Abstract
OBJECTIVE:
Interest in using bedside C-reactive protein (CRP) and ferritin levels to identify patients with hyperinflammatory sepsis who might benefit from anti-inflammatory therapies has piqued with the COVID-19 pandemic experience. Our first objective was to identify patterns in CRP and ferritin trajectory among critically ill pediatric sepsis patients. We then examined the association between these different groups of patients in their inflammatory cytokine responses, systemic inflammation, and mortality risks.
DESIGN:
A prospective, observational cohort study. Plasma CRP (mg/dL), ferritin (ng/mL), and 31 cytokine levels were measured at two timepoints during sepsis (median Day 2 and Day 5). Group-based multi-trajectory models (GBMTM) identified groups of children with distict patterns of CRP and ferritin.
SETTING:
Nine pediatric intensive care units in the United States.
PATIENTS:
Children with sepsis and organ failure.
MEASUREMENTS AND MAIN RESULTS:
Two hundred and fifty-five children were enrolled. Five distinct clinical multi-trajectory groups were identified. Group 1 had normal CRP and ferritin levels (n = 8; 0% mortality); Group 2 had high CRP levels that became normal, with normal ferritin levels throughout (n = 80; 5% mortality); Group 3 had high ferritin levels alone (n=16; 6% mortality); Group 4 had very high CRP levels, and high ferritin levels (n = 121; 11% mortality); and, Group 5 had very high CRP and very high ferritin levels (n = 30; 40% mortality). Cytokine responses differed across the 5 groups, with ferritin levels correlated with macrophage inflammatory protein 1α levels and CRP levels reflective of many cytokines.
CONCLUSIONS:
Bedside CRP and ferritin levels can be used together to distinguish groups of children with sepsis who have different systemic inflammation cytokine responses and mortality risks. These data suggest future potential value in personalized clinical trials with specific targets for anti-inflammatory therapies.
Keywords: Sepsis, Immunomodulation, C-reactive protein, Ferritin, Children, Multiple Organ Failure
A recent audit of 109 million death records found that sepsis contributes to 1 in 5 deaths globally with one third occurring in the pediatric population, predominantly in resource poor settings.1 Current management of pediatric sepsis includes early use of appropriate antibiotics and cardiovascular support with little attention to inflammation. Interest in therapeutic targeting of systemic inflammation in sepsis has been piqued by the SARS CoV2 pandemic. Two inexpensive, readily available, non-specific biomarkers, C-reactive protein (CRP) and ferritin, are commonly used to identify systemic inflammation and track therapies directed to systemic inflammation unrelated to infection.2 In two single center studies we reported that hospitalized children overall, and septic children in particular, have: 1) little to no risk of mortality when neither biomarker is elevated; 2) increased mortality risk when peak CRP alone or peak ferritin alone are high; and 3) very high mortality risk when both peak CRP and peak ferritin levels are high.3,4 These observations raise the possibility that paired measurement of CRP and ferritin levels could provide a novel metric for systemic inflammation mortality risk.
In this manuscript we provide the first multicenter assessment using the combination of CRP and ferritin levels to identify systemic inflammation mortality risk in pediatric sepsis. We also use a novel statistical method termed group-based multi-trajectory modeling (GBMTM) to take full advantage of our multivariable longitudinal data.5 The primary purpose of this study is to identify independently distinct systemic inflammation groups based on their trajectory in CRP and ferritin level during sepsis. We also test the hypothesis that designation by group is also associated with certain discriminate inflammatory cytokine profiles, and mortality risks.
Methods
Study protocol and definitions
The present manuscript employs multi-trajectory modeling of CRP and ferritin to derive novel pediatric sepsis groups in children previously enrolled and reported in the 9 center Eunice Kennedy Shriver National Institutes of Child Health and Development Collaborative Pediatric Critical Care Research Network Phenotyping Pediatric Sepsis induced Multiple Organ Failure (PHENOMS) study who had two sequential CRP and ferritin measured in a week.6,7 The study was approved by the central Institutional Review Board (IRB) of the University of Utah, IRB #70976. Written informed consent was obtained from one or more parents/guardians for each child. Assent was garnered when the child was able. Patients were enrolled from 2015–2017.
The details of the clinical study protocol have been previously published. 6, 7 In brief, the children qualified for enrollment in the study if they 1) were between the ages of 44 weeks gestation to 18 years of age; 2) were suspected of having infection with two or more of four systemic inflammatory response criteria;8 3) had one or more organ failures as defined by modified criteria of Proulx et al;9 and 4) had an indwelling arterial line or central venous catheter for blood drawing. The children were excluded from enrollment if there was lack of commitment to aggressive care.
Clinical data were assessed daily until 28 days or discharge from the PICU. These included demographic variables (age, sex, previously healthy status, post-op status), pediatric risk of mortality score 3 (PRISM-3) to assess severity of illness at admission, and organ failures (Central Nervous System = Glasgow Coma Scale score < 12 not explained by use of sedation; Cardiovascular = requirement for vasoactive agents for systolic blood pressure < 5th percentile for age; Respiratory = paO2/FiO2 ratio < 300 requiring mechanical ventilation; Renal = oliguria and serum creatinine > 1mg/dL; Hepatic = alanine aminotransferase > 100 and bilirubin > 1 mg/dL; Hematologic = platelet count < 100K and international normalized ratio > 1.5) to assess organ failure index or the number of organ failures.4,6,7 An organ failure index was generated as an integer score with 1 point assigned for each of the above organ failures. PRISM-3 was calculated using the worst physiologic values within a modified six-hour window ranging from two hours prior ICU admission to four hours post ICU admission.
Blood samples were obtained after 24 hours of sepsis associated organ failure (because CRP and ferritin levels peak at 12 – 24 hours)2 and twice a week thereafter. The blood samples were analyzed for measurement of CRP and ferritin as well as 31 additional cytokine biomarkers, including an assay for ADAMTS13 activity which is a marker of thrombotic microangiopathy when < 57% of control, and an assessment of immunoparalysis defined as whole blood ex vivo TNFα response to endotoxin < 200 pg/mL.5–7,10,11 Plasma for cytokine measurement was divided into three assays. IL-18, IL-18BP, and CXCL9 were measured at 25-fold dilution.12 IFNβ, sCD163, and IL-22 were measured by Bioplex inflammatory flex-set assay per manufacturer’s instructions (Bio-Rad). The remainder were measured by Bioplex Group I/II flex-set assay (Bio-Rad). All cytokines were measured on a BioPlex 200 System (Bio-Rad). The functional assays were measured as previously described.4,6,7,10,11 The biomarkers were used to generate cytokine response profile heat maps and also to confirm multiple organ failure (MOF) outcomes characterized as immunoparalysis associated MOF (immunoparalysis beyond three days with two or more organ failures),4,6,7,10,11 and macrophage activation syndrome (MAS, defined by ferritin > 500 ng/mL with platelet count < 100×109/L, INR > 1.5, ALT > 100 IU/L and bilirubin > 1 mg/dL).4,6,7,13,14
Statistical Methods
Study sample collection occurred twice weekly with over 90% of the first two scheduled samples occurring within the first week of sepsis induced organ failure. Recognizing that a significant number of patients were discharged, died, or met blood draw limits before later collection periods, we chose to build early models across subjects’ first two sampling time points. A modern statistical approach to finite mixture modeling, GBMTM, was used to categorize pediatric sepsis patients into distinct groups or latent classes that demonstrated similar CRP and ferritin trajectories over the first two study sampling periods.5 Multi-trajectories for a different number of specified groups (1 through 5) were then constructed and evaluated. Statistical measures of model fit included Bayesian Information Criteria (BIC) and Akaike Information Criteria (AIC) and were evaluated alongside clinical interpretations for each set of biomarker trajectories to determine the most appropriate number of trajectory groups to present and analyze (Supplemental Table 1). The set of five-group multi-trajectories appeared to offer the most clinical information, as well as the best AIC and BIC values. Five groups were accordingly chosen for further analysis with patients’ clinical outcomes and other clinically meaningful biological measures. GBMTM was performed using the SAS based software implementation found at https://www.andrew.cmu.edu/user/bjones/index.htm. Analyses were performed using SAS 9.4 (SAS Institute; Cary, NC) software with p-values < 0.05 deemed statistically significant unless stated otherwise.
Distributions of combined CRP and ferritin measurements at the first two sampling periods were explored by constructing a two-dimensional bagplot. A bagplot is analogous to a bivariate boxplot.15 A center point is calculated based on determination of the Tukey depth, through which any biplane will split the data approximately in half, and a darker-shaded, inner “bag” is plotted which encloses approximately 50% of data points. An outer “fence” encompassing a distribution of points is then constructed by inflating the bag by a specified factor. Lines connect the edge of the inner bag to points between the bag and outer fence. A factor of 3.0 was selected for this analysis such that the fence would encompass approximately 97% of points if the data were of a normal distribution. Both variables were converted to log scale for data exploration given high skew of the untransformed distributions.
All cytokine values are grouped based on multi-trajectory groups (1–5) and sampling time points (the first two samples collected according to the PHENOMS sampling schedule and reported as Sample Day 1 and Sample Day 2) to compare between groups and sampling days. In the heatmap, the color of each cell represents the log ratio of the median biomarker value comparing each group to the entire cohort study. Red represents a greater median biomarker value for that group compared with the median for the entire study cohort, whereas blue represents lower median biomarker values compared with the median for the entire study cohort. Cytokines were clustered by the hierarchical clustering method. Heatmap analyses were performed with R version 3.6.2.
The primary outcome was death in the PICU (PICU mortality). Secondary outcomes included development of organ failure(s); length of stay in the PICU; and as defined previously in this methods section subsequent development of immunoparalysis,4,6,7,11,16 and MAS.4,6,7,13,14,16 For exploratory outcomes related to the multi-trajectory groups, we also evaluated use of invasive organ support therapies including mechanical ventilation (MV), continuous renal replacement therapies (CRRT), and extracorporeal membrane oxygenation (ECMO). CRRT and ECMO were used according to local site convention and protocols. Exploratory multivariable models were also constructed to examine associations between trajectory groups and mortality, adjusting for age, PRISM-3, and immunocompromised status.
Results
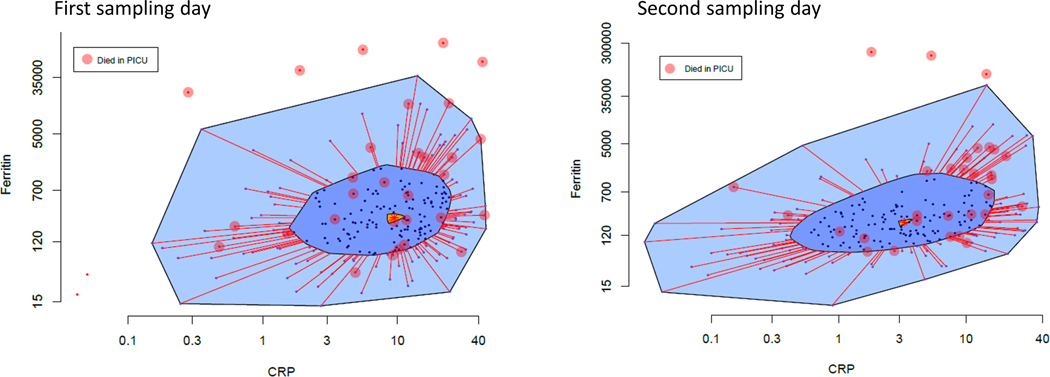
Two hundred and fifty five of the 401 patients originally enrolled in the PHENOMS study had paired CRP and ferritin twice in the first week and were selected for analysis. One hundred and forty-six patients were missing the first or second samples in the first week of enrollment, 82 of which left the PICU alive (n=76) or died (n=6) before or on the same study day the second sample was scheduled to be collected (Supplemental Figure 1). Thirty of the 255 children with two samples died in the PICU (12% mortality). Figure 1 shows the bagplot of combined CRP and ferritin measurements on the day of first sampling (Figure 1a - median day 2 of sepsis; [IQR day 1 - day 3]) and the day of second sampling (Figure 1b - median day 5 of sepsis; IQR day 5 - day 6]). The median patient on the first sampling day (Tukey Median) had a CRP of 9.44 mg/dL and ferritin of 274 ng/mL. The median patient (Tukey Median) on the second sampling day had a CRP of 3.27 mg/dL and ferritin of 206 ng mL. All five patients exhibiting extreme hyperinflammation, indicated by being outside the outer bag and therefore beyond the 97th percentile of systemic inflammation distribution who had CRP or ferritin levels greater than the Tukey Median CRP or ferritin levels, died in the PICU. Patients representing the lower half of combined CRP and ferritin inflammation (inner bag) at first sampling had 8% mortality compared to 16% mortality in those with higher combined CRP and ferritin inflammation (outside the inner bag) (Figure 1 top panel).
Figure 1 –
Bagplot of combined CRP and ferritin levels at first and second sampling (median day 2 and day 5 of sepsis). Figure 1a – Bagplot on first sampling day shows Tukey median CRP 9.44 mg/dL and ferritin 273.61 ng/mL. Figure 1b – Bagplot on second sampling day shows Tukey median decreased to CRP 3.27 mg/dL and ferritin 206 ng/ mL. All children with CRP and/or ferritin levels beyond the outer bag (> 97% inflammation) and > Tukey median CRP and ferritin levels died (circles).
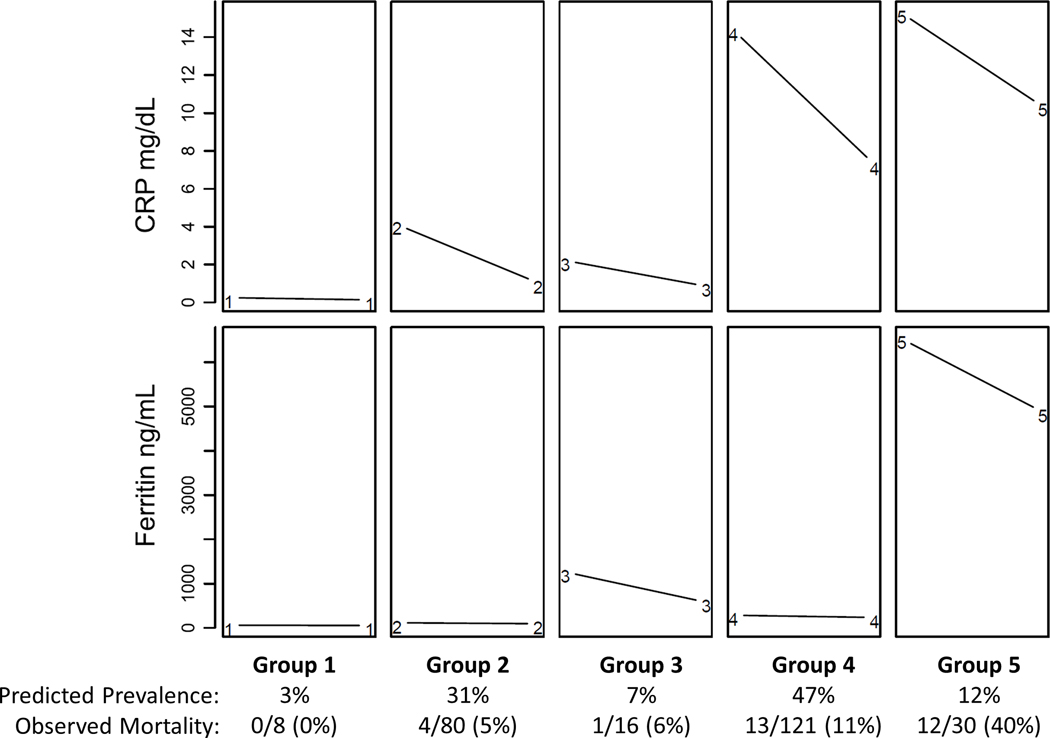
Figure 2 shows the results obtained using group-based trajectory modeling of multiple outcomes to assess the paired CRP and ferritin levels. The results presented in Figure 2 are model generated estimates and are therefore representative but not identical to the values of CRP and ferritin observed in the cohort. Five distinct multiple trajectory groups were derived based on BIC and AIC criteria (Supplemental Table 1) with probability modeling and clinical relevance assessment: Group 1 showed normal CRP and ferritin levels (n = 8); Group 2 had high CRP alone (median 4 mg/dL) that normalized to < 1 mg/dL (n = 80); Group 3 had high ferritin alone (median 908 ng/mL) that remained increased (median 605 ng/mL) (n = 16); Group 4 had very high CRP alone (median 16 mg/dL) that remained high (median 8 mg/dL) (n = 121); and Group 5 had both very high CRP levels (median 17 mg/dL) and very high ferritin levels (median 4,058 ng/mL) that remained very high at 11.12 mg/dL and 3,926 ng /mL, respectively (n = 30).
Figure 2 –
Predicted prevalence of five distinct CRP-ferritin Multi-trajectory Groups – CRP and Ferritin levels both low and normal in Group 1, CRP levels alone increased in Group 2, Ferritin levels alone increased in Group 3, only CRP levels very high in Group 4, both CRP and ferritin levels very high in Group 5. Group 1 = 3% (0% mortality); Group 2 = 31% (5 % mortality); Group 3 = 7% (6 % mortality); Group 4 = 47% (11% mortality); and Group 5 = 12% (40% mortality).
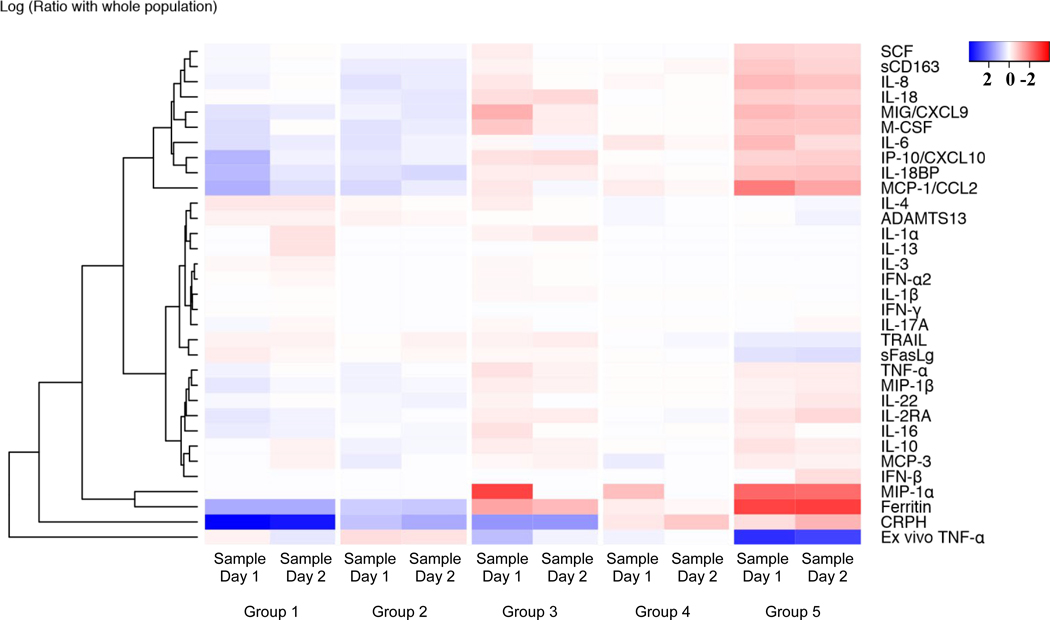
Table 1 shows the clinical characteristics of Groups 1 through 5. Age, physiological severity of illness, cancer diagnosis, and transplantation diagnosis differed across the groups (p < 0.05; Table 1). Figure 3 shows the heatmap of 29 inflammatory cytokines, CRP and ferritin, and two functional assays, ADAMTS13 activity and assessment of immunoparalysis. Hierarchical cluster analysis shows that ferritin levels correlated with MIP-1-α whereas, CRP reflected many cytokines (y – axis Figure 3). Supplemental Tables 2 and 3 show the median [IQR] values of these tests at sampling day 1 and sampling day 2.
Table 1.
Clinical characteristics by CRP-ferritin multi- trajectory group1
| CRP-ferritin Multi- Trajectory Group | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| Group 1 (N = 8) | Group 2 (N = 80) | Group 3 (N = 16) | Group 4 (N = 121) | Group 5 (N = 30) | P-value | |
| Age at onset of organ failure (in years) | 1 [0, 2] | 3 [1, 8] | 4 [0, 11] | 8 [3, 14] | 10 [6, 13] | <.0012 |
|
| ||||||
| Male | 3 (38%) | 42 (53%) | 9 (56%) | 75 (62%) | 14 (47%) | 0.3533 |
|
| ||||||
| PRISM | 6 [0, 12] | 7 [3, 12] | 15 [8, 22] | 9 [5, 15] | 9 [5, 13] | 0.0192 |
|
| ||||||
| OFI on Day 0 | 1 [1, 2] | 2 [1, 2] | 3 [1, 3] | 2 [1, 2] | 2 [1, 3] | 0.0992 |
|
| ||||||
| Cancer | 0 (0%) | 2 (3%) | 3 (19%) | 6 (5%) | 15 (50%) | <.0013 |
|
| ||||||
| Transplant | 0 (0%) | 2 (3%) | 2 (13%) | 5 (4%) | 11 (37%) | <.0013 |
|
| ||||||
| Chronic illness | 4 (50%) | 41 (51%) | 11 (69%) | 64 (53%) | 23 (77%) | 0.1043 |
|
| ||||||
| INFECTIONS AT ELIGIBILITY4 | ||||||
|
| ||||||
| Bacterial | 3 (38%) | 22 (28%) | 4 (25%) | 55 (45%) | 13 (43%) | 0.2753 |
|
| ||||||
| Viral | 4 (50%) | 35 (44%) | 4 (25%) | 25 (21%) | 8 (27%) | 0.0883 |
|
| ||||||
| Fungal | 0 (0%) | 0 (0%) | 0 (0%) | 2 (2%) | 1 (3%) | 0.6273 |
Only subjects with the first two samples were included in biomarker trajectory analyses.
Kruskal-Wallis test.
Fisher’s exact test (Monte Carlo approximation).
Documented infections at the time of study eligibility are reported. The statistical tests compare documented vs. suspected infections.
Figure 3 –
Heatmap of CRP, ferritin and 31 inflammatory biomarkers among CRP-ferritin multi-trajectory Groups 1–5 on sampling days 1 and 2 (median day 2 and day 5 of sepsis). Legend shows color coding for log odds ratio at −2, 0, and +2 comparing group median to overall median, blue median less than whole cohort median, red median greater than whole cohort median (eg. Highest inflammation and lowest ADAMTS13, TRAIL, and whole blood ex vivo TNFα response observed in Group 5). Ferritin co-regulates with MIP-1-a and CRP is reflective of many cytokines according to hierarchical cluster analysis on Y axis.
The cytokines SCF, sCD163, IL-8, IL-18, CXCL9, M-CSF, IL-6, IP10/XCXCL10, IL-18BP, MCP-1/CCL2, TNFα, MIP-1-β, IL-22, IL2RA, IL-16, IL-10, MCP-3, IFNβ, MIP-1-α and the non-specific systemic inflammation biomarkers CRP and ferritin differed across the five groups and were increased in Group 5 (red) compared to Group 1 (blue). The macrophage activation inhibitor TRAIL response (TNF receptor apoptosis inducing ligand), the presence of immunoparalysis, and the ADAMTS13 activity response also differed across the five groups but in the opposite direction being decreased in Group 5 (blue) compared to Group 1 (red). The different cytokine patterns across the groups demonstrate that the five distinct multiple trajectories of CRP and ferritin are characterized by different immune activation profiles in children with sepsis.
Patients with the highest ferritin levels (Group 3 and Group 5) had the highest MIP-1-α response and accompanying immunoparalysis evident at first sampling. Pairwise comparison indicated that Group 5 had a significantly greater prevalence of immunoparalysis of 67% compared to 25% in Group 3 (Fisher’s Exact Test P = 0.01). This increased MIP-1-α response and immunoparalysis resolved by second sampling in patients with high ferritin and low CRP (Group 3); however, the increased MIP-1-α response and immunoparalysis persisted at second sampling in patients with very high CRP and very high ferritin (Group 5) as did increased IL-6, IL-8, MCP-1/CCL2, and IFNβ. Patients with very high CRP levels and very high ferritin levels (Group 5) also had higher sCD163, IL-8, IL-18, CXCL9, M-CSF, IL-18BP, MCP1. IL-10, MIP-1-α, IL-16, IP-10, TNFα, and IL2RA levels than patients with very high CRP levels alone (Group 4). Patients with very high CRP levels alone (Group 4) had higher sCD163, IL-8, MCSF, IL-6, IL-18, MCP1, TNFα, MIP-1-β, IL-22, IL-10, MIP-1-α, CRP, and ferritin levels than those with normal or high CRP levels alone (Groups 1 and 2).
Table 2 shows outcomes according to multi-trajectory group. Development of maximum organ failures (median [IQR]) and mortality (%) increased across the groups: Group 1 – maximum organ failures = 2 [1, 2], 0% (0/8) mortality; Group 2 – maximum organ failures = 2 [1, 2], 5% (4/80) mortality; Group 3 – maximum organ failures = 3 [2, 3], 6% (1/16) mortality; Group 4 - maximum organ failures = 2 [2, 3], 11% (13/121) mortality; and Group 5 – maximum organ failures = 3 [2, 4], 40% (12/30) mortality. Development of MAS, and immunoparalysis differed across the groups: Group 1 - MAS 0% (0/8), immunoparalysis 25% (2/8); Group 2 – MAS 1% (1/80), immunoparalysis 16% (13/80); Group 3 – MAS 25% (4/16), immunoparalysis 25% (4/16); Group 4 – MAS 7% (8/121), immunoparalysis 27% (33/121); and Group 5 – MAS 20% (6/30), immunoparalysis 67% (20/30). Requirements for CRRT also differed across the groups: Group 1 - CRRT 13% (1/8); Group 2 – CRRT 5% (4/80); Group 3 – CRRT 19% (3/16); Group 4 CRRT 11% (13/121); and Group 5 – CRRT 57% (17/30). In exploratory multivariable models, Group 5 was significantly associated with mortality compared with Groups 1–4 after adjusting for age, PRISM III and immunocompromised status both concomitantly (Table 3) and individually (all P<0.01; Supplemental Tables 4–7).
Table 2.
Outcomes by CRP-ferritin multi- trajectory group1
| CRP-ferritin multi-trajectory groups | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| Group 1 (N = 8) | Group 2 (N = 80) | Group 3 (N = 16) | Group 4 (N = 121) | Group 5 (N = 30) | P-value | |
| PICU mortality | <.0013 | |||||
|
| ||||||
| Survived | 8 (100%) | 76 (95%) | 15 (94%) | 108 (89%) | 18 (60%) | |
|
| ||||||
| Died | 0 (0%) | 4 (5%) | 1 (6%) | 13 (11%) | 12 (40%) | |
|
| ||||||
| Hospital Length of Stay (days) | 17 [13, 46] | 19 [13, 33] | 24 [13, 38] | 22 [15, 35] | 39 [20, 74] | 0.1082 |
|
| ||||||
| ICU Length of Stay (days) | 14 [11, 19] | 13 [9, 22] | 9 [6, 19] | 16 [10, 25] | 18 [11, 35] | 0.0782 |
|
| ||||||
| MAS | 0 (0%) | 1 (1%) | 4 (25%) | 8 (7%) | 6 (20%) | <.0013 |
|
| ||||||
| IPMOF | 2 (25%) | 13 (16%) | 4 (25%) | 33 (27%) | 20 (67%) | <.0013 |
|
| ||||||
| Maximum OFI | 2 [1, 2] | 2 [1, 2] | 3 [2, 3] | 2 [2, 3] | 3 [2, 4] | <.0012 |
|
| ||||||
| ECMO on study | 0 (0%) | 3 (4%) | 1 (6%) | 14 (12%) | 6 (20%) | 0.0803 |
|
| ||||||
| CRRT on study | 1 (13%) | 4 (5%) | 3 (19%) | 13 (11%) | 17 (57%) | <.0013 |
|
| ||||||
| Plasma exchange on study | 0 (0%) | 3 (4%) | 3 (19%) | 6 (5%) | 6 (20%) | 0.0143 |
|
| ||||||
| Mechanical ventilation (days) | 11 [9, 14] | 10 [8, 16] | 8 [3, 10] | 13 [7, 21] | 13 [8, 29] | 0.0592 |
Only subjects with the first two samples were included in biomarker trajectory analyses.
Kruskal-Wallis test.
Fisher’s exact test (Monte Carlo approximation).
Table 3.
Biomarker trajectory multivariable model
| PICU mortality | |||||
|---|---|---|---|---|---|
|
|
|||||
| Overall (N = 255) | Survived (N = 225) | Died (N = 30) | Odds ratio (95% CI) | P-value | |
| Biomarker trajectory | <.001 | ||||
|
| |||||
| Groups 1–4 | 225 (88%) | 207 (92%) | 18 (60%) | Reference | |
|
| |||||
| Group 5 | 30 (12%) | 18 (8%) | 12 (40%) | 7.03 (2.52, 20.15) | |
|
| |||||
| Age | 6 [2, 12] | 6 [2, 12] | 9 [2, 15] | 1.03 (0.95, 1.11) | 0.469 |
|
| |||||
| PRISM-3 | 8 [3, 15] | 8 [3, 14] | 11 [3, 19] | 1.04 (0.98, 1.09) | 0.186 |
|
| |||||
| Immunocompromised | 66 (26%) | 53 (24%) | 13 (43%) | 1.08 (0.39, 2.76) | 0.872 |
Estimates are based on a multivariable model, adjusting for each of the predictors in this table.
Abbreviations: PRISM, pediatric risk of mortality score.
Discussion
This multicenter study should inform planning of future clinical trials of anti-inflammatory therapies in pediatric sepsis in two ways.17 First, it corroborates our two, previous single center reports showing that systemic inflammation mortality risk increases in children with high CRP or ferritin alone, with even greater mortality risk when CRP and ferritin are both elevated.3,4 These findings also confirm feasibility of using inexpensive and readily available CRP and ferritin bedside measurements for mortality risk stratification in pediatric sepsis. Second, this multicenter study provides information about different cytokine response profiles in five distinct CRP and ferritin multi-trajectory groups. These data may better enable our ability to rationalize personalized testing of specific therapies that target cytokines with the purpose of reducing systemic inflammation and mortality risk.
Repeated assessment of inflammatory biomarkers and cytokine-targeted therapies are mainstays of rheumatic disease management. The recent COVID-19 pandemic has brought more attention to the interplay between infectious syndromes and detrimental host immune response. Relatedly, sepsis has long been broadly defined as infection leading to life-threatening organ dysfunction, though the threshold at which dysregulated systemic inflammation becomes the predominant threat versus an invading pathogen is poorly understood. Antibiotics and infection source control remain the foundation of sepsis management. However, we have now found that CRP and ferritin – two widely available biomarkers -- hold promise for individualizing treatments directed at sepsis-related systemic inflammation. As an example, two anti-cytokine therapies, IL-6 monoclonal antibody (inhibits IL-6 effects) and interleukin-1 receptor antagonist protein (inhibits production of IL-8 and MIP-1-α in mixed lymphocyte reactions without affecting lymphocyte proliferation),18 have been reported effective in reducing mortality in adults with hyperinflammatory COVID-19 sepsis, with the former showing biologic efficacy only in patients with elevated CRP, and the latter showing biologic efficacy in all patients.19 In our multicenter study we observed elevated IL-6 levels in the two multi-trajectory groups of children with very high CRP levels; elevated IL-8 and MIP-1-α levels in the three groups of children with highest ferritin levels; and the highest IL-6, IL-8, and MIP-1-α levels in the one group of children with very high CRP and very high ferritin levels (all potentially targetable with IL-1 receptor antagonist protein). These observations have therefore provided a rationale for our research network to design an adaptive placebo-controlled trial of interleukin-1 receptor antagonist protein to target CRP and ferritin based systemic inflammation mortality risk in pediatric sepsis (NCT05267821).
Interaction with the CRP receptor on macrophages in the reticuloendothelial system leads to phagocytosis, antigen processing, and clearance. In the context of sepsis, CRP can orchestrate reticuloendothelial system activation. Groups 2 and 4 demonstrated persistently elevated CRP values and greater mortality than Group 1, which had persistently low CRP measurements. These findings are compatible with previous reports associating persistently high levels of CRP despite antibiotic administration with mortality due to unremitting bacterial infection and organ failure.3,20 Notably, ferritin was also low in Group 1 and the cytokine profile for this group identified elevated levels of IL-1α, IL-4 and IL-13. IL-4 and IL-13 are mediators of allergic inflammation, while IL-1α rises in response to sterile injury, raising the possibility that this group may represent patients with aseptic or allergic systemic inflammatory response syndrome.21
More recently, very high ferritin levels in the absence of iron overload have been recognized as indicative of macrophage activation and hyperinflammation. During viral infection, toll like receptor 9 is activated by viral antigens leading to macrophage inflammasome activation with feed forward IL-1 and IL-18 production leading to synthesis and release of iron-poor ferritin.22–24 We observe high ferritin, IL-18 and MIP-1-α with low CRP levels in patients in Group 3. Wang and colleagues have shown in rodents that sequential exposure to viral antigen/infection and then bacterial antigen/infection (but not viral followed by viral, or bacterial followed by bacterial, or bacterial followed by viral infection) induces the hyperinflammatory, hyperferritinemic, sepsis response with high CRP and ferritin analogous to the biomarker trajectories evident in Group 5 in our analysis.25 Under this sequential antigen challenge, rodents reach ferritin levels of 6,000 ng/mL with concomitantly increased TNFα, IL-6 and sCD25 levels. Patients with very high CRP and very high ferritin in our present study (Group 5) similarly exhibit increased TNFα, IL-6, and sCD25 (IL2RA) levels, as well as very high levels of IL-8 and MIP-1-α (co-regulates with ferritin) indicative of hyperinflammatory hyperferritinemic sepsis-driven macrophage activation. Immunoparalysis paralleled the hyperferritinemic inflammation profile.
Children with low CRP and high ferritin (Group 3) had high MIP-1-α, and immunoparalysis at first sampling that resolved and recovered respectively by second sampling; whereas, children with very high CRP and very high ferritin (Group 5), had high MIP-1-α (co-regulates with ferritin), and immunoparalysis that persisted along with elevated IL-6 and IL-8 levels. On comparing group 5 with group 3, immunoparalysis differed significantly, occurring in two-thirds of patients in group 5 (both very high CRP and ferritin), but only one-quarter of patients in group 3 (low CRP and high ferritin). Persistent immunoparalysis may be related, in part, to profound iatrogenic myeloablation or immunosuppression, occult viral DNAemia, or pathogenic variants of immunity.26–28 Interestingly, circulating levels of the macrophage activation inhibitor TRAIL (TNF receptor apoptosis inducing ligand), which also has anti-cancer properties,29 were decreased in patients with very high CRP levels with or without very high ferritin levels (Groups 4 and 5). This observation supports further study of therapeutic use of TRAIL to decrease inflammation in these two groups of patients who had the highest systemic inflammation mortality risks.
There are several limitations to consider in our study. First, this cohort was enrolled prior to the COVID-19 pandemic and included children with sepsis, the leading global killer of children from 1998–2017. Second, the cohort is limited to sepsis patients who were in the PICU for two blood draws in a week. This small cohort size with a relatively rare primary outcome of mortality precluded the construction of robust multivariable models due to limited statistical degrees of freedom. Third, although CRP and ferritin monitoring is feasible in resource-poor and resource-rich settings alike, our study was performed in 9 resource-rich PICUs, therefore findings will require validation in resource-poor centers. Fourth, CRP and ferritin are non-specific biomarkers. Established risk stratification tools including the Pediatric Sepsis Biomarker Risk Model II (PERSERVERE II)30,31 and transcriptomics32 are superior but not yet practicable in resource-poor settings where most global deaths occur. Fifth we only assessed 31 cytokines and functional assays associated with hyperinflammation syndromes in children limiting clinical insights into other cytokines.
Conclusions
Our multi-center study validates our previous two single center studies showing that systemic inflammation mortality risk can be assessed with combined measurement of CRP and ferritin levels. The present study further reveals different cytokine response profiles according to CRP and ferritin level multi-trajectory groups. In the future, such phenotypic characterization, and targeting with specific biologic therapies and anti-inflammatory strategies to reduce systemic inflammation may improve outcomes in pediatric sepsis in resource rich and resource poor settings alike.11,14
Supplementary Material
RESEARCH IN CONTEXT
Interpretable and readily available diagnostics are needed to guide targeted anti-inflammatory therapies for children with sepsis.
Emerging evidence is identifying unique sepsis phenotypes.
This study examines the utility of C-reactive protein and ferritin for distinguishing sepsis inflammation phenotypes.
AT THE BEDSIDE
C-reactive protein and ferritin are low-cost, widely available tests that are associated with specific inflammatory profiles.
Distinct sepsis inflammation phenotypes may offer novel therapeutic targets among children with sepsis.
Prospective trials are needed to examine biomarker-guided sepsis treatments.
ACKNOWLEDGMENTS
Clinical Research Investigation and Systems Modeling of Acute illness center: Ali Smith, BS; Octavia Palmer, MD; Vanessa Jackson, AA; Renee Anderko, BS, MS. Children’s Hospital of Pittsburgh: Jennifer Jones, RN; Luther Springs. Children’s Hospital of Philadelphia: Carolanne Twelves, RN, BSN, CCRC; Mary Ann Diliberto, BS, RN, CCRC; Martha Sisko, BSN, RN, CCRC, MS; Pamela Diehl, BSN, RN; Janice Prodell, RN, BSN, CCRC; Jenny Bush, RNC, BSN; Kathryn Graham, BA; Kerry Costlow, BS; Sara Sanchez. Children’s National Medical Center: Elyse Tomanio, BSN, RN; Diane Hession, MSN, RN; Katherine Burke, BS. Children’s Hospital of Michigan: Ann Pawluszka, RN, BSN; Melanie Lulic, BS. Nationwide Children’s Hospital: Lisa Steele, RN, CCRC; Andrew R. Yates, MD; Josey Hensley, RN; Janet Cihla, RN; Jill Popelka, RN; Lisa Hanson-Huber, BS. Children’s Hospital of Los Angeles and Mattel Children’s Hospital: Jeni Kwok, JD; Amy Yamakawa, BS. Children’s Hospital of Washington University of Saint Louis: Michelle Eaton, RN. Mott Children’s Hospital: Frank Moler, MD; Chaandini Jayachandran, MS, CCRP. University of Utah Data Coordinating Center: Teresa Liu, MPH, CCRP; Jeri Burr, MS, RN-BC, CCRC, FACRP; Missy Ringwood, BS, CMC; Nael Abdelsamad, MD, CCRC; Whit Coleman, MSRA, BSN, RN, CCRC.
Supported, in part, by grant R01GM108618 (to Dr. Carcillo PI, Dr Park, and Dr Canna) from the National Institutes of General Medical Sciences, and by 5U01HD049934-10S1 (to Dr Carcillo), 1K23HD099331-01A1 (to Dr Horvat) and K12HD047349 (to Dr Kernan) from the Eunice Kennedy Shriver National Institutes of Child Health and Human Development, National Institutes of Health, Department of Health and Human Services and the following cooperative agreements: U10HD049983, U10HD050096, U10HD049981, U10HD063108, U10HD63106, U10HD063114, U10HD050012, and U01HD049934.
Footnotes
Authors - Carcillo’s, Berg’s, Wessel’s, Pollack’s, Meert’s, Hall’s, Doctor’s, Cornell’s, Harrison’s, Zuppa’s, Reeder’s, Banks’s, and Holubkov’s institutions received funding from the National Institutes of Health (NIH). Drs. Carcillo’s, Newth’s, Shanley’s, and Dean’s institutions received funding from the National Institutes of Child Health and Human Development. Authors Carcillo, Berg, Wessel, Pollack, Meert, Hall, Newth, Doctor, Shanley, Cornell, Harrison, Zuppa, Reeder, Banks, Holubkov, Notterman, and Dean received support for article research from the NIH. Dr. Carcillo’s institution also received funding from the National Institutes of General Medical Sciences. Dr. Pollack disclosed that his research is supported by philanthropy from Mallinckrodt Pharmaceuticals. Dr. Hall received funding from Bristol Myers-Squibb (for service on an advisory board) and LaJolla Pharmaceuticals (service as a consultant), both unrelated to the current submission. Dr. Newth received funding from Philips Research North America. Dr. Doctor’s institution received funding from the Department of Defense and Kalocyte. Dr. Shanley received funding from Springer publishing, International Pediatric Research Foundation, and Pediatric Academic Societies. Dr. Cornell disclosed he is co-founder of Pre-Dixon Bio. Dr. Holubkov received funding from Pfizer (Data Safety Monitoring Board [DSMB] member), Medimmune (DSMB member), Physicians Committee for Responsible Medicine (biostatistical consulting), DURECT Corporation (biostatistical consulting), Armaron Bio (DSMB past member), and St Jude Medical (DSMB past member). The remaining authors have disclosed that they do not have any potential conflicts of interest.
Supplemental digital content is available for this manuscript
Copyright Form Disclosure: Drs. Horvat, Banks, Zuppa, Sward, and Carcillo’s institutions received funding from the National Institute of Child Health and Human Development (NICHD). Drs. Horvat, Banks, Park, Kernan, Canna, Berg, Wessel, Pollack, Meert, Hall, Newth, Doctor, Shanley, Harrison, Zuppa, Reeder, Sward, Holubkov, Dean, and Carcillo received support for article research from the National Institutes of Health (NIH). Drs. Horvat and Carcillo disclosed the off-label product use of TNF Receptor Apoptosis Inducing Ligand Interleukin Receptor Antagonist Protein. Dr. Kernan’s institution received funding from the NICHD (K12HD047349). Dr. Canna’s institution received funding from from InnVention Therapeutix; he received funding from Simcha Therapeutics. Drs. Berg, Wessel, Pollack, Meert, Hall, Newth, Doctor, Shanley, Harrison, Reeder, Holubkov, and Dean’s institutions received funding from the NIH. Dr. Hall received funding from La Jolla Pharmaceuticals, Abbvie, and Kiadis. Dr. Newth received funding from Philips Research North America, Hamilton Medical AG, and Nihon Kohden Orange Med. Dr. Doctor’s institution received funding from the Department of Defense and KaloCyte. Dr. Holubkov’s institution received funding from AltaThera Pharmaceuticals; he received funding from Pfizer, the Physicians Committee for Responsible Medicine, and DURECT corporation. Dr. Carcillo’s institution received funding from the National Institute of General Medical Sciences. The remaining authors have disclosed that they do not have any potential conflicts of interest.
References
- 1.Rudd KE, Johnson SC, Agesa KM, Shackelford KA, Tsoi D, Kievlan DR, et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: analysis for the Global Burden of Disease Study. Lancet Lond Engl. 2020. Jan 18;395(10219):200–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor MD, Allada V, Moritz ML, Nowalk AJ, Sindhi R, Aneja RK, et al. Use of C-Reactive Protein and Ferritin Biomarkers in Daily Pediatric Practice. Pediatr Rev. 2020. Apr;41(4):172–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horvat CM, Bell J, Kantawala S, Au AK, Clark RSB, Carcillo JA. C-Reactive Protein and Ferritin Are Associated With Organ Dysfunction and Mortality in Hospitalized Children. Clin Pediatr (Phila). 2019. Jun;58(7):752–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carcillo JA, Sward K, Halstead ES, Telford R, Jimenez-Bacardi A, Shakoory B, et al. A Systemic Inflammation Mortality Risk Assessment Contingency Table for Severe Sepsis. Pediatr Crit Care Med J Soc Crit Care Med World Fed Pediatr Intensive Crit Care Soc. 2017. Feb;18(2):143–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagin DS, Jones BL, Passos VL, Tremblay RE. Group-based multi-trajectory modeling. Stat Methods Med Res. 2018. Jul;27(7):2015–23. [DOI] [PubMed] [Google Scholar]
- 6.Carcillo JA, Berg RA, Wessel D, Pollack M, Meert K, Hall M, et al. A Multicenter Network Assessment of Three Inflammation Phenotypes in Pediatric Sepsis-Induced Multiple Organ Failure. Pediatr Crit Care Med J Soc Crit Care Med World Fed Pediatr Intensive Crit Care Soc. 2019. Dec;20(12):1137–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carcillo JA, Halstead ES, Hall MW, Nguyen TC, Reeder R, Aneja R, et al. Three Hypothetical Inflammation Pathobiology Phenotypes and Pediatric Sepsis-Induced Multiple Organ Failure Outcome. Pediatr Crit Care Med J Soc Crit Care Med World Fed Pediatr Intensive Crit Care Soc. 2017. Apr 13; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldstein B, Giroir B, Randolph A, International Consensus Conference on Pediatric Sepsis. International pediatric sepsis consensus conference: definitions for sepsis and organ dysfunction in pediatrics. Pediatr Crit Care Med J Soc Crit Care Med World Fed Pediatr Intensive Crit Care Soc. 2005. Jan;6(1):2–8. [DOI] [PubMed] [Google Scholar]
- 9.Villeneuve A, Joyal JS, Proulx F, Ducruet T, Poitras N, Lacroix J. Multiple organ dysfunction syndrome in critically ill children: clinical value of two lists of diagnostic criteria. Ann Intensive Care. 2016. Dec;6(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muszynski JA, Nofziger R, Moore-Clingenpeel M, Greathouse K, Anglim L, Steele L, et al. Early Immune Function and Duration of Organ Dysfunction in Critically III Children with Sepsis. Am J Respir Crit Care Med. 2018. Aug 1;198(3):361–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall MW, Knatz NL, Vetterly C, Tomarello S, Wewers MD, Volk HD, et al. Immunoparalysis and nosocomial infection in children with multiple organ dysfunction syndrome. Intensive Care Med. 2011. Mar;37(3):525–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiss ES, Girard-Guyonvarc’h C, Holzinger D, de Jesus AA, Tariq Z, Picarsic J, et al. Interleukin-18 diagnostically distinguishes and pathogenically promotes human and murine macrophage activation syndrome. Blood. 2018. Mar 29;131(13):1442–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kyriazopoulou E, Leventogiannis K, Norrby-Teglund A, Dimopoulos G, Pantazi A, Orfanos SE, et al. Macrophage activation-like syndrome: an immunological entity associated with rapid progression to death in sepsis. BMC Med. 2017. Sep 18;15(1):172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shakoory B, Carcillo JA, Chatham WW, Amdur RL, Zhao H, Dinarello CA, et al. Interleukin-1 Receptor Blockade Is Associated With Reduced Mortality in Sepsis Patients With Features of Macrophage Activation Syndrome: Reanalysis of a Prior Phase III Trial. Crit Care Med. 2016. Feb;44(2):275–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rousseeuw PJ, Ruts I, Tukey JW. The Bagplot: A Bivariate Boxplot. Am Stat. 1999. Nov 1;53(4):382–7. [Google Scholar]
- 16.Carcillo JA, Podd B, Aneja R, Weiss SL, Hall MW, Cornell TT, et al. Pathophysiology of Pediatric Multiple Organ Dysfunction Syndrome. Pediatr Crit Care Med J Soc Crit Care Med World Fed Pediatr Intensive Crit Care Soc. 2017. Mar;18(3_suppl Suppl 1):S32–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall M. Targeted Reversal of Inflammation in Pediatric Sepsis-induced MODS (TRIPS) [Internet]. clinicaltrials.gov; 2022. May [cited 2022 May 30]. Report No.: NCT05267821. Available from: https://clinicaltrials.gov/ct2/show/NCT05267821
- 18.Lukacs NW, Kunkel SL, Burdick MD, Lincoln PM, Strieter RM. Interleukin-1 receptor antagonist blocks chemokine production in the mixed lymphocyte reaction. Blood. 1993. Dec 15;82(12):3668–74. [PubMed] [Google Scholar]
- 19.Cavalli G, Larcher A, Tomelleri A, Campochiaro C, Della-Torre E, De Luca G, et al. Interleukin-1 and interleukin-6 inhibition compared with standard management in patients with COVID-19 and hyperinflammation: a cohort study. Lancet Rheumatol. 2021. Feb 3; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmit X, Vincent JL. The time course of blood C-reactive protein concentrations in relation to the response to initial antimicrobial therapy in patients with sepsis. Infection. 2008. Jun;36(3):213–9. [DOI] [PubMed] [Google Scholar]
- 21.Edwards MR, Strong K, Cameron A, Walton RP, Jackson DJ, Johnston SL. Viral infections in allergy and immunology: How allergic inflammation influences viral infections and illness. J Allergy Clin Immunol. 2017. Oct;140(4):909–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shrivastava G, Valenzuela Leon PC, Calvo E. Inflammasome Fuels Dengue Severity. Front Cell Infect Microbiol. 2020;10:489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai JH, Wang MY, Huang CY, Wu CH, Hung LF, Yang CY, et al. Infection with the dengue RNA virus activates TLR9 signaling in human dendritic cells. EMBO Rep. 2018. Aug;19(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simon DW, Halstead ES, Davila S, Kernan KF, Clark RSB, Storch G, et al. DNA Viremia Is Associated with Hyperferritinemia in Pediatric Sepsis. J Pediatr. 2019. Oct;213:82–87.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang A, Pope SD, Weinstein JS, Yu S, Zhang C, Booth CJ, et al. Specific sequences of infectious challenge lead to secondary hemophagocytic lymphohistiocytosis-like disease in mice. Proc Natl Acad Sci U S A. 2019. Feb 5;116(6):2200–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kernan KF, Carcillo JA. Hyperferritinemia and inflammation. Int Immunol. 2017. Nov 1;29(9):401–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kernan KF, Ghaloul-Gonzalez L, Shakoory B, Kellum JA, Angus DC, Carcillo JA. Adults with septic shock and extreme hyperferritinemia exhibit pathogenic immune variation. Genes Immun. 2019. Jul;20(6):520–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kernan KF, Ghaloul-Gonzalez L, Vockley J, Lamb J, Hollingshead D, Chandran U, et al. Prevalence of Pathogenic and Potentially Pathogenic Inborn Error of Immunity Associated Variants in Children with Severe Sepsis. J Clin Immunol. 2022. Feb;42(2):350–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schenck EJ, Ma KC, Price DR, Nicholson T, Oromendia C, Gentzler ER, et al. Circulating cell death biomarker TRAIL is associated with increased organ dysfunction in sepsis. JCI Insight. 2019. May 2;4(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanski NL, Stenson EK, Cvijanovich NZ, Weiss SL, Fitzgerald JC, Bigham MT, et al. PERSEVERE Biomarkers Predict Severe Acute Kidney Injury and Renal Recovery in Pediatric Septic Shock. Am J Respir Crit Care Med. 2020. Apr 1;201(7):848–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong HR, Caldwell JT, Cvijanovich NZ, Weiss SL, Fitzgerald JC, Bigham MT, et al. Prospective clinical testing and experimental validation of the Pediatric Sepsis Biomarker Risk Model. Sci Transl Med. 2019. Nov 13;11(518). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sweeney TE, Azad TD, Donato M, Haynes WA, Perumal TM, Henao R, et al. Unsupervised Analysis of Transcriptomics in Bacterial Sepsis Across Multiple Datasets Reveals Three Robust Clusters. Crit Care Med. 2018. Jun;46(6):915–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.