Abstract
Mycoplasma gallisepticum, the cause of chronic respiratory infections in the avian host, possesses a family of M9/pMGA genes encoding an adhesin(s) associated with hemagglutination. Nucleotide sequences of M9/pMGA gene family members indicate extensive sequence similarity in the promoter regions of both the transcribed and silent genes. The mechanism that regulates M9/pMGA gene expression is unknown, but studies have revealed an apparent correlation between gene expression and the number of tandem GAA repeat motifs located upstream of the putative promoter. In this study, transposon Tn4001 was used as a vector with the Escherichia coli lacZ gene as the reporter system to examine the role of the GAA repeats in M9/pMGA gene expression in M. gallisepticum. A 336-bp M9 gene fragment (containing the GAA repeat region, the promoter, and the translation start codon) was amplified by PCR, ligated with a lacZ gene from E. coli, and inserted into the Tn4001-containing plasmid pISM2062. This construct was transformed into M. gallisepticum PG31. Transformants were filter cloned on agar supplemented with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) to monitor lacZ gene expression on the basis of blue/white color selection. Several cycles of filter cloning resulted in cell lineages in which lacZ gene expression alternated between the On and Off states in successive generations of progeny clones. The promoter regions of the M9-lacZ hybrid genes of individual progeny clones were amplified by PCR and sequenced. The only differences between the promoter regions of the blue and white colonies were in the number of GAA repeats. Clones that expressed lacZ had exactly 12 tandem copies of the GAA repeat. Clones that did not express lacZ invariably had either more than 12 (14 to 16) or fewer than 12 (5 to 11) GAA repeats. Southern analysis of M. gallisepticum chromosomal DNA confirmed that the phase-variable expression of the lacZ reporter gene was not caused by Tn4001 transposition. These data strongly indicate that changes in the length of the GAA repeat region are responsible for regulating M9/pMGA gene expression.
A family of pMGA genes with a potential to code for hemagglutinins has been identified in the avian chronic respiratory disease agent Mycoplasma gallisepticum (14). Recently, we demonstrated that the 62-kDa M9 protein associated with monoclonal antibody-mediated agglutination of M. gallisepticum is encoded by a member of the pMGA family (12). The tandemly arranged multiple pMGA genes in M. gallisepticum S6 account for as much as 8% of the 1,030-kb total genome (2). Delineation of a significant amount of DNA for the synthesis of pMGA gene products denotes an important dynamic function for this gene cluster in the morphogenesis and survival of the organism. Among the 32 to 70 pMGA genes that are estimated to be present in the genomes of different strains of M. gallisepticum, there is appreciable sequence similarity both within and across strains. It is thought that only one of the pMGA genes is predominately expressed in a given strain as a result of transcriptional regulation (7). A notable similarity in the intergenic regions of members of the M9/pMGA family is the occurrence of tandem GAA trinucleotide repeats located upstream of the putative promoter. While both the pMGA1.1 gene expressed in strain S6 and the M9 gene expressed in strain PG31 have exactly 12 copies of the GAA repeat, none of the silent genes of the M9/pMGA family for which nucleotide sequence data are known have 12 repeats (8, 12). In vitro growth of M. gallisepticum S6 in the presence of specific antibodies to pMGA1.1 (the expressed protein in strain S6) negates the production of pMGA1.1 and leads to the production of a related pMGA family member, pMGA1.9 (15). The change in pMGA protein production resulting from the incubation of cells with pMGA-specific antibody was accompanied by changes in the number of GAA repeats upstream of pMGA genes, consistent with a mechanism by which oscillation of pMGA gene expression between the On and Off states is regulated by the length of the GAA repeat region (8).
In this study we demonstrate for the first time the use of a lacZ reporter to investigate Mycoplasma gene regulation. A lacZ-M9 fusion gene was constructed and inserted into the M. gallisepticum PG31 chromosome by using the Staphylococcus aureus transposon Tn4001 as the delivery vehicle. The fusion gene consisted of a 336-bp PCR product containing the M9 GAA repeat region, M9 transcription and translation start sites, and a promoterless lacZ gene from Escherichia coli. Expression of the fusion gene in M. gallisepticum was monitored by observing the blue/white color of colonies on agar supplemented with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Pedigree analysis of several generations of subclones demonstrated that colonies that were predominately blue had 12 copies of the GAA repeat sequence upstream of the lacZ gene while white colonies had either more or fewer than 12 repeats but never exactly 12. The change in the length of the GAA repeat region was the only difference in the nucleotide sequence of the M9 portion of the fusion genes from blue and white colonies. These data indicate that the 336-bp region contains all the sequences necessary to drive GAA-dependent M9 gene expression.
MATERIALS AND METHODS
Bacteria, plasmids, and culture conditions.
M. gallisepticum PG31 (ATCC 19610) was cultured in modified Frey (6) broth or agar supplemented with 10% swine serum as described previously (10). Broth cultures were grown at 37°C to mid-log phase as observed by a change in the phenol red pH indicator to orange (ca. 2 to 4 days). After transformation, mycoplasmas were grown in modified Frey medium supplemented with 80 μg of gentamicin per ml. Plasmids pISM2062 containing Tn4001 and pISM2062.2lac (11) containing the promoterless lacZ gene were provided by F. C. Minion, University of Iowa, Ames, Iowa. E. coli INVαF′ and the TA cloning vector pCR2.1 were obtained from Invitrogen, Carlsbad, Calif., and E. coli XL1-Blue MRF′ was obtained from Stratagene, La Jolla, Calif. The E. coli strains were cultured in Luria-Bertani medium.
Construction of the lacZ reporter.
A 336-bp M9 gene fragment of M. gallisepticum (containing the GAA region, the putative promoter, and the translation start codon) was amplified by PCR with the 5′ M9 primer CGAAGCTTAGTCCAGAACCCATAAAACCG and the 3′ M9 primer CCAGGATCCGCTAACATTACAAACGAACC (Fig. 1), designed on the basis of previously published M9 gene sequences (GenBank accession no. AF032890 [12]). Plasmid VSP #3 DNA containing the whole M9 gene of M. gallisepticum PG31 (12) was used as the template. PCR was performed in a 50-μl reaction mixture containing 1.25 U of Taq DNA polymerase (Perkin-Elmer Cetus, Norwalk, Conn.), deoxynucleoside triphosphates (0.2 mM each), and 1.5 mM MgCl2. The PCR amplification was done with a GeneAmp 9600 thermal cycler under the following conditions: denaturation at 95°C for 1 min, three cycles of annealing (90 s) at 49°C, extension (30 s) at 72°C, and denaturation (20 s) at 95°C, and then 25 cycles with the same extension and denaturation conditions but with annealing at 58°C.
FIG. 1.
Nucleotide sequence of the 5′-end region of the M9-lacZ fusion gene. Sequences derived from the oligonucleotide primers used for PCR amplification of the M9 promoter region and the trinucleotide GAA repeat region are underlined, as is the sequence complementary to the lacZ primer used to amplify the promoter region of the reporter gene in transformants. The putative transcription start site (arrow) of the M9 gene was identified based on sequence similarity to the start site previously determined for pMGA (7).
The amplified M9 gene fragment was ligated into the pCR2.1 cloning vector and transformed into E. coli INVαF′, selecting for ampicillin resistance. The resulting plasmid was linearized by digestion with BamHI and ligated with the 3-kb BamHI fragment containing the promoterless lacZ gene from plasmid pISM2062.2lac (Fig. 2A). The ligation mixture was used to transform E. coli XL1-Blue MRF′, which was assayed on Luria-Bertani plates containing 100 μg of ampicillin per ml, 80 μg of isopropyl-d-thiogalactopyranoside (IPTG) per ml, and 150 μg of X-Gal per ml. Restriction enzyme analysis confirmed that the plasmid (pCR2.1-M9.lacZ), obtained from blue colonies, contained the 336-bp M9 gene fragment and the lacZ gene in the correct orientation.
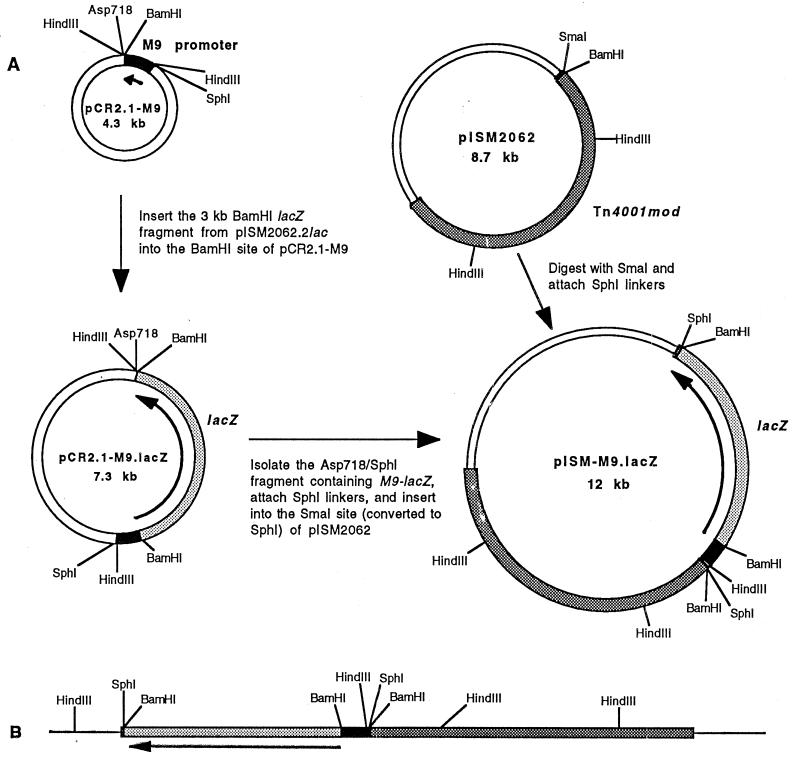
FIG. 2.
Schematic diagram of the construction of the M9-lacZ reporter and its insertion into the chromosome of M. gallisepticum. (A) A 336-bp PCR product containing the 5′ end of the M9 gene was inserted in frame with a promoterless lacZ gene and inserted into Tn4001 as illustrated. White regions denote sequences derived from E. coli plasmid vectors. Regions shaded in black, dark gray, and light gray denote the M9 gene fragment, sequences originating from Tn4001mod, and lacZ, respectively. In pISM2062 and pISM-M9.lacZ, the transposon portion of the plasmid is denoted by a thick line. Arrows marking plasmid regions illustrate the direction of transcription. (B) A representation of Tn4001 containing M9-lacZ inserted into the chromosome of M. gallisepticum. Thin lines denote mycoplasmal chromosomal DNA flanking the transposon insert. Shaded regions are as indicated in panel A. The HindIII site shown in chromosomal DNA sequences on the left represents the mycoplasmal HindIII site most proximal to the left end of the transposon.
The strategy for transferring the M9-lacZ gene from pCR2.1-M9.lacZ to pISM2062, containing Tn4001, is illustrated in Fig. 2A. The M9-lacZ gene was excised as a 3.5-kb fragment from pCR2.1-M9.lacZ by digestion with Asp718 and SphI. The ends of the fragment were made flush by digestion with T4 DNA polymerase (New England BioLabs, Beverly, Mass.), and SphI linkers were attached. To prepare the pISM2062 vector, the SmaI cloning site (located within Tn4001) was converted to a SphI site by digestion of the plasmid with SmaI followed by attachment of SphI linkers. The 3.5-kb fragment containing the M9-lacZ fusion gene was ligated into the SphI site of the modified pISM2062 plasmid to generate plasmid pISM-M9.lacZ. Restriction analysis of pISM-M9.lacZ demonstrated that the M9-lacZ gene is oriented within Tn4001, as shown in Fig. 2A.
Transformation of M. gallisepticum by electroporation.
Cells from log-phase cultures of M. gallisepticum PG31 were harvested by centrifugation at 8,000 × g, washed, and suspended in HEPES-sucrose buffer (HSB) (8 mM HEPES, 272 mM sucrose [pH 7.4]) at 108 to 109 cells/100 μl of buffer. The mycoplasma cells and plasmid pISM-M9.lacZ DNA (20 μg in 10 μl of HSB) were mixed in a prechilled 1.5-ml microtube and incubated on ice for 10 min. The mixture was transferred to a prechilled cuvette with an electrode gap of 0.2 cm and electroporated with a Gene Pulser II (Bio-Rad, Hercules, Calif.) at 2.5 kV, 100 Ω, and 25 μF (9). The cells were immediately transferred to 1 ml of Frey medium in a prechilled 15-ml polypropylene tube, placed on ice for 7 min, incubated at 37°C for 2 h, and assayed on agar supplemented with gentamicin (80 μg/ml) and X-Gal (150 μg/ml) (X-Gal plates). Representative blue and white colonies (transformants) were picked, initially cultured in 2 ml of Frey broth containing gentamicin, and stored frozen at −80°C for future use.
Pedigree analysis and monitoring of blue/white color selection.
To perform pedigree analysis, mycoplasmas were passed through a 0.2-μm-pore-size filter before being assayed for CFU on X-Gal plates. This filter-cloning procedure was used to eliminate cell aggregates. Thus, the resulting colonies arose from individual cells and are as homogeneous as possible. Individual blue and white colonies were picked and initially cultured in 2 ml of Frey broth containing gentamicin, and aliquots were stored frozen at −80°C for future use. To assess the activity of the lacZ reporter in the cell population of each subclone that was stored at −80°C, cells from the aliquot were passed through a 0.2-μm-pore-size filter and assayed on X-Gal plates. The numbers of blue colonies and white colonies were determined, and the percentage of colonies that were blue was calculated. For this enumeration, colonies that were intensely blue were recorded as blue and colonies that were either completely white or only moderately blue were recorded as white. This approach was taken because we reasoned that colonies arising from cells that failed to produce LacZ would develop some blue color as lacZ was switched from Off to On in some cells within the developing population in the colony.
DNA analysis of subclones.
Genomic DNA was isolated from 50-ml cultures of representative subclones that had been stored at −80°C (16). The promoter region of the lacZ reporter was amplified by PCR with the 5′ M9 primer (see above) and the lacZ primer, TTCCCAGTCACGACGTTGTAAAAC (Fig. 1). PCR products were directly sequenced (without cloning) with the 3′ M9 primer at the Iowa State University DNA Sequencing and Synthesis Facility, Ames, Iowa. The sequences were aligned and analyzed by using the MacVector nucleotide and protein sequence analysis software package (version 6.01; Oxford Molecular Group Inc., Beaverton, Oreg.). For Southern analysis, HindIII- digested genomic DNA was electrophoresed on a 0.7% agarose gel, transferred to a nylon membrane, and hybridized with the lacZ gene probe under normal (high-stringency) hybridization conditions. The lacZ probe consisted of the 3-kb BamHI fragment described above, radiolabeled with 32P by the random primer method as described previously (5).
RESULTS
Phase-variable expression of the M9-lacZ reporter in M. gallisepticum.
The M9-lacZ gene was constructed such that the DNA fragment containing the promoter region and coding for the first 22 amino acids of the M9 protein would be fused in frame with the lacZ gene. When transformed into E. coli, plasmid pCR2.1-M9.lacZ containing the M9-lacZ reporter resulted in transformants that were blue when assayed on agar supplemented with X-Gal. Nucleotide sequence analysis of the M9-lacZ gene confirmed that no mutations had been introduced during construction. From the nucleotide sequence, the reporter was predicted to encode an M9-LacZ fusion protein of 1,039 amino acids.
The initial transformation of strain PG31 with pISM-M9.lacZ resulted in gentamicin-resistant transformants at a frequency of 10−5 transformants per CFU. pISM-M9.lacZ does not replicate in mycoplasmas, and transformants could be obtained only if the Tn4001 had inserted into the mycoplasma chromosome, as described previously (4). The M9-lacZ fusion gene that had been incorporated into the Tn4001 portion of pISM-M9.lacZ consisted of a lacZ gene combined in frame with a 336-bp M9 gene fragment containing the GAA repeat region, the transcription start site, and the translation start codon. Because the lacZ gene expression is controlled by the transcriptional regulatory regions of the M9 gene fragment to which it was fused, β-galactosidase activity in the transformants reflected regulation of lacZ gene expression by the M9 promoter. Representative examples of blue (lacZ gene expressed) and white (lacZ gene not expressed) colonies of M. gallisepticum containing the M9-lacZ fusion gene assayed on X-Gal plates are shown in Fig. 3. From the initial transformation of PG31 with pISM-M9.lacZ, 20 transformants were selected for further study. The CFU counts from each of these 20 parent transformants were determined on X-Gal plates to find whether the M9-lacZ gene underwent phase variation resulting in cell populations that gave rise to both blue colonies expressing lacZ and white colonies not expressing lacZ. CFU derived from each of the initial transformants exhibited a mixture of blue and white colonies on X-Gal plates. Transformant MGT-6 yielded a nearly equal mixture of blue (60%) and white (40%) colonies and was chosen for further study by pedigree analysis.
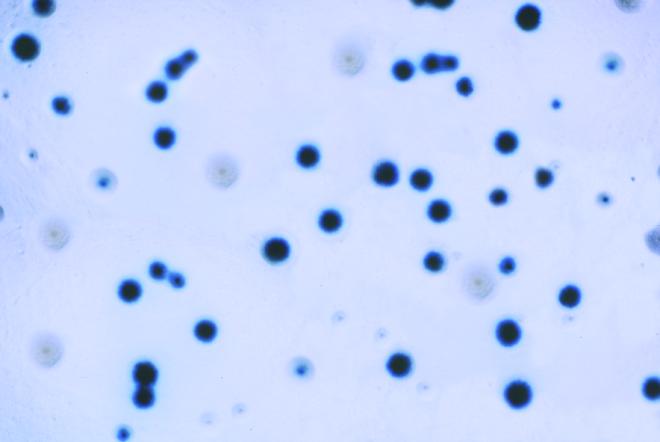
FIG. 3.
Representative photograph illustrating blue and white colonies of M. gallisepticum assayed on X-Gal agar.
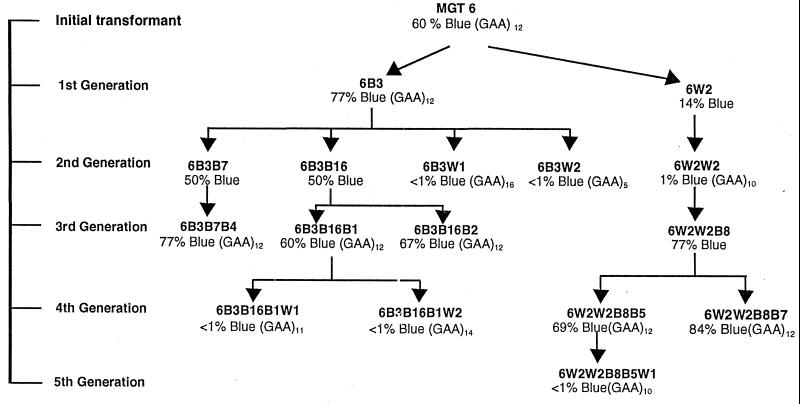
Subclones of MGT-6 were analyzed for up to five generations (Fig. 4). A first-generation blue (LacZ+) colony (6B3) was subcloned and found to give rise to 77% blue-colony progeny. A first-generation white (LacZ−) colony (6W2) was subcloned and gave rise to only 14% blue colonies. Thus, the blue/white phenotype was somewhat unstable but for the most part bred true, indicative of phase variation. Subclones of 6B3 and 6W2 were isolated, and these second-generation subclones also gave rise to a mixture of blue and white colonies, as diagrammed in Fig. 4. One lineage that was studied switched from LacZ+ (MGT-6) to LacZ− (6W2 and 6W2W2) and back to LacZ+ (6W2W2B8 and 6W2W2B8B5) and back to LacZ− again (6W2W2B8B5W1). Similarly, several LacZ− subclones (6B3W1, 6B3W2, 6B3B16B1W1, and 6B3B16B1W2) were independently isolated from the LacZ+ subclones MGT-6 and 6B3. The M9-lacZ gene from subclones derived from these lineages was studied to investigate the mechanism of variation in LacZ activity.
FIG. 4.
Pedigree analysis of M. gallisepticum containing the M9-lacZ reporter. For each of the analyzed subclones, the percentage of progeny colonies that were scored as blue are indicated. For subclones in which the nucleotide sequence of the M9 promoter region was determined, the number of trinucleotide GAA repeats is indicated by (GAA)n.
Phase variation in M9-lacZ gene expression correlates with variation in the GAA repeat region.
The promoter region of the M9-lacZ reporter gene was PCR amplified as described in Materials and Methods to generate a 376-bp product containing the GAA tandem repeat region and the beginning of the lacZ coding region. The primers used for PCR amplification would not amplify the native M9 gene of PG31, because one of the primers was specific for lacZ. The nucleotide sequence of each PCR product was determined, and no sequence variation was observed except for the length of the trinucleotide GAA repeat region. Subclones that gave rise to predominantly blue colonies (e.g., the LacZ+ subclones MGT-6, 6B3, 6B3B16B1, 6W2W2B8B5, and 6W2W2B8B7) invariably contained 12 copies of the GAA repeat. The LacZ− subclones never had 12 GAA repeats, sometimes having more than 12 repeats (16 repeats in 6B3W1 and 14 repeats in 6B3B16B1W2) and sometimes fewer (11 repeats in 6B3B16B1W1, 10 repeats in 6W2W2 and in 6W2W2B8B5W1, and only 5 repeats in 6B3W2). Therefore, it appears that exactly 12 GAA repeats are required for expression.
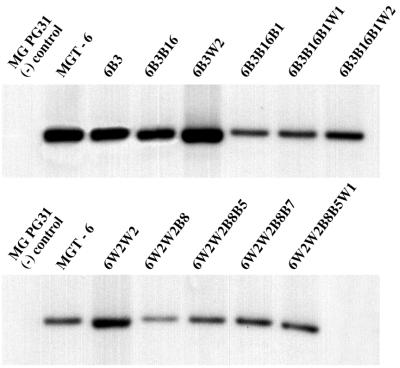
Because the M9-lacZ gene is located on a transposable element, it is possible that PG31 DNA sequences flanking the transposon influence the expression of the reporter. Therefore, we determined whether change in the expression status of the reporter gene was associated with transposition to a new site in the genome. Genomic DNAs from transformant MGT-6 and representative subclones thereof were analyzed on Southern blots probed with lacZ. A single HindIII fragment of 4 kb hybridized with the probe (Fig. 5). The HindIII fragment that hybridized with lacZ contains one of the Tn4001-mycoplasmal DNA junction regions (Fig. 2B), and the size of this fragment would vary if Tn4001 were to transpose to a different site in the chromosome. Because the size of the hybridizing DNA fragment was invariant, transposition of Tn4001 from its initial insertion site in the MGT-6 chromosome to an alternative site did not occur in the subclones that were analyzed. Thus, the phase-variable expression of the lacZ reporter did not occur as a consequence of Tn4001 transposition. We conclude that the phase-variable expression of M9-lacZ gene is regulated by the length of the GAA repeat region.
FIG. 5.
Southern analysis of mycoplasmal genomic DNAs probed with lacZ. HindIII-digested chromosomal DNAs were probed with the 3-kb BamHI lacZ fragment used to construct pCR2.1-M9.lacZ. The DNA fragment from each transformant that hybridized with the probe was 4 kb (the DNA fragment from 6W2W2B8B5W1 had an apparent mobility of slightly less than 4 kb due to smiling effects evident from the ethidium bromide-stained gel). Analysis of the parent strain PG31 served as a control to verify that the lacZ probe did not hybridize to mycoplasmal DNA from cells that had not been transformed.
DISCUSSION
Colonies of M. gallisepticum containing the M9-lacZ reporter appear intensely blue, weakly blue, or white on X-Gal plates. We have shown that intensely blue colonies are composed predominantly of cells containing 12 GAA repeats in the reporter gene while white colonies contain either more or fewer than 12 repeats. One might propose that colonies possessing a weakly blue phenotype are composed of cells that produce a low level of LacZ due to an intermediate level of M9-lacZ gene expression. This does not appear to be the case. The subcloning of weakly blue colonies did not result in progeny with a weakly blue phenotype. Subclone analysis of weakly blue colonies revealed a high degree of heterogeneity of cell phenotypes consisting of some progeny that produced intensely blue colonies and other progeny that produced white colonies. Our interpretation is that M9-lacZ gene transcription is all-or-nothing. With 12 GAA repeats, the gene is fully transcribed. Without 12 repeats, we suggest that no transcription occurs. Therefore, we disagree with the interpretation of experiments in which Northern and reverse transcription-PCR data were used to argue that multiple pMGA genes are simultaneously transcribed, with most genes (pMGA1.2, pMGA1.4, and pMGA1.8) being expressed at low levels and only a single gene (pMGA1.1), containing 12 GAA repeats, being transcribed at a high level (7). A more likely interpretation for the data is that the only M9/pMGA genes that are transcribed are those that contain 12 GAA repeats. The low level of expression of pMGA1.2, pMGA1.4, and pMGA1.8 as described by Glew et al. (7) would result from subpopulations of cells in which these alternative pMGA genes happen to contain 12 GAA repeats.
The GAA repeat region appears to act as a transcriptional switch regulating the expression of each member of the M9/pMGA gene family. The pMGA gene family is very large, and the phase-variable expression of each family member may play an important role in immune avoidance. The incorporation of antibodies specific for pMGA protein into the growth medium results in cultures containing cells that fail to produce pMGA (8, 15). The most likely explanation is that pMGA-specific antibodies inhibit cell growth, conferring a growth advantage on cells that fail to produce pMGA. Regulation of M9/pMGA gene expression by the GAA repeat permits cell cultures to contain cell subpopulations producing alternative pMGA proteins or in some cases lacking pMGA protein altogether. Thus, an antibody response could not effectively attack all cells within the population.
What is the mechanism by which the GAA repeats regulate pMGA gene expression? Mycoplasmas are thought to possess only a single ς factor analogous to the eubacterial general factor ςA. However, the region upstream of the transcription start site of the M9/pMGA genes lacks significant sequence similarity to the well-defined consensus −10 (TATAAT) and −35 (TTGACA) ςA recognition sequences (3). Thus, the failure of most pMGA genes (those lacking 12 GAA repeats) to be transcribed is understandable. The mechanism by which genes containing 12 GAA repeats are transcribed remains to be elucidated. Perhaps M9/pMGA transcription uses an activator similar to the TRAP system of Bacillus subtilis, in which tryptophan biosynthesis is regulated by 11 copies of a (G/U)AG repeat (1). However, the (G/U)AG repeats are in the trp leader transcript, and TRAP acts by binding to this RNA. The GAA repeats of the M9/pMGA genes are upstream of the putative transcription start site (7). M9/pMGA RNA transcripts would lack the GAA repeat region, and regulation of gene expression is predicted to occur by binding of a positive regulator to the GAA repeat region of the DNA. This putative activator might bind to single-stranded GAA repeat DNA, because it has recently been shown that GAA tandem repeats near the physiological pH will form a triple-helix structure in which the TTC strand folds onto either side of the same GAA strand, leaving the remaining portion of the GAA strand not base paired (13). A single-stranded GAA repeat region may be able to wrap around an activator protein similarly to how the repeat portion of the trp leader transcript binds to TRAP.
ACKNOWLEDGMENTS
This work was supported by the U.S. Department of Agriculture National Research Initiative Competitive Grants Program (grant 93-37204-9113) and by Auburn University College of Veterinary Medicine (grant ALAV 304).
REFERENCES
- 1.Babitzke P. Regulation of tryptophan biosynthesis: Trp-ing the TRAP or how Bacillus subtilis reinvented the wheel. Mol Microbiol. 1997;26:1–9. doi: 10.1046/j.1365-2958.1997.5541915.x. [DOI] [PubMed] [Google Scholar]
- 2.Baseggio N, Glew M D, Markham P F, Whithear K G, Browning G F. Size and genomic location of the pMGA multigene family of Mycoplasma gallisepticum. Microbiology. 1996;142:1429–1435. doi: 10.1099/13500872-142-6-1429. [DOI] [PubMed] [Google Scholar]
- 3.Busby S, Ebright R H. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell. 1994;79:743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 4.Cao J, Kapke P A, Minion F C. Transformation of Mycoplasma gallisepticum with Tn916, Tn4001, and integrative plasmid vectors. J Bacteriol. 1994;176:4459–4462. doi: 10.1128/jb.176.14.4459-4462.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dybvig K, Sitaraman R, French C T. A family of phase-variable restriction enzymes with differing specificities generated by high-frequency gene rearrangements. Proc Natl Acad Sci USA. 1998;95:13923–13928. doi: 10.1073/pnas.95.23.13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frey M L, Hanson R P, Anderson D P. A medium for isolation of avian mycoplasmas. Am J Vet Res. 1968;29:2163–2171. [PubMed] [Google Scholar]
- 7.Glew M D, Markham P F, Browning G F, Walker I D. Expression studies on four members of the pMGA multigene family in Mycoplasma gallisepticum S6. Microbiology. 1995;141:3005–3014. doi: 10.1099/13500872-141-11-3005. [DOI] [PubMed] [Google Scholar]
- 8.Glew M D, Baseggio N, Markham P F, Browning G F, Walker I D. Expression of the pMGA genes of Mycoplasma gallisepticum is controlled by variation in the GAA trinucleotide repeat lengths within the 5′ noncoding regions. Infect Immun. 1998;66:5833–5841. doi: 10.1128/iai.66.12.5833-5841.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heydreyda C T, Lee K K, Krause D C. Transformation of Mycoplasma pneumoniae with Tn4001 by electroporation. Plasmid. 1993;30:170–175. doi: 10.1006/plas.1993.1047. [DOI] [PubMed] [Google Scholar]
- 10.Hwang Y S, Panangala V S, Rossi C R, Giambrone J J, Lauerman L H. Monoclonal antibodies that recognize specific antigens of Mycoplasma gallisepticum and Mycoplasma synoviae. Avian Dis. 1989;33:42–52. [PubMed] [Google Scholar]
- 11.Knudtson K L, Minion F C. Construction of Tn 4001 lac derivatives to be used as promoter probe vectors in mycoplasmas. Gene. 1993;137:217–222. doi: 10.1016/0378-1119(93)90009-r. [DOI] [PubMed] [Google Scholar]
- 12.Liu L, Payne D M, van Santen V L, Dybvig K, Panangala V S. A protein associated with monoclonal antibody-mediated agglutination of Mycoplasma gallisepticum is a member of the pMGA family. Infect Immun. 1998;66:5570–5575. doi: 10.1128/iai.66.11.5570-5575.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mariappan S V S, Catasti P, Silks III L A, Bradbury E M, Gupta G. The high-resolution structure of the triplex formed by the GAA/TTC triplet repeat associated with Friedreich's ataxia. J Mol Biol. 1999;285:2035–2052. doi: 10.1006/jmbi.1998.2435. [DOI] [PubMed] [Google Scholar]
- 14.Markham P F, Glew M D, Whithear K G, Walker I D. Molecular cloning of a member of the gene family that encodes pMGA, a hemagglutinin of Mycoplasma gallisepticum. Infect Immun. 1993;61:903–909. doi: 10.1128/iai.61.3.903-909.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Markham P F, Glew M D, Browning G F, Whithear K G, Walker I D. Expression of two members of the pMGA gene family of Mycoplasma gallisepticum oscillates and is influenced by pMGA-specific antibodies. Infect Immun. 1998;66:2845–2853. doi: 10.1128/iai.66.6.2845-2853.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voelker L L, Weaver K E, Ehle L J, Washburn L R. Association of lysogenic bacteriophage MAV1 with virulence in Mycoplasma arthritidis. Infect Immun. 1995;63:4016–4023. doi: 10.1128/iai.63.10.4016-4023.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]