Abstract
Transposon mutagenesis and marker rescue were used to isolate and identify an 8.5-kb contiguous region containing six open reading frames constituting the operon for the sorbitol P-enolpyruvate phosphotransferase transport system (PTS) of Streptococcus mutans LT11. The first gene, srlD, codes for sorbitol-6-phosphate dehydrogenase, followed downstream by srlR, coding for a transcriptional regulator; srlM, coding for a putative activator; and the srlA, srlE, and srlB genes, coding for the EIIC, EIIBC, and EIIA components of the sorbitol PTS, respectively. Among all sorbitol PTS operons characterized to date, the srlD gene is found after the genes coding for the EII components; thus, the location of the gene in S. mutans is unique. The SrlR protein is similar to several transcriptional regulators found in Bacillus spp. that contain PTS regulator domains (J. Stülke, M. Arnaud, G. Rapoport, and I. Martin-Verstraete, Mol. Microbiol. 28:865–874, 1998), and its gene overlaps the srlM gene by 1 bp. The arrangement of these two regulatory genes is unique, having not been reported for other bacteria.
Oral streptococci, particularly aciduric species such as Streptococcus mutans, contribute to dental caries by degrading dietary sugars and sugar alcohols to metabolic acid end products, resulting in the demineralization of tooth mineral (4). Caries formation in the presence of readily fermentable carbohydrates, such as sucrose, has led to the use of low-cariogenic sugar substitutes, such as sorbitol (glucitol), in sugar-free gums and lozenges (3). More recently, however, the frequent use of sorbitol-containing products has been shown to result in increased levels of sorbitol-utilizing bacteria due to adaptation to sorbitol. The major sugar transport process in S. mutans is via the phosphoenolpyruvate: sugar phosphotransferase system (PTS) (17, 28), a group translocation process utilizing phosphoenolpyruvate as a substrate in phosphoryl transfer involving the general, non-sugar-specific proteins enzyme I and HPr and ultimately the sugar-specific, membrane-bound enzyme II (EII) complex, resulting in the transport and phosphorylation of the specific sugar being transported. The EII complexes are normally comprised of three functional domains, fused either within a single protein or on separate proteins, with domains IIA (formerly enzyme III) and IIB possessing the first and second phosphorylation sites, respectively, while the IIC domain forms the transmembrane channel and the sugar-binding site (17).
Early work with S. mutans revealed that sorbitol transport by glucose-grown cells required the concomitant induction of the sorbitol-PTS and sorbitol-6-phosphate dehydrogenase (SDH), resulting in the formation of fructose-6-phosphate (8, 19). Sorbitol-PTS and SDH activities were repressed by low concentrations of glucose (8, 19) by a mechanism that was at least in part due to inducer exclusion, a mechanism not observed with glucose-PTS-negative mutants. Sorbitol transport by Streptococcus sanguis also occurs via an inducible sorbitol-PTS (10, 21); however, unlike S. mutans, S. sanguis is not subject to catabolite repression by glucose, being capable of growth on glucose and sorbitol concurrently, with sorbitol utilized at a slightly lower rate than glucose. The first sorbitol-PTS to be genetically characterized was from Escherichia coli L163sr (30, 31). Sequence and expression analysis revealed the presence of the genes gutA, gutB, gutD, gutM, and gutR, coding for EIIBC, EIIA, SDH, an activator, and a repressor, respectively. Subsequent reanalysis of the E. coli L163sr sequence has revealed that the EIIC domain is encoded by two distinct genes, one half by gutA and one half by gutE, which also encodes the EIIB domain (16). The genetic designations gut, for glucitol, and srl, for sorbitol, have both been used by different groups in designating the genes from characterized sorbitol operons.
There is currently a single report of a genetically defined sorbitol mutant of S. mutans (32). This strain failed to ferment sorbitol anaerobically but did so aerobically, and it was determined that the defect was due to a chromosomal deletion that included the pfl gene, coding for pyruvate formate-lyase. Consequently, the specific aim of the present study was to clone, sequence, and identify the genes involved in sorbitol metabolism by S. mutans. A mutant strain of S. mutans LT11 defective in sorbitol metabolism was generated via transposon mutagenesis, and this strain led to the recovery of the genes coding for the sorbitol-PTS, as well as the gene coding for SDH. In addition, the identification of two regulatory genes within the sorbitol operon has led to a better understanding of the mechanism involved in the regulation of sorbitol-related enzymes.
Bacterial strains, plasmids, and growth conditions.
Table 1 lists the bacteria and plasmids used in this study. The plasmids pα, pΩ, and pΩIS were a kind gift of R. Lunsford. S. mutans strains were maintained anaerobically at 37°C on Todd-Hewitt (TH) plates (Difco or BBL) with antibiotics as appropriate and grown for DNA isolation in TH broth supplemented with 0.3% yeast extract. Where appropriate, antibiotics were used at the indicated concentrations: erythromycin at 500 μg/ml for E. coli and 10 μg/ml for S. mutans, kanamycin (KM) at 30 μg/ml for E. coli and 300 μg/ml for S. mutans, ampicillin at 100 μg/ml, and tetracycline at 10 μg/ml. Growth studies were carried out in tryptone (1%)-yeast extract (0.5%) broth (TYE) with the appropriate carbon source.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Genetic markers or description | Source or reference |
|---|---|---|
| S. mutans strains | ||
| LT11 | Highly transformable mutant of UA159 | 23 |
| BH96SR | LT11 carrying Tn4001 in srlR; Sorb− | This work |
| BH96SRΩIS | BH96SR with pΩIS integrated at one end of Tn4001 | This work |
| BH97SRT+ and BH97SRT− | LT11 with pSR(+) and pSR(−), respectively, integrated in srlR; Sorb− | This work |
| BH98SDH | LT11 with Emr gene inserted into srlD; Sorb− | This work |
| E. coli strains | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIq ZΔM15 Tn10 (Tetr)] | Stratagene |
| JWL163 | fhuA2 glnV44 λ− gatC49 gat50 upp-33 srlD50 rpsL104 malT1 xylA7 mtlA2 metB1 | 12 |
| Plasmids | ||
| pBluescript | Apr ColE1 ori f1 ori multipurpose cloning vector | Stratagene |
| pΩ | Emr; pBluescript-based streptococcal integration vector | 13 |
| pα | pΩ carrying Tn4001 | 13 |
| pΩIS | pΩ carrying a portion of IS256 | R. Lunsford |
| pΩIS-SR | Contains 1 kb of LT11 DNA rescued from BH96SRΩIS | This work |
| pSR(+) and pSR(−) | Contain an 0.66-kb NspV fragment from pΩIS-SR cloned into pΩ in both orientations | This work |
| pSR-EcDN | Contains 3 kb of LT11 DNA rescued from BH97SRT+ | This work |
| pSR-EcUP | Contains 5 kb of LT11 DNA rescued from BH97SRT− | This work |
| pSDH1.6 | 1.6-kb BglII/MunI fragment from pSR-EcUP cloned into pBluescript | This work |
| pSDH-Em | Emr gene cloned into the BamHI site of pSDH1.6 | This work |
DNA methodology.
S. mutans DNA isolation, plasmid isolation, agarose gel electrophoresis, Southern hybridizations, DNA ligations, and transformation of E. coli were performed as previously described (5, 6). Transformation of S. mutans was essentially done as described by Perry et al. (15). Sequencing was carried out manually using Sequenase version 2.0 (Amersham) with the modifications described by Mytelka and Chamberlin (14) or the CircumVent Thermal Cycle DNA sequencing kit (New England Biolabs), and automatic sequencing was carried out using fluorescent dye terminators by the University of Florida (Gainesville) DNA Sequencing Core Laboratory. Custom-made primers for manual sequencing or for PCR were synthesized by the University of Calgary, University Core DNA Services, or the University of Florida DNA Sequencing Core Laboratory. The DNA sequences to complete the sequence of the srlB gene and the additional sequence downstream were obtained by directly sequencing genomic DNA using primers designed based on previously sequenced DNA. Searches for homologous proteins were carried out against the GenBank database using the BLAST suite of programs (2) at the National Center for Biotechnology Information via their World Wide Web interface (http://www.nih.nlm.ncbi/BLAST). Multiple alignment of proteins was carried out using CLUSTAL W (25).
Isolation of S. mutans BH96SR.
S. mutans LT11 was transformed with pα (13), and dilutions of the culture were plated out on TH-KM plates to allow growth of individual colonies. Approximately 600 transformants were picked onto TYE-sorbitol indicator plates containing KM and incubated overnight. One non-acid-producing colony (BH96SR) was tested for its ability to ferment PTS sugars on TYE-sugar indicator plates and shown to be unable to metabolize sorbitol. A marker rescue strategy involved the use of plasmid pΩIS (Table 1) for Tn4001 junction rescue in BH96SR as follows. Plasmid pΩIS was transformed into S. mutans BH96SR, and transformants were selected on TH-erythromycin plates. Integration of pΩIS via Campbell-type recombination at the Tn4001 copy was confirmed for six transformants by Southern hybridization analysis. The genomic DNA from one isolate, BH96SRΩIS, was cut with SstI, ligated at a dilute DNA concentration, and used to transform E. coli to Emr. Fifteen E. coli transformants were screened, and all appeared to carry a plasmid with ∼1 kb of DNA flanking the transposon junction. One plasmid was selected for further analysis and was named pΩIS-SR. The transposon sequences were removed from pΩIS-SR by HindIII digestion-religation to form pΩ-SR. An 0.66-kb NspV fragment from pΩ-SR was cloned into the ClaI site of pΩ in both orientations to give pSR(+) and pSR(−). Rescue of DNA from the sorbitol locus of LT11 was essentially performed as described above, except that EcoRI was used to cut the genomic DNA prior to rescue in E. coli after the integration of pSR(+) and pSR(−) to give BH97SRT+ and BH97SRT−, respectively. The plasmids recovered were named pSR-EcUP and pSR-EcDN.
Inactivation of the srlD gene.
A 1.1-kb BamHI fragment containing an Emr gene was isolated from pGh9:ISS1 (22) and cloned into the BamHI site of pSDH1.6 to give pSDH-Em (Fig. 1). Linearized pSDH-Em was used to transform LT11 to Emr, and six colonies were analyzed by Southern hybridization for the presence of the disrupted srlD gene. All six contained the Emr gene in srlD, and one was picked for further analysis and named BH98SDH.
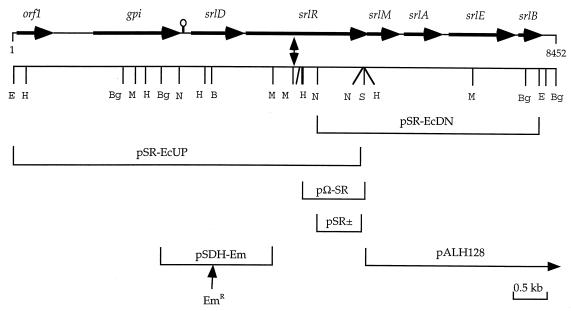
FIG. 1.
Schematic representation of the genetic characterization of the sorbitol transport operon in S. mutans LT11 obtained by sequencing. Inserts of plasmids used for sequencing, marker rescue, and/or complementation are indicated under the restriction map. A double-headed arrow represents the point of insertion of Tn4001 in S. mutans BH96SR. A putative hairpin structure between gpi and srlD is indicated. Restriction enzyme sites are BamHI (B), BglII (Bg), EcoRI (E), HindIII (H), MunI (M), NspV (N), and SstI (S). Gene symbols: orf1, unknown; gpi, glucose-6-phosphate isomerase; srlD, sorbitol-6-phosphate dehydrogenase; srlR and srlM, regulatory proteins; srlA to srlE, EIIBC; and srlB, EIIA.
Characterization of S. mutans BH96SR and rescue of the sorbitol locus from S. mutans LT11.
The Tn4001 system (13) was employed with S. mutans LT11 to successfully isolate a strain, BH96SR, that did not ferment sorbitol but did ferment all other PTS sugars tested, indicating that the transposon was not located in the ptsHI operon. We used a marker rescue strategy to isolate approximately 1 kb of genomic DNA flanking one end of the transposon insertion in BH96SR(pΩ-SR) and determined its nucleotide sequence. Analysis of the sequence revealed that, in BH96SR, Tn4001 had inserted itself into an open reading frame (ORF) whose putative translation product showed homology to several transcriptional regulators from Bacillus subtilis, including the LicR protein from a β-glucoside PTS operon that transports lichenan (β-1,3-1,4-glucan) degradation products, including cellobiose (26). Thus, it appeared that the transposon insertion inactivated a putative regulator of the sorbitol operon of LT11, causing the sorbitol-negative phenotype of BH96SR. To further characterize the upstream and downstream regions, the marker rescue plasmids pSR(+) and pSR(−) were used to obtain two overlapping clones (pSR-EcUP and pSR-EcDN) whose inserts represent approximately 8.5 kb of contiguous sequence from the LT11 genome (Fig. 1).
DNA sequence analysis.
The inserts from pSR-EcUP and pSR-EcDN were completely sequenced to give a total of 7,957 bp defining an EcoRI fragment from this region (data not shown). A further 495 bp of sequence was obtained by direct genomic sequencing, PCR product sequencing, and sequencing from an additional overlapping clone, pALH128 (Fig. 1). Analysis of the 8,452 bp of total sequence obtained revealed a total of eight ORFs, all in the same orientation, whose putative translation products were used as query sequences in BLAST searches of the GenBank database to identify the corresponding genes (Fig. 1). ORF1 (564 bp) begins with an ATG codon and encodes a putative protein of 187 amino acids that produced no significant matches with any entries in GenBank. After an intergenic region of 556 bp begins an ORF (1,350 bp, ATG start codon) whose product (449 amino acids) shares between 59 and 66% identity with the glucose-6-phosphate isomerase A and B isozymes, respectively, from Bacillus stearothermophilus and 62% identity to the enzyme from B. subtilis. We, therefore, designated this ORF as the gpi gene of S. mutans LT11. We have detected a region of dyad symmetry (ΔG = −71 kJ) beginning 18 bp downstream of the gpi stop codon and ending 113 bp upstream of the srlD start codon. This may play a role in transcriptional termination of gpi and/or in regulation of the sorbitol operon expression. The remaining six ORFs appear to constitute the genetic components of the sorbitol-PTS operon of S. mutans. Separated from the gpi gene by 164 bp is the srlD gene (801 bp, ATG start codon) coding for a protein of 266 residues sharing 58% identity with the gutD gene coding for a SDH from the sorbitol-PTS operon of Clostridium beijerinckii (22) and 57% identity to the sorD gene encoded by the l-sorbose PTS operon of Klebsiella pneumoniae (29). This system encodes components to transport and phosphorylate sorbose to sorbose-1-phosphate and to reduce it to sorbitol-6-phosphate followed by conversion to fructose-6-phosphate by the SDH. Interestingly, the S. mutans SDH shares only 28% identity with the SDHs from the sorbitol-PTS operons of E. coli and Erwinia amylovora (1, 30). Twenty base pairs downstream of the stop codon of the srlD gene is a GTG codon, which we believe signifies the start of a gene that we have designated srlR, coding for a putative transcriptional regulator protein. There is a ribosomal binding site (RBS) motif, AGAGGG, located 7 bp upstream of the GTG codon, whereas there is no suitable RBS upstream of the first or second ATG codons of the ORF. The ORF (1,866 bp) codes for a protein of 621 residues (SrlR), which shares 22% identity with the LicR regulator (641 residues) from the β-glucoside PTS operon from B. subtilis (26) and 19.5% identity with the MtlR regulator (697 residues) from the mannitol-PTS operon from B. stearothermophilus (11). As a consequence of the B. subtilis genome sequencing project, two other similar putative regulators have been identified, YjdC (648 residues), sharing 21.5% identity with SrlR, and YdaA (694 residues), sharing 18% identity with SrlR. All five proteins share between 18 and 27% identity with each other, except for the MltR and YdaA proteins, which are 40.5% identical, indicating a closer relationship (data not shown). Overlapping the stop codon of srlR by 1 nucleotide is srlM (489 bp, ATG start codon), coding for a homolog of the GutM activator protein from the E. coli sorbitol operon (31). There is another ATG codon 7 nucleotides downstream; however, the spacing between the putative RBS (AAGGAG) and the first ATG is more optimal (7 versus 16 bp). An alignment between SrlM (162 residues) and GutM (119 residues) reveals that they share 23.5% identity and that the additional amino acids in SrlM are located at the C terminus (data not shown). The next three ORFs code for the EII components of the system as identified by their similarity to their E. coli and C. beijerinckii homologs. Beginning 79 bp downstream of srlM is srlA (543 bp, ATG start codon) coding for one half of the EIIC domain (180 residues), which shares 58 and 52% identity with the EIICs from C. beijerinckii and E. coli, respectively. Beginning 90 bp after the end of srlA is srlE (1,011 bp, ATG start codon), coding for a fusion protein (336 residues) consisting of the other half of the EIIC domain fused to an EIIB domain and sharing 65 and 57% identity with the EIIBCs from C. beijerinckii and E. coli, respectively. Beginning 42 bp downstream of srlE is srlB (369 bp, ATG start codon) coding for the EIIA subunit (121 residues), which shares 41 and 33% identity with the EIIAs from C. beijerinckii and E. coli, respectively. We could detect no other ORFs downstream of srlB or a structure resembling a possible transcriptional terminator. However, we did find evidence of a nonfunctional transaldolase-like-protein-encoding gene similar to the one in the sorbitol operon of C. beijerinckii (22).
Growth characteristics of S. mutans LT11 and sorbitol mutants.
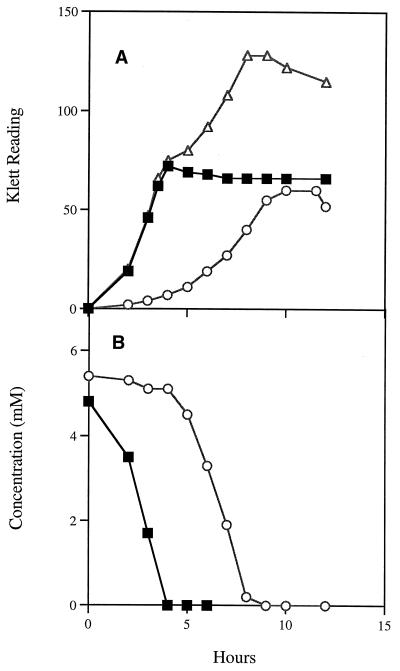
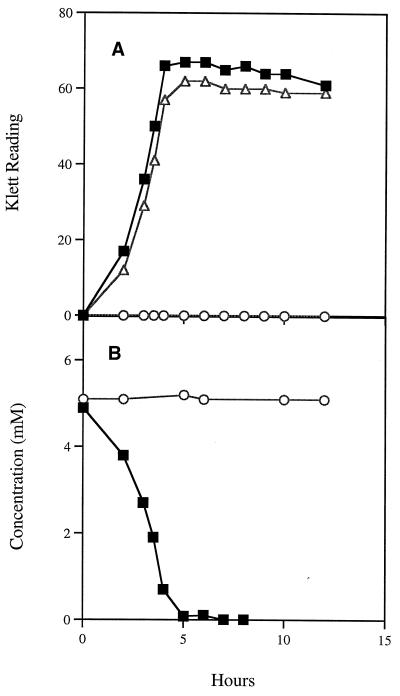
When S. mutans LT11 was grown in medium with sorbitol as the sole carbon source, there was a long lag period (5 to 6 h), indicating that the proteins involved in sorbitol transport and metabolism are inducible (Fig. 2A). In medium containing glucose or glucose-sorbitol, a shorter lag period of 1.5 to 2 h was observed (Fig. 2A). Analysis of the glucose-sorbitol culture medium showed that glucose was utilized to near exhaustion before sorbitol was consumed (Fig. 2B). A very short lag period was observed during transition from growth on glucose to growth on sorbitol, indicating that the enzymes required for sorbitol utilization were induced prior to complete exhaustion of glucose. These observations indicate that sorbitol metabolism in S. mutans is inducible and subject to catabolite repression in the presence of glucose, confirming earlier research (8, 19). In similar experiments with S. mutans BH96SR, growth was unimpaired in medium containing glucose or glucose-sorbitol, and as expected, this S. mutans strain failed to grow when sorbitol was the sole carbon source (Fig. 3). Strains BH97SRT+ and BH97SRT− (Table 1) failed to utilize sorbitol when grown on indicator plates, as did BH98SDH, containing an insertionally inactivated srlD gene. Analysis of the sequence would seem to indicate that, in both cases, it is likely that there is a polar effect by the insertions in these strains on the transcription of the downstream genes. Transcript analysis, however, would be needed to confirm this hypothesis. We transformed pSR-EcUP and pSDH1.6 into E. coli JWL163, which contains a mutation in its srlD gene (12). When plated on MacConkey-sorbitol plates, the plasmid-containing colonies were dark red, indicating sorbitol utilization. Thus, the expression of the srlD genes from these plasmids in E. coli is not dependent on the presence of the S. mutans SrlR or SrlM protein.
FIG. 2.
(A) Growth of the wild-type S. mutans LT11 on glucose (■), sorbitol (○), and glucose-sorbitol (▵). (B) Glucose (■) and sorbitol (○) utilization in the glucose-sorbitol culture.
FIG. 3.
(A) Growth of the mutant S. mutans BH96SR on glucose (■), sorbitol (○), and glucose-sorbitol (▵). (B) Glucose (■) and sorbitol (○) utilization in the glucose-sorbitol culture.
The results of this study support the earlier biochemical and physiological studies that indicated that sorbitol-fermenting S. mutans possesses an inducible sorbitol-PTS that is subject to catabolite repression in the presence of glucose (8, 19). Thus, it is not surprising to find a regulatory region as an integral part of the operon. The available evidence suggests that sorbitol transport via the PTS in S. mutans is likely the only route for sorbitol uptake by the organism, since, unlike Bacillus (33, 34), the organism is devoid of SDH activity (19). Furthermore, a mutant defective in the general PTS protein, enzyme I, failed to ferment sorbitol (6), and sorbitol is not a substrate of the multiple-sugar metabolism transport system first reported by Tao and coworkers (24). As sorbitol metabolism in S. sanguis appears not to be subject to catabolite repression (10, 21), it will be interesting to characterize the genetics of the sorbitol-PTS in this related oral bacterium.
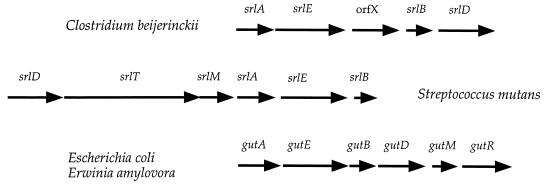
It is obvious from the results, however, that there has been extensive divergence between the S. mutans operon and other sorbitol operons in terms of the gene order and in the mode of regulation. Although the EII genes are found in the same order in S. mutans, C. beijerinckii, E. coli, and E. amylovora (srlA, srlE, and srlB), there are several differences between locations of other genes in the operons (Fig. 4). The srlD gene is found after srlB in other sorbitol operons, while it is likely the first gene in the S. mutans operon. We have detected a region of dyad symmetry upstream of srlD, which may act as a transcriptional terminator of the preceding gene, gpi, and we have also shown that sequences upstream of the gpi gene are not necessary for expression of srlD in E. coli. It is interesting to note, however, that two genes whose products both catalyze reactions that form fructose-6-phosphate are found adjacent to each other in the genome. We also note that, whereas the SDH proteins of S. mutans, C. beijerinckii, and K. pneumoniae share about 57% identity, this group shares only about 26% identity with the SDH proteins of E. coli and E. amylovora. A multiple alignment of the SDHs (data not shown) reveals that overall the proteins have identical residues in 34 positions and similar residues in another 48 positions, producing an overall similarity of about 30%. Thus, we believe that the SDHs form two distinct but distantly related families with a common ancestor. This is in contrast to the work of Wehmeier and Lengeler (29), who noted no significant similarity between K. pneumoniae SDH and E. coli SDH and speculated that their srlD genes evolved convergently rather than by gene duplication. In C. beijerinckii, there is a gene (orfX) coding for a putative transaldolase-like protein found between the srlE and srlB genes (22). No significant ORFs were found downstream of the srlB gene of S. mutans; however, when we used this sequence in BLAST searches of the GenBank database, several short stretches of DNA were identified that when translated had significant similarity to segments from the orfX gene product of C. beijerinckii (data not shown). Point mutations and/or small insertions-deletions appear to prevent this region from encoding a functional orfX gene product. It is tempting to speculate that, in S. mutans, the transaldolase-like product of the orfX gene became dispensable and the gene was allowed to mutate to nonfunctionality. The E. coli and E. amylovora sorbitol operons contain two genes downstream of gutD, gutM and gutR, encoding regulatory proteins with activator and repressor functions, respectively. It will be interesting to determine by further sequence analysis if the C. beijerinckii sorbitol operon contains a regulatory region.
FIG. 4.
The structure of the sorbitol-PTS operons from S. mutans, C. beijerinckii, E. coli, and E. amylovora. The operons are shown arbritrarily aligned by their respective srlA and srlE genes.
The srlR and srlM genes constitute the regulatory region of the S. mutans sorbitol operon and overlap by 1 nucleotide, indicating a translational coupling. The SrlM protein shares low but significant homology with the GutM protein of E. coli, which is postulated to be a DNA-binding protein and has been shown to be necessary for full activation of the operon (31). The SrlM protein does, however, contain an additional 41 residues at its C terminus compared to GutM, and it is likely that this region contributes to the activity of the protein in S. mutans. The SrlR protein is similar to several transcriptional regulators from Bacillus, all which exhibit a multidomain structure. In the N-terminal region, the proteins contain two helix-turn-helix motifs, the first similar to that from the DeoR family and the second similar to that from the LysR family of transcriptional regulators (7). The middle parts of the proteins are similar to a region from the BglG-SacT-SacY family of transcriptional antiterminators (18, 27). This region has been shown to consist of two homologous domains, PRD-I and PRD-II (PTS regulation domain), that presumably arose by duplication (20, 27). Figure 5 shows an alignment of the PRD domains of the S. mutans SrlR protein with some of the PRD-containing proteins found in Bacillus. An arginine (position 14 [Fig. 5]) and a glutamate (position 61 [Fig. 5]) are conserved in all the proteins shown and in most other PRD-containing proteins (20). More interesting are the highly conserved histidine residues (positions 7 and 69 [Fig. 5]), which have been shown to be phosphorylated in some PRD-containing proteins either by EIIBs or by the general PTS protein HPr and are involved in either negative or positive regulation of the regulators (reviewed in reference 20). The first type of PRD phosphorylation occurs in the PRD-I of antiterminator-type proteins in the absence of inducer and leads to negative regulation of the regulator. The second type occurs only in gram-positive bacterial regulators, where, in the absence of PTS substrates, phosphorylated HPr (His-15) phosphorylates in turn PRD-containing regulators and stimulates their activity (9). Interestingly, the first histidine (position 7 [Fig. 5]) is not conserved in the S. mutans SrlR PRD-I and the second histidine (position 69 [Fig. 5]) is not conserved in either the SrlR PRD-I or the SrlR PRD-II. We note, however, that there is a histidine found six residues downstream of the tyrosine at position 69 in the S. mutans SrlR PRD-II (data not shown). The S. mutans SrlR protein contains a third domain in the C-terminal region which is similar to the EIIAFru family of PTS proteins, especially around a site that is the phosphorylation signature sequence of these proteins (data not shown). The Bacillus transcriptional regulators LicR, MtlR, YjdC, and YdaA also contain a similar region (11, 26).
FIG. 5.
Alignment of the PRD-I and -II domains from the YdaA, LicR, and YjdC proteins from B. subtilis (Bs); the MtlR protein from B. stearothermophilus (Bt); and the SrlR proteins from S. mutans LT11 (Sm). Identical residues present in all domains at positions 14 and 61 are boxed, positions with similar residues in at least five domains are indicated by a period, and positions with identical residues in at least five domains are indicated by an asterisk. Positions 7, 14, 61, and 69 of the alignment (see text) are indicated at the top.
Nucleotide sequence accession number.
The sequence of the region depicted in Fig. 1 has been submitted to the GenBank database under accession no. AF132127.
Acknowledgments
This study was supported by grants to I.R.H. from the Medical Research Council of Canada (MT-3546) and to A.L.H. from NIH/NIDCR (DE10890).
We thank Christopher Cote for his excellent technical assistance.
REFERENCES
- 1.Aldridge P, Metzger M, Geider K. Genetics of sorbitol metabolism in Erwinia amylovora and its influence on bacterial virulence. Mol Gen Genet. 1997;256:611–619. doi: 10.1007/s004380050609. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birkhed D, Edwardsson S, Kalfas S, Svensäter G. Cariogenicity of sorbitol. Swed Dent J. 1984;8:147–154. [PubMed] [Google Scholar]
- 4.Bowden G H W. Which bacteria are cariogenic in humans? In: Johnson N M, editor. Dental caries. 1. Markers of high and low risk groups and individuals. Cambridge, United Kingdom: Cambridge University Press; 1991. pp. 266–286. [Google Scholar]
- 5.Boyd D, Cvitkovitch D G, Hamilton I. Sequence and expression of the genes for HPr (ptsH) and enzyme I (ptsI) of the phosphoenolpyruvate-dependent phosphotransferase transport system from Streptococcus mutans. Infect Immun. 1994;62:1156–1165. doi: 10.1128/iai.62.4.1156-1165.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cvitkovitch D G, Boyd D, Thevenot T, Hamilton I. Glucose transport by a mutant of Streptococcus mutans unable to accumulate sugars via the phosphoenolpyruvate phosphotransferase system. J Bacteriol. 1995;177:2251–2258. doi: 10.1128/jb.177.9.2251-2258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Débarbouille M, Martin-Verstraete I, Klier A, Rapoport G. The transcriptional regulator LevR of Bacillus subtilus has domains homologous to both s54- and phosphotransferase system-dependent regulators. Proc Natl Acad Sci USA. 1991;88:2212–2216. doi: 10.1073/pnas.88.6.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dills S S, Seno S. Regulation of hexitol catabolism in Streptococcus mutans. J Bacteriol. 1983;153:861–866. doi: 10.1128/jb.153.2.861-866.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorke B, Rak B. Catabolite control of Escherichia coli regulatory protein BblG activity by antagonistically acting phosphorylations. EMBO J. 1999;18:3370–3379. doi: 10.1093/emboj/18.12.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamilton I R, Svensater G. Sorbitol inhibition of glucose metabolism by Streptococcus sanguis 160. Oral Microbiol Immunol. 1991;6:151–159. doi: 10.1111/j.1399-302x.1991.tb00470.x. [DOI] [PubMed] [Google Scholar]
- 11.Henstra S A, Tolner B, ten Hoeve Duurkens R H, Konings W N, Robillard G T. Cloning, expression, and isolation of the mannitol transport protein from the thermophilic bacterium Bacillus stearothermophilus. J Bacteriol. 1996;178:5586–5591. doi: 10.1128/jb.178.19.5586-5591.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lengeler J. Mutations affecting transport of the hexitols, d-mannitol, d-glucitol, and galacitol in Escherichia coli K-12: isolation and mapping. J Bacteriol. 1973;124:26–47. doi: 10.1128/jb.124.1.26-38.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lunsford R. A Tn4001 delivery system for Streptococcus gordonii (Challis) Plasmid. 1995;33:153–157. doi: 10.1006/plas.1995.1016. [DOI] [PubMed] [Google Scholar]
- 14.Mytelka D S, Chamberlin M J. Analysis and supression of DNA pauses associated with a trinucleotide consensus. Nucleic Acids Res. 1996;24:2774–2781. doi: 10.1093/nar/24.14.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perry D, Wondrack L M, Kuramitsu H K. Genetic transformation of putative cariogenic properties in Streptococcus mutans. Infect Immun. 1983;41:722–727. doi: 10.1128/iai.41.2.722-727.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reizer J, Reizer A, Yamada M, Saier M H., Jr The glucitol permease of Escherichia coli: a tripartite permease of the phosphotransferase system. Microbiology. 1998;144:1463–1464. doi: 10.1099/00221287-144-6-1463. [DOI] [PubMed] [Google Scholar]
- 17.Saier M H, Jr, Reizer J. The bacterial phosphotransferase system: new frontiers 30 years later. Mol Microbiol. 1994;13:755–764. doi: 10.1111/j.1365-2958.1994.tb00468.x. [DOI] [PubMed] [Google Scholar]
- 18.Schnetz K, Stülke J, Gertz S, Krüger S, Kreig M, Hecker M, Rak B. LicT, a Bacillus subtilis transcriptional antiterminator protein of the BlgG family. J Bacteriol. 1996;178:1971–1979. doi: 10.1128/jb.178.7.1971-1979.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slee A M, Tanzer J M. The repressible metabolism of sorbitol (d-glucitol) by intact cells of the oral plaque-forming bacterium Streptococcus mutans. Arch Oral Biol. 1983;28:830–845. doi: 10.1016/0003-9969(83)90041-9. [DOI] [PubMed] [Google Scholar]
- 20.Stülke J, Arnaud M, Rapoport G, Martin-Verstraete I. PRD—a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol Microbiol. 1998;28:865–874. doi: 10.1046/j.1365-2958.1998.00839.x. [DOI] [PubMed] [Google Scholar]
- 21.Svensater G, Hamilton I R. Sorbitol transport by Streptococcus sanguis 160. Oral Microbiol Immunol. 1991;6:160–168. doi: 10.1111/j.1399-302x.1991.tb00471.x. [DOI] [PubMed] [Google Scholar]
- 22.Tangney M, Brehm J K, Minton N P, Mitchell W J. A gene system for glucitol transport and metabolism in Clostridium beijerinckii NCIMB 8052. Appl Environ Microbiol. 1998;64:1612–1619. doi: 10.1128/aem.64.5.1612-1619.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tao L, MacAlister T J, Tanzer J M. Transformation efficiency of EMS-induced mutants of Streptococcus mutans of altered cell shape. J Dent Res. 1993;72:1032–1039. doi: 10.1177/00220345930720060701. [DOI] [PubMed] [Google Scholar]
- 24.Tao L, Sutcliffe I C, Russell R R B, Feretti J J. Transport of sugars, including sucrose, by the msm transport system of Streptococcus mutans. J Dent Res. 1993;72:L1386–1390. doi: 10.1177/00220345930720100701. [DOI] [PubMed] [Google Scholar]
- 25.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tobisch S, Glaser G, Krüger S, Hecker M. Identification and characterization of a new β-glucoside utilization system in Bacillus subtilis. J Bacteriol. 1997;179:496–506. doi: 10.1128/jb.179.2.496-506.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tortosa P, Aymerich S, Lindner C, Saier M H, Jr, Reizer J, Le Coq D. Multiple phosphorylation of SacY, a Bacillus subtilis transcriptional antiterminator negatively controlled by the phosphotransferase system. J Biol Chem. 1997;272:17230–17237. doi: 10.1074/jbc.272.27.17230. [DOI] [PubMed] [Google Scholar]
- 28.Vadeboncoeur C, Pelleter M. The phosphoenolpyruvate:sugar phosphotransferase system of oral streptococci and its role in the control of sugar metabolism. FEMS Microbiol Rev. 1997;19:187–201. doi: 10.1111/j.1574-6976.1997.tb00297.x. [DOI] [PubMed] [Google Scholar]
- 29.Wehmeier U F, Lengeler J W. Sequence of the sor-operon for l-sorbose utilization from Klebsiella pneumoniae KAY2026. Biochim Biophys Acta. 1994;1208:348–351. doi: 10.1016/0167-4838(94)90124-4. [DOI] [PubMed] [Google Scholar]
- 30.Yamada M, Saier M H., Jr Glucitol-specific enzymes of the phosphotransferase system in Escherichia coli. Nucleotide sequence of the gut operon. J Biol Chem. 1987;262:5455–5463. [PubMed] [Google Scholar]
- 31.Yamada M, Saier M H., Jr Positive and negative regulators for glucitol (gut) operon expression in Escherichia coli. J Mol Biol. 1988;203:569–583. doi: 10.1016/0022-2836(88)90193-3. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto Y, Sato Y, Takahashi-Abbe S, Abbe K, Yamada T, Kizaki H. Cloning and sequence analysis of the pfl gene encoding pyruvate formate-lyase from Streptococcus mutans. Infect Immun. 1996;64:385–391. doi: 10.1128/iai.64.2.385-391.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye R, Rehemtulla S N, Wong S-L. Glucitol induction in Bacillus subtilis is mediated by a regulatory factor, GutR. J Bacteriol. 1994;176:3321–3327. doi: 10.1128/jb.176.11.3321-3327.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye R, Wong S-L. Transcriptional regulation of the Bacillus subtilis glucitol dehydrogenase gene. J Bacteriol. 1994;176:3314–3320. doi: 10.1128/jb.176.11.3314-3320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]