Abstract
Glypicans are proteoglycans that are bound to the outer surface of the plasma membrane by a glycosylphosphatidylinositol anchor. The mammalian genome contains six members of the glypican family (GPC1 to GPC6). Although the degree of sequence homology within the family is rather low, the three-dimensional structure of these proteoglycans is highly conserved. Glypicans are predominantly expressed during embryonic development. Genetic and biochemical studies have shown that glypicans can stimulate or inhibit the signaling pathways triggered by Wnts, hedgehogs, fibroblast growth factors, and bone morphogenetic proteins. The study of mutant mouse strains demonstrated that glypicans have important functions in the developmental morphogenesis of various organs. In addition, a role of glypicans in synapsis formation has been established. Notably, glypican loss-of-function mutations are the cause of three human inherited syndromes. Recent analysis of glypican compound mutant mice has demonstrated that members of this protein family display redundant functions during embryonic development.
Keywords: embryo, glypican, proteoglycan
THE GLYPICAN FAMILY
Glypicans are proteoglycans that are anchored in the cell membrane by a glycosylphosphatidylinositol (GPI) linker. Glypican-encoding genes have been found in lower organisms, including flies and nematodes (1, 2). Like all proteoglycans, glypicans can carry glycosaminoglycan (GAG) chains. In most cases, these chains consist of heparan sulfate (3). The mammalian glypican family is composed of six members: GPC1 to GPC6 (4). Although the sequence homology shared by all mammalian glypicans is not very high, the localization of 14 cysteines and of the GAG chain insertion sites is conserved across the family, indicating that the overall protein structure of mammalian glypicans is similar (3). The crystal structure of GPC1 shows a densely packed protein that displays an elongated cylindrical shape containing 14 α-helices and three major loops. The C-terminal domain containing the GPI anchor and the GAG insertion sites seems to be unstructured (5). The cylindrical core protein appears to be parallel to the membrane, with a surface that is evolutionary conserved facing the GAG chains. This suggests that GPC1-binding proteins could interact with the GAG chains, with the core protein, or with both simultaneously (5, 6). Based on their sequence homology, glypicans can be classified into two subfamilies. One includes GPC1, GPC2, GPC4, and GPC6, and the other GPC3 and GPC5. The members of the first subfamily display a high degree of homology with Dally-like, one of the two Drosophila glypicans (1). The members of the second subfamily share a high degree of similarity with Dally, the other Drosophila glypican (1). Notably, the mouse and human genomes display two glypican clusters: GPC3 and GPC4 on the X chromosome, and GPC5 and GPC6 on human chromosome 13 (mouse chromosome 14). Moreover, there is a high degree of homology between GPC4 and GPC6, and between GPC3 and GPC5, suggesting that this chromosomal arrangement is the result of gene duplication events during evolution (1).
FUNCTION OF GLYPICANS
Genetic and biochemical studies have shown that glypicans regulate the activity of several growth factors/morphogens, including Wnts, hedgehogs (Hhs), fibroblast growth factors (FGFs), and bone morphogenetic proteins (BMPs; 1, 7–12). This regulatory activity is based on the ability of glypicans to stimulate or inhibit the interaction of these growth factors with their signaling receptors (3). Because the growth factors regulated by glypicans are known to bind to heparan sulfate, it was initially thought that these glycan polymers mediate the glypican-growth factor interaction. However, it is now well established that in many cases the glypican core protein is the main mediator of this interaction (7, 13).
Based on experiments performed in Drosophila, it has been proposed that glypicans are also involved in the transport and formation of morphogen gradients in the embryo (14). However, the involvement of glypicans in a similar function in mammals has not been reported so far.
Glypicans also have specific functions in the nervous system. GPC4 and GPC6, for example, play a role in the formation of excitatory synapses in the central nervous system (15–17). This function is based on their ability to form part of the synapse-organizing protein complexes. The heparan sulfate chains mediate the interaction of glypicans with the proteins involved in synapse formation (15).
THE ROLE OF GLYPICANS IN THE EMBRYO
All glypicans are expressed in the embryo in a stage- and tissue-specific manner (18). Given the fact that the growth factors/morphogens regulated by these proteoglycans play key roles in embryonic development, the discovery that glypicans have important functions in the embryo, and that glypican mutations are involved in inherited human syndromes, was not surprising. We describe below some of the main findings regarding the role of each mammalian glypican in embryonic morphogenesis, and their involvement in genetic syndromes. Some of this information is summarized in Table 1.
Table 1.
Glypicans: signaling and congenital diseases
| Glypican | Signaling Pathway Altered in Knockout Mice | Human Congenital Syndrome |
|---|---|---|
| GPC1 | FGF | |
| GPC2 | ND | |
| GPC3 | Hh, Wnt, BMP | Simpson–Golabi–Behmel |
| GPC4 | ND | Keipert |
| GPC5 | ND | |
| GPC6 | Hh, Wnt | Recessive omodysplasia |
BMP, bone morphogenetic protein; FGF, fibroblast growth factor; Hh, hedgehog; ND, not determined.
Glypican-3
This glypican is the one that has been most extensively studied. This is probably due to the fact that GPC3 was the first member of the family that was found to be associated with a genetic syndrome. Loss-of-function mutations of GPC3 are the cause of the Simpson–Golabi–Behmel syndrome (SGBS; 19). This syndrome is characterized by pre- and postnatal overgrowth, dysmorphic facial features including cleft palate, polydactyly, syndactyly, supernumerary nipples, cystic and dysplastic kidneys, cardiac defects, rib and vertebral fusions, and umbilical and inguinal hernias (20, 21). The multiple developmental abnormalities of patients with SGBS indicate that GPC3 plays a critical role in the morphogenesis of various organs. The causal role of the GPC3 mutations in SGBS was confirmed by the generation of GPC3-null mice, which display many of the abnormalities described in patients with SGBS, including significant prenatal overgrowth and kidney dysplasia (22). Given the critical role that insulin-like growth factor II (IGF-II) plays in the regulation of embryonic growth, it was initially proposed that GPC3 acts as an IGF-II inhibitor (19). However, it was later demonstrated that GPC3 does not interact with the IGF-II signaling pathway (22, 23). Instead, a series of genetic and biochemical experiments demonstrated that GPC3 inhibits Hh signaling during embryonic development (7, 24). Notably, organs that present developmental abnormalities in GPC3-null mice display a significant increase in Hh signaling activity (24). The generation of GPC3/Ihh double knockout mice demonstrated that the overgrowth caused by the lack of GPC3 is partially mediated by Indian Hh (Ihh) signaling (24). The Hh-inhibitory activity of GPC3 is based on the ability of this glypican to bind with high affinity to Sonic Hh (Shh) and Indian Hh (Ihh) and to act as a competitive inhibitor of the binding of Hh to patched, the Hh signaling receptor. Furthermore, the binding of Hh to GPC3 induces endocytosis and degradation of the GPC3/Hh complex (7).
GPC3-null embryos also display an inhibition of the noncanonical Wnt signaling pathway. Tissue culture experiments confirmed that GPC3 stimulates noncanonical Wnt activity (25). Finally, reduced BMP signaling has been reported in the kidneys and bones of the GPC3-null embryos, indicating that this glypican promotes BMP signaling in these organs (26, 27).
Glypican-6
Autosomal recessive omodysplasia (OMOD1) is a rare inherited condition caused by loss-of-function mutations of GPC6 (28). Patients with OMOD1 display short stature, facial dysmorphism, and proximal limb shortening. GPC6-null mice, which die at birth, show many of the abnormalities observed in patients with OMOD1, including significantly shorter long bones and face dysmorphisms (29). Notably, the growth plate of femurs from GPC6-mutant embryos display reduced cell proliferation and a significant inhibition of Hh signaling (30). Tissue culture experiments demonstrated that GPC6 stimulates the binding of Hh to patched. This was consistent with the observation that GPC6 interacts with Hh through its core protein, and with patched via its GAG chains (30). These results led to the conclusion that GPC6 promotes the embryonic growth of long bones by stimulating Hh signaling (30).
Unexpectedly, GPC6-null embryos display dramatically shortened small intestines (75% shorter at birth) and significantly smaller stomachs (50% smaller at birth; 31). This phenotype has not been reported in patients with OMOD1 so far. However, newborn babies usually do not survive with intestines that are very short (32). It is possible, therefore, that OMOD1 babies with short intestines have eluded characterization due to their early demise. It is also possible that the few patients that survive after birth display a genetic background that reduces the impact of the lack of GPC6 in intestinal lengthening. The elongation of the embryonic intestine is mostly under the control of Hh and noncanonical Wnt signaling pathways (33, 34). Notably, the analysis of the GPC6-null embryos showed that this glypican stimulates intestinal lengthening by simultaneously regulating Hh and noncanonical Wnt signaling (31). As expected, given the capacity of GPC6 to stimulate Hh signaling, the GPC6-null embryonic intestine shows reduced Hh activity (31). In addition, the mutant gut displays increased noncanonical Wnt signaling, indicating that GPC6 inhibits this signaling pathway in the developing intestine (31). In this regard, it should be noted that it had been previously demonstrated that both an increase and a decrease in noncanonical Wnt signaling significantly reduce intestinal length (33, 35). Tissue culture studies confirmed that GPC6 can act as an inhibitor of noncanonical Wnt (31).
The study of the GPC3 and GPC6 knockout embryos has shown that in the context of Hh and noncanonical Wnt signaling, these two glypicans display opposite activities. With regard to the impact of GPC3 and GPC6 on noncanonical Wnt activity, it should be noted that it has recently been reported that GPC6 can bind to the palmitoleate moiety of Wnt (13). This binding occurs by the insertion of the palmitoleate group into a tunnel-like hydrophobic grove of GPC6, which shields this lipid moiety from interacting with other proteins. GPC3, on the other hand, binds to two small loops located in the middle region of Wnt (36). Because the palmitic group is essential for the binding of Wnt to its signaling receptor Frizzled, whereas the Wnt middle region is not involved in the interaction with this receptor, it is reasonable to propose that GPC6 inhibits Wnt activity by interfering with the binding of Wnt to Frizzled, whereas GPC3 does not inhibit this interaction. Moreover, in addition to interacting with Wnt, GPC3 can also bind to Frizzled through its GAG chains (37). Thus, the ability of GPC3 to simultaneously interact with Wnt and Frizzled could facilitate/stabilize the Wnt/Frizzled interaction and increase their signaling activity.
The opposite effect of GPC6 and GPC3 on Hh signaling could be explained, at least in part, by the fact that only GPC6 can interact with patched (30). The simultaneous binding of GPC6 to Hh and its receptor may facilitate/stabilize the ligand/receptor interaction.
Glypican-4
The Keipert syndrome is a rare X-linked disorder characterized by a prominent forehead, a flat midface, hypertelorism, and digital abnormalities. Cognitive impairment and deafness are variable features (38). It has been recently reported that this genetic syndrome is caused by loss-of-function mutations of GPC4 (38). The study of GPC4-null mice showed evidence of craniofacial and digital abnormalities, the two main features of the Keipert syndrome (38). The molecular mechanism by which GPC4 regulates facial and digital development has not been identified yet.
In addition to the craniofacial and digital abnormalities, GPC4-null mice display defective excitatory synapse formation and less synaptic GluA1 AMPA receptors in the developing hippocampus (16). This deficit in synapsis formation results in behaviors associated with autism spectrum disorder (39). Biochemical studies have shown that GPC4 can interact with several components of the synaptic-organizing protein complexes, including leucine-rich repeat transmembrane neural proteins (LRRTMs), receptor protein tyrosine phosphatases (RPTPs), and G-protein-coupled receptor 158 (GPR158; 15).
Glypican-1
GPC1-null mice display brains that are significantly smaller than their wild-type littermates (40). The difference in brain size emerges very early during embryogenesis (between E8.5 and E9.5) and remains roughly constant after that. The smaller size of the GPC1-knockout brain is due to reduced cell proliferation at the early stages of neurogenesis. Only subtle differences on brain pattern can be identified in the GPC1 mutants, although they display premature differentiation of postmitotic neurons (38). Compound mutants between GPC1-null and a mouse strain with a gene-trap allele of GPC4 generated mice with a synergistically reduced brain size. Thus, GPC1 appears to act redundantly with GPC4 in controlling brain size (40). Analysis of GPC1 mutants indicated that FGF17 mediates the role of GPC1 in brain development (40).
So far, no inherited syndromes implicating GPC1 have been reported.
Glypican-2 and Glypican-5
No detailed studies of GPC2-null and GPC5-null mice have been published to date, and so far, these two glypicans have not been associated with any genetic syndrome. GPC5 has been linked to the acquired nephrotic syndrome (41). The role of GPC5 in this syndrome seems to be mediated by the FGF signaling pathway (41). To study the role of GPC5 in this disease, podocyte-specific GPC5 knockdown mice were generated. These mice are fertile and develop normally up to adulthood with no obvious phenotype (41). In the case of GPC2, it was reported that GPC2-null mice are phenotypically normal (40).
GPC6 and GPC4 Compound Mutants
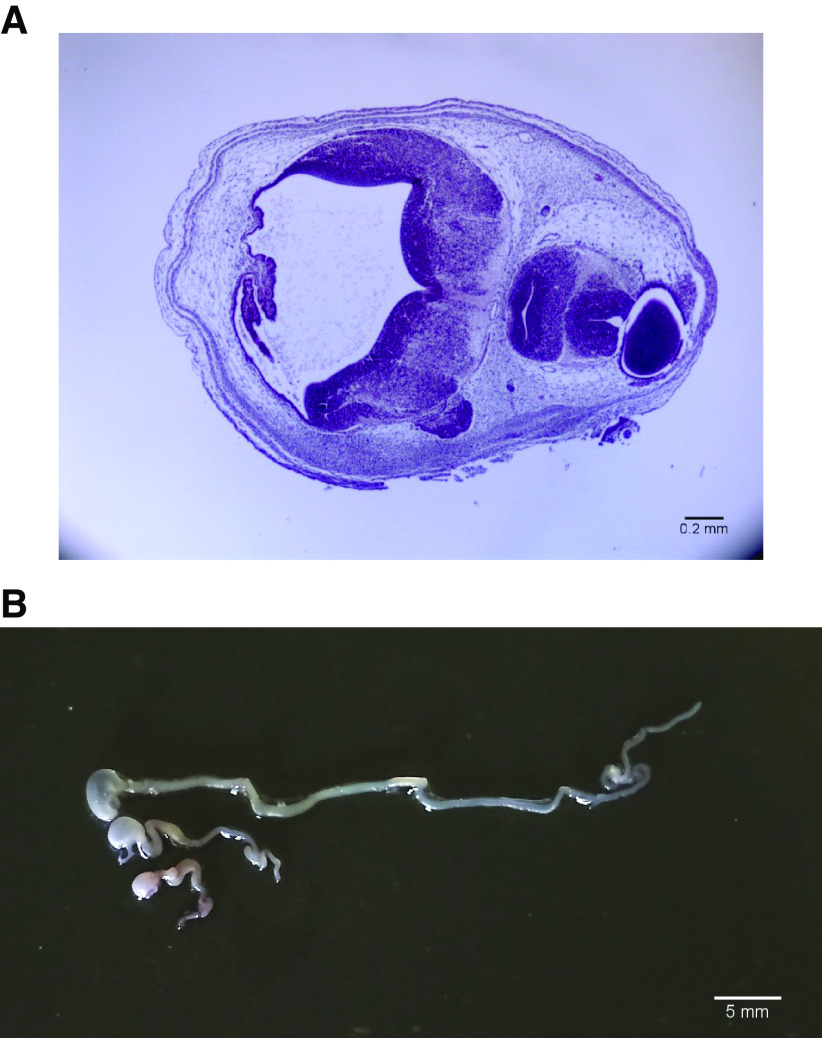
GPC6 and GPC4 are the pair of glypicans that show the highest degree of homology within the glypican family. As described earlier, mutations of these two glypicans are the cause of two inherited human syndromes that include bone abnormalities and facial dysmorphisms. Moreover, these abnormalities have been replicated in the corresponding mouse knockouts. Both glypicans are expressed in several organs, including the bones in the face, the brain, and the gastrointestinal system. In our laboratory we decided, therefore, to investigate whether these two genes display complementary functions during development by generating GPC6 and GPC4 double knockouts. To this end, we bred GPC6 and GPC4 heterozygotes and genotyped the litters at different embryonic stages. However, we were not able to identify any double knockouts, even at early stages of development. We concluded therefore that the simultaneous lack of GPC6 and GPC4 induces early lethality. Notably, during the genotyping of the litters, we noticed that ∼10% of the GPC6-null/GPC4 heterozygous embryos display obvious cranial and facial abnormalities that are not seen in the single mutants. Analysis of embryo sections showed that the compound mutants display various degrees of cyclopia and brain malformations (Fig. 1A). GPC4-null embryos have a normal gastrointestinal system. However, all the compound GPC6/GPC4 double mutants display a small intestine that is significantly shorter than that of the GPC6-null embryos. In addition, the stomach of all the double mutants was clearly smaller than the one from the GPC6-null embryos (Fig. 1B). From these results, we conclude that GPC6 and GPC4 display functional redundancy during embryonic development.
Figure 1.
A: tissue section of the head of E13.5 GPC6-null/GPC4-heterozygous double mutant embryo. The tissue was stained with hematoxylin-eosin. B: guts dissected from E17.5 embryos. Top: wild-type, middle: GPC6-null/GPC4 wild type, bottom: GPC6-null/GPC4-heterozygous.
FUTURE WORK
The analysis of mutant mouse strains and the involvement of glypicans in various inherited human syndromes clearly demonstrate that this family of proteoglycans play a critical role in mammalian embryonic development. Genetic and biochemical studies have shown that the signaling pathways triggered by Wnts, Hhs, FGFs, and BMPs mediate the role of glypicans in the morphogenesis of specific tissues, including bone, brain, kidney, and the gastrointestinal system. Based on findings obtained in lower organisms and the expression pattern of glypicans (18, 42, 43), it is expected that further analysis of mutant mice will reveal other tissues that are impacted by glypican activity in the mammalian embryo. In addition to their role in developmental morphogenesis, glypicans regulate synapsis formation by interacting with protein complexes that regulate trans-synaptic signaling. GPC4 has already been implicated in behavioral disorders. Future studies could unveil the impact of additional glypican mutations in other neurological disorders. Analysis of GPC1/GPC4 and GPC6/GPC4 double mutants indicates that glypicans can display redundant functions. It is expected, therefore, that the study of other glypican compound mutant mouse strains will uncover additional roles of these proteoglycans in mammalian embryonic development.
GRANTS
This work was supported by Canadian Institutes of Health Research (CIHR) Grant MOP-142344.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
This article is part of the special collection “Deciphering the Role of Proteoglycans and Glycosaminoglycans in Health and Disease.” Liliana Schaefer, MD, served as Guest Editor of this collection.
AUTHOR CONTRIBUTIONS
J.F. drafted manuscript; edited and revised manuscript; approved final version of manuscript.
REFERENCES
- 1.Filmus J, Capurro M, Rast J. Glypicans. Genome Biol 9: 224, 2008. doi: 10.1186/gb-2008-9-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fico A, Maina F, Dono R. Fine-tuning of cell signaling by glypicans. Cell Mol Life Sci 68: 923–929, 2011. doi: 10.1007/s00018-007-7471-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filmus J, Capurro M. The glypican family. In: Extracellular Matrix: Pathobiology and Signaling, edited by Nikos K. Karamanos. Berlin/Boston: De Gruyter, 2012, p. 209–220. [Google Scholar]
- 4.Veugelers M, De Cat B, Ceulemans H, Bruystens AM, Coomans C, Dürr J, Vermeesch J, Marynen P, David G. Glypican-6, a new member of the glypican family of cell surface proteoglycans. J Biol Chem 274: 26968–26977, 1999. doi: 10.1074/jbc.274.38.26968. [DOI] [PubMed] [Google Scholar]
- 5.Svensson G, Awad W, Håkansson M, Mani K, Logan DT. Crystal structure of N-glycosylated human glypican-1 core protein: structure of two loops evolutionary conserved in vertebrate glypican-1. J Biol Chem 287: 14040–14051, 2012. doi: 10.1074/jbc.M111.322487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Awad W, Adamczyk B, Örnros J, Karlsson NG, Mani K, Logan DT. Structural aspects of N-glycosylations and the C-terminal region in human glypican-1. J Biol Chem 290: 22991–23008, 2015. doi: 10.1074/jbc.M115.660878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capurro MI, Xu P, Shi W, Li F, Jia A, Filmus J. Glypican-3 inhibits hedgehog signaling during development by competing with patched for hedgehog binding. Dev Cell 14: 700–711, 2008. doi: 10.1016/j.devcel.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Lum L, Yao S, Mozer B, Rovescalli A, Von Kessler D, Nirenberg M, Beachy PA. Identification of hedgehog pathway components by RNAi in Drosophila cultured cells. Science 299: 2039–2045, 2003. doi: 10.1126/science.1081403. [DOI] [PubMed] [Google Scholar]
- 9.Shiau CE, Hu N, Bronner-Fraser M. Altering glypican-1 levels modulates canonical Wnt signaling during trigeminal placode development. Dev Biol 348: 107–118, 2010. doi: 10.1016/j.ydbio.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galli A, Roure A, Zeller R, Dono R. Glypican 4 modulates FGF signalling and regulates dorsoventral forebrain patterning in Xenopus embryos. Development 130: 4919–4929, 2003. doi: 10.1242/dev.00706. [DOI] [PubMed] [Google Scholar]
- 11.Akiyama T, Kamimura K, Firkus C, Takeo S, Shimmi O, Nakato H. Dally regulates Dpp morphogen gradient formation by stabilizing Dpp on the cell surface. Dev Biol 313: 408–419, 2008. doi: 10.1016/j.ydbio.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan J, Ho M. The role of glypican-1 in regulating multiple cellular signaling pathways. Am J Physiol Cell Physiol 321: C846–C858, 2021. doi: 10.1152/ajpcell.00290.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGough IJ, Vecchia L, Bishop B, Malinauskas T, Beckett K, Joshi D, O'Reilly N, Siebold C, Jones EY, Vincent JP. Glypicans shield the Wnt lipid moiety to enable signalling at a distance. Nature 585: 85–90, 2020. doi: 10.1038/s41586-020-2498-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan D, Wu Y, Yang Y, Belenkaya TY, Tang X, Lin X. The cell-surface proteins Dally-like and Ihog differentially regulate hedgehog signaling strength and range during development. Development 137: 2033–2044, 2010. doi: 10.1242/dev.045740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamimura K, Maeda N. Glypicans and heparan sulfate in synaptic development, neural plasticity, and neurological disorders. Front Neural Circuits 15: 595596, 2021. doi: 10.3389/fncir.2021.595596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allen NJ, Bennett ML, Foo LC, Wang GX, Chakraborty C, Smith SJ, Barres BA. Astrocyte glypicans 4 and 6 promote formation of excitatory synapses via GluA1 AMPA receptors. Nature 486: 410–414, 2012. doi: 10.1038/nature11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Wit J, O’Sullivan ML, Savas JN, Condomitti G, Caccese MC, Vennekens KM, Yates JR, Ghosh A. Unbiased discovery of glypican as a receptor for LRRTM4 in regulating excitatory synapse development. Neuron 79: 696–616, 2013. doi: 10.1016/j.neuron.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song HH, Filmus J. The role of glypicans in mammalian development. Biochim Biophys Acta 1573: 241–246, 2002. doi: 10.1016/s0304-4165(02)00390-2. [DOI] [PubMed] [Google Scholar]
- 19.Pilia G, Hughes-Benzie RM, MacKenzie A, Baybayan P, Chen EY, Huber R, Neri G, Cao A, Forabosco A, Schlessinger D. Mutations in GPC3, a glypican gene, cause the Simpson-Golabi-Behmel overgrowth syndrome. Nat Genet 12: 241–247, 1996. doi: 10.1038/ng0396-241. [DOI] [PubMed] [Google Scholar]
- 20.Chen E, Johnson JP, Cox VA, Golabi M. Simpson-Golabi-Behmel syndrome: congenital diaphragmatic hernia and radiologic findings in two patients and folow-up of a previously reported case. Am J Med Genet 46: 574–578, 1993. doi: 10.1002/ajmg.1320460523. [DOI] [PubMed] [Google Scholar]
- 21.Vuillaume ML, Moizard MP, Rossignol S, Cottereau E, Vonwill S, Alessandri JL, Busa T, Colin E, Gérard M, Giuliano F, Lambert L, Lefevre M, Kotecha U, Nampoothiri S, Netchine I, Raynaud M, Brioude F, Toutain A. Mutation update for the GPC3 gene involved in Simpson-Golabi-Behmel syndrome and review of the literature. Hum Mutat 39: 790–805, 2018. [Erratum in Hum Mutat 39: 2110–2112, 2018]. doi: 10.1002/humu.23428. [DOI] [PubMed] [Google Scholar]
- 22.Cano-Gauci DF, Song H, Yang H, McKerlie C, Choo B, Shi W, Pullano R, Piscione TD, Grisaru S, Soon S, Sedlackova L, Tanswell AK, Mak TW, Yeger H, Lockwood GA, Rosenblum N, Filmus J. Glypican-3-deficient mice exhibit the overgrowth and renal abnormalities typical of the Simpson-Golabi-Behmel syndrome. J Cell Biol 146: 255–264, 1999. doi: 10.1083/jcb.146.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiao E, Fisher P, Crisponi L, Deiana M, Dragatsis I, Schlessinger D, Pilia G, Efstratiadis A. Overgrowth of a mouse model of the Simpson-Golabi-Behmel syndrome is independent of IGF signaling. Dev Biol 243: 185–206, 2002. doi: 10.1006/dbio.2001.0554. [DOI] [PubMed] [Google Scholar]
- 24.Capurro MI, Li F, Filmus J. Overgrowth of a mouse model of Simpson-Golabi-Behmel syndrome is partly mediated by Indian Hedgehog. EMBO Rep 10: 901–907, 2009. doi: 10.1038/embor.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song HH, Shi W, Xiang Y, Filmus J. The loss of glypican-3 induces alterations in Wnt signaling. J Biol Chem 280: 2116–2125, 2005. doi: 10.1074/jbc.M410090200. [DOI] [PubMed] [Google Scholar]
- 26.Hartwig S, Hu MC, Cella C, Piscione T, Filmus J, Rosenblum ND. Glypican-3 modulates inhibitory BMP2-SMAD signaling to control renal development in vivo. Mech Dev 122: 928–938, 2005. doi: 10.1016/j.mod.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 27.Paine-Saunders S, Viviano BL, Zupicich J, Skarnes WC, Saunders S. Glypican-3 controls cellular responses to Bmp4 in limb patterning and skeletal development. Dev Biol 225: 179–187, 2000. doi: 10.1006/dbio.2000.9831. [DOI] [PubMed] [Google Scholar]
- 28.Campos-Xavier AB, Martinet D, Bateman J, Belluoccio D, Rowley L, Tan TY, Baxová A, Gustavson K-H, Borochowitz ZU, Innes AM, Unger S, Beckmann JS, Mittaz L, Ballhausen D, Superti-Furga A, Savarirayan R, Bonafé L. Mutations in the heparan-sulfate proteoglycan glypican-6 (GPC6) impair endochondral ossification and cause recessive omodysplasia. Am J Hum Genet 84: 760–770, 2009. doi: 10.1016/j.ajhg.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borochowitz Z, Sabo E, Misselevitch I, Boss JH. Autosomal-recessive omodysplasia: prenatal diagnosis and histomorphometric assessment of the physeal plates of the long bones. Am J Med Genet 76: 238–244, 1998. . [DOI] [PubMed] [Google Scholar]
- 30.Capurro M, Izumikawa T, Suarez P, Shi W, Cydzik M, Kaneiwa T, Gariepy J, Bonafe L, Filmus J. Glypican-6 promotes the growth of developing long bones by stimulating hedgehog signaling. J Cell Biol 216: 2911–2926, 2017. doi: 10.1083/jcb.201605119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi W, Kaneiwa T, Cydzik M, Gariepy J, Filmus J. Glypican-6 stimulates intestinal elongation by simultaneously regulating hedgehog and non-canonical Wnt signaling. Matrix Biol 88: 19–32, 2020. doi: 10.1016/j.matbio.2019.11.002. [DOI] [PubMed] [Google Scholar]
- 32.Walton KD, Freddo AM, Wang S, Gumucio DL. Generation of intestinal surface: an absorbing tale. Development 143: 2261–2272, 2016. doi: 10.1242/dev.135400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cervantes S, Yamaguchi TP, Hebrok M. Wnt5a is essential for intestinal elongation in mice. Dev Biol 326: 285–294, 2009. doi: 10.1016/j.ydbio.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao J, Kim BM, Rajurkar M, Shivdasani RA, McMahon AP. Hedgehog signaling controls mesenchymal growth in the developing mammalian digestive tract. Development 137: 1721–1729, 2010. doi: 10.1242/dev.044586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bakker ERM, Raghoebir L, Franken PF, Helvensteijn W, van Gurp L, Meijlink F, van der Valk MA, Rottier RJ, Kuipers EJ, van Veelen W, Smits R. Induced Wnt5a expression perturbs embryonic outgrowth and intestinal elongation, but is well tolerated in adult mice. Dev Biol 369: 91–100, 2012. doi: 10.1016/j.ydbio.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 36.Li N, Wei L, Liu X, Bai H, Ye Y, Li D, Li N, Baxa U, Wang Q, Lv L, Chen Y, Feng M, Lee B, Gao W, Ho M. A frizzled-like cysteine rich domain in glypican-3 mediates Wnt binding and regulates hepatocellular carcinoma tumor growth in mice. Hepatology 70: 1231–1245, 2019. doi: 10.1002/hep.30646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Capurro M, Martin T, Shi W, Filmus J. Glypican-3 binds to Frizzled and plays a direct role in the stimulation of canonical Wnt signaling. J Cell Sci 127: 1565–1575, 2014. doi: 10.1242/jcs.140871. [DOI] [PubMed] [Google Scholar]
- 38.Amor DJ, Stephenson SEM, Mustapha M, Mensah MA, Ockeloen CW, Lee WS, Tankard RM, Phelan DG, Shinawi M, de Brouwer APM, Pfundt R, Dowling C, Toler TL, Sutton VR, Agolini E, Rinelli M, Capolino R, Martinelli D, Zampino G, Dumic M, Reardon W, Shaw-Smith C, Leventer RJ, Delatycki MB, Kleefstra T, Mundlos S, Mortier G, Bahlo M, Allen NJ, Lockhart PJ. Pathogenic variants in GPC4 cause Keipert syndrome. Am J Hum Genet 104: 914–924, 2019. doi: 10.1016/j.ajhg.2019.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dowling C, Allen NJ. Mice lacking glypican 4 display juvenile hyperactivity and adult social interaction. Brain Plast 4: 197–209, 2018. doi: 10.3233/BPL-180079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jen YHL, Musacchio M, Lander AD. Glypican-1 controls brain size through regulation of fibroblast growth factor signaling in early neurogenesis. Neur Dev 4: 33, 2009. doi: 10.1186/1749-8104-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okamoto K, Tokunaga K, Doi K, Fujita T, Suzuki H, Katoh T, Watanabe T, Nishida N, Mabuchi A, Takahashi A, Kubo M, Maeda S, Nakamura Y, Noiri E. Common variation in GPC5 is associated with acquired nephrotic syndrome. Nat Genet 43: 459–463, 2011. doi: 10.1038/ng.792. [DOI] [PubMed] [Google Scholar]
- 42.Iglesias BV, Centeno G, Pascuccelli H, Ward F, Peters MG, Filmus J, Puricelli L, de Kier Joffé EB. Expression pattern of glypican-3 (GPC3) during human embryonic and fetal development. Histol Histopathol 23: 1333–1340, 2008. doi: 10.14670/HH-23.1333. [DOI] [PubMed] [Google Scholar]
- 43.Dwivedi PP, Lam N, Powell BC. Boning up on glypicans - opportunities for new insights into bone biology. Cell Biochem Funct 31: 91–114, 2013. doi: 10.1002/cbf.2939. [DOI] [PubMed] [Google Scholar]