A pharmacogenomics-enriched comprehensive medication management program that includes pharmacists empowered with pharmacogenomic data and a clinical decision support tool can detect risks in prescription regimens that are prevalent in an employee population and offers opportunities to improve medication management.
Keywords: adverse drug reactions, employee health, employer health costs, medication management, medication safety, personalized medicine, pharmacogenetics, polypharmacy
Objectives
The aims of the study are to assess adoption of a pharmacogenomic-enriched comprehensive medication management program in a self-insured employer setting and to better understand medication risks that affect employees.
Methods
Employees were identified to be at high risk of medication mismanagement and were subsequently provided with a program and process to improve their health. DNA testing, a clinical decision support system, and pharmacists were used to identify medication safety and effectiveness issues and to recommend appropriate changes.
Results
A total of 10.6% of the invited employees enrolled in the program. Actionable recommendations were suggested by pharmacists for 85.8% of employees who completed the program, averaging 5.2 recommendations per person.
Conclusions
Implementation of a PGx + CMM program in a self-insured employer setting is feasible, detects risks in prescription regimens, and offers opportunities to improve medication management and reduce the burden of healthcare expenses.
The burden of high and rising medical and pharmacy costs is of great concern for both consumers and policymakers.1 Healthcare costs are primarily borne by employees and employers in the form of insurance expenses, premiums, and copays—in the case of the US work force—and Medicare, Medicaid, and retiree/pension plans for the remainder of the insured population.2 Medications are associated with 80% of treatment strategies,3 and most US adults (82%) take at least one medication, and more than 25% take 5 or more.4 Individual responses to medication may vary considerably, and medications are ineffective or cause adverse effects in greater than 50% of patients.5 Tools that help predict an individual’s response to specific medications may enable the shift of current medical practice from reactive to proactive. Pharmacogenomics (PGx) may provide such a tool.
Pharmacogenomics integrates genomic information to inform a person’s response to medications and is personalized to the individual. As certain inherited genetic differences are known to influence the effectiveness and safety of pharmacological treatments, PGx may enable providers to choose more appropriate medications based on a patient’s genetic traits.6–8 Valuable clinical information related to drug response and/or adverse drug reactions can be obtained through genomic sequencing.9 Pharmacogenetic variants are highly prevalent with 99% of individuals carrying at least one variant that could influence medication-related outcomes.10 Pharmacogenomic research has identified a multitude of gene-drug response associations, which have resulted in genetically guided treatment and dosing decisions to yield higher success rates of pharmacological treatments.11 The list of medications that are clinically impacted by genetic biomarkers continues to expand and includes various therapeutic areas.12,13 In fact, the Food and Drug Administration (FDA) Table of Pharmacogenomic Biomarkers in Drug Labeling lists approximately 130 biomarkers found in drug labeling and more than 340 medications approved by the US FDA contain pharmacogenetic information in the drug label.12 Pharmacogenomics-guided medication management has been shown to be cost saving in various therapeutic areas and promises to be an effective tool for healthcare improvement.14–16
Comprehensive medication management (CMM) services represent a practical method to integrate PGx into clinical care. These services are designed to individually assess each patient’s medications to determine the most effective therapeutic regimen that is also safe. Similar to PGx, CMM has also shown positive impact in healthcare cost and outcomes.17 The combination of CMM with PGx testing could provide clinicians with more information relevant to medication selection and dosing. In fact, it has been demonstrated that PGx + CMM would be more useful than either approach alone.18,19
A real-world implementation of the Corigen® PGx + CMM program, using a clinical decision support system (CDSS), in a state-run, retiree health plan demonstrated the ability to decrease healthcare costs and improve health outcomes in patients 65 years and older.20 We hypothesized that broader populations, such as a self-insured employer group, could exhibit similar impactful economic and clinical outcomes in the presence of clinical factors where more appropriate medication selection is guided by pharmacists using a CDSS that incorporates comprehensive medication management and pharmacogenomics.
In this retrospective program review for early insights, we document the feasibility of implementing the Corigen® PGx + CMM program in an employee cohort as evidenced by enrollment and participant completion of the program. In addition, we describe the identification and potential implications of medication risks in the employee cohort in relation to other employee populations and the effects that increased medication safety and effectiveness may have on improving healthcare, medication management, and healthcare resource utilization.
METHODS
In an effort to improve employee health in a self-insured employer group by reducing medication-related adverse events, increasing the success of medication therapies, and reducing the burden of healthcare expenses, Coriell Life Sciences partnered with a national health services company to implement a PGx + CMM program. The Corigen® program, offered by Coriell Life Sciences, supported employee eligibility, enrollment, education, DNA testing, medication appropriateness and risk determination, PGx-CMM review, alternative medication evaluation, telehealth interactions, ongoing participant outreach, and finally the development of a comprehensive medication action plan (MAP).
Eligibility and Enrollment
Employees and eligible adult dependents enrolled in the employer-sponsored health plan from February 2021 were eligible for participation if they met the following criteria: at least 18 years old, taking at least one prescribed medication, and were identified with potential medication risk. Individuals were identified as eligible by a risk score based on potential drug-drug interactions, anticholinergic burden, contraindications, and medications impacted by genetics in pharmacy and medical claims records in the 12 months before the start of the program. The risk score is a relative risk score that ranks individuals in the population based on a weighted aggregation of the calculated risks for the previously mentioned categories. The risk score was used to identify a subset of high-risk individuals in the entire employee population. A cohort of 7500 single employer employees were invited to enroll in an employer-sponsored PGx + CMM program via an email and letter containing educational material. They were not currently enrolled nor had previously been offered a comprehensive medication management program through their employer. Individuals enrolled in the program between February and August 2021 were included in the analysis.
Enrollment was subsequently completed via a web-based system that included eligibility verification, an informational video, frequently asked questions, and the ability to document the employee’s current medication list. Upon enrollment, saliva sample collection kits were shipped to the participant and upon return, subsequently genotyped by a Clinical Laboratory Improvement Amendments and College of American Pathologists-licensed laboratory (Quest Diagnostics Nichols Institute, San Juan Capistrano, CA). All costs of the program were covered by the employer.
Clinical Decision Support System
Genomic DNA is extracted and amplified by both whole-genome amplification and multiplex PCR to overcome complexities associated with genotyping homologous pharmacogenetic genes. Genotype analysis includes allele translation and copy number analysis (from zero to 3 copies) for clinically applicable genes. Results from the genotyping and copy number assays were posted to GeneDose LIVE™ (GDL; Coriell Life Sciences, Philadelphia, PA), a comprehensive CDSS empowered by a purpose-built medication risk assessment engine.21 This risk assessment engine incorporates a genetic knowledge repository containing reputable, publicly available pharmacogenomics information curated according to a modified PhAESIS process.22 In addition, the risk assessment engine contains a nongenetic medication knowledge base compiled from US FDA drug labels, professional guidelines (eg, AGS Beers Criteria), medical associations, and drug costing sources. Upon the addition of patient-specific information, the risk assessment engine analyzed, and the CDSS subsequently reported, the magnitude of medication risks across distinct but not mutually exclusive factors: pharmacogenomics, drug-drug interactions (including phenoconversions), contraindications, anticholinergic burden, lifestyle factors (eg, foods, alcohol, and smoking), pregnancy and lactation warnings, AGS Beers Criteria, FDA black box warnings, and pediatric-specific risks. Pharmacists, trained by Coriell Life Sciences, used the CDSS to evaluate the risks—genetic and nongenetic—associated with an employee’s medication regimen, choose alternative medications, and communicate the suggested changes to the prescribing clinician via a report.
Pharmacist Evaluation and Results Delivery
A pharmacist interviewed the patient, reviewed the risk chart, and used the full-complement of GeneDose LIVE™ tools, including the alternative medication selector, to choose and subsequently document recommendations for medication adjustment with the goal of safer and more effective regimen. Recommendations were discussed during the pharmacist consultation and participants were advised to discuss the recommendations with their healthcare providers who could implement changes where appropriate to reduce medication risks. Patients and clinicians also received a MAP outlining current medication regimens and recommended changes. In addition, patients and clinicians received a medication compatibility report outlining future, potential medications with an associated risk based on the patient’s genetic information.
Analysis and Patient Consent
Evaluation was conducted at 6 months after the start of the PGx + CMM program and was approved by the Biomedical Research Alliance of New York Institutional Review Board. Written consent for the use of employee’s deidentified sample, and information for research was requested, although it was determined that the retrospective chart review did not constitute research involving human subjects. Data from employees not consenting to research use were excluded from the retrospective chart review of the MAP. A univariate data analysis was performed to describe baseline medication regimens for consenting individuals, including distributions of specific medications, number of medications, and number of PGx impacted medications. In addition, the number of types of actionable recommendations provided in the MAPs was examined and tabulated.
RESULTS
Of 7500 individuals invited to the program, 802 enrolled (10.7%). Of those enrolled, 594 (77.3%) returned a sample for analysis; 469 (61%) had completed the pharmacist consultation and received a MAP by 180 days after program initiation (Fig. 1). Employees who enrolled or samples that arrived at the laboratory after the 180-day time point were excluded from this analysis. Employees who were invited to participate averaged 51.9 years old and were mostly female (69%). Those who enrolled were slightly older (54.4 years old) and included a higher proportion of females (75%) than the overall invitee group. Of the 469 individuals who completed a MAP, 437 consented to research inclusion.
FIGURE 1.
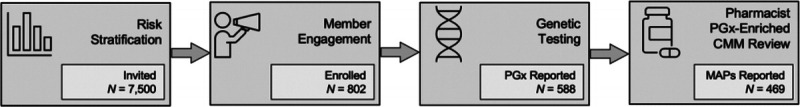
Program workflow and participant count for the Corigen® program implementation in the employee population. The participant count is as of the 180-day time point cutoff after program inception. Of the 7500 individuals identified to be at high risk of medication management and invited to the program, 802 enrolled (10.7%). Of the 802 who enrolled, 469 (61%) completed the pharmacist consultation and received a MAP by the 180-day time point. The GeneDose LIVE™ CDSS facilitated both the unification of PGx with CMM and the development of pharmacist-authored MAPs that were observed to identify potential medication safety and effectiveness issues in program participants.
Participants who completed the program (n = 437) in the first 6 months were prescribed an average of 6.9 medications (SD = 4.2) before the start of the program (Fig. 2B). Most participants (n = 252/437, 57.7%) were prescribed more than 5 medications at the time of the pharmacist consultation. The most commonly prescribed medications with pharmacogenomic implications were atorvastatin (25.4%), metformin (24.3%), metoprolol (13.5%), and bupropion (12.8%; Fig. 2A, notated with star), whereas vitamins (48.5%), hydrochlorothiazide (21.1%), and levothyroxine (20.8%) were among the most commonly prescribed medications lacking pharmacogenomic clinical utility (Fig. 2A).
FIGURE 2.
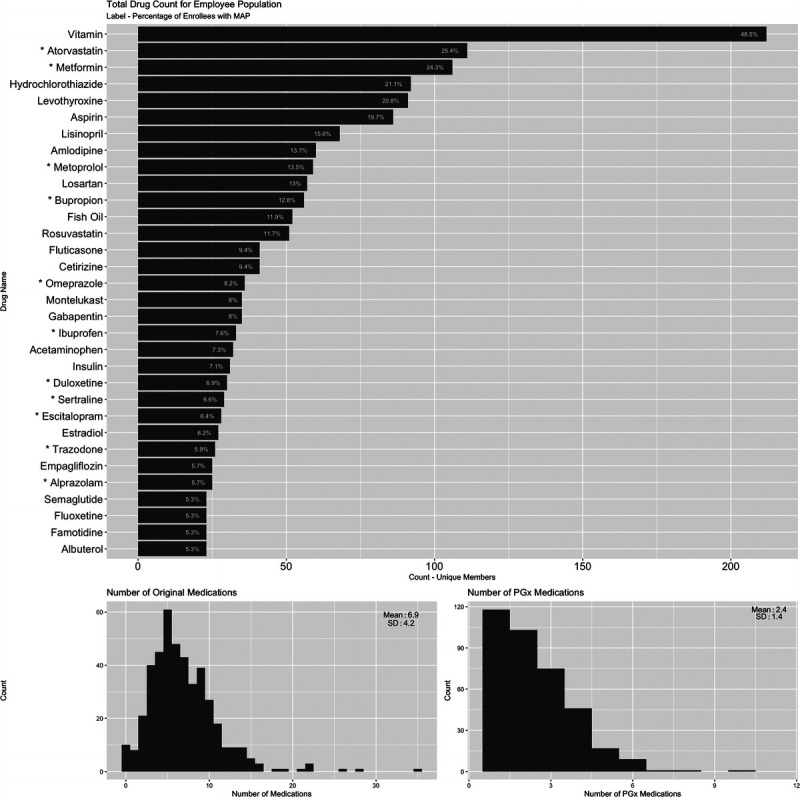
Summary of the original medication regimen for Corigen® program participants. A, The most common medications are listed with the unique member count for medications included in the original regimen. The label indicates the percentage of enrollees with a MAP who were currently taking each medication. Medications with pharmacogenomic implications are designated with an asterisk (*). B, Histogram of the number of original medications per participants who completed the program. The mean is 6.9 and the standard deviation is 4.2. C, Histogram of the number of original medications with pharmacogenomic implication per participants who completed the program. The mean is 2.4 and the SD is 1.4.
Of the 437 consented participants who received a MAP, 375 (85.8%) had at least one actionable recommendation suggested by the pharmacists. Participants received 5.2 (SD = 4.2) recommendations per participant (n = 437) on average or 6.0 (SD = 4.0) recommendations for those who received at least one actionable recommendation (n = 375). Of the 375 who received an actionable recommendation, 165 participants (44.0%) received a recommendation to initiate alternative medications and 161 (42.9%) participants received a recommendation to discontinue a current medication (Table 1). Most pharmacist recommendations (n = 322, 85.9%) were categorized as “monitor” with MAPs containing additional notes to the prescribing physician intended as a meaningful, substantive intervention (Table 1). These actionable notes contained specific details including physiological metrics, adverse effects, and distinct laboratory values to monitor, written with the intent to decrease the likelihood of additional medication related adverse events.
TABLE 1.
Recommendations Provided in Medication Action Plans (n = 375 of 437) Who Received an Actionable Recommendation
| Recommendation | Count | Percent of Total |
|---|---|---|
| Initiate new medication | 165 | 44.0% |
| Discontinue medication | 161 | 42.9% |
| Modify prescription | 3 | 0.8% |
| Monitor (ADR, efficacy, laboratories, physiology) | 322 | 85.9% |
DISCUSSION
We implemented a PGx + CMM program that incorporated member engagement and education, genetic testing, and pharmacist consultation to identify medication risks for the improvement of medication therapy outcomes in the employee population. The adoption of the PGx + CMM program in this employee population resulted in 802 of the 7500 invited employees (10.6%) enrolling in the program in the first 6 months. Completing the program, 469 (61%) employees had a pharmacist consultation and had a MAP created. Of the 437 participants consenting to research, 375 (85.8%) had MAPs with actionable recommendations that were suggested and documented by the pharmacists, with more than 5 actionable recommendations per participant on average.
The successful enrollment, risk evaluation, and communication of suggested medication changes associated with this real-world implementation provides evidence that the Corigen® program is feasible in this employee population and suggests that it may be widely applied to general workforce populations. Specifically, we identified no unique challenges that seem different from those of similar program implementations in other employee populations. However, we acknowledge that it may be difficult to understand differences that may exist between this employee population and others.
The utilization of educational materials, virtual enrollment forms, and at-home DNA collection kit resulted in more than 10% of the invited population enrolling in the program. This rate is in line with expected participation in similar program implementations at the 6-month time point and thus suggests common limitations and barriers to enrollment in wellness programs. Higher enrollment and completion rates may be expected in future analysis for this population because of expanded outreach and extended invitation and analysis period. Still, these findings show that a group previously naive to pharmacogenomics testing and comprehensive medication management took an action to improve their health as a direct result of the program invitation. After enrollment and DNA testing, employees had their medication regimen reviewed by a pharmacist trained in pharmacogenomics and in using the GeneDose LIVE™ CDSS.
The program’s pharmacists, empowered with the GeneDose LIVE™ CDSS, identified actionable medication recommendations in most participants. Most of the participants (57.7%) who completed the program were defined as having polypharmacy (taking greater than 5 medications). Excessive or unnecessary use of medications—inappropriate polypharmacy—generally increases the risks of drug-drug, drug-disease, and drug-gene interactions.23 The presence of these risk factors was confirmed by the risk assessment tool for this specific population. Participants were also taking medications that are common in adults with many having a pharmacogenomic implication, suggesting that both pharmacogenomics testing and comprehensive medication management can be drivers of change in this program. The combination of polypharmacy and common medications, along with the number of actionable recommendations and high proportion of medications flagged for removal or monitoring, provides evidence for the risk of medication mismanagement in this and likely other workforce populations. Not only did the GeneDose Live CDSS identify potential issues, but a pharmacist verified the issue, recommended a change, and then communicated this information via a consultation and report to both the employee and their prescribing physician. Together, these findings validate that medication issues do exist, were previously not identified, and increase the likelihood that an employee’s prescription will be changed to a more safe and effective medication. The prevalence of medication issues in this population can be extended to general workforce populations and suggests a significant opportunity to improve care by adopting a PGx + CMM program.
Adoption of the PGx + CMM program has been shown to produce positive economic and healthcare resource utilization changes for a retiree population 65 years old and older.20 Altogether, the findings presented in this report suggest similar opportunities to improve healthcare outcomes and medication management in broad employee, adult populations, and even in those who may not be part of a unified health system. In the retiree population, greater than 76% of MAPs were identified as actionable,20 similar to 85.8% observed in this employee population, supporting similar opportunities to address medication risks. Similar populations may include many adults with a therapeutic regimen that puts them at risk for negative health outcomes and increased medical costs for the employer sponsoring the health plan. A combined PGx + CMM program in such populations holds potential to facilitate both reactive correction and proactive mitigation of medication issues.
This study presents the potential for realization of positive health outcomes resulting from the PGx + CMM program. Further analysis is necessary to examine the economic, clinical (including medication changes), and healthcare resource utilization impacts of this specific program implementation. However, it is reasonable to suggest that the program benefits employees, employers, and healthcare providers.
CONCLUSIONS
This report documents early insights of a real-world implementation of the Corigen® PGx + CMM (PGx + CMM) program in a workforce population. The findings demonstrate that the implementation is feasible; the CDSS, GeneDose LIVE™, detects risks in prescription regimens that are prevalent in the population, and the program offers opportunities to improve care and medication management when compared with existing standard of care models.
Footnotes
Funding sources: None to disclose.
Conflict of interest: M.S.F., R.A.L., and S.E.G. are employed by and receive a salary from Quest Diagnostics. M.S.F. and S.E.G. have stock ownership in Quest Diagnostics. J.A.S., A.P.P., and M.K. are employed by and receive a salary from Coriell Life Sciences. J.A.S. and M.K. have an equity interest in Coriell Life Sciences.
Ethical Considerations and Disclosures: This study was conducted in accordance with the Declaration of Helsinki and approved by the Biomedical Research Alliance of New York Institutional Review Board, date of approval: December 23, 2020.
Contributor Information
Murray Keogh, Email: mkeogh@coriell.com.
Maren S. Fragala, Email: Maren.S.Fragala@QuestDiagnostics.com.
Arul Prakasam Peter, Email: aprakasam@coriell.com.
Raymond A. Lorenz, Email: Raymond.A.Lorenz@QuestDiagnostics.com.
Steven E. Goldberg, Email: Steven.E.Goldberg@questdiagnostics.com.
REFERENCES
- 1.Fragala MS, Shaman JA, Lorenz RA, Goldberg SE. Role of pharmacogenomics in comprehensive medication management: considerations for employers. 2022. [DOI] [PubMed]
- 2.Cubanski J, Rae M, Young K, ADHow does prescription drug spending and use compare across large employer plans, Medicare part D, and Medicaid? Kaiser Family Foundation; 2019. Available at: https://www.kff.org/medicare/issue-brief/how-does-prescription-drug-spending-and-use-compare-across-large-employer-plans-medicare-part-d-and-medicaid/. Accessed March 21, 2022. [Google Scholar]
- 3.Pharmacy ACoC . Comprehensive Medication Management in Team-Based Care. 2017. 2019. Available at: https://www.pcpcc.org/sites/default/files/event-attachments/CMM%20Brief.pdf. Accessed March 22, 2022.
- 4.Slone Epidemiology Center at Boston University . Patterns of medication use in the United States. 2006. Available at: https://www.bu.edu/slone/files/2012/11/SloneSurveyReport2006.pdf. Accessed March 22, 2022.
- 5.Spear BB, Heath-Chiozzi M, Huff J. Clinical application of pharmacogenetics. Trends Mol Med. 2001;7:201–204. [DOI] [PubMed] [Google Scholar]
- 6.Roden DM McLeod HL Relling MV, et al. Pharmacogenomics. Lancet. 2019;394:521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roden DM, Wilke RA, Kroemer HK, Stein CM. Pharmacogenomics: the genetics of variable drug responses. Circulation. 2011;123:1661–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrod AE. The Croonian lectures on inborn errors of metabolism. Lancet. 1908;172:1–7. [Google Scholar]
- 9.Wright GEB, Carleton B, Hayden MR, Ross CJD. The global spectrum of protein-coding pharmacogenomic diversity. Pharmacogenomics J. 2018;18:187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chanfreau-Coffinier C Hull LE Lynch JA, et al. Projected prevalence of actionable pharmacogenetic variants and level a drugs prescribed among US Veterans Health Administration pharmacy users. JAMA Netw Open. 2019;2:e195345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lauschke VM, Milani L, Ingelman-Sundberg M. Pharmacogenomic biomarkers for improved drug therapy-recent progress and future developments. AAPS J. 2017;20:4. [DOI] [PubMed] [Google Scholar]
- 12.US FDA . Table of Pharmacogenomic Biomarkers in Drug Labeling. Washington, DC: U.S. Food & Drug Administration; 2022. [Google Scholar]
- 13.Kim JA, Ceccarelli R, Lu CY. Pharmacogenomic biomarkers in US FDA-approved drug labels (2000–2020). J Pers Med. 2021;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Y Swanson KM Rojas RL, et al. Systematic review of the evidence on the cost-effectiveness of pharmacogenomics-guided treatment for cardiovascular diseases. Genet Med. 2020;22:475–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winner JG Carhart JM Altar CA, et al. Combinatorial pharmacogenomic guidance for psychiatric medications reduces overall pharmacy costs in a 1 year prospective evaluation. Curr Med Res Opin. 2015;31:1633–1643. [DOI] [PubMed] [Google Scholar]
- 16.Brown LC, Lorenz RA, Li J, Dechairo BM. Economic utility: combinatorial pharmacogenomics and medication cost savings for mental health care in a primary care setting. Clin Ther. 2017;39:592–602.e591. [DOI] [PubMed] [Google Scholar]
- 17.Chung TH Hernandez RJ Libaud-Moal A, et al. The evaluation of comprehensive medication management for chronic diseases in primary care clinics, a Texas delivery system reform incentive payment program. BMC Health Serv Res. 2020;20:671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown JT, Bishop JR, Schneiderhan ME. Using pharmacogenomics and therapeutic drug monitoring to guide drug selection and dosing in outpatient mental health comprehensive medication management. Ment Health Clin. 2020;10:254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez-Escudero I Cedeno JA Rodriguez-Nazario I, et al. Assessment of the clinical utility of pharmacogenetic guidance in a comprehensive medication management service. J Am Coll Clin Pharm. 2020;3:1028–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarvis JP Peter AP Keogh M, et al. Real-world impact of a pharmacogenomics-enriched comprehensive medication management program. J Pers Med. 2022;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaman JA, Chernin PH, Megill SE. Coriell Life Sciences: empowering the most precise medical care for a healthier world. Pharmacogenomics. 2022;23:457–462. [DOI] [PubMed] [Google Scholar]
- 22.Gharani N Keller MA Stack CB, et al. The Coriell personalized medicine collaborative pharmacogenomics appraisal, evidence scoring and interpretation system. Genome Med. 2013;5:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maher RL, Hanlon J, Hajjar ER. Clinical consequences of polypharmacy in elderly. Expert Opin Drug Saf. 2014;13:57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]


