Abstract
WbpM is a highly conserved protein involved in synthesis of the O antigens of Pseudomonas aeruginosa. Homologues of this protein have been identified in a large number of bacteria, and they can be divided into two subfamilies: subfamily 1, including WbpM, contains large proteins (∼600 amino acids), while subfamily 2, typified by HP0840 (FlaA1) of Helicobacter pylori, contains smaller proteins (∼350 amino acids) homologous to the C termini of proteins in subfamily 1. Analysis of knockout mutants of wbpM in P. aeruginosa serotypes O3, O10, O15, and O17 showed that although all 20 serotypes of P. aeruginosa possess wbpM, it is not universally required for O-antigen biosynthesis. Homologous genes from Bordetella pertussis (wlbL), Staphylococcus aureus (cap8D), and H. pylori (flaA1) complemented a P. aeruginosa O5 wbpM mutant to various degrees. These conserved proteins may represent interesting targets for the design of inhibitors of bacterial exopolysaccharide biosynthesis.
Exopolysaccharides of pathogenic bacteria, including lipopolysaccharides (LPS), lipooligosaccharides, and capsules, play a major role in virulence (reviewed in references 9, 11, 29, and 32). We are interested in identifying enzymes that are conserved amongst various exopolysaccharide biosynthetic pathways in order to provide targets for the rational design of inhibitor molecules. WbpM was previously identified as being a highly conserved and essential protein for the biosynthesis of the O antigens of serotypes O5 and O6 of Pseudomonas aeruginosa, although these molecules have different sugar compositions (5, 7). A gapped BLASTP search (3) of the GenBank database revealed a large number of WbpM homologues in both gram-negative and gram-positive bacteria, as well as in the Archeae (Table 1). These homologues could be divided into two subfamilies. Subfamily 1, including WbpM, contains large (approximately 600 amino acids) proteins with two distinct domains, with an NAD+ or NADP+ binding motif in the C terminus, and often a second motif in the N terminus (7). The other subfamily, embodied by the product of the Helicobacter pylori HP0840 gene (FlaA1) (36), consists of smaller (between 300 and 400 amino acids) proteins that are homologous to the C-terminal half of the larger proteins in the other subfamily (7) (Table 1). Interestingly, some species of bacteria contain examples of both subfamilies. In the capsular biosynthestic clusters of Staphylococcus aureus serotypes 5 and 8, contiguous large (capD) and small (capE) wbpM homologues are present in the same operon (29, 30, 31), whereas S. aureus serotype 1 has only capD (21). P. aeruginosa serotype O11 has recently been shown to contain both WbpM and a subfamily 2 homologue, WbjB (10).
TABLE 1.
WbpM and its homologues from other bacteria
| Subfamily and protein | Organism | Length (aa)c | % Similarity to WbpMO5d | Putative function | Accession no. (reference or source) |
|---|---|---|---|---|---|
| 1a | |||||
| WbpM | Pseudomonas aeruginosa serotype O5 | 665 | 100 | Fuc2NAc biosynthesis | U50396 (7) |
| WbpMO6 | Pseudomonas aeruginosa serotype O6 | 665 | 98 | Fuc2NAc biosynthesis | AF035937 (5) |
| WbpMO11 | Pseudomonas aeruginosa serotype O11 | 665 | 97 | Fuc2NAc biosynthesis | U44089, AF147795 (10) |
| WbcP (TrsG) | Yersinia enterocolitica serotype O:3 | 638 | 65 | Galactose modification | S51266 (34) |
| WlbL | Bordetella bronchiseptica | 624 | 61 | Nucleotide sugar dehydratase or epimerase | AJ007747 (direct submission) |
| WlbL (BplL) | Bordetella pertussis | 624 | 61 | FucNAcMe biosynthesis | S70683 (1) |
| WlaL, PglF | Campylobacter jejuni | 590 | 52 | Unknown; protein glycosylation | Y11648 (15), AF108897 (direct submission) |
| RfbV | Vibrio cholerae serotype O1 | 621 | 64 | Unknown | Y07788 (13) |
| ORF22-30 | Vibrio cholerae serotype O22 | 646 | 65 | Unknown | AB012957 (direct submission) |
| ORF10 | Vibrio cholerae serotype O139 | 646 | 66 | Epimerase or dehydratase | U47057 (8) |
| CapD | Staphylococcus aureus serotype 1 | 599 | 59 | Type 1 capsule synthesis | U10927 (22) |
| Cap5D | Staphylococcus aureus serotype 5 | 607 | 56 | Unknown | U81973 (31) |
| Cap8D | Staphylococcus aureus serotype 8 | 607 | 56 | Unknown | U73374 (29) |
| LpsB | Rhizobium etli | 683 | 65 | dTDP-glucose-4,6-dehydratase | U56723 (direct submission) |
| PglD | Neisseria meningitidis | 636 | 61 | Pilin glycosylation | AF014804 (direct submission) |
| WbiI | Burkholderia pseudomallei | 637 | 54 | Epimerase/dehydratase | AF0064070 (12) |
| TP0077 | Treponema pallidum | 538 | 55 | Capsular polysaccharide biosynthesis protein | AE001192 (14) |
| TM1548 | Thermotoga maritima | 605 | 53 | Unknown | AE001801 (26) |
| YveM | Bacillus subtilis | 598 | 56 | Unknown | Z99121 (20) |
| 2b | |||||
| WbjB | Pseudomonas aeruginosa | 344 | 50 | O-antigen biosynthesis | AF147795 (10) |
| HP0840 (FlaA1) | Helicobacter pylori strain 26695 | 333 | 52 | Unknown | AE000595 (36) |
| JHP0778 | Helicobacter pylori strain J99 | 333 | 51 | Sugar nucleotide biosynthesis | AE001508 (2) |
| FlaA1 | Caulobacter crescentus | 331 | 51 | Unknown | U27301 (direct submission) |
| Cap5E | Staphylococcus aureus serotype 5 | 342 | 50 | Unknown | U81973 (31) |
| Cap8E | Staphylococcus aureus serotype 8 | 342 | 50 | Unknown | U73374 (31) |
| Protein D | Methanococcus jannaschii | 333 | 62 | Capsular biosynthetic protein | U67549 (6) |
| CapD | Rickettsia prowazekii | 341 | 51 | Unknown | AJ235271 (4) |
| KasD | Streptomyces kasugaensis | 329 | 42 | NDP-hexose 4,6-dehydratase | AB005901 (18) |
| Gdh | Saccharopolyspora erythraea | 329 | 42 | dTDP-d-glucose-4,6-dehydratase | L37354 (23) |
| OrfJ06 | Leptospira interrogans | 336 | 51 | Unknown | AF144879 (11) |
| OrfH10 | Leptospira borgpetersenii | 336 | 52 | Unknown | AF078135 (11) |
Subfamily 1 contains proteins of approximately 600 amino acids in length.
Subfamily 2 contains proteins under 360 amino acids in length.
aa, amino acids.
Similarity was the sum of identical and conserved amino acids, as determined by nonfiltered Gapped BLASTP searches (3). The similarities of subfamily 1 are to WbpMO5 in its entirety, while the similarities of subfamily 2 are to the 3′ half of WbpMO5, from approximately amino acid 290 to the end.
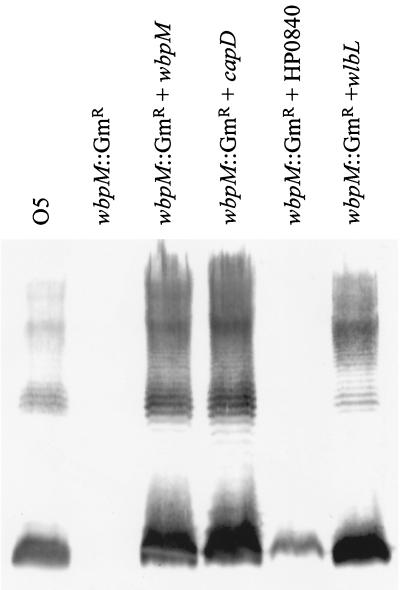
Although the specific function of these proteins has not yet been demonstrated at the biochemical level, they can be shown by complementation to be functionally homologous. The wbpM homologues from Bordetella pertussis strain BP536 (1), S. aureus serotype 8 (29), and H. pylori strain 26695 (36) were individually cloned into the broad-host-range vector, pUCP26 (37) (Table 2) and used to transform a P. aeruginosa O5 wbpM::Gmr mutant, which cannot synthesize B-band LPS (7). Silver-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western immunoblot analyses showed that the P. aeruginosa wbpM::Gmr mutant was complemented for O-antigen production by wlbL from B. pertussis (1), cap8D of S. aureus (29), and, partially, by HP0840 of H. pylori (36) (Fig. 1). The ability of HP0840 to partly complement a wbpM mutation demonstrates that the subfamily 2 homologues have activities related to those of subfamily 1. These data support the hypothesis previously stated by members of our group (7) that the larger proteins may have arisen through a fusion between two contiguous open reading frames, with the carboxy-terminal region, corresponding to subfamily 2, containing the putative active site. Further characterization of the larger proteins will permit the identification of the minimum structure required for function in P. aeruginosa.
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype, phenotype, or propertiesa | Source and/or reference |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| O3 | Wild type, serotype reference strain, B-band positive | ATCC 33350 (24) |
| O3 wbpM::Gmr | wbpM knockout mutant, B-band negative | This study |
| O10 | Wild type, serotype reference strain, B-band positive | ATCC 33357 (24) |
| O10 wbpM::Gmr | wbpM knockout mutant, B-band negative | This study |
| O15 | Wild type, serotype reference strain, B-band positive | ATCC 33362 (24) |
| O15 wbpM::Gmr | wbpM knockout mutant, B-band positive | This study |
| O17 | Wild type, serotype reference strain, B-band positive | ATCC 33364 (24) |
| O17 wbpM::Gmr | wbpM knockout mutant, B-band positive | This study |
| E. coli | ||
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δlac-proAB F′[tra D36 proAB+ lacIqlacZΔM15]; used as host strain for recombinant plasmids | 38 |
| SM10 | thi-1 thr leu tonA lacY supE recA RP4-2-Tcr::Mu, Kmr; mobilizes plasmids into P. aeruginosa strains via conjugation | 34 |
| Plasmids | ||
| pUCP26 | 4.9-kb pUC18-based broad-host-range vector; Tcr | 37 |
| pCRII-TOPO | T/A topoisomerase PCR cloning vector | Invitrogen |
| pFV163-26 | pUCP26 containing wbpMO5 on a 3.6-kb XbaI-SalI fragment from pFV115 (Burrows et al. [7]) | This study |
| pCAPCD-26 | pUCP26 containing the S. aureus cap8CD genes on a 4.0-kb SalI-PstI fragment from pCL7815 (Sau and Lee [29]) | This study |
| pWLBL-26 | B. pertussis wlbL gene amplified by PCR from a pLAFR1 clone containing the entire B. pertussis band A coding region (Allen and Maskell [1]) using primers with the following sequences: upstream, 5′ GCC CCG TCC CGT CCA GCC GC 3′; and downstream, 5′ CAT CGA CCC CAC CAA GGC ATC 3′, and then cloned into pCRII-TOPO and subcloned as a 2.1-kb BamHI-XbaI fragment into pUCP26 | This study |
| pHP0840-26 | pUCP26 containing the H. pylori 26695 ORF HP0840 gene (encodes putative protein FlaA1) subcloned as a 1.3-kb BamHI-SstI fragment from pUC18; the DNA fragment containing HP0840 was purchased from the ATCC/TIGR collection | This study |
| pEX18Ap | pEX100T-based suicide construct containing bla and sacB | 17 |
| pUCGM | Source of gentamicin resistance cassette; Apr, Gmr | 33 |
| pFV169-18Ap | pEX18Ap containing wbpMO5 on a 2.3-kb XbaI-HindIII fragment, with an ∼1-kb nonpolar gentamicin cassette (excised from pUCGM with SalI) inserted at a unique XhoI site with wbpM | This study |
ATCC, American Type Culture Collection; TIGR, The Institute for Genomic Research.
FIG. 1.
Complementation of a serotype O5 B-band-deficient mutant. Plasmids (see Table 2) containing wbpM or its homologues from S. aureus serotype 8 (capD), H. pylori 26695 (HP0840) and B. pertussis (wlbL) were used to complement a wbpM::Gmr mutant of P. aeruginosa PAO1 (7). B-band LPS prepared by the proteinase K digestion method of Hitchcock and Brown (15) was separated on SDS-PAGE, transferred to nitrocellulose, and detected using monoclonal antibody 18-19, which recognizes the O5 O unit (18). The fastest-migrating band represents the core plus one O unit, the most abundant species.
All 20 serotypes of P. aeruginosa contain wbpM, despite the fact that they produce structurally distinct O antigens (7, 25). Therefore, it was of interest to understand the role of WbpM in O-antigen synthesis in P. aeruginosa and to determine whether it is essential in serotypes other than O5 and O6 (5, 7). Members of our group proposed previously (7) that WbpM could be a C4 epimerase required for the conversion of UDP-N-acetyl-d-glucosamine (UDP-d-GlcNAc) to UDP-N-acetyl-d-galactosamine (UDP-d-GalNAc). However, results from studies on the biosynthesis of the UDP-d-GalNAc residues required for serotype O6 O-antigen synthesis showed that among the proteins encoded by the O-antigen biosynthesis genes, WbpP, and not WbpM, is the C4 epimerase involved in conversion of UDP-d-GlcNAc to UDP-d-GalNAc (5). Instead, WbpM is likely involved in the formation of the 6-deoxyaminohexose UDP-N-acetyl-d-quinovosamine (UDP–d-QuiNAc) (or UDP-6-deoxy-d-GlcNAc) residues of the serotype O6 O antigen. In serotype O5, the UDP-d-QuiNAc product of WbpM activity could be epimerized at C4 to form UDP-N-acetyl-d-fucosamine (UDP-d-Fuc2NAc) (or UDP-6-deoxy-d-GalNAc) by the UDP-galactose-4-epimerase (GalE) homologue, WbpK. This hypothesis is supported by the observation that WbpK is able to partly complement a Salmonella enterica serovar Typhimurium galE mutant (M. Matewish, H. L. Rocchetta, L. L. Burrows, R. Rahim, K. Pigeon, and J. S. Lam, Abstr. 60th N. Am. Cyst. Fibros. Meet., abstr. 374, 1998), suggesting it possesses C4 epimerase activity.
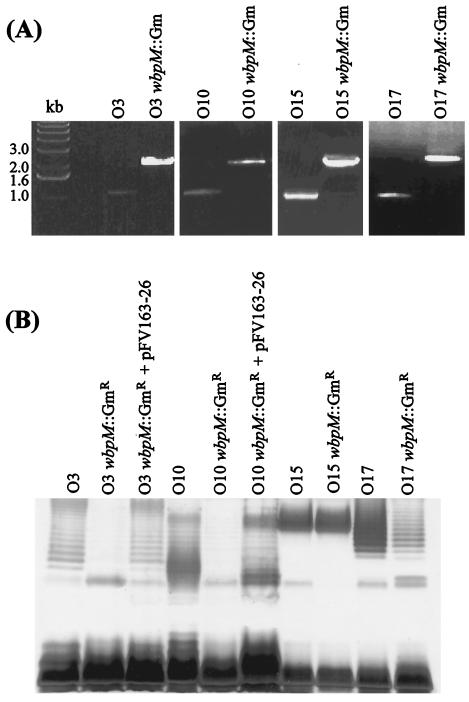
To further investigate the role of WbpM in synthesis of P. aeruginosa O antigens containing d-QuiNAc and its derivatives, we generated nonpolar chromosomal wbpM::Gmr mutations, using previously described methodology (7), in serotypes O3, O10, O15, and O17. These serotypes have primary O-antigen structures that are distinct from those of serotypes O5 and O6 (Table 3). The O antigen of serotype O10 contains d-QuiNAc, while serotype O3 contains a d-QuiNAc derivative, bacillosamine (4-amino-d-QuiNAc; shown in bold in Table 3). In contrast, the O antigens of O15 and O17 contain neither d-QuiNAc nor d-FucNAc. Briefly, a gentamicin-resistance cassette containing its own promoter sequence was inserted within a unique XhoI site approximately in the middle of the 1.9-kb wbpM gene of serotype O5, and the mutated gene was introduced first into the chromosome of serotype O5 to demonstrate that the mutation abrogated O-antigen synthesis (data not shown). The wbpM::Gmr construct was then introduced in the chromosomes of serotypes O3, O10, O15, and O17 by homologous recombination. To demonstrate the correct replacement of wild-type wbpM with the knockout construct, PCR using wbpM-specific primers flanking the XhoI site was performed. Upon agarose gel analysis, amplicons from the mutants were approximately 1 kb larger than those of the parental serotype strains, corresponding to the size of the Gmr cassette (Fig. 2A). Silver-stained SDS-PAGE analysis of LPS from the O3, O10, O15, and O17 parent strains and their isogenic wbpM mutants showed that synthesis of the serotype O3 and O10 O antigens containing 4-amino-d-QuiNAc {4-amino-2,4,6-trideoxy-N-acetylglucosamine [Bac(2NAc4N)]} and d-QuiNAc, respectively (Table 3), was abrogated by the loss of WbpM (Fig. 2B). The deficiency in B-band O-antigen production was restored in the serotype O3 and O10 wbpM::Gmr mutants by complementation with the wbpMO5 gene in trans (Fig. 2B). In contrast, synthesis of the serotype O15 and O17 O antigens that contain neither d-QuiNAc nor d-Fuc2NAc was not affected by the wbpM mutation (Fig. 2B). These data are consistent with our previous results (5, 7) showing wbpM is essential for the synthesis of the serotype O6 and O5 O antigens, containing d-QuiNAc and its C4 epimer d-FucNAc, respectively.
TABLE 3.
P. aeruginosa O-antigen structures
| P. aeruginosa IATS serotypea | O antigen structureb |
|---|---|
| O3c | [-6)-α-d-GlcNAc-(1-4)-α-l-GalNAcA-(1-3)-β-d-Bac(2NAc4N)-(1-2)-α-l-Rha-(1-] |
| 4 | | |
| (S)-CH3CH(OH)CH2CO | |
| O5 | [-4)-β-d-Man(2NAc3N)A-(1-4)-β-d-Man(2NAc3NAc)A-(1-3)-α-d-FucNAc-(1-] |
| 3| | |
| CH3C⩵NH | |
| O6 | [3)-α-l-Rha-(1-4)-α-d-GalNAcA-(1-4)-α-d-GalNFmA-(1-3)-α-d-QuiNAc-(1-] |
| 3 | 6| | |
| OAc NH2 | |
| O10 | [-3)-α-l-Rha-(1-4)-α-l-GalNAcA-(1-3)-α-d-QuiNAc-(1-] |
| O15 | [-4)-α-d-GalNAc-(1-2)-β-d-Ribf-(1-] |
| O17 | [-3)-β-d-ManNAc-(1-4)-α-l-Rha-(1-] |
IATS, International Antigenic Typing Scheme (23).
Based on reference 19. Abbreviations: Bac(2NAc4N), 4-amino-2,4,6-trideoxy-N-acetylglucosamine; FucNAc, 6-deoxy-N-acetylgalactosamine; GalNAcA, N-acetylgalactosaminuronic acid; GalNFmA, N-formylgalactosaminuronic acid; GlcNAc, N-acetylglucosamine; Man(2NAc3NAc)A, 2,3-dideoxy-2,3-di-N-acetylmannosaminuronic acid; QuiNAc, 6-deoxy-N-acetylglucosamine; Rha, rhamnose; Ribf, ribofuranose.
The Bac(2NAc4N) residue in the serotype O3 O antigen is substituted with an (S)-3-hydroxybutyryl group.
FIG. 2.
Analysis of wbpM::Gmr mutants of selected P. aeruginosa serotype strains. (A) Correct insertion of the gentamicin resistance cassette within wbpM was ascertained by PCR using the wbpM-specific primers flanking the unique XhoI site (upstream primer, 5′ AGGGTGGCTATCTATGGCGCGGGG 3′; downstream primer, 5′ AACGGGTGATGCTCGGGTGGGTGA 3′). The mutants showed a shift in the size of the amplicon of approximately 1 kb, corresponding to the size of the Gmr cassette. (B) LPS prepared from selected serotype strains, their wbpM::Gmr mutants, and their complemented mutants was analyzed by silver-stained SDS-PAGE. Mutants lacking B-band O antigen were complemented with the serotype O5 wbpM gene on pFV163-26. It is not clear why the O17 wbpM mutant produces somewhat less B-band LPS than the wild-type strain, but the banding pattern is very similar and its O antigen is genuine O17 as determined by slide agglutination with O17-specific antiserum (data not shown).
In summary, we have shown that WbpM is a member of a large and growing family of functionally homologous proteins that are involved in synthesis of exopolysaccharides in both gram-positive and gram-negative bacteria. WbpM is thought to play a role in the synthesis of the deoxyhexose, UDP-d-QuiNAc, and its derivatives, including UDP-d-Bac(2NAc4N) and UDP-d-FucNAc. Work is underway in our laboratory to provide biochemical evidence for the role of WbpM in the biosynthesis of these unusual sugars. Since homologues of WbpM occur in a number of medically significant bacteria, these proteins may be of interest as therapeutic targets.
Acknowledgments
This work was supported by grants to J.S.L. from the Medical Research Council of Canada (MT18647) and the Canadian Bacterial Diseases Network (a federal Network of Centres of Excellence). L.L.B. is the recipient of a Canadian Cystic Fibrosis Foundation Fellowship.
We thank D. Maskell and C. Y. Lee for providing clones containing wlbL and cap8D, respectively, and A. Clarke, M. Bélanger and C. Creuzenet for helpful discussions.
REFERENCES
- 1.Allen A, Maskell D. The identification, cloning and mutagenesis of a genetic locus required for lipopolysaccharide biosynthesis in Bordetella pertussis. Mol Microbiol. 1996;19:37–52. doi: 10.1046/j.1365-2958.1996.354877.x. [DOI] [PubMed] [Google Scholar]
- 2.Alm R A, Ling L S, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersson S G, Zomorodipour A, Andersson J O, Sicheritz-Ponten T, Alsmark U C, Podowski R M, Naslund A K, Eriksson A S, Winkler H H, Kurland C G. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 5.Bélanger M, Burrows L L, Lam J S. Functional analysis of genes responsible for synthesis of the B-band O antigen of Pseudomonas aeruginosa serotype O6 lipopolysaccharide. Microbiology. 1999;145:3505–3521. doi: 10.1099/00221287-145-12-3505. [DOI] [PubMed] [Google Scholar]
- 6.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 7.Burrows L L, Charter D F, Lam J S. Molecular characterization of the Pseudomonas aeruginosa serotype O5 (PAO1) B-band lipopolysaccharide gene cluster. Mol Microbiol. 1996;22:481–495. doi: 10.1046/j.1365-2958.1996.1351503.x. [DOI] [PubMed] [Google Scholar]
- 8.Comstock L E, Johnson J A, Michalski J M, Morris J G, Jr, Kaper J B. Cloning and sequence of a region encoding a surface polysaccharide of Vibrio cholerae O139 and characterization of the insertion site in the chromosome of Vibrio cholerae O1. Mol Microbiol. 1996;19:815–826. doi: 10.1046/j.1365-2958.1996.407928.x. [DOI] [PubMed] [Google Scholar]
- 9.Cryz S J, Jr, Pitt T L, Furer E, Germanier R. Role of lipopolysaccharide in virulence of Pseudomonas aeruginosa. Infect Immun. 1984;44:508–513. doi: 10.1128/iai.44.2.508-513.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dean C R, Franklund C V, Retief J D, Coyne M J, Jr, Hatano K, Evans D J, Pier G B, Goldberg J B. Characterization of the serogroup O11 O-antigen locus of Pseudomonas aeruginosa PA103. J Bacteriol. 1999;181:4275–4284. doi: 10.1128/jb.181.14.4275-4284.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Pena-Moctezuma A, Bulach D M, Kalambaheti T, Adler B. Comparative analysis of the LPS biosynthetic loci of the genetic subtypes of serovar Hardjo: Leptosprira interrogans subtype Hardjoprajitno and Leptospira borgpetersenii subtype Hardjobovis. FEMS Microbiol Lett. 1999;177:319–326. doi: 10.1111/j.1574-6968.1999.tb13749.x. [DOI] [PubMed] [Google Scholar]
- 12.DeShazer D, Brett P J, Woods D E. The type II O-antigenic polysaccharide moiety of Burkholderia pseudomallei lipopolysaccharide is required for serum resistance and virulence. Mol Microbiol. 1998;30:1081–1100. doi: 10.1046/j.1365-2958.1998.01139.x. [DOI] [PubMed] [Google Scholar]
- 13.Fallarino A, Mavrangelos C, Stroeher U H, Manning P A. Identification of additional genes required for O-antigen biosynthesis in Vibrio cholerae O1. J Bacteriol. 1997;179:2147–2153. doi: 10.1128/jb.179.7.2147-2153.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fraser C M, Norris S J, Weinstock G M, White O, Sutton G G, Dodson R, Gwinn M, Hickey E K, Clayton R, Ketchum K A, Sodergren E, Hardham J M, McLeod M P, Salzberg S, Peterson J, Khalak H, Richardson D, Howell J K, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton M D, Venter J C, et al. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 15.Fry B N, Korolik V, ten Brinke J A, Pennings M T, Zalm R, Teunis B J, Coloe P J, van der Zeijst B A. The lipopolysaccharide biosynthesis locus of Campylobacter jejuni 81116. Microbiology. 1998;144:2049–2061. doi: 10.1099/00221287-144-8-2049. [DOI] [PubMed] [Google Scholar]
- 16.Hitchcock P J, Brown T M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoang T T, Karkhoff-Schweizer R R, Kutchma A J, Schweizer H P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 18.Ikeno S, Tsuji T, Higashide K, Kinoshita N, Hamada M, Hori M. A 7.6 kb DNA region from Streptomyces kasugaensis M338-M1 includes some genes responsible for kasugamycin biosynthesis. J Antibiot (Tokyo) 1998;51:341–352. doi: 10.7164/antibiotics.51.341. [DOI] [PubMed] [Google Scholar]
- 19.Knirel Yu A, Kochetkov N K. Structure of lipopolysaccharides from gram-negative bacteria. III. Structure of O-specific polysaccharides. Biokhimiya. 1994;59:1784–1851. [PubMed] [Google Scholar]
- 20.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 21.Lam J S, Handelsman M Y C, Chivers T R, MacDonald L A. Monoclonal antibodies as probes to examine serotype-specific and cross-reactive epitopes of lipopolysaccharides from serotypes O2, O5, and O16 of Pseudomonas aeruginosa. J Bacteriol. 1992;174:2178–2184. doi: 10.1128/jb.174.7.2178-2184.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin W S, Cunneen T, Lee C Y. Sequence analysis and molecular characterization of genes required for the biosynthesis of type 1 capsular polysaccharide in Staphylococcus aureus. J Bacteriol. 1994;176:7005–7016. doi: 10.1128/jb.176.22.7005-7016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linton K J, Jarvis B W, Hutchinson C R. Cloning of the genes encoding thymidine diphosphoglucose 4,6- dehydratase and thymidine diphospho-4-keto-6-deoxyglucose 3,5-epimerase from the erythromycin-producing Saccharopolyspora erythraea. Gene. 1995;153:33–40. doi: 10.1016/0378-1119(94)00809-7. [DOI] [PubMed] [Google Scholar]
- 24.Liu P V, Wang S. Three new major somatic antigens of Pseudomonas aeruginosa. J Clin Microbiol. 1990;28:922–925. doi: 10.1128/jcm.28.5.922-925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu P V, Matsumoto H, Kusama H, Bergan T. Survey of heat-stable major somatic antigens of Pseudomonas aeruginosa. Int J Syst Bacteriol. 1983;33:256–264. [Google Scholar]
- 26.Nelson K E, Clayton R A, Gill S R, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Nelson W C, Ketchum K A, McDonald L, Utterback T R, Malek J A, Linher K D, Garrett M M, Stewart A M, Cotton M D, Pratt M S, Phillips C A, Richardson D, Heidelberg J, Sutton G G, Fleischmann R D, Eisen J A, Fraser C M, et al. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 27.Roberts I S. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu Rev Microbiol. 1996;50:285–315. doi: 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- 28.Roche R J, Moxon E R. Phenotypic variation of carbohydrate surface antigens and the pathogenesis of Haemophilus influenzae infections. Trends Microbiol. 1995;3:304–309. doi: 10.1016/s0966-842x(00)88959-3. [DOI] [PubMed] [Google Scholar]
- 29.Sau S, Lee C Y. Cloning of type 8 capsule genes and analysis of gene clusters for the production of different capsular polysaccharides in Staphylococcus aureus. J Bacteriol. 1996;178:2118–2126. doi: 10.1128/jb.178.7.2118-2126.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sau S, Sun J, Lee C Y. Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. J Bacteriol. 1997;179:1614–1621. doi: 10.1128/jb.179.5.1614-1621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sau S, Bhasin N, Wann E R, Lee J C, Foster T J, Lee C Y. The Staphylococcus aureus allelic genetic loci for serotype 5 and 8 capsule expression contain the type-specific genes flanked by common genes. Microbiology. 1997;143:2395–2405. doi: 10.1099/00221287-143-7-2395. [DOI] [PubMed] [Google Scholar]
- 32.Schnaitman C A, Klena J D. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol Rev. 1993;57:655–682. doi: 10.1128/mr.57.3.655-682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schweizer H P. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques. 1993;15:831–834. [PubMed] [Google Scholar]
- 34.Simon R, Priefer U, Pühler A. A broad-host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 35.Skurnik M, Venho R, Toivanen P, al-Hendy A. A novel locus of Yersinia enterocolitica serotype O:3 involved in lipopolysaccharide outer core biosynthesis. Mol Microbiol. 1995;17:575–594. doi: 10.1111/j.1365-2958.1995.mmi_17030575.x. [DOI] [PubMed] [Google Scholar]
- 36.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D, Venter J C, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 37.West S E H, Schweizer H P, Dall C, Sample A K, Runyen-Janecky L J. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and the sequence of the region required for their replication in Pseudomonas aeruginosa. Gene. 1994;128:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 38.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]