n=8) |
|||||||
|
Aspergillus niger |
33023 NT |
74.13 |
b |
128.56 |
d |
150.0 |
a |
125.44 |
b |
98.13 |
a |
|
A. brasiliensis |
105650 |
84.0 |
b |
105.13 |
b |
99.88 |
b |
104.88 |
d |
102.88 |
b |
|
A. luchuensis |
4281 T |
37.38 |
b |
82.13 |
b |
109.13 |
b |
100.31 |
c |
102.0 |
c |
|
A. neoniger |
4068 |
101.0 |
a |
143.69 |
b |
136.38 |
a |
109.44 |
c |
101.88 |
b |
|
A. tubingensis |
4050 |
57.63 |
b |
126.56 |
b |
100.50 |
b |
101.38 |
b |
136.88 |
b |
|
A. carbonarius |
5864 |
71.0 |
a |
106.44 |
c |
104.0 |
b |
85.94 |
c |
80.38 |
b |
|
A. japonicus |
32856 |
47.63 |
b |
65.56 |
b |
88.75 |
b |
88.50 |
b |
105.0 |
b |
|
Mean |
67.54 |
108.30 |
112.66 |
102.27 |
103.88 |
||||||
|
Standard deviation |
20.18 |
25.41 |
20.45 |
12.28 |
15.51 |
||||||
a Stability index was classified as follows: a, 100—90.0%; b, 89.9—80.0%; c, 79.9—70%; d, less than 69.9% similarity of mass spectra within repeated data in the same strain and incubation period. Unstable results (stability index c and d) are highlighted by gray shading.
Fig. 1 -Examples of variation in MALDI-TOF MS spectra with incubation periods (Aspergillus niger NBRC 33023 grown on potato dextrose agar medium). The vertical axis represents relative intensity (%) with absolute value of maximum mV, and horizontal axis represents mass-to-charge ratio (m/z).
3.2. Selection of mass profiles of each strain
The phylogenetic tree constructed from the calmodulin gene sequences constructed in this study was not in conflict with the tree generated by using a dataset combining four genes (Houbraken et al., 2020). The studied strains were classified into nine phylogenetic clades (I-1 to VII; Supplementary Fig. S1).
We next carried out cluster analyses based on supermasses from two selected incubation periods (Fig. 2). The resulting dendrograms were not expected to reflect evolutionary relationships but instead simply illustrate MS data similarities among samples. Although the branching order was different, the clusters in the two dendrograms were roughly equivalent to each other and to those in the phylogenetic tree (Supplementary Fig. S1). The exception was the position of a 10-d culture of one strain (NBRC 9455) of A. brasiliensis Varga, Frisvad & Samson. The highest similarity between NBRC 9455 and other strains of A. brasiliensis was 61.7% at 5 d. Likewise, the similarity of A. japonicus Saito NBRC 32856 with other strains of the same species was low (47.9% at 5 d and 59.9% at 10 d). To establish a superspectral library, a supermass similarity of at least 70.0% is preferable within a given species. These two strains were considered separately for subsequent data manipulation. Supermasses of A. tubingensis (clade IV-2) were approximately separated from A. neoniger Varga et al. (clade IV-1) in 5-d cultures (Fig. 2A), but not in 10-d cultures (Fig. 2B, clade IV). This was because raw MS data from cultures of the same strain frequently had shared similarities less than 70.0% in both species. Raw MS data selection and supermass calculations were repeated until within-strain similarities increased up to 70.0%. In cases where similarities remained under 70.0%, such as 10-d cultures of NBRC 109442 and NBRC 8872, the unstable MS data were excluded from subsequent experiments.
Fig. 2 -Dendrogram of Aspergillus sect. Nigri constructed from the supermasses per strain by the SARAMIS using single-linkage agglomerative cluster analysis. Cultures incubated at 5 d (A) and 10 d (B) on potato dextrose agar medium were used in the analysis. The scale bar below indicates the similarity (%). Broken vertical line means the position of 70% similarity in the dendrogram. Broken horizonal line shows separation of phylogenetic clades. Accumulated number of peaks showed as datacount.
Aspergillus welwitschiae (Bres.) Henn. is morphologically indistinguishable from A. niger (Hong et al., 2013), but eleven to twelve sites in their nucleotide sequences differ between the two species by amino acid sequence of calmodulin and β-tubulin. Five strains, NBRC 4043, 6649, 6650, 31012 and 31638 assigned to A. welwitschiae (clade I-2) in the phylogenetic tree (Supplementary Fig. S1) were not separated from A. niger in the supermass-based dendrograms (Clade I in Fig. 2). The ex-neotype strain of A. foetidus Thom & Raper NBRC 4031 was grouped with A. niger both in the calmodulin phylogenetic tree and in the supermass dendrograms. The distributions of varieties and strains of these three species were scattered throughout clades I-1 and I-2 in Supplementary Fig. S1, and not separable using proteins of metabolites; these two clades were thus analyzed together, as clade I in Fig. 2.
3.3 Construction and verification of superspectra
Characteristic masses that were retained for creation of the final version of superspectra of Aspergillus sect. Nigri are shown in Supplementary Table S2. The superspectra were constructed using 36 to 48 specific peaks without score adjustment. Masses at m/z 7,027.7–7,033.7 (13/15 superspectra), 4,683.9–4,689 (10/15), 5,244–5,246.9 (10/15), and 6,010–6,015 (10/15) were common to almost all superspectra (Supplementary Table S2). A dendrogram including these superspectra along with those from the supplier is shown in Fig. 3. Compared with the dendrograms in Fig. 2, this dendrogram was more deeply branched because only selected peaks were used. To verify the constructed superspectral library, we carried out rapid identification of 61 strains of Aspergillus sect. Nigri based on raw MS data using the SARAMIS system (Supplementary Table S3). The number of raw MS matching entries in the supplier’s SARAMIS library, and that in the library constructed in this study, at a confidence level higher than 75.0% for each clade is shown in Table 3. The overall sensitivity with specificity of the supplier’s library was 36.41%, whereas that using the expanded library was 86.64% (n = 217).
Table 3. Comparison of the performance of the SARAMIS supplier’s data library and the library constructed in this study for identification of Aspergillus sect. Nigri. The respective numbers of selected superspectra above 75.0% of confidence level are shown.
|
Species |
Aspergillus niger / A. welwitschiae |
A. brasiliensis |
A. luchuensis / A. kawachii |
A. neoniger |
A. tubingensis |
A. carbonarius |
A. japonicus |
A. aculeatus (negative control) |
total b |
|
|
Numbers of used strains |
28 |
3 |
10 |
4 |
6 |
3 |
6 |
1 |
60 |
|
|
Used spectra (a) |
101 |
12 |
30 |
16 |
24 |
12 |
22 |
2 |
217 |
|
|
Library: supplier’s superspectra | ||||||||||
|
Confidence level of correct answer (b) |
99.90% |
16 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
19 |
|
99.8 — 90.0% |
19 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
21 |
|
|
89.9 — 80.0% |
23 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
25 |
|
|
79.9 — 75.0% |
12 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
14 |
|
|
Un-identified (c) |
31 |
3 |
30 |
16 |
24 |
12 |
22 |
2 |
138 |
|
|
Correctly identified spectra [(b) / (a)] |
69.31% |
75.00% |
0% |
0% |
0% |
0% |
0% |
0% |
36.41% |
|
|
Discordant answer a |
2 |
0 |
1 |
4 |
5 |
0 |
0 |
0 |
12 |
|
|
Unexpected taxa |
Yersinia pestis, Staphylococcus homini |
- |
A. niger complex |
A. niger complex |
A. niger complex |
- |
- |
- |
||
|
Library: superspectra constructed in this study | ||||||||||
|
Confidence level of correct answer (b) |
99.90% |
52 |
12 |
14 |
14 |
11 |
12 |
17 |
0 |
132 |
|
99.8 — 90.0% |
15 |
0 |
5 |
1 |
6 |
0 |
4 |
0 |
31 |
|
|
89.9 — 80.0% |
10 |
0 |
5 |
0 |
3 |
0 |
0 |
0 |
18 |
|
|
79.9 — 75.0% |
4 |
0 |
1 |
1 |
0 |
0 |
1 |
0 |
7 |
|
|
Un-identified (c) |
20 |
0 |
5 |
0 |
4 |
0 |
0 |
2 |
29 |
|
|
Correctly identified spectra [(b) / (a)] |
80.20% |
100.00% |
83.33% |
100.00% |
83.33% |
100.00% |
100.00% |
0% |
86.64% |
|
|
Discordant answer a |
0 |
0 |
0 |
5 |
1 |
0 |
0 |
0 |
6 |
|
|
Unexpected taxa |
- |
- |
- |
A. tubingensis |
A. luchuensis / A. kawachii |
- |
- |
- |
||
a Discordant answer represents the number of returned candidate names greater than 80.0% confidence level (see Supplementary Table S2), and are included in the total number for (b) and (c).
b Two spectra for the negative control were excluded.
Fig. 3 -Dendrogram of Aspergillus sect. Nigri constructed from the supplier’s and newly constructed superspectra library by the SARAMIS using single-linkage agglomerative cluster analysis. The scale bar above indicates the similarity (%). The superspectra newly constructed in the present study in bold, those provided by the supplier are shown in regular font.

Although most strains in clade I (A. niger/A. welwitschiae, Supplementary Fig. S1) were correctly identified by supermasses, the confidence levels were generally low when MS data from 10 d cultures were used. In a few cases, the bacteria names were returned as the second or third matching candidates (Supplementary Table S3). Approximately 20 superspectra of A. niger already installed in the SARAMIS database were also correctly returned as matches to the tested spectra, but the confidence levels were even lower than those from our new library, possibly because of differences in culture conditions. Similar results were observed for A. luchuensis and A. kawachii, supplier-provided superspectra of A. niger was returned in low confidence level only at once. Because similarities among MS from A. brasiliensis strains NBRC 6341, NBRC 105650, and NBRC 9455 were low, we separated the superspectra into two groups. Aspergillus brasiliensis (clade II) was correctly identified, with or without supplier’s superspectra. In A. brasiliensis and A. carbonarius (Bainier) Thom, the spectral waveforms of these species were highly dependent on culturing time, with the two superspectra showing less than 70.0% similarity to each other (Fig. 3). In other words, MS from 5-d culture were highly similar to the 5-d superspectrum, and MS from 10-d culture were identifiable using either the 10-d or cumulative (5- and 10-d) superspectrum. Accurate identification of A. carbonarius in this case required the incorporation of both 5-d and cumulative superspectra into the library.
Aspergillus foetidus var. acidus (Nakaz. et al.) Raper & Fennell is now considered to be a synonym of A. luchuensis (Hong et al., 2013). In the phylogenetic tree, clade III included A. foetidus NBRC 4338, A. foetidus var. pallidus (Nakaz., Simo & A. Watan.) Raper & Fennell NBRC 4118 and 4123, and A. foetidus var. acidus NBRC 4121 along with A. luchuensis. The final clade-III superspectrum was constructed only from strains of A. luchuensis and A. kawachii, but raw MS from 10-d cultures of these strains of A. foetidus varieties were slightly different from the superspectrum of A. luchuensis. The most highly supported match returned by the SARAMIS system was A. luchuensis, but confidence levels were relatively low. In contrast, the MS of A. kawachii NBRC 4308, actually an albino mutant and a synonym of A. luchuensis, was highly similar to black strains of A. luchuensis.
Although all strains of A. japonicus occupied the same phylogenetic position with ex-type CBS 114.51, the MS of A. japonicus NBRC 32856 was distinct from other strains of this species. Aspergillus aculeatus Iizuka NBRC 5330 (ex-type strain of A. yezoensis Y. Sasaki) was placed in clade VI (Supplementary Fig. S1), and its MS was more similar to A. japonicus than A. aculeatus NBRC 31348 (Supplementary Table S3).
Strains belonging to clades IV-1 (A. neoniger) and VI-2 (A. tubingensis), and several strains of A. niger and A. brasiliensis could not be clearly discriminated using some pre-supplied superspectra, although identifications at the 95.0% confidence level were made, some pre-supplied superspectra of A. niger and A. brasiliensis were assigned with low support; the source strains of these superspectra are unknown. The MS of A. neoniger NBRC 113385 (derived from the ex-type strain NRRL 62634 = CBS 115656; Supplementary Fig. S1) was slightly different from spectra of the other three strains of A. neoniger examined by us. Likewise, the MS of A. tubingensis NBRC 113384 (derived from the ex-type strain NRRL 4875) was distinct from Japanese strains belonging to the same clade in the calmodulin phylogenetic tree. This problem was solved by establishing a superspectrum encompassing both species (A. tubingensis + A. neoniger).
4. Discussion
In MALDI-TOF MS identification of filamentous fungi, extraction protocols have likewise been designed to use young mycelia (very short incubation, less than 3 d) before miscellaneous proteins are expressed. Alternative methods that require purification of ribosomal proteins have also been used (Nakamura et al., 2017). Becker et al. (2015) found that the performance of MALDI-TOF MS for identification of general environmental species was 84%, which was lower than the values of 89% to 100% reported for clinical strains (Sanguinetti & Posteraro, 2014). Data for species from the general environment are obviously lacking. The greater the numbers of mass peaks collected, the better the capability to accurately identify closely related species. In the present study, we aimed to construct a mass spectral dataset for quality control at food manufacturing sites and examined over 3,600 MS data and 900 culture samples from 61 strains of Aspergillus sect. Nigri. The established superspectral library have been published from the NBRC website (https://www.nite.go.jp/nbrc/industry/maldi/maldi.html, in Japanese). Given that only four harmful species, namely A. niger, A. brasiliensis, A. welwitschiae, and A. japonicus were registered in the pre-installed library (Fig. 3), the ability to identify A. luchuensis, which is used for food fermentation, is of industrially importance. In the current study, we analyzed two selections—5-d and 10-d samples—because of the large difference in their waveforms.
Sulc, Peslova, Zabka, Hajduch, and Havlicek (2009) have suggested that the number of peaks of black Aspergillus detected by MALDI-TOF MS tend to be reduced because black pigments may interfere with ionization. Similar observations made in the present study may explain the stability of MS data on 3-d cultures, which preceded production of conidia and black pigments. The smaller number of peaks in Aspergillus sect. Nigri compared with other sections of the genus hindered the selection of key peaks for identification.
In a previous study used to construct superspectra of Trichoderma, De Respinis et al. (2010) detected possible hydrophobins between m/z 6,000 and 8,000. Hydrophobins were found to be major proteins in intact-cell mass spectrometry, and showed that it was a candidate of effective biomarker for the identification of 29 species of Trichoderma (Neuhof et al., 2007). For example, among superspectra selected from strains of the clade I (A. niger and A. welwitschiae), several dominant peaks were observed at approximately m/z 3,500, 4,500, 6,280 and 7,140 that were independent of the culture period. However, the peak stability was poor and it was necessary to re-verify whether the peaks were genuinely common in the species (data not shown). Those molecules might be not ribosomes because they were smaller than that reported before (Nakamura et al., 2017). The proteome is a phenotypic reflection of an evolutionary lineage. As confirmed by the results, the use of total proteins for MALDI-TOF MS identification is very reasonable. Topics for future exploration include the identity of these proteins and their establishment as a chemical classification index.
More recently, D'hooge et al. (2019) tested the identification by MALDI Biotyper v3.0 software (Bruker Daltonics, Bremen, Germany) using 18 species and 175 strains of Aspergillus sect. Nigri preserved at the BCCM/IHEM collection, Mycology and Aerobiology, Belgium. These authors demonstrated that MALDI-TOF MS could correctly identify all strains at least clade level (e.g., A. niger and A. welwitschiae, A. luchuensis and A. piperis Samson & Frisvad). However, the methodology to create the main spectrum profiles (MSP), adopted in the Biotyper’s database and involving superposition of multiple measurements of a single strain, differs from that of the SARAMIS’s superspectra used in the present study. There are two different concepts for fingerprinting method using MALDI-TOF MS. The MSP is constructed from about 70 peaks, which are selected in order from the highest frequency per the strain, and installed into a customized library. Then the homology score is calculated from the matching rate to both mass and intensity of MSPs (as described in the Biotyper user manual). It shows high discrimination performance at the strain level. On the other hand, only the presence or absence of the molecule is evaluated, regardless of the strength of the detected signal in the case of superspectra. This approach is considered to be advantageous for identification of organisms that show uneven growth patterns, such as filamentous fungi, because peaks with high frequency are preferentially used even if the detection rate and strength of those peaks are low (De Respinis et al., 2010). Conversely, strain-level discriminative ability might be less reliable than that of the MSP library. The need to revise ‘unique’ peaks after each species addition is a disadvantageous because the system must make subtractions from the entire superspectral library (e.g., including bacteria) to determine specific peaks in the grouped peak list. As more species are added, it is expected that the number of unique peaks will decrease and finally attain zero. Thus, at least to avoid truncation of the peaks in common with bacterial species, the approach can use more peaks for superspectra of filamentous fungi. The authors suggested the switching and use of datasets designed for a specific target microbial group rather than reliance on the current comprehensive superspectral library of all microbes.
Because rapid identification based on MALDI-TOF MS depends on an abundance of databases, the collection of numerous ubiquitous and environmental fungal species is urgently needed. Given the existence of regional differences among strains in the same species (e.g., A. tubingensis and A. neoniger), the inclusion of strains derived from a user’s own region is desirable. Public and private culture collections, which possess many isolates from local areas, can play a key role in breakthroughs associated with this identification method through mutual cooperation and the expansion of mass spectral data of their own strains.
Disclosures
The authors declare no conflicts of interest. All the experiments undertaken in this study comply with the current laws of the country where they were performed.
Supplementary Material
Acknowledgments
We thank Dr. Vit Hubka, an assistant professor of Department of Botany, Charles University in Prague, and Edanz Group (en-author-services.edanzgroup.com) for carefully English proofreading the manuscript.
References
- Américo, F. M., Machado Siqueira, L. P., Del Negro, G. M. B., Favero Gimenes, V. M., Trindade, M. R. S., Motta, A. L., Santos de Freitas, R., Rossi, F., Colombo, A. L., Benard, G., & de Almeida Júnior, J. N. (2019). Evaluating VITEK MS for the identification of clinically relevant Aspergillus species. Medical Mycology, 58, 322-327. https://doi.org/10.1093/mmy/myz066 [DOI] [PubMed] [Google Scholar]
- Becker, P. T., Stubbe, D., Claessens, J., Roesems, S., Bastin, Y., Planard, C., Cassagne, C., Piarroux, R., & Hendrickx, M. (2015). Quality contorol in culture collection: Confirming indentity of filamentous fungi by MALDI-TOF MS. Mycoscience, 56, 273–279. https://doi.org/10.1016/j.myc.2014.08.002 [Google Scholar]
- De Respinis, S., Monnin, V., Girard, V., Welker, M., Arsac, M., Cellière, B., Durand, G., Bosshard, P. P., Farina, C., Passera, M., Van Belkum, A., Petrini, O., & Tonolla, M. (2014). Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry using the Vitek MS system for rapid and accurate identification of dermatophytes on solid cultures. Journal of Clinical Microbiology, 52, 4286–4292. https://doi.org/10.1128/JCM.02199-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Respinis, S., Tonolla, M., Pranghofer, S., Petrini, L., Petrini, O., & Bosshard, P. P. (2013). Identification of dermatophytes by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Mediccal Mycology, 51, 514–521. https://doi.org/10.3109/13693786.2012.746476 [DOI] [PubMed] [Google Scholar]
- De Respinis, S., Vogel, G., Benagli, C., Tonolla, M., Petrini, O., & Samuels, G. J. (2010). MALDI-TOF MS of Trichoderma: a model system for the identification of microfungi. Mycological Progress, 9, 79–100. https://doi.org/10.1007/s11557-009-0621-5. [Google Scholar]
- D’hooge, E., Becker, P., Stubbe, D., Normand, A. C., Piarroux, R., & Hendrickx, M. (2019). Black aspergilli: A remaining challenge in fungal taxonomy? Medical Mycology, 57, 773–780. https://doi.org/10.1093/mmy/myy124 [DOI] [PubMed] [Google Scholar]
- Emonet, S., Shah, H., Cherkaoui, A., & Schrenzel, J. (2010). Application and use of various mass spectrometry methods in clinical microbiology. Clinical Microbiology and Infection, 16, 1604–1613. https://doi.org/10.1111/j.1469-0691.2010.03368.x [DOI] [PubMed] [Google Scholar]
- Erhard, M., Hipler, U. C., Burmester, A., Brakhage, A. A., & Wöstemeyer, J. (2008). Identification of dermatophyte species causing onychomycosis and tinea pedis by MALDI-TOF mass spectrometry. Experimental Dermatology, 17, 356–361. https://doi.org/10.1111/j.1600-0625.2007.00649.x [DOI] [PubMed] [Google Scholar]
- Gams, W., Christensen, M., Onions, A. H. S., Pitt, J. I., & Samson, R. A. (1986) [“1985”] Infrageneric taxa of Aspergillus. In: Samson R. A. & Pitt J. I. (Eds.), Advances in Penicillium and Aspergillus systematics (pp. 55–62). New York: Plenum Press. [Google Scholar]
- Gautier, M., Normand, A. C., L'Ollivier, C., Cassagne, C., Reynaud-Gaubert, M., Dubus, J.-C., Brégeon, F., Hendrickx, M., Gomez, C., Ranque, S., & Piarroux, R. (2016). Aspergillus tubingensis: a major filamentous fungus found in the airways of patients with lung disease. Medical Mycology, 54, 459–470. https://doi.org/10.1093/mmy/myv118. [DOI] [PubMed] [Google Scholar]
- Hong, S. B., Lee, M., Kim, D. H., Varga, J., Frisvad, J. C., Perrone, G., Gomi, K., Yamada, O., Machida, M., Houbraken, J., & Samson, R. A. (2013). Aspergillus luchuensis, an industrially important black Aspergillus in East Asia. PLoS One, 8, e63769. https://doi.org/10.1371/journal.pone.0063769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken, J., Kocsube, S., Visagie, C. M., Yilmaz, N., Wang, X. C., Meijer, M., Kraak, B., Hubka, V., Bensch, K., Samson, R. A., & Frisvad, J. C. (2020). Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Studies in Mycology, 95, 5–169. https://doi.org/10.1016/j.simyco.2020.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallow, W., Erhard, M., Dieckmann, R., & Sauermann, S. (2004). Process for identifying microoraganisms by means of mass spectrometry. US Patent, 10/752,224
- Kumar, S., Stecher, G., & Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870–1874. https://doi.org/10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima-Neto, R., Santos, C., Lima, N., Sampaio, P., Pais, C., & Neves, R. P. (2014). Application of MALDI-TOF MS for requalification of a Candida clinical isolates culture collection. Brazilian Journal of Microbiology, 45, 515–522. https://doi.org/10.1590/S1517-83822014005000044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirhendi, H., Zarei, F., Motamedi, M., & Nouripour-Sisakht, S. (2016). Aspergillus tubingensis and Aspergillus niger as the dominant black Aspergillus, use of simple PCR-RFLP for preliminary differentiation. Journal de Mycologie Médicale, 26, 9–16. https://doi.org/10.1016/j.mycmed.2015.12.004 [DOI] [PubMed] [Google Scholar]
- Nakamura, S., Sato, H., Tanaka, R., Kusuya, Y., Takahashi, H., & Yaguchi, T. (2017). Ribosomal subunit protein typing using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) for the identification and discrimination of Aspergillus species. BMC Microbiology, 17, 100. https://doi.org/10.1186/s12866-017-1009-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nenoff, P., Erhard, M., Simon, J. C., Muylowa, G. K., Herrmann, J., Rataj, W., & Gräser, Y. (2013). MALDI-TOF mass spectrometry – a rapid method for the identification of dermatophyte species. Medical Mycology, 51, 17–24. https://doi.org/10.3109/13693786.2012.685186. [DOI] [PubMed] [Google Scholar]
- Neuhof, T., Dieckmann, R., Druzhinina, I. S., Kubicek, C. P., Nakari-Setälä, T., Penttilä, M., & von Döhren, H. (2007). Direct identification of hydrophobins and their processing in Trichoderma using intact-cell MALDI-TOF MS. The FEBS Journal, 274, 841–852. https://doi.org/10.1111/j.1742-4658.2007.05636.x [DOI] [PubMed] [Google Scholar]
- Normand, A. C., Cassagne, C., Gautier, M., Becker, P., Ranque, S., Hendrickx, M., & Piarroux, R. (2017). Decision criteria for MALDI-TOF MS-based identification of filamentous fungi using commercial and in-house reference databases. BMC Microbiology, 17, 25. https://doi.org/10.1186/s12866-017-0937-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahi, P., Prakash, O., & Shouche, Y. S. (2016). Matrix-assisted laser desorption/ionization time-of-flight mass-spectrometry (MALDI-TOF MS) based microbial identifications: Challenges and scopes for microbial ecologists. Frontiers in Microbiology, 7, 1359. https://doi.org/10.3389/fmicb.2016.01359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raper, K. B., & Fennell, D. I. (1965). The genus Aspergillus. Baltimore: The Williams & Wilkins. [Google Scholar]
- Rodrigues, P., Santos, C., Venâncio, A., & Lima, N. (2011). Species identification of Aspergillus section Flavi isolates from Portuguese almonds using phenotypic, including MALDI-TOF ICMS, and molecular approaches. Journal of Applied Microbiology, 111, 877–892. https://doi.org/10.1111/j.1365-2672.2011.05116.x [DOI] [PubMed] [Google Scholar]
- Ryzhov, V., & Fenselau, C. (2001). Characterization of the protein subset desorbed by MALDI from whole bacterial cells. Analytical chemistry, 73, 746–750. https://doi.org/10.1021/ac0008791 [DOI] [PubMed] [Google Scholar]
- Samson, R. A., Visagie, C. M., Houbraken, J., Hong, S. B., Hubka, V., Klaassen, C. H. W., Perrone, G., Seifert, K. A., Susca, A., Tanney, J. B., Varga, J., Kocsubé, S., Szigeti, G., Yaguchi, T. & Frisvad, J. C. (2014). Phylogeny, identification and nomenclature of the genus Aspergillus. Studies in Mycology, 78, 141–173. https://doi.org/10.1016/j.simyco.2014.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanguinetti, M., & Posteraro, B. (2014). MALDI-TOF mass spectrometry: any use for Aspergilli? Mycopathologia, 178, 417–426. https://doi.org/10.1007/s11046-014-9757-1 [DOI] [PubMed] [Google Scholar]
- Schuster, E., Dunn-Coleman, N., Frisvad, J. C., & Van Dijck, P. W. M. (2002). On the safety of Aspergillus niger – a review. Applied Microbiology and Biotechnology, 59, 426–435. https://doi.org/10.1007/s00253-002-1032-6 [DOI] [PubMed] [Google Scholar]
- Stephan, R., Ziegler, D., Pfluger, V., Vogel, G., & Lehner, A. (2010). Rapid genus- and species-specific identification of Cronobacter spp. by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Journal of Clinical Microbiology, 48, 2846–2851. https://doi.org/10.1128/JCM.00156-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulc, M., Peslova, K., Zabka, M., Hajduch, M., & Havlicek, V. (2009). Biomarkers of Aspergillus spores: Strain typing and protein identification. International Journal of Mass Spectrometry, 280, 162–168. https://doi.org/10.1016/j.ijms.2008.08.012. [Google Scholar]
- Varga, J., Frisvad, J. C., Kocsubé, S., Brankovics, B., Töth, B., Szigeti, G., & Samson, R. A. (2011). New and revisited species in Aspergillus section Nigri. Studies in Mycology, 69, 1–17. https://doi.org/10.3114/sim.2011.69.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga, J., Kocsubé, S., Töth, B., Frisvad, J. C., Perrone, G., Susca, A., Meijer, M., & Samson, R. A. (2007). Aspergillus brasiliensis sp. nov., a biseriate black Aspergillus species with world-wide distribution. International Journal of Systematic and Evolutionary Microbiology, 57, 1925–1932. https://doi.org/10.1099/ijs.0.65021-0 [DOI] [PubMed] [Google Scholar]
- Yamazaki, A., & Kawasaki, H. (2014). Lipomyces chichibuensis sp. nov., isolated in Japan, and reidentification of the type strains of Lipomyces kononenkoae and Lipomyces spencermartinsiae. International Journal of Systematic and Evolutionary Microbiology, 64, 2566–2572. https://doi.org/10.1099/ijs.0.059972-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.