Abstract
Pleurotus tuber-regium is a sclerotium-producing mushroom with pharmacological and nutritional value. Many researches applied molecular biological strategies that allow analyzing numerous genes expressed in response to different growth stresses or tissues. In order to capture accurate and reliable results in gene expression studies, it is necessary to select appropriate internal reference genes. They must have relatively uniform expression in the majority of tested samples, thereby reducing the impact of the sample itself on the RT-qPCR results. The selection and validation of 14 candidate reference genes, including α-tub, β-tub, γ-tub, GAPDH, Tif-5a, Tef-1α, ATPase, acyl-CoA, U2ribo, S/G, Wlp, VtpAsE, E3upl, and Sdh, were carried out for gene expression analysis in P. tuber-regium mycelia in response to different temperatures, drought levels, and salinity shifts, fruitbody, and sclerotium. Four statistical algorithms, NormFinder, geNorm, BestKeeper, and RefFinder, were recruited to evaluate the transcription stability of candidate reference genes. The RT-qPCR expression stability analysis indicated that the E3upl and Tif-5a were the most stable expressed genes among all experimental samples, so the combination of these two reference genes is suitable for the analysis of gene expression patterns in P. tuber-regium.
Keywords: reference gene, stability evaluation, transcription abundance
1. Introduction
The basidiomycete Pleurotus tuber-regium (Fr.) Singer, known as tiger milk mushroom, is a tropical and subtropical sclerotium-producing mushroom (Oso, 1977). Pleurotus tuber-regium is of popularity and economic importance due to the nutritional value and bioactive compounds around the world especially in China, Africa, and the United States (Wong & Cheung, 2008; Liu et al., 2018a). Previous studies have focused on the research of P. tuber-regium reproduction (Fasidi & Ekuere, 1993; Okhuoya & Etugo, 1993; Bamigboye, Oloke, & Dames, 2019), nutrition (Oyetayo & Akindahunsi, 2005; Ohiri, 2018), pharmacological value (Wu, Lu, Jiang, Li, & Huang, 2014; Okolo, Orisakwe, & Siminialayi, 2018), and molecular biology (Huang, Lin, Cheung, & Wu, 2011; Liu et al., 2018b). With the completion of the diploid genome sequencing (Lam et al., 2018) and the construction of Agrobacterium tumefaciens-mediated transformation system (Liu et al., 2018b) in P. tuber-regium, there is an increasing attention on the molecular biological research of important functional genes as it provides necessary information about fungal development and metabolite synthesis.
Currently, gene expression analysis is one of the most important fields in molecular functional research. One of the most widely used techniques to determine gene expression is reverse transcription quantitative polymerase chain reaction (RT-qPCR), due to its high accuracy, specificity and reproducibility (Derveaux, Vandesompele, & Hellemans, 2010). Thereinto, appropriate reference genes are necessary to standardize RT-qPCR results, and they must be validated in analyses to minimize changes in the quantity and quality of RNA, reverse transcription efficiency in the PCR steps (Fleige & Pfaffl, 2006; Derveaux et al., 2010). Reference genes, used to normalize mRNA fractions, are often referred to as housekeeping genes which should not vary their expression during development, among tissues or cells, or in response to experimental treatments (de Oliveira et al., 2012).
Although several genes have been routinely used and reported as reference genes in mushroom molecular biology studies, the reference genes for P. tuber-regium have not yet been reported. Many studies make use of the housekeeping genes without proper validation of their presumed stability, based on the assumption that they would be constitutively expressed due to their roles in basic cellular processes (de Oliveira et al., 2012). To obtain a precise quantification of gene expression, it is necessary to catch hold of stabilized reference genes for the different conditions being researched in P. tuber-regium. In this study, 14 candidate reference genes from the transcriptomic data and commonly used reference genes were screened and verified for gene expression research in P. tuber-regium different tissues and mycelia which grown under stress treatments of different temperatures, water deficit, and salinity shifts. Four different statistical algorithms were used to obtain a combination of reference genes that can be used for prospective P. tuber-regium gene expression studies. To our knowledge, this is the first report on selection and validation of reference genes for RT-qPCR data normalization in P. tuber-regium.
2. Materials and methods
2.1. Strain and culture situations
The P. tuber-regium strain ACCC 50657, provided by the Laboratory of Food Microbiology, Huazhong Agricultural University, was maintained and sub-cultured on Petri dishes with PDA medium (potato 200 g/L, glucose 20 g/L, agar 20 g/L) in the darkness at room temperature and 32 °C, respectively.
To prepare sclerotia and fruitbodies samples, the culture substrates composed of cottonseed shell 500 g, bran 15 g, gypsum 2.5 g, lime 2.5 g, and H2O 700 mL were made. The 300 g sterilized culture bags inoculated with 5–10 g matrix covered with vigorous hyphae were incubated at 32 °C for 15 d to make the hyphae overgrown with culture substrates. Then, the sclerotia and fruitbodies were induced on culture substrate covered with nutrient soil (approximately 2.5 L) in the plastic vase (approximately 5.5 L) with round bottom at room temperature with proper humidity. The sclerotia and fruitbodies were grown randomly. The sclerotia were harvested when the diameter reached about 1 cm. The fruitbodies of P. tuber-regium were divided into pilei and stipes. To prepare mycelia samples, the PDA medium supplemented with mannitol or NaCl were made. The hyphae grown on PDA medium covered with cellophane in advance were harvested when they were 8 d old under different experimental conditions. 12 samples were selected for this assay, including sclerotia, the stipes of fruitbodies, the pilei of fruitbodies, and 9 mycelia samples grown on PDA medium under different temperatures (25 °C, 28 °C, 32 °C), water deficit (mannitol: 0.1 M, 0.2 M, 0.3 M, 0.4 M), and salinity shifts (NaCl: 0.086 M, 0.171 M).
2.2. Extraction of RNA isolation and cDNA synthesis
The RNA of P. tuber-regium was extracted by means of KKFast Plant RNApure Kit (Zomanbio, Beijing, China) according to the manufacturer’s instructions. The quantity and purity of RNA were measured in a NanoPhotometer-N60 (Implen, Munich, German), meanwhile the quality and integrity of the RNA were verified using electrophoresis with 1.0% agarose gel. The OD260/OD280 of the extracted RNA ranged from 1.8 to 2.2, and agarose gel electrophoresis had clear three bands that could be used for subsequent experiments. The cDNA was synthesized by reverse transcription reaction from 1 µg of RNA isolation according to the PrimerScriptTM RT reagent Kit with gDNA Eraser (TaKaRa, Dalian, China).
2.3. Candidate reference gene selection and primer design
Novel candidate reference genes were selected based on differentially expressed genes (DEGs) profiles between newly formed sclerotia (approximately 1 cm diam) and mycelia grown on culture substrates for 12 d in P. tuber-regium. The RNA-seq data used in this study were acquired by the Illumina Hiseq and three biological replicates were performed (unpublished data). The Cufflinks software was employed to estimate gene expression levels by using FPKM values (fragments per kilobase of exon per million fragments mapped) (Trapnell et al., 2010). In addition, six genes commonly used as reference genes mentioned in other literatures were also employed in this study.
Primers used in this study were designed using Primer 3 plus online website (http://primer3plus.com/cgi-bin/dev/primer3plusPackage.cgi), and the specificity of the product was verified using 1.0% agarose gel electrophoresis and RT-qPCR melting curves. The PCR products were within 80–150 bp range and each primer pair was anchored within exon-exon junction. The relevant information of all primers was listed in Table 1.
Table 1. Candidate genes, primers, product lengths, and amplification efficiencies in the RT-qPCR analysis.
| Gene | Gene symbol |
Orientation Sequence (5’→3’) | Amplification length (bp) |
Amplification efficiency (%) |
R2 | |
| α-tubulin | α-tub | Forward | TCACGGCTTCGCTTCGTT | 114 | 103.98 | 0.99445 |
| Reverse | TGATAGGGGCAAAGGTCGC | |||||
| β-tubulin | β-tub | Forward | TACACGCAGGAGGGTATGGA | 137 | 83.59 | 0.99865 |
| Reverse | TCGACAGGCTCTTCCTCGTA | |||||
| γ-tubulin | γ-tub | Forward | CGGTACAGCTGGGCCAAT | 116 | 90.98 | 0.99945 |
| Reverse | TCTCCTCCCTCAGTCGCC | |||||
| Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | Forward | AGGCTGTTGGCAAGGTCA | 147 | 97.17 | 0.9996 |
| Reverse | CCTTGACGACGGCCTTGAT | |||||
| Translation initiation factor 5a | Tif-5a | Forward | CTGCCCTTCCACCCACAAT | 106 | 85.57 | 0.9981 |
| Reverse | ATTGCCATCCGCGCTCAT | |||||
| Translation elongation factor 1α | Tef-1α | Forward | CGAGCAGGGTGTTCCAGG | 95 | 86.12 | 0.99875 |
| Reverse | TCTTGGAGTCGGAGGCGA | |||||
| Transitional endoplasmic reticulum ATPase | ATPase | Forward | CACGCCGTTCTGTATCCGA | 85 | 86.53 | 0.9993 |
| Reverse | TTGTTGCCGAAGCCACGA | |||||
| Short/branched chain specific acyl-CoA dehydrogenase | acyl-CoA | Forward | TGGATCCCTCAGTGTCCGT | 137 | 79.14 | 0.99785 |
| Reverse | CAAGCAAAACGACCCGAGC | |||||
| U2 small nuclear ribonucleoprotein A&apos | U2ribo | Forward | ATCGCGAGTGGCTTGTGT | 140 | 82.02 | 0.99955 |
| Reverse | TGATAGCGTCGTAGCGAGC | |||||
| Succinate/glutarate-semialdehyde dehydrogenase | S/G | Forward | TGGCGGCTTCGACTTTGA | 111 | 88.5 | 0.9978 |
| Reverse | TGCATATGGCCGCTCCTT | |||||
| WHI2-like protein | Wlp | Forward | ATTTTCACGGCGCTGCAG | 131 | 84.17 | 0.9989 |
| Reverse | CTTGAAGGCTCCAATGCGC | |||||
| V-type proton ATPase subunit E | VtpAsE | Forward | AGAAGAAGCACAAGGGCGT | 95 | 94.17 | 0.9994 |
| Reverse | TTCTTCGCGGCGGTGAA | |||||
| E3 ubiquitin-protein ligase RBX1 | E3upl | Forward | AGCAACCAGTGAAGAGTGCA | 88 | 87.44 | 0.99805 |
| Reverse | CCTGGTCTTGAGCCAACGT | |||||
| Succinate dehydrogenase (ubiquinone) cytochrome b560 subunit | Sdh | Forward | TTCCTGATGCCGCCAAGT | 123 | 93.35 | 0.99965 |
| Reverse | AAGCCCCTTTGACCGTGA | |||||
2.4. RT-qPCR
The total volume of RT-qPCR reactions was 10 μL, containing 5 μL of TB GreenTM Premix Ex TaqTM II (TaKaRa), 0.2 μL (10 μM) of each primer (forward and reverse), 1 μL of cDNA (dilution 1:10 previously defined) and 3.6 μL of ultrapure water. The reactions were performed in QuantStudioTM 6 Flex System (Thermo Fisher Scientific, Waltham, USA) using the following amplification parameters: 50 °C for 2 min, 95 °C for 10 min, 40 cycles of 95 °C for 15 s, 60 °C for 1 min, with insertion of the melting curve from 72 to 95 °C, with a heating rate of 1.6 °C/s and a continuous fluorescence measurement. All reactions were performed with three technical repetitions and two biological replications with negative controls.
2.5. Amplification efficiencies
To acquire the PCR amplification efficiencies (E) of each primer pair, standard curves were calculated using a 5-fold serial diluted pMD18-T plasmid (TaKaRa). Every candidate reference gene amplified by PCR was constructed into the plasmid. The E values was ranged (Radonić et al., 2004) and listed in Table 1.
2.6. Data analyses
To select suitable reference genes, three software was recruited to calculate the stability: geNorm (Vandesompele et al., 2002), NormFinder (Andersen, Jensen, & Ørntoft, 2004), and BestKeeper (Pfaffl, Tichopad, Prgomet, & Neuvians, 2004). In addition, RefFinder (https://www.heartcure.com.au/reffinder) which is a comprehensive web-based tool that integrates the three applets was also applied to produce a comprehensive ranking of the candidate reference genes.
3. Results
3.1. The selection of candidate reference genes
In this study, the constitutively expressed candidate genes were screened using the following standards: (1) the FPKM values range from 200 to 5000 in all of the tested samples; (2) FPKM value in the sclerotia sample/ FPKM in the mycelia sample approximately equal to 1; (3) P value<0.5 for each candidate gene; (4) the genes can be annotated in the databases including NR, Swiss-Prot, KOG, KEGG, GO, Pfam, TrEMBL. We filtrated and evaluated the candidate genes of interest with fold-change close to one, indicating stability of transcript levels under those conditions. They are Transitional endoplasmic reticulum ATPase (ATPase), Short/branched chain specific acyl-CoA dehydrogenase (acyl-CoA), U2 small nuclear ribonucleoprotein A & apos (U2ribo), Succinate/glutarate-semialdehyde dehydrogenase (S/G), WHI2-like protein (Wlp), V-type proton ATPase subunit E (VtpAsE), E3 ubiquitin-protein ligase RBX1 (E3upl), and Succinate dehydrogenase (ubiquinone) cytochrome b560 subunit (Sdh). In addition, 6 genes used as reference genes in reported literatures, including α-tubulin (α-tub), β-tubulin (β-tub), γ-tubulin (γ-tub), Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), Translation initiation factor 5a (Tif-5a), and Translation elongation factor 1α (Tef-1α), were also recruited to normalize RT-qPCR data in this research (Zampieri, Nora, Basso, Camassola, & Dillon, 2014; Castanera, López-Varas, Pisabarro, & Ramírez, 2015). The gene name, gene ID, accession number, function, and FPKM in transcriptome samples of each candidate gene were described in Table 2.
Table 2. The gene name, gene ID, FPKM in transcriptome samples, and KOG description of each candidate gene used in this study.
| Gene | Gene ID | Accession number |
FPKM value in RNA-seq mycelia samples | FPKM value in RNA-seq sclerotia samples | Function | ||||
| Mycelium 1 | Mycelium 2 | Mycelium 3 | Sclerotium1 | Sclerotium2 | Sclerotium3 | ||||
| α-tub | evm.TU.contig_3_pilon.401 | MW504066 | 1,637.05 | 1,981.61 | 2,022.09 | 1,391.35 | 1,424.07 | 1,786.9 | Cytoskeleton |
| β-tub | evm.TU.contig_38_pilon.186 | MW579480 | 2,089.67 | 2,061.52 | 2,212.61 | 1,735.01 | 1,698.92 | 1,954.91 | Cytoskeleton |
| γ-tub | evm.TU.contig_32_pilon.312 | MW579481 | 40.267 | 50.9676 | 43.6822 | 36.9135 | 38.2139 | 29.1037 | Cytoskeleton |
| GAPDH | evm.TU.contig_13_pilon.60 | MW579482 | 1,902.2 | 2,558.2 | 2,833.49 | 946.535 | 604.03 | 1,577.32 | Carbohydrate transport and metabolism |
| Tif-5a | evm.TU.contig_13_pilon.46 | MW625948 | 2,778.9 | 2,904.14 | 3,921.95 | 1,925.2 | 1,914.82 | 2,169.98 | Translation, ribosomal structure and biogenesis |
| Tef-1α | maker-contig_3_pilon-exonerate_protein2genome-gene-52.4 | MW625949 | 7,981.22 | 7,587.76 | 8,554.87 | 3,889.41 | 2,804.54 | 5,091.9 | Translation, ribosomal structure and biogenesis |
| ATPase | evm.TU.contig_15_pilon.85 | MW625950 | 452.421 | 532.817 | 546.236 | 486.035 | 464.005 | 583.112 | Posttranslational modification, protein turnover, chaperones |
| acyl-CoA | evm.TU.contig_3_pilon.882 | MW625951 | 444.297 | 432.19 | 441.913 | 449.624 | 416.271 | 449.464 | Lipid transport and metabolism |
| U2ribo | maker-contig_3_pilon-exonerate_protein2genome-gene-45.6 | MW625952 | 253.134 | 262.828 | 347.344 | 260.039 | 309.41 | 270.064 | RNA processing and modification |
| S/G | evm.TU.contig_3_pilon.218 | MW625953 | 277.18 | 235.686 | 269.645 | 294.402 | 252.925 | 258.03 | Energy production and conversion |
| Wlp | maker-contig_25_pilon-exonerate_protein2genome-gene-42.2 | MW625954 | 211.321 | 222.871 | 225.605 | 208.97 | 212.471 | 220.273 | NA |
| VtpAsE | evm.TU.contig_27_pilon.329 | MW625956 | 299.824 | 302.765 | 374.97 | 348.827 | 327.401 | 297.42 | Energy production and conversion |
| E3upl | maker-contig_32_pilon-exonerate_protein2genome-gene-33.2 | MW625957 | 1,105.04 | 1,059.68 | 1,866.21 | 1,392.85 | 1,356.21 | 1,173.37 | Posttranslational modification, protein turnover, chaperones |
| Sdh | evm.TU.contig_6_pilon.48 | MW625955 | 428.991 | 407.464 | 572.122 | 336.315 | 653.744 | 378.491 | Carbohydrate transport and metabolism |
3.2. The specificity and amplification efficiency of each primer pair of candidate reference genes
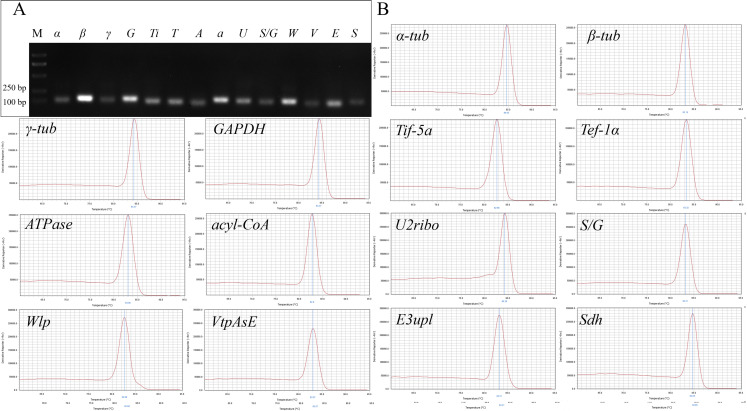
The expression levels of the 14 candidate reference genes were measured under each of the 12 experimental conditions (see Materials and methods). 14 primer pairs were designed for the selected candidate genes, respectively (Table 1). The agarose gel electrophoresis and a melting curve analysis by RT-qPCR were performed to verify the specificity of the primers used in this study (Fig. 1A, B). The PCR amplification efficiencies for the 14 candidate genes varied from 79.14% (acyl-CoA) to 103.98% (α-tub), and the correlation coefficients ranged from 0.99445 for α-tub to 0.99965 for Sdh (Table 1).
Fig. 1. The specificity verification of pair-wise primers used in this study. A: 1% agarose gel electrophoresis demonstrated the amplification of a single band of the expected size for the 14 reference genes. B: Melting curves of the 14 candidate reference genes showed single peaks.
3.3. The variances of Ct values among 14 candidate reference genes
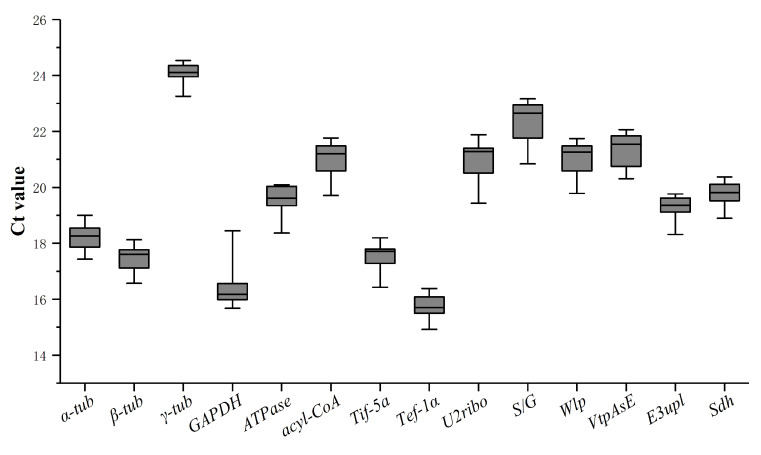
For visual comparisons of different RNA transcription abundances, the Ct value was analyzed in the samples directly. The variances of Ct values demonstrated a compact distribution in Fig. 2. According to the fluctuation of Ct value, each of candidate gene showed moderate abundance with a limited range between 14.92 and 24.53 in the tested P. tuber-regium samples. γ-tub demonstrated equivalent levels of gene expression, while the expression of GAPDH seemed to be significantly fluctuated by the treatments. The main programs, including geNorm (Vandesompele et al., 2002), NormFinder (Andersen et al., 2004), and BestKeeper (Pfaffl et al., 2004) were employed to analyze the 14 candidate reference genes.
Fig. 2. Variations in cycle threshold (Ct) values of 14 selected candidate reference genes under the 12 experimental conditions.
3.4. The prediction of the expression stability of candidate reference genes by geNorm, NormFinder, BestKeeper software
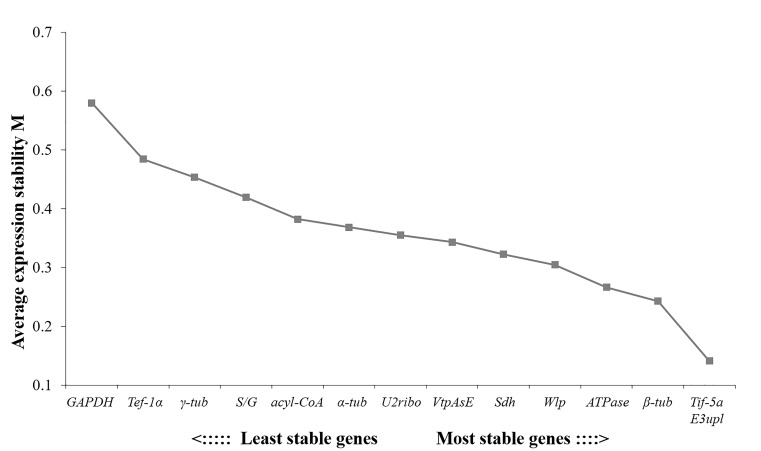
The geNorm tool defined the internal control gene-stability measure M as the average pairwise variation of a particular gene with all other control genes (Vandesompele et al., 2002). For all 12 samples tested, Tif-5a and E3upl were the most stable gene pair, followed by β-tub, ATPase and Wlp. In contrast, GAPDH was the least stable candidate gene (Fig. 3). At the same time, the Vn/(n+1) value was calculated by geNorm software, and the optimal number of internal reference genes was obtained. In the treatments, data acquired from all 12 samples were analyzed and demonstrated that the V value with the inclusion of two genes (V2/3) was 0.095, lower than the arbitrary cut off value of 0.15. Therefore, it was enough to select 2 candidate genes as optimal reference gene combination, and the third one was not required for further correction.
Fig. 3. Average expression stability of the 14 candidate reference genes calculated by the geNorm.
The NormFinder analysis strategy provides a direct measure for the estimated expression variation, enabling the user to evaluate the systematic error introduced when using the gene (Andersen et al., 2004). In this analysis, E3upl was the most stable internal reference gene, consistent with geNorm analysis, followed by Sdh, and the most unstable gene was GAPDH.
BestKeeper was the third method used to evaluate the expression stability of the 14 candidate reference genes. Because it can only analyze no more than 10 genes, we chose the top 10 candidates from the geNorm classification as input data. The selected genes were also within the top 10 in the NormFinder ranking besides acyl-CoA. The applet determines the optimal housekeeping gene employing the pair-wise correlation analysis of all pairs of candidate genes and calculates the geometric mean of the best suited ones (Pfaffl et al., 2004), and it is usually employed for preliminary filtration. Similar with the results of the previous two methods, the most consistently expressed gene was E3upL, followed by Sdh, the gene with the most erratic expression was U2ribo.
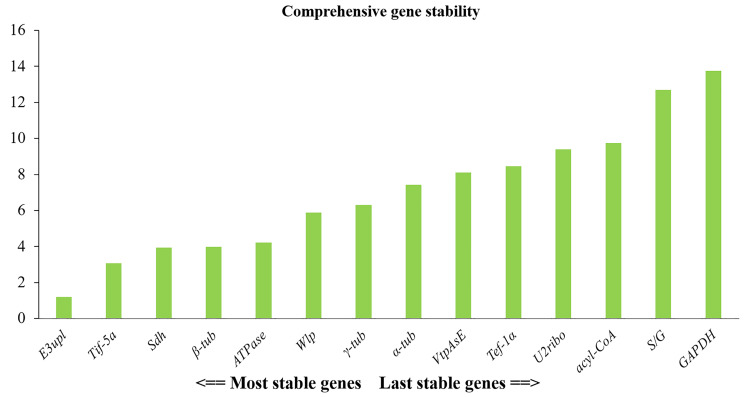
3.5. The prediction of best reference gene by online RefFinder analysis
RefFinder compares and ranks the tested candidate reference genes integrating the currently available major computational programs (Xie, Xiao, Chen, Xu, & Zhang 2012). It was demonstrated that E3upl was the best reference gene among all tested samples (Fig. 4). In gene expression analysis, using two or more reference genes in the meantime can reduce systematic bias and help to obtain more accurate quantitative data of gene expression. Comprehensive analysis of the gene stability revealed that E3upl and Tif-5a could be used as the optimal reference gene combination among mycelia under different stress responses and different tissues.
Fig. 4. Comprehensive expression stability of the 14 candidate reference genes calculated by the RefFinder.
3.6. The expression stabilities of candidate reference genes under different conditions
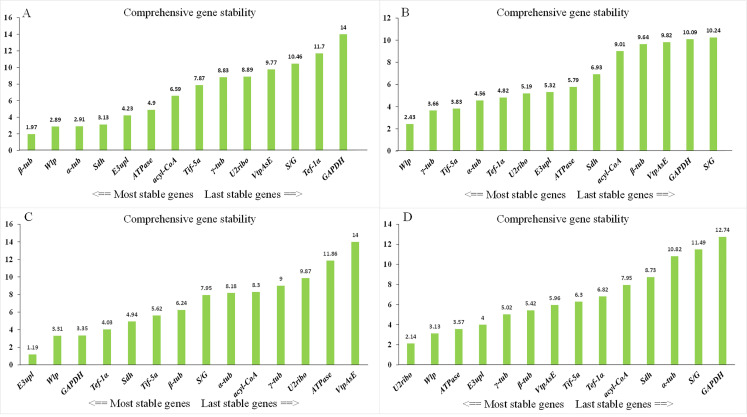
When all 12 samples were considered together, the combination of E3upl and Tif-5a genes was the most stable for serving as endogenous reference genes by the analyses of four statistical algorithms. Due to RefFinder integrates the currently available major computational programs (geNorm, NormFinder, BestKeeper, and the comparative Delta-Ct method) to compare and rank the tested candidate reference genes (Xie et al., 2012), we selected it to detect the expression stabilities of candidate reference genes under four different conditions (Fig. 5). Under different development stages, β-tub was the most stable reference gene, while GAPDH was considered as the most unstable one (Fig. 5A). For the mycelia samples, Wlp showed the most stability under different temperatures (Fig. 5B), E3upl showed the most stability under water deficit (Fig. 5C), while U2ribo demonstrated the most stability under salinity shifts (Fig. 5D). The S/G, VtpAsE, and GAPDH exhibited the lowest stability, respectively (Fig. 5B–D).
Fig. 5. Expression stability and ranking of candidate reference genes determined by RefFinder. A: Different developmental stages. B: Mycelia under different temperatures. C: Mycelia under water deficit. D: Mycelia under salinity shifts.
4. Discussion
Among various technologies, RT-qPCR is considered to be the most appropriated method for gene expression analyses (Xiang et al., 2018). The expression stability of the same reference gene under different conditions is differential among various species. In order to acquire suitable genes for data normalization in P. tuber-regium, 14 candidate reference genes were selected from transcriptomic data and internal reference genes commonly used in other fungal species, and measured their expression levels by RT-qPCR among 12 samples. The four statistical algorithms (NormFinder, geNorm, BestKeeper, and RefFinder) have been recruited to select and assess candidate reference genes for RT-qPCR data normalization from different tissues, developmental stages, and stress conditions in various filamentous fungi like Lentinula edodes (Xiang et al., 2018; Zhao et al., 2018), P. ostreatus (Castanera et al., 2015), and Ganoderma lucidum (Xu et al., 2014).
In the current study, the selected 14 candidate reference genes exhibited a wide range of expression profiles under experimental conditions (Fig. 2), demonstrating that no single gene had a consistent expression profile. The expression stability of each candidate reference gene is different according to different test conditions. Each reference gene was ranked with respect to stability using the four tests (Table 3). In general, E3upl and Tif-5a were identified as the stable gene combination for various stress conditions and different samples by comprehensive statistical analysis, and was also recommended as the best reference gene combination for RT-qPCR data normalization in the tested samples in P. tuber-regium. The results demonstrated that there was a relatively good correlation among the results obtained from the four analysis approaches, despite the fact that the fundamental difference in the algorithms implemented by the four software. Particularly, E3upl, Wlp, and U2ribo were the most consistently expressed genes in mycelia under water deficit, different temperatures, and salinity shifts, respectively. While β-tub was the most stably expressed gene in different tissues. GAPDH was the most unstable gene in different tissues. S/G, VtpAsE, and GAPDH were the most unstable reference genes in mycelia under different temperatures, water deficit, and salinity shifts, respectively. In general, E3upl and Tif-5a were identified as the stable gene combination for various stress conditions and different samples by comprehensive statistical analysis, and was also recommended as the best reference gene combination for RT-qPCR data normalization in the all samples in P. tuber-regium.
Table 3. Ranking of Pleurotus tuber-regium candidate reference genes according to their expression stability, analyzed by geNorm, NormFinder, BestKeeper, and RefFinder for 12 experimental conditions.
| Rank | Gene expression stability | |||
| GeNorm | NormFinder | BestKeeper | RefFinder | |
| 1 | E3upl/Tif-5a | E3upl | E3upl | E3upl |
| 2 | β-tub | Sdh | Sdh | Tif-5a |
| 3 | ATPase | Tif-5a | Tif-5a | Sdh |
| 4 | Wlp | β-tub | α-tub | β-tub |
| 5 | Sdh | ATPase | β-tub | ATPase |
| 6 | VtpAsE | Wlp | ATPase | Wlp |
| 7 | U2ribo | α-tub | acyl-CoA | γ-tub |
| 8 | α-tub | ATPase | Wlp | α-tub |
| 9 | acyl-CoA | VtpAsE | VtpAsE | VtpAsE |
| 10 | S/G | U2ribo | U2ribo | Tef-1α |
| 11 | γ-tub | acyl-CoA | U2ribo | |
| 12 | Tef-1α | γ-tub | acyl-CoA | |
| 13 | GAPDH | Tef-1α | S/G | |
| 14 | GAPDH | GAPDH | ||
5.Conclusions
In summary, the E3upl and Tif-5a were recommended as the best reference gene combination for RT-qPCR data analyses among the research samples in P. tuber-regium. This research provides basic background information with respect to reference gene selection and utilization for RT-qPCR studies in P. tuber-regium. The study will contribute to sequencing data and gene function analyses.
Conflicts of Interest
No conflicts of interest among researchers.
Author’s contribution
All authors contributed to this study. XS and AM had the ideas for the article. AM guided the experiments. XS preformed the experiments, and wrote the manuscript. JW, CM and AM critically reviewed the manuscript.
Acknowledgements
This work was supported by a grant from the National Natural Science Foundation of China (NSFC) (No. 31772375) to Aimin Ma.
References
- Andersen, C. L., Jensen, J. L., & Ørntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research, 64, 5245–5250. https://doi.org/10.1158/0008-5472.CAN-04-0496 [DOI] [PubMed] [Google Scholar]
- Bamigboye, C. O., Oloke, J. K., & Dames, J. F., (2019). Development of high yielding strain of Pleurotus tuber-regium: fructification, nutritional and phylogenetic studies. Journal of Food Science and Technology, 56, 3597–3608. https://doi.org/10.1007/s13197-019-03786-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanera, R., López-Varas, L., Pisabarro, A. G., & Ramírez, L. (2015). Validation of reference genes for transcriptional analyses in Pleurotus ostreatus by using reverse transcription-quantitative PCR. Applied and Environmental and Microbiology, 81, 4120–4129. https://doi.org/10.1128/AEM.00402-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira, L. A., Breton, M. C., Bastolla, F. M., Camargo, S. D. S., Margis, R., Frazzon, J., & Pasquali, G. (2012).Reference genes for the normalization of gene expression in Eucalyptus species. Plant and Cell Physiology, 53, 405–422. https://doi.org/10.1093/pcp/pcr187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derveaux, S., Vandesompele, J., & Hellemans, J. (2010). How to do successful gene expression analysis using real-time PCR, Methods, 50, 227–230. https://doi.org/10.1016/j.ymeth.2009.11.001 [DOI] [PubMed] [Google Scholar]
- Fasidi, I. O., & Ekuere, U. U. (1993). Studies on Pleurotus tuber-regium (Fries) Singer: cultivation, proximate composition and mineral contents of sclerotia. Food Chemistry, 48, 255–258. https://doi.org/10.1016/0308-8146(93)90136-4 [Google Scholar]
- Fleige, S., & Pfaffl, M. W. (2006). RNA integrity and the effect on the real-time qRT-PCR performance. Molecular Aspects of Medicine, 27, 126–139. https://doi.org/10.1016/j.mam.2005.12.003 [DOI] [PubMed] [Google Scholar]
- Huang, B., Lin, W., Cheung, P. C. K., & Wu, J.(2011). Differential proteomic analysis of temperature-induced autolysis in mycelium of Pleurotus tuber-regium. Current Microbiology, 62, 1160–1167. https://doi.org/10.1007/s00284-010-9838-4 [DOI] [PubMed] [Google Scholar]
- Lam, K. L., Si, K., Wu, X., Tang, S., Sun, X., Kwan, H. S., & Cheung, P. C. K. (2018). The diploid genome of the only sclerotia-forming wild-type species in the genus Pleurotus–Pleurotus tuber-regium–provides insights into the mechanism of its biomass conversion from lignocellulose substrates. Journal of Biotechnology, 283, 22–27. https://doi.org/10.1016/j.jbiotec.2018.07.009 [DOI] [PubMed] [Google Scholar]
- Liu, D., Zhu, H., Chen, Y., Zheng, L., Chen, L., & Ma, A. (2018. a). Agrobacterium tumefaciens-mediated transformation of the King Tuber medicinal mushroomLentinus tuber-regium (Agaricomycetes). International Journal of Medicinal Mushrooms, 20, 791–796. https://doi.org/10.1615/IntJMedMushrooms.2018026991 [DOI] [PubMed] [Google Scholar]
- Liu, D., Zhu, H., Chen, Y., Zheng, L., Chen, L., & Ma, A. (2018. b). Cloning and heterologous expression of a hydrophobin gene Ltr.hyd from the tiger milk mushroom Lentinus tuber-regium in yeast-like cells of Tremella fuciformis. Electronic Journal of Biotechnology, 32, 6–12. https://doi.org/10.1016/j.ejbt.2017.12.003 [Google Scholar]
- Ohiri, R. C. (2018). Nutriceutical potential of Pleurotus tuber-regium sclerotium. The Ukrainian Biochemical Journal, 90, 84–93. https://doi.org/10.15407/ubj90.03.084 [Google Scholar]
- Okhuoya, J. A., & Etugo, J. E. (1993). Studies of the cultivation of Pleurotus tuber-regium (Fr) Sing. an edible mushroom. Bioresource Technology, 44, 1–3. https://doi.org/10.1016/0960-8524(93)90198-K [Google Scholar]
- Okolo, K. O., Orisakwe, O. E., & Siminialayi, I. M. (2018). Nephroprotective and antioxidant effects of king tuber oyster medicinal mushroom, Pleurotus tuber-regium (Agaricomycetes), on carbon tetrachloride-induced nephrotoxicity in male sprague dawley rats. International Journal of Medicinal Mushrooms, 20, 419–429. https://doi.org/10.1615/IntJMedMushrooms.2018026044 [DOI] [PubMed] [Google Scholar]
- Oso, B. A. (1977). Pleurotus tuber-regium from Nigeria. Mycologia, 69, 271–279. https://doi.org/10.2307/3758652 [PubMed] [Google Scholar]
- Oyetayo, F. L., & Akindahunsi, A. A. (2005). Nutrient and antinutrient distribution of edible mushroom, Pleurotus tuber-regium (fries) singer. LWT–Food Science and Technology, 39, 548–553. https://doi.org/10.1016/j.lwt.2005.04.005 [Google Scholar]
- Pfaffl, M. W., Tichopad, A., Prgomet, C., & Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper – Excel-based tool using pair-wise correlations. Biotechnology Letters, 26, 509–515. https://doi.org/10.1023/b:bile.0000019559.84305.47 [DOI] [PubMed] [Google Scholar]
- Radonić, A., Thulke, S., Mackay, I. M., Landt, O., Siegert, W., & Nitsche, A. (2004).Guideline to reference gene selection for quantitative real‑time PCR. Biochemical and Biophysical Research Communications, 313, 856–862. https://doi.org/10.1016/j.bbrc.2003.11.177 [DOI] [PubMed] [Google Scholar]
- Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., Salzberg, S. L., Wold, B. J., & Pachter, L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology, 28, 511–515. https://doi.org/10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele, J., de Preter, K., Pattyn, F., Poppe, B., van Roy, N., de Paepe, A., & Speleman, F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology, 3, research0034.1–0034.11. https://doi.org/10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, K. H., & Cheung, P. C. K. (2008).Sclerotium of culinary-medicinal king tuber oyster mushroom, Pleurotus tuberregium (Fr.) Singer (Agaricomycetideae): its cultivation, biochemical composition, and biopharmacological effects (review). International Journal of Medicinal Mushrooms, 10, 303–313. https://doi.org/10.1615/IntJMedMushr.v10.i4.20 [Google Scholar]
- Wu, G. H., Lu, C. L., Jiang, J. G., Li, Z. Y., & Huang, Z. L. (2014). Regulation effect of polysaccharides from Pleurotus tuber-regium (Fr.) on the immune activity of mice macrophages. Food & Function, 5, 337–344. https://doi.org/10.1039/c3fo60410a [DOI] [PubMed] [Google Scholar]
- Xiang, Q., Li, J., Qin, P., He, M., Yu, X., Zhao, K., Zhang, X., Ma, M., Chen, Q., Chen, X., Zeng, X., & Gu, Y. (2018). Identification and evaluation of reference genes for qRT-PCR studies in Lentinula edodes. PLoS One, 13, e0190226. https://doi.org/10.1371/journal.pone.0190226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, F., Xiao, P., Chen, D., Xu, L., & Zhang, B. (2012). miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Molecular Biology, 80, 75–84. https://doi.org/10.1007/s11103-012-9885-2 [DOI] [PubMed] [Google Scholar]
- Xu, J., Xu, Z., Zhu, Y., Luo, H., Qian, J., Ji, A., Hu, Y., Sun, W., Wang, B., Song, J., Sun, C., & Chen, S.(2014). Identification and evaluation of reference genes for qRT-PCR normalization in Ganoderma lucidum. Current Microbiology, 68, 120–126. https://doi.org/10.1007/s00284-013-0442-2 [DOI] [PubMed] [Google Scholar]
- Zampieri, D., Nora, L. C., Basso, V., Camassola, M., & Dillon, A. J. P. (2014). Validation of reference genes in Penicillium echinulatum to enable gene expression study using real-time quantitative RT-PCR. Current Genetics, 60, 231–236. https://doi.org/10.1007/s00294-014-0421-6 [DOI] [PubMed] [Google Scholar]
- Zhao, X., Yang, H., Chen, M., Song, X., Yu, C., Zhao, Y., & Wu, Y. (2018). Reference gene selection for quantitative real-time PCR of mycelia from Lentinula edodes under high-temperature stress. Biomed Research International, 2018, 1670328. https://doi.org/10.1155/2018/1670328 [DOI] [PMC free article] [PubMed] [Google Scholar]