Abstract
Jumbo phages such as Pseudomonas aeruginosa ФKZ have potential as antimicrobials and as a model for uncovering basic phage biology. Both pursuits are currently limited by a lack of genetic engineering tools due to a proteinaceous “phage nucleus” structure that protects from DNA-targeting CRISPR-Cas tools. To provide reverse-genetics tools for DNA jumbo phages, we combined homologous recombination with an RNA-targeting CRISPR-Cas13a enzyme, and used an anti-CRISPR gene (acrVIA1) as a selectable marker. We show that this process can insert foreign genes, delete genes and add fluorescent tags to the ФKZ genome. Fluorescent tagging of endogenous gp93 revealed that it is ejected with the phage DNA while the deletion of tubulin-like protein PhuZ surprisingly had only a modest impact on phage burst size. Editing of two other phages that resist DNA-targeting CRISPR-Cas systems was also achieved. RNA-targeting Cas13a holds great promise for becoming a universal genetic editing tool for intractable phages, enabling the systematic study of phage genes of unknown function.
Editor summary:
Phage genetic engineering platform will enable a better understanding of phage biology, and engineering of phage therapies.
INTRODUCTION
Bacteriophages infect bacteria and can cause cell lysis after replication. In recent decades, the rapid emergence of multi-drug resistant bacterial pathogens and simultaneous decline in the discovery of antibiotics has rekindled interest in the use of phages as alternative antimicrobial therapeutics (phage therapy) [1, 2]. Phages offer many advantages over antibiotics, including high specificity and efficient self-propagation in the presence of their bacterial host [3–5]. However, host range limitations, and the rapid emergence of phage resistance in clinical strains, present barriers to adoption of phage therapy [1, 3, 6]. Phage genome engineering might help to overcome these hurdles [7, 8]. Robust phage engineering tools could aid fundamental discoveries, broaden host range, enhance evasion of host antiviral defense systems, and reduce phage immunogenicity [9–12]. Phage engineering techniques often include homologous recombination (HR) with a template plasmid [13, 14], coupled with a selective pressure, such as CRISPR-Cas targeting. CRISPR-Cas systems (clustered regularly interspaced short palindromic repeats and CRISPR-associated proteins) are adaptive anti-phage immune systems in prokaryotes [15, 16]. CRISPR-Cas programmable targeting enables effective enrichment for phage recombinants by removing wild-type phages from the population and has been coupled with the integration of an anti-CRISPR gene as a selectable marker [17].
To date, all CRISPR-based screening tools used in phage engineering recognize and target phage genomic DNA. However, phages have evolved a multitude of strategies to circumvent DNA-targeting immunity, including anti-CRISPR proteins, DNA base modifications, and genome segregation [18, 19]. P. aeruginosa jumbo phage ФKZ is resistant to a broad range of DNA-targeting immune systems via assembly of a proteinaceous “phage nucleus” structure that shields phage DNA during replication [20, 21]. This phage family is therefore a good candidate for use as a phage therapeutic, but no genetic tools are available for jumbo phages and most studies have relied on plasmid-based over-expression of jumbo phage genes [22].
Although the phage nucleus protects phage DNA from being targeted by bacterial immune defenses, the mRNA-targeting CRISPR-Cas13a system (type VI-A) [23] effectively inhibits ФKZ replication by degrading phage mRNA that is exported from the phage nucleus to the cytoplasm [20]. The promiscuous cleavage of bacterial and phage transcripts by activated Cas13a limits the emergence of CRISPR-resistant escape phages [24], and may aid the identification of engineered mutant phages.
We report the development of a CRISPR-Cas13a system as a genetic engineering method for ФKZ. Using Cas13a to target an essential transcript, we select for phages that have undergone homologous recombination resulting in a desired genetic change along with the acquisition of an anti-Cas13a trans gene, acrVIA1 (derived from Listeriophage ФLS46 [25]), as a selectable marker. This approach allows us to precisely insert foreign gene fragments into the ФKZ genome, knock out non-essential genes, and fuse fluorescent tags to individual genes. Importantly, the same guide can be used for any genomic manipulation as engineered phages are identified based on the acquisition of the Cas13a inhibitor, not a change in the target sequence. Our work establishes a Cas13a-based phage engineering strategy that could be a universally applied for engineering phages.
RESULTS
Optimization of CRISPR-Cas13a for efficient phage targeting
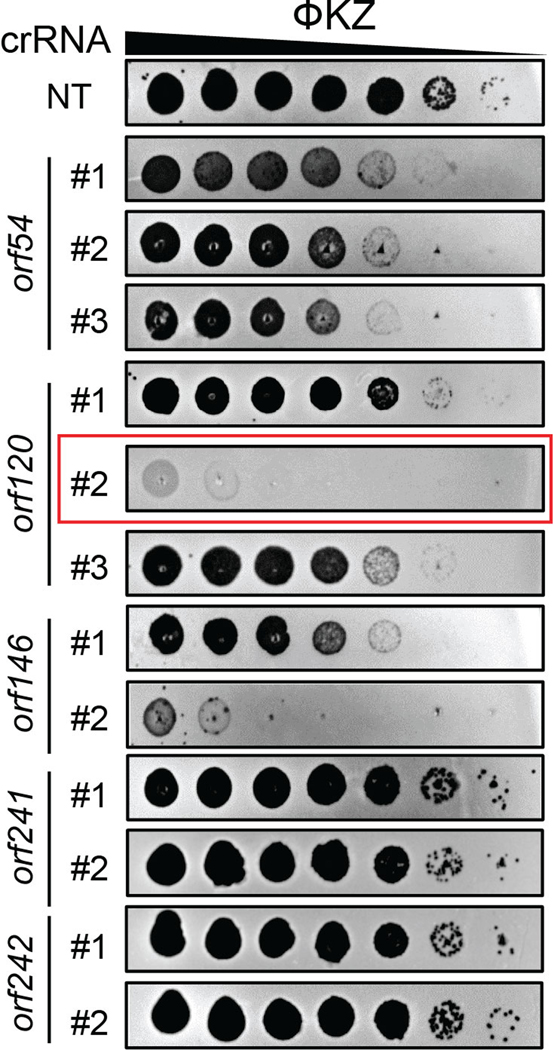
Cas13a is an RNA-guided RNA nuclease that can block ΦKZ replication in P. aeruginosa PAO1. We previously targeted ΦKZ by expressing LseCas13a (derived from Listeria seeligeri) from the PAO1 chromosome under a pLac inducible promoter and crRNA guides from a plasmid (Fig. 1A, V1) [20]. We selected LseCas13a because of its prior thorough biochemical characterization [23, 26, 27]. For the effective elimination of WT phages in the population, we first sought to enhance activity of crRNA guides. We designed Version 2 (V2) with the repeat-spacer-repeat unit moved to the +1 transcription start site of PBAD promoter and the second direct repeat (DR) mutated to remove repeat homology (Fig. 1A). To further stabilize the crRNA cassette, we next omitted the second DR and generated V3 (Fig. 1A), which could prevent recombination between the repeats. Using the same spacer, both V2 and V3 provided more robust defense against phage JBD30 and ΦKZ compared with V1 (Fig. 1A). For simplicity, we selected the V3 cassette that has a single repeat to express crRNAs against ΦKZ. We designed guides with 24 nucleotide spacers complementary to randomly selected regions of various ΦKZ gene transcripts. Strong targeting was observed for some crRNAs, to the point that mutant escape phages could be isolated, such as the two spacers matching orf120 and orf146 transcripts, but not all crRNAs were efficacious (Extended Data Fig. 1). Given the variability of targeting efficiency, for the remainder of this report, we use the crRNA targeting ΦKZ orf120 transcripts (Extended Data Fig. 1) as our primary guide to screen for engineered phages. We refer the PAO1 strain simultaneously expressing Cas13a and orf120-crRNA to as Cas13a counter-selection strain. We describe below how the same guide can be used to facilitate the engineering of distinct genomic loci.
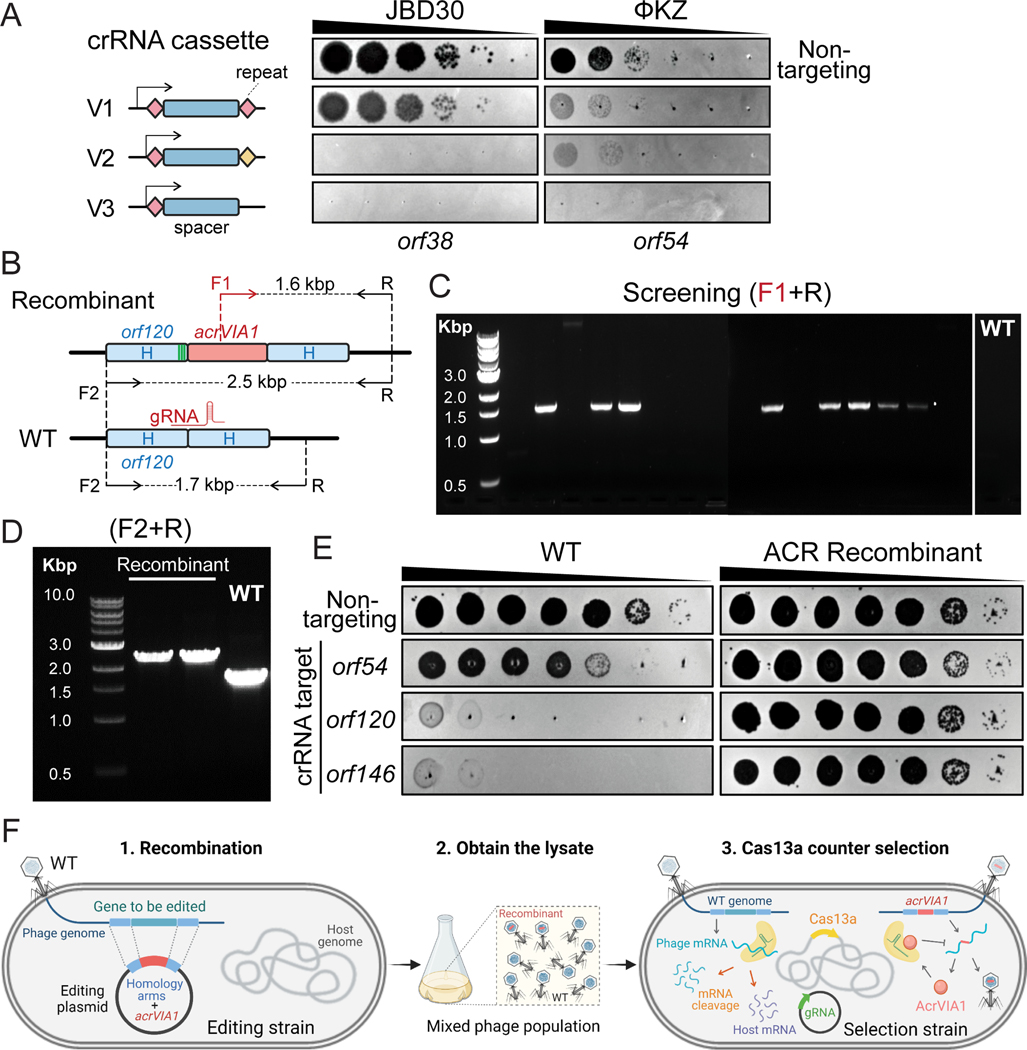
Fig. 1: Screening for ΦKZ recombinants by CRISPR-Cas13a counter selection.
(A) Schematic of three versions of CRISPR-Cas13a crRNA cassette and their efficiency of plating assays targeting two unrelated phage strains: JBD30 and ΦKZ. Spacers are represented by dark blue rectangles. Repeats are shown as diamonds with different colors (pink and yellow) indicating different repeat sequences. Arrows mean transcription start sites of PBAD promoter. The cassettes carried the same spacer sequences targeting transcripts of orf38 of JBD30 and orf54 of ΦKZ, respectively. (B) Schematic of WT ΦKZ and recombinant genomes at the editing site. The acrVIA1 gene, shown as a red rectangle, was inserted downstream of orf120, with up-and downstream of homology arms (H) indicated by light blue rectangles. Green stripes represent synonymous mutations that were introduced to the homology region in only this case. Synonymous mutations were not necessary due to the effectiveness of the AcrVIA1 marker on inhibition of Cas13a activities. F and R indicate forward and reverse primers, respectively, being used to confirm the insertion of acrVIA1. (C) Recombinant phages were screened by PCR using primer F1 and R. 8 out of the 16 tested phage plaques generated the expected 1.6 kbp band, which was not detectable in a WT plaque. (D) Recombinant phage plaques underwent 3 rounds of purification and were further confirmed by PCR using primer F2 and R. The PCR products revealed the expected size increase, from ~1.7 kbp of WT to ~2.5 kbp of recombinants (E) Plaque assays showing robust anti-CRISPR activities acquired by recombinant ΦKZ against distinct crRNAs, owing to the successful expression and execution of the incorporated AcrVIA1. PCR-based mutant screening, as well as the subsequent plaque assays, have been independently repeated three times yielding similar results. (F) Workflow of phage genome engineering using CRISPR-Cas13a. 1. an editing plasmid introduces desired genetic modifications via recombination, 2. a mixed phage lysate is generated, 3. the lysate is plated on the selection strain harboring Cas13a and a crRNA targeting WT phages.
Isolation of ΦKZ recombinants by CRISPR-Cas13a selection
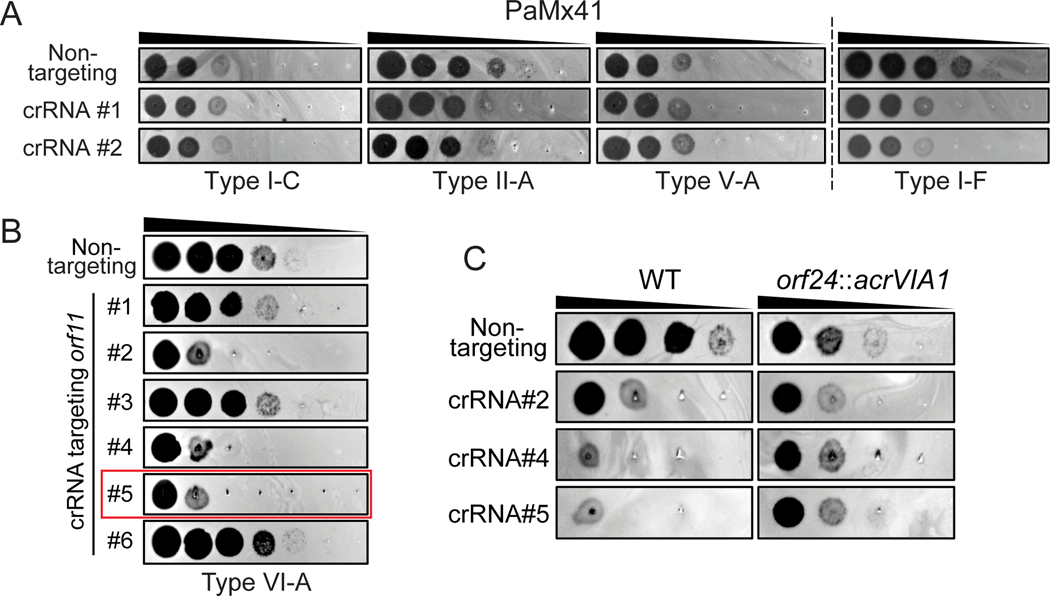
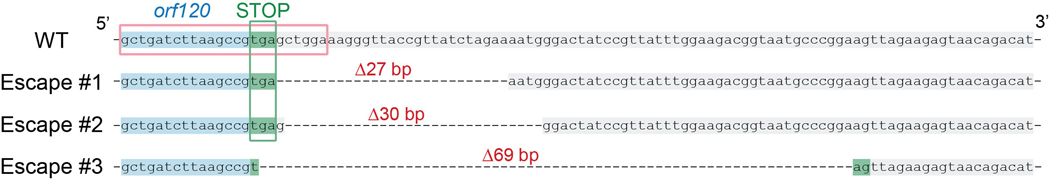
To avoid disrupting any essential genes that are required for phage replication, we first attempted to insert acrVIA1 immediately downstream of the ΦKZ major capsid gene (orf120). A template DNA substrate for homologous recombination, composed of ~600 bp homology arms flanking acrVIA1 (669 bp) plus a 30 bp sequence including Shine-Dalgarno site was cloned into a plasmid, referred to as an editing plasmid (Fig. 1B). After infecting a PAO1 strain possessing the editing plasmid to allow recombination, the phage lysate was then titrated on a lawn of the Cas13a counter-selection strain to eliminate WT phages. To screen for recombinants, individual plaques were examined for acrVIA1 integration via PCR using two sets of primers to confirm integration of acrVIA1 at the correct locus (Fig. 1B). This approach resulted in the isolation of recombinants (Fig. 1C and 1D). Sanger sequencing further confirmed the correct genomic integration junction. Recombinant phages propagated well on hosts expressing crRNAs targeting other transcripts due to the expression of acrVIA1 (Fig. 1E), which abolishes Cas13a immunity regardless of the crRNA sequence. Recombinant phages formed plaques on PAO1 with a similar size and efficiency compared to WT phages and phage latent periods and burst sizes were unaffected (Extended Data Fig. 2). This demonstrated that the insertion did not cause a fitness defect. Three randomly selected plaques that escaped Cas13a targeting but screened negative for the acrVIA1 integration contained genomic deletions ranging from 27 bp to 69 bp starting immediately downstream of the orf120 stop codon, disrupting the protospacer (Extended Data Fig. 3). While the crRNA used is therefore not inescapable, the recombination efficiency to insert the selectable marker is clearly efficient enough to enable facile identification of the desired mutants.
To test the flexibility of this nascent genetic technology (Fig. 1F) and generate new biological insights of phage ΦKZ, we next replaced (or attempted to replace) multiple genes with acrVIA1 (results summarized in Table 1); phuZ (orf39, tubulin homolog), orf54 (major shell/nucleus protein), orf89-orf93 (inner body proteins), orf93, orf146 (tail protein), orf241, and orf241-orf242 (small accessory proteins), in addition to attempting to add fluorescent tags onto phuZ, orf54 and orf93 in the phage genome. The successes, failures, and new insights gained are discussed below. Whole genome sequencing of two deletion mutants, phuZ::acrVIA1 and orf93::acrVIA1, revealed no other mutations, highlighting the accuracy of this RNA-targeting system to select for the desired changes in DNA phage genomes.
Table 1.
Summary of phage mutants engineered by CRISPR-Cas13a.
| Phage | Gene No./Genomic site | Identified protein | Modification | % plaques acrVIA1+1 (n2) | Desired mutant isolated? |
|---|---|---|---|---|---|
| ФKZ | Upstream of orf54 | - | Insertion | 41.2% (17) | Yes |
| Downstream of orf120 | - | Insertion | 50.0% (16) | ||
| orf39 | PhuZ | Deletion | 52.9% (17) | ||
| mNeonGreen-PhuZ fusion | 25.0% (16) | ||||
| mCherry-PhuZ fusion |
25.0% (16) | ||||
| orf93 | Inner body protein | Deletion | 20.0% (20) | ||
| gp93-mNeonGreen fusion | 18.2% (22) | ||||
| orf241 | Hypothetical | Deletion | 26.1% (23) | ||
| orf241, orf242 | Hypothetical | Double deletions | 41.7% (12) | ||
| orf54 | Shell | Deletion | 20.0% (15) | No | |
| mCherry-gp54 fusion |
8.3% (24) | ||||
| gp54-mCherry fusion |
9.1% (11) | ||||
| gfp11-gp54 fusion | 7.0% (57) | ||||
|
orf89, orf90,
orf91, orf92, orf93 |
Inner body proteins | Deletion | 11.8% (17) | ||
| orf146 | Structural protein | Deletion | 0% (89) | ||
| OMKO1 | Downstream of the capsid gene | - | Insertion with a barcode | 70.8% (24) | Yes |
| Upstream of the shell gene | - | Insertion | 50.0% (12) | ||
| PaMx41 | orf24 | Hypothetical | Deletion | 100.0% (8) | Yes |
Percentage of PCR-identified recombinants out of all tested plaques.
Number of plaques analyzed by PCR to screen for recombinants.
Characterization of PhuZ and gp93 using engineered ΦKZ
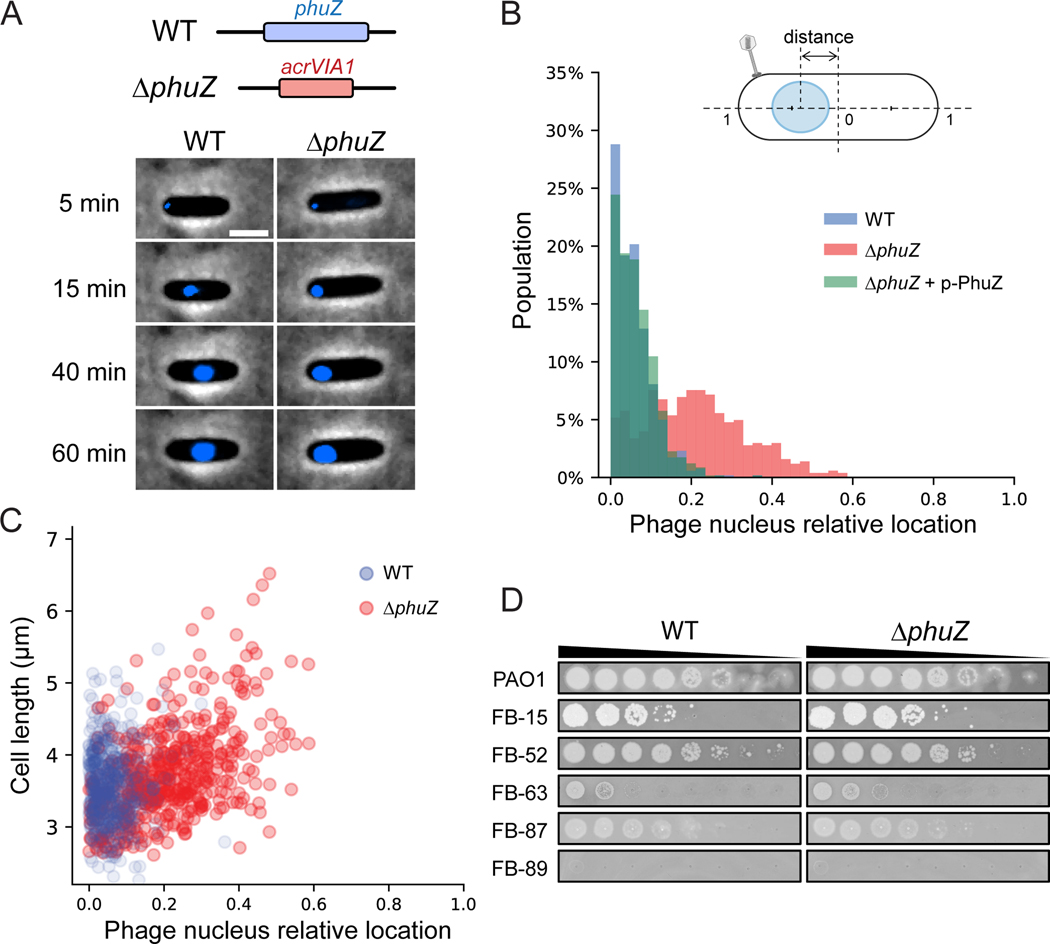
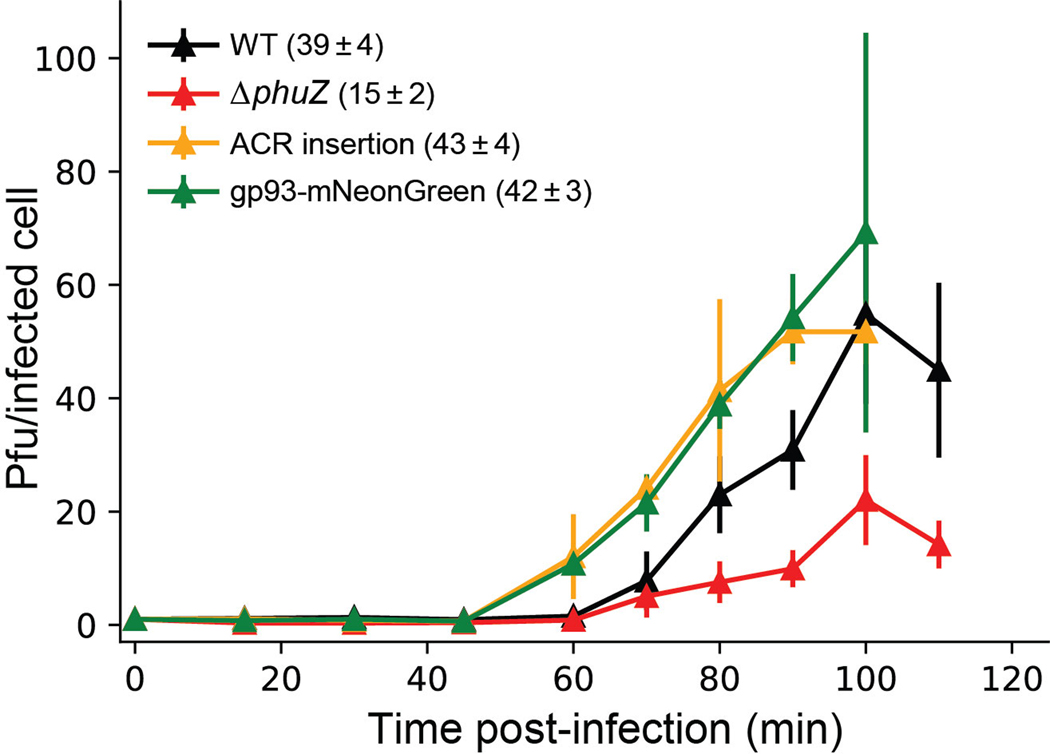
PhuZ (gp39) is a tubulin homolog conserved across many jumbo phages and some megaphages [28, 29]. It assembles a bipolar spindle to center the phage nucleus during phage intracellular development [30, 31], and “treadmill” newly synthesized phage capsids from the cell inner membrane to the phage nucleus for DNA packaging [32]. These roles made us speculate that PhuZ might be essential for phage growth, however, this is not the case. phuZ::acrVIA1 (herein, ΔphuZ) mutants formed plaques on a PAO1 lawn that were indistinguishable from WT plaques and exhibited a similar latent period (~60 min) compared with WT phages (Extended Data Fig. 2). However, the burst size significantly decreased (15 phage particles per infected bacterial cell on average vs. 39 of WT phage) (Extended Data Fig. 2). While cells infected with WT phages or ΔphuZ mutants complemented in trans had phage nuclei in the center of the cell ~80% of the time, the localization of the phage nucleus showed a wide distribution in cells infected by ΔphuZ mutants (Fig. 2A and 2B, Movie S1). This is consistent with the previous findings that trans over-expression of catalytic mutant PhuZ resulted in mispositioning of the phage nucleus [31, 33]. ~25% of mutant-infected cells still positioned the phage nucleus at the cell center (Fig. 2B), a phenotype most commonly seen in shorter cells (Pearson correlation coefficient = 0.486, p < 0.001), in contrast with WT infection where no correlation was observed with cell size (Pearson correlation coefficient = 0.029, p = 0.505) (Fig. 2C). Considering that PhuZ is proposed to traffic phage capsids from the cell inner membrane to the phage nucleus [32, 34], we speculate that this is only required under unidentified conditions or not at all. Taken together, these data confirm that PhuZ is required for the consistent positioning of the phage nucleus at the cell center during infection, but its removal, and the subsequent mislocalization of the phage nucleus, does not abolish phage replication under laboratory conditions.
Fig. 2: Absence of PhuZ causes mispositioning of the phage nucleus.
(A) Top: schematic of ΦKZ WT and phuZ::acrVIA1 (ΔphuZ) mutant genomes at the editing site. Bottom: Representative cells infected by WT (left) and ΔphuZ mutant (right). Phage DNA is stained by DAPI and shown as blue signals. The phage nucleus is mispositioned near the cell polar region upon infection by ΔphuZ, in contrast to the WT infection where the phage nucleus is centered. Scale bar denotes 2 μm. (B) Distribution of subcellular location of the phage nucleus in PAO1 cells infected by WT (blue, N = 521) and ΔphuZ (red, N = 503), and a PAO1 strain expressing wild-type PhuZ in trans (p-PhuZ) and infected by ΔphuZ (light green, N = 573). The diagram of an infected cell is shown on the top. The phage nucleus location is defined as the relative distance between the cell center and the nucleus center. (C) The phage nucleus location is plotted against the cell length for WT and ΔphuZ. The phage nucleus position in ΔphuZ-infected cells positively correlates with the cell length with a Pearson correlation coefficient of 0.486, p < 0.001 (two-sided), whereas there is no such correlation for WT infection with a Pearson correlation coefficient of 0.029, p = 0.505 (two-sided). (D) Efficiency of plating of WT and ΔphuZ on representative P. aeruginosa clinical strains. (The full panel of plaque assays is presented in Extended Data Fig. 4)
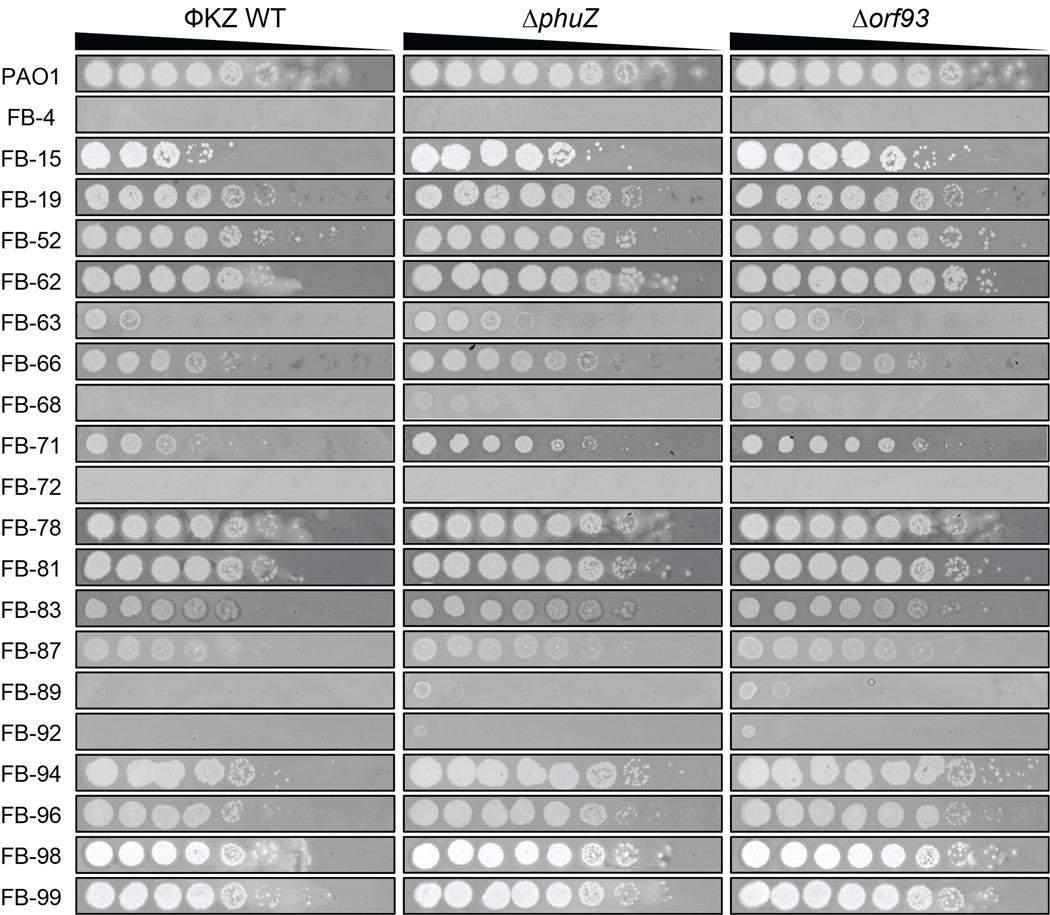
orf93 encodes gp93, a high copy number “inner body (IB)” protein that is packaged in the phage head [35, 36]. Replacement of orf93 with acrVIA1 (orf93::acrVIA1, herein Δorf93) also yielded viable phage that generated plaques with similar size and efficiency. To assess whether the loss of phuZ or orf93 impact growth in a strain-dependent manner, we challenged a panel of 21 P. aeruginosa clinical strains with the mutant phages. Plaque assays showed that host ranges and plaque morphologies of both mutants were quite similar to WT (Fig. 2D, Extended Data Fig. 4), suggesting that these knockouts, and Cas13a-mediated genetic engineering in general, has no impact on the ΦKZ host range.
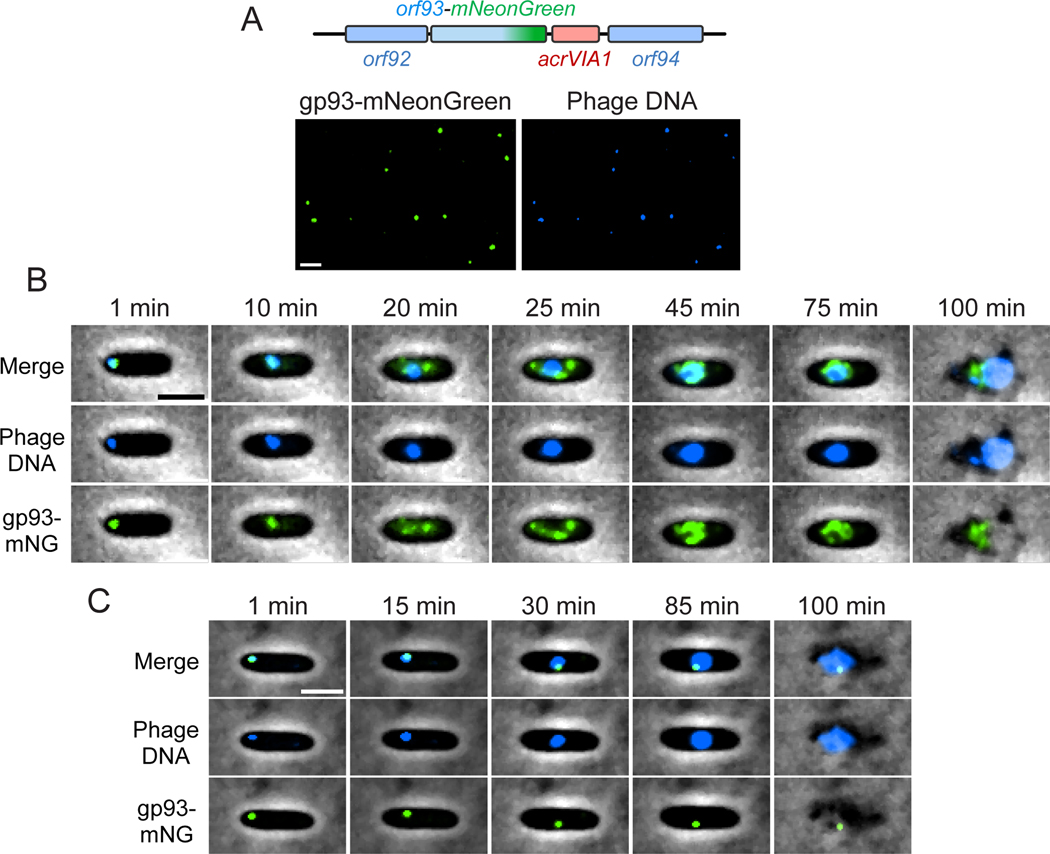
We next inserted a fluorescent label at the C-terminus of gp93 (Fig. 3A), which is notable as the first genomic protein tag in this phage family. This was again achieved with the acrVIA1 selectable marker (Fig. 3A). Labeled gp93 was observed in the mature virion (Fig. 3A), as predicted by previous mass spectrometry studies [36]. Excitingly, time-lapse movies revealed the fluorescently labeled protein being injected with the phage DNA at the cell pole (Fig. 3B, Movie S2) and subsequently translocating to the cell center where it remained bound to the phage nucleus. As new gp93-mNeonGreen was expressed from the phage genome, more and more green signals concentrated on the surface of the phage nucleus, while some foci appeared near the cell inner membrane. Finally, cells lysed and released fluorescent phage progeny. To confirm that the protein that appears to be injected was not rapidly synthesized de novo, we monitored the infection behaviors of WT phages loaded with gp93-mNeonGreen expressed from a plasmid during phage production, but where no new fluorescent protein could be made during infection (Fig. 3C, Movie S3). Similar to the engineered phage, the phage particles were fluorescent, injected the labeled protein, and the green focus migrated from the cell pole to the cell center along with phage DNA on the surface of the phage nucleus until cell lysis. No new protein synthesis was observed however, as expected. Therefore, the IB protein gp93 is not only packaged in the phage head, but may also play a role during phage injection and subsequent maturation to the phage nucleus. Our new ability to endogenously label phage proteins, as demonstrated here, will be beneficial for characterizing ΦKZ virion and cell biology in the future. Using a similar approach, mNeonGreen or mCherry were fused to PhuZ in the phage genome (Table 1), however, no fluorescent PhuZ filaments were visible upon infection. These fusions formed fluorescent filaments when expressed from a plasmid, but not from the phage genome, perhaps due to low expression levels or other factors.
Fig. 3: Fluorescent labeling of an inner body protein of ΦKZ.
(A) Top: schematic of ΦKZ orf93-mNeonGreen mutant at the editing site. The acrVIA1 gene was inserted downstream of the orf93-mNeonGreen fusion cassette. Bottom: visualization of individual phage particles under the fluorescence microscope. Each mutant phage particle is visible as a green focus (left), due to the packaging of gp93-mNeonGreen in the capsid. ΦKZ genomic DNA is stained by DAPI (right). mNeonGreen and DAPI signals colocalize very well and individual virions are easily distinguishable. Only ~1.4% of the fluorescent phage particles examined (6 out of 438) lacked the mNeonGreen signal. (B) Overlay images (phase-contrast and fluorescent channels) from a time-lapse movie depicting a representative PAO1 cell being infected by a gp93-mNeonGreen mutant phage. gp93-mNeonGreen and phage DNA are shown as green and blue signals, respectively. The representative cell was chosen out of 1093 infected cells from two independent infection experiments. (C) Overlay images from a time-lapse movie showing a PAO1 cell being infected by a WT ΦKZ loaded with gp93-mNeonGreen fusions. Packaged gp93-mNeonGreen appears as a green focus and remains bound to phage DNA throughout the infection cycle. The representative cell was chosen out of 1045 infected cells from two independent infection experiments. Scale bar denotes 2 μm.
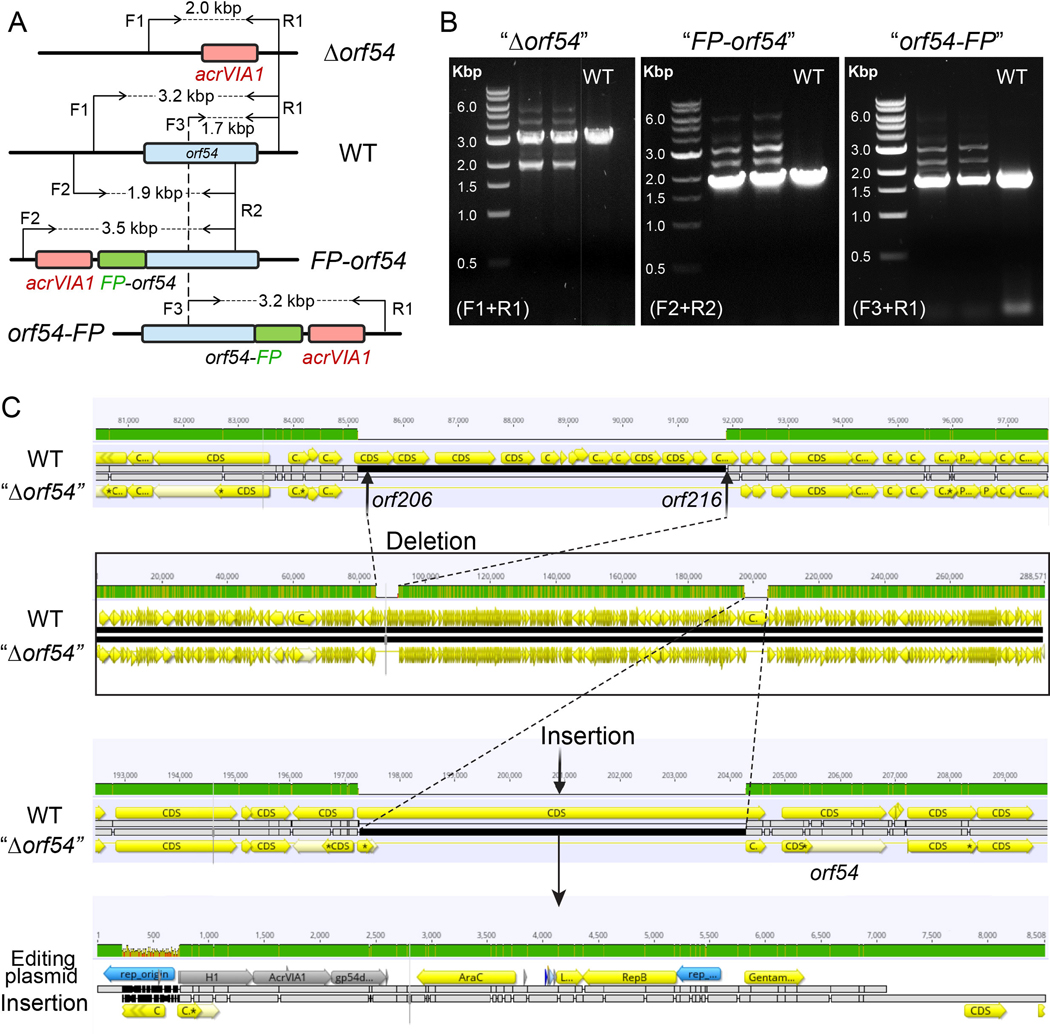
The phage nucleus is primarily composed of gp54 [31]. We were unable to knockout or fluorescently label orf54, even when wild-type gp54 was provided by expressing from a plasmid in trans. The primers used to amplify the region of editing generated multiple bands for both deletion and tag-addition mutant variants (Extended Data Fig. 5A and 5B). N- or C-terminal fusion of gp54 with mCherry tags yielded similar results. Whole genome sequencing of an isolated orf54 “pseudo knock-out” (i.e. a plaque that grew under Cas13a selection) strain revealed that part of the editing plasmid was integrated upstream of orf54, while the orf54 gene was left intact (Extended Data Fig. 5C). Interestingly, a gene cluster of nearly the same length as the integrated plasmid (~ 7 kbp, orf206–216) was missing in the mutant, likely due to a limitation of phage packaging capacity. These data suggested that natural gp54 produced from phages must be present to ensure phage viability. A similar attempt to delete the structural gene (orf146) and a cluster of IB genes (orf89-orf93) also failed, while deletion of small, hypervariable accessory genes orf241 and orf241–242 succeeded but yielded no change in plaque size or efficiency. These results highlighted that the CRISPR-Cas13a counter selection system is a strong and efficient phage genome engineering tool, but the modification of phage essential genes remains challenging.
Precise genome engineering of clinical phage OMKO1
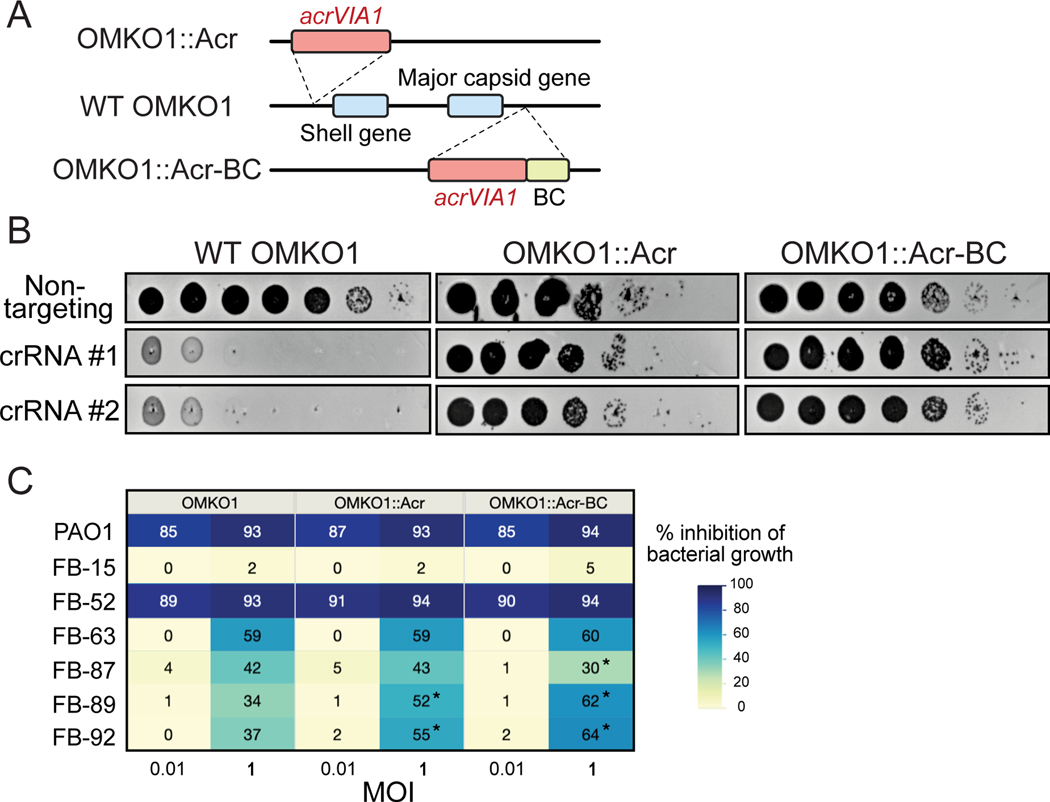
We next explored the versatility of our phage engineering platform by editing the genome of a clinical jumbo phage. We selected OMKO1, a P. aeruginosa phage with a ~280 kbp genome that has 90.1% nucleotide sequence identity to ΦKZ. OMKO1 is a potentially ideally therapeutic phage as P. aeruginosa strains that evolve resistance to infection become sensitized to small-molecule antibiotics [37]. This phage has been used for phage therapy as emergency treatment for chronic infections caused by antibiotic resistant P. aeruginosa [38], and it is currently being tested in a phase I/II clinical trial (CYPHY, https://clinicaltrials.gov/ct2/show/NCT04684641). With this phage, we tested whether we could insert “DNA barcodes” without impacting host range, for downstream clinical applications. Insertion of a DNA tracking signature into clinical phages would enable differentiation from naturally occurring phages during the manufacturing process and following administration to patients.
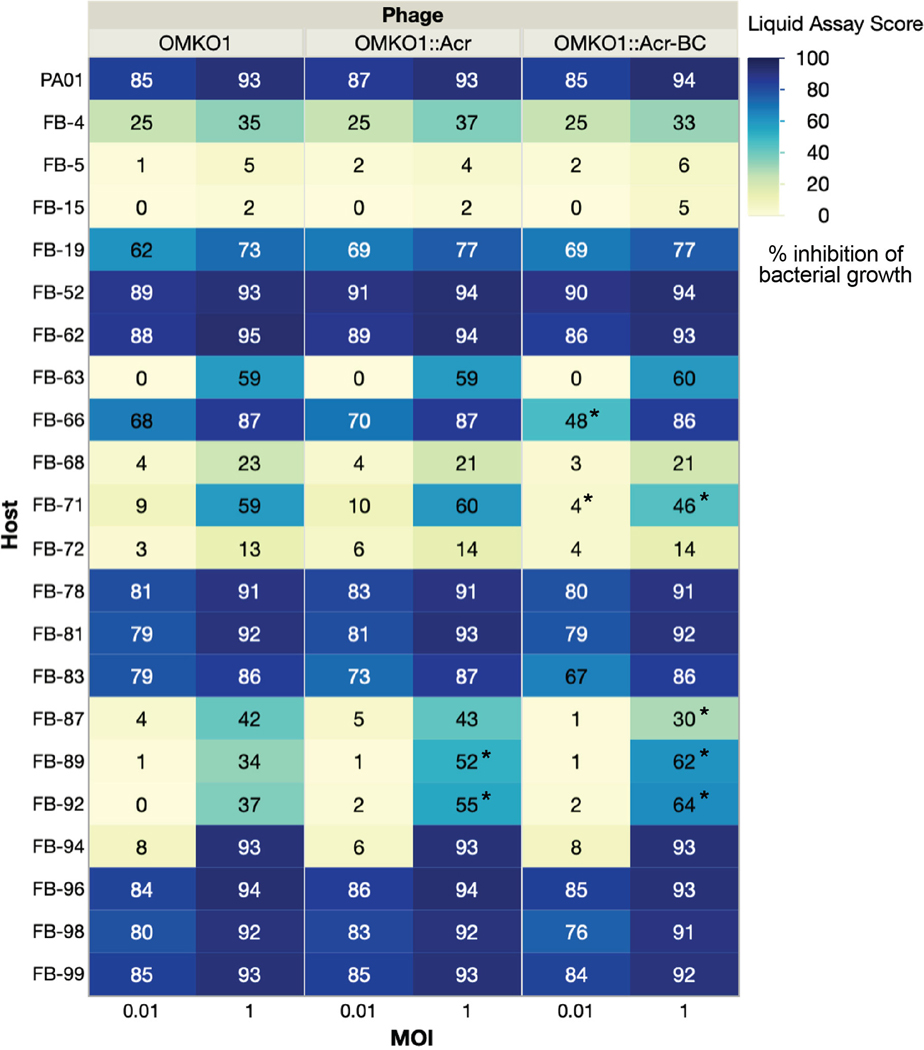
Two engineered OMKO1 strains were generated, one with acrVIA1 and a 120-nt barcode inserted downstream of the capsid gene, and another with acrVIA1 integrated upstream of the shell gene (orf54 homolog, Fig. 4A, Table 1). The presence of the desired inserts was confirmed with whole genome sequencing of both OMKO1 engineered strains, and no unintended genetic changes occurred. Moreover, both strains exhibited strong resistance to Cas13a targeting (Fig. 4B), owing to the expression of acrVIA1 from the phage genomes. The host range and virulence of the two engineered OMKO1 variants together with the parental phage was then assessed on 22 P. aeruginosa clinical strains (including PAO1). The experiment was performed in a microplate liquid assay, where phage variants were individually mixed with each host strain at a MOI~1 and MOI~0.01. All three phages displayed the same host range (Fig. 4C, Extended Data Fig. 6) and were capable of infecting and suppressing growth of 20/22 (91%) clinical strains tested. Infections at high MOI (MOI = 1) resulted in a broader host range and greater bacterial growth suppression, while low MOI (MOI = 0.01) infections were able to suppress cell growth of 12/22 (55%) hosts. All phages exhibited similar virulence across all hosts with small differences in 5/22 strains (marked with * in Extended Data Fig. 6). Altogether, these results indicate that OMKO1’s host range was not affected, and virulence was impacted only modestly by inserting acrVIA1 or acrVIA1 and a barcode in the two selected genome locations under the tested conditions.
Fig. 4: Genetic engineering of therapeutic jumbo phage OMKO1.
(A) Schematic of genomes of two engineered OMKO1 variants where indicated gene fragments are integrated into the WT genome. OMKO1::Acr, acrVIA1 is inserted upstream of the shell gene. OMKO1::Acr-BC, acrVIA1 is inserted together with a barcode sequence (BC) downstream of the major capsid gene. (B) Plaque assays of WT OMKO1 and mutants. Both engineered variants exhibit robust resistance against distinct CRISPR-Cas13a crRNAs, suggesting that the incorporated acrVIA1 is successfully functioning. crRNA #1 and #2 target OMKO1 homologs of ФKZ orf120 and orf146 transcripts, respectively. (C) Determination of host range of WT OMKO1 and mutants on representative P. aeruginosa clinical strains by microplate liquid assay at MOI of 0.01 and 1. Data are presented as the mean liquid assay scores across three independent experiments. Asterisks (*) indicate significant difference between WT and mutants as determined by two-sided Students’ T-tests (p < 0.05). The color intensity of each phage-host combination reflects the liquid assay score, with the darker color the stronger intensity displaying a greater score. Liquid assay score represents how well the phage strain can repress the growth of a given bacterial host. No inhibition of bacterial growth is reflected by a liquid assay score of 0, while complete suppression would result in a score of 100. (The full table of plaque assays is shown in Extended Data Fig. 6)
CRISPR-Cas13a engineering of a lytic Podophage
To evaluate the applicability of the CRISPR-Cas13a-mediated genome editing approach to other virulent phages, we selected P. aeruginosa phage PaMx41. PaMx41 is a Podophage (Jamesmcgillvirus) isolated from environmental and sewage water samples in Central Mexico [39]. Its genome is approximately 43.5 kbp long and harbors 55 open reading frames (ORFs), ~70% of which have unknown function [40]. Remarkably, we discovered that PaMx41 appears to be resistant to many DNA-targeting CRISPR-Cas systems (Type I-C, II-A, and V-A) and showed partial sensitivity (~10-fold reduction in efficiency of plating) to Type I-F to a degree that is not sufficient for counter selection (Fig. 5A). In contrast, when the transcripts of the major capsid gene (orf11) were targeted by CRISPR-Cas13a, PaMx41 exhibited strong sensitivities to specific crRNAs (Fig. 5B). Following the same approach as we developed to engineer ФKZ, we successfully substituted one hypothetical gene (orf24) and its downstream non-coding region with acrVIA1 and isolated a pure mutant strain using an efficient crRNA (#5, ~100-fold reduction in efficiency of plating) (Fig. 5B, Table 1). The mutant showed expected anti-CRISPR activities against different orf11-targeting crRNAs, indicating that the incorporated acrVIA1 was expressed and properly functioning (Fig. 5C). No other significant change in plaque size or efficiency was observed for the PaMx41 orf24::acrVIA1 phage on strain PAO1. This mutant did display decreased plaquing ability in another P. aeruginosa strain however, which has now been more thoroughly characterized in a separate publication [41]. Notably, initial PCR screening for recombinant plaques showed that 100% of surviving phages were desired recombinants, with no spontaneous escape plaques. The data suggest that the RNA-targeting Cas13a system holds great promise for becoming a universal genetic editing tool to deal with previously intractable phages.
Fig. 5: Genetic engineering of Podophage PaMx41.
(A) Plaque assays showing PaMx41 resisting a broad variety of DNA-targeting CRISPR-Cas systems, while being (B) targeted by CRISPR-Cas13a. The crRNA sequences of different CRISPR-Cas systems were listed in Table S3. crRNA#5 highlighted in the red frame has been used for PaMx41 genome engineering. (C) PaMx41 orf24::acrVIA1 mutant strain is resistant to diverse crRNAs of CRISPR-Cas13a, due to the expression of AcrVIA1 from the mutant genome. The mutant phages propagated better than WT on the cell lawns of all three crRNAs, though they appeared to exhibit slightly reduced EOP.
CONCLUSIONS
We applied the RNA-targeting CRISPR-Cas13a system, in conjunction with homologous recombination, to achieve genetic modification of jumbo phages ΦKZ, OMKO1, and the small podophage PaMx41, all of which are resistant to DNA-targeting CRISPR-Cas systems. CRISPR-Cas13a-mediated counter-selection recovered rare (~10−5) phage recombinants from a large pool of wild-type phages. It is notable that despite the phage nucleus present in jumbo phage ΦKZ, which renders the ΦKZ genome inaccessible to many bacterial proteins, the editing plasmid DNA can interact with the ΦKZ genome to recombine with expected frequencies.
Many studies have reported that phages can hamper CRISPR-Cas activities, for example, by repressing transcription of endogenous CRISPR-Cas components [42, 43], possessing covalent DNA modifications [44–47], or encoding anti-CRISPR proteins (recently reviewed in [48]). Furthermore, the assembly of a proteinaceous nucleus-like structure that shields phage genomes from attack by distinct DNA-targeting nucleases [20, 21] represents the ultimate “anti-CRISPR/anti-RM” mechanism. Therefore, development of phage genomic manipulation approaches that target mRNA, which is a relatively consistent and exposed molecule, may provide a near-universal approach. Moreover, Cas13 is rarely encoded in bacteria [49, 50] and most phages are therefore not expected to encode anti-Cas13a proteins.
Applying gene editing to ΦKZ allowed us to query endogenous gene function and essentiality for the first time. We observed that ΔphuZ mutant phages mispositioned the phage nucleus during viral intracellular development. Previous studies revealed that newly assembled phage capsids trafficked along PhuZ filaments towards the phage nucleus for viral DNA packaging [32]. However, our work demonstrates that successful DNA loading into capsids is not dependent on PhuZ. Moreover, loss of PhuZ appeared to lower burst size, but did not impact host range or plaque morphology. Overall, phuZ seems to be a bona fide nonessential gene for ΦKZ. The evolutionary advantage of encoding tubulin in this jumbo phage and many others requires further investigation. Furthermore, a fluorescent label on gp93 demonstrated that it is packaged in the phage head, injected with the genome, and massively synthesized later in infection, with peri-nuclear localization. The labelling not only allows us to visualize individual virions under the microscope, but also to observe the injection of this inner body protein into the host cell, which had been previously suggested with little evidence [36, 51].
One major challenge of using Cas13a, in our experience, has been the wide variability of crRNA efficacy. Future studies focusing on the optimization of crRNA design for phage targeting or perhaps the implementation of other RNA-targeting enzymes, such as Cas13 orthologues or Cas7–11 [52, 53] will be important. However, we can circumvent this problem by implementing an anti-CRISPR selectable marker [17] to ensure that the same strong guide can be used for all genetic manipulations for a given phage strain. The downside is that this limits the user to a single perturbation and leaves the acr gene in the phage genome. However, double and triple mutants are possible in principle if one uses crRNAs specific to the site of editing [50], negating the need for an acr gene. One could also remove the acr gene from the genome, for example with the introduction of FLP recombination sites and a phage nucleus-localized Flp recombinase.
In summary, the RNA-targeting CRISPR-Cas13a counter-selection tool should be applicable to a broad range of phages and enable downstream high throughput phage engineering. In addition to the dsDNA phage efficacy demonstrated here, this approach could also be suitable, in principle, to engineer ssDNA phages. The ability to generate synthetic phages precisely and efficiently with desired features will not only benefit phage therapeutic applications but will also advance our understanding of fundamental phage biology and phage-bacteria interactions.
ONLINE METHODS
Strains, DNA oligonucleotides and plasmid constructions
All bacterial and phage strains, plasmids, spacer sequences, and primers used in this study are listed in Tables S1, S2, S3, and S4, respectively.
The crRNAs designed for CRISPR-Cas13 targeting were constructed in the pHERD30T backbone. The pHERD30T-crRNA Version 2 was constructed by thermal annealing of oligonucleotides oSDM465 and oSDM466 and phosphorylation by polynucleotide kinase (PNK). The annealed product was introduced by Gibson assembly into pHERD30T linearized by PCR using oligonucleotides oSDM457 and oSDM458. Proper construction of the expression vector was verified by Sanger sequencing. The pHERD30T-crRNA Version 3 was constructed just as for Version 2, but the crRNA-coding insert was instead composed of oligonucleotides oSDM455 and oSDM456. Both Version 2 and Version 3 of this plasmid were designed such that cleavage by BsaI would generate a linear plasmid that would accept annealed oligonucleotide spacers via ligation. Oligonucleotide pairs with repeat-specific overhangs encoding spacer sequences were annealed and phosphorylated using T4 polynucleotide kinase and then cloned into the BsaI-digested empty vectors. Cloning procedures were performed in commercial E. coli DH5α cells (New England Biolabs) according to the manufacturer’s protocols. The resulting crRNA plasmids were electroporated into P. aeruginosa PAO1 strain harboring the tn7::cas13aLse (SDM084) on the chromosome as described previously [20]. Gene expression was induced by the addition of L-arabinose at a final concentration of 0.3% and isopropyl β-D-1-thiogalactopyranoside (IPTG) at a final concentration of 1 mM.
To construct editing plasmids for homologous recombination, homology arms of >500 bp in length were amplified by PCR using the ΦKZ genomic DNA as the template. To prevent Cas13a cleavage, several synonymous mutations were introduced into the crRNA-targeting site of the left orf120 homology arm by designing the reverse primer (JG064) to contain appropriate mismatches. The acrVIA1 gene was amplified from plasmid pAM383 [25], a gift from Luciano Marraffini, The Rockefeller University. PCR products were purified and assembled as a recombineering substrate and then inserted into the NheI site of the pHERD30T backbone by Gibson Assembly (New England Biolabs) following the manufacturer’s protocols. The resulting plasmids were electroporated into PAO1.
To construct plasmids for complementing wild-type PhuZ, gp54, and gp146 of ФKZ in trans, phuZ, orf54 or operon#24 (containing both orf53 and orf54), and orf146 were amplified by PCR using the ΦKZ genomic DNA as the template. The PCR products were then cloned into the the pHERD30T backbone at either the NheI recognition site or between two different restriction sites. The resulting plasmids were introduced into PAO1 by electroporation. Thereafter, mutant phages were used to infect the appropriate PAO1 strains carrying the corresponding plasmid for complementation.
Isolation of phage recombinants
Host strains bearing editing plasmids were grown in LB supplemented with 10 mM MgSO4 and 50 μg/ml gentamicin, at 37°C with aeration at 250 rpm. When OD600 is around 2, Wild type ΦKZ was added into the culture at a MOI (multiplicity of infection) of 1 to allow infection to occur for ~18 hours. 2% volume of chloroform was added into the infection culture and left to shake gently on an orbital shaker at room temperature for 15 min, followed by centrifugation at 4,000 x g for 15 min to remove cell debris. The supernatant lysate was further treated with 2% of chloroform for 15 min and centrifuged again under the same conditions, followed by a 30-min treatment with DNase I (New England Biolabs) at 37°C. The resulting phage lysate containing both WT phages and recombinants are tittered on PAO1 strains bearing the CRISPR-Cas13a system with the most efficient crRNA (orf120 crRNA#2) to screen for recombinants. Individual phage plaques were picked from top agar and purified for three rounds using the CRISPR counter-selection strain to ensure thorough removal of any remaining WT or escape mutants. Whether or not they are recombinant phages or Cas13a escape phages were determined by PCR using appropriate pairs of primers amplifying the modified regions of the phage genome. Identified phages were further confirmed and analyzed by sequencing the PCR products or the whole genomes and then stored at 4°C.
Phage plaque assay
Host strains were grown in LBM (LB supplemented with 10 mM MgSO4), 50 μg/ml gentamicin, 1 mM IPTG and 0.3% arabinose inducers for gene expression, at 37°C with aeration at 250 rpm for overnight. Phage spotting assays were performed using 1.5% LB agar plates and 0.42% LB top agar, both of which contained 10 mM MgSO4 and inducers. 100 μl of appropriate overnight culture was suspended in 3.5 ml of molten top agar and then poured onto an LB+10 mM MgSO4 agar plate, leading to the growth of a bacterial lawn. After 10–15 min at room temperature, 2 μl of ten-fold serial dilutions of phages was spotted onto the solidified top agar. Plates were incubated overnight at 37°C. Plate images were obtained using Gel Doc EZ Gel Documentation System (BioRad) and Image Lab (BioRad) software version 6.0.1.
Microplate Liquid Assay
Fresh overnight cultures were diluted to a cell concentration of 1×108 cfu/ml in LB media supplemented with 10 mM MgSO4. Phage lysates were added to reach a MOI of ~1 and ~0.01 in a Corning Costar 96-well clear flat-bottom microplate (Thermo Fisher Scientific) sealed with a Breathe-Easy® sealing membrane (Merck KGaA). After the infection cultures were incubated at room temperature for 20 min, plates were incubated at 37°C, 800 rpm for 8 hours in a BioTek LogPhase 600 plate reader (Agilent Technologies, Inc.). Cell growth was monitored by measuring OD600 every 20 min. Each phage-host combination was performed in three biological replicates.
Data Analysis
Growth curves for each phage-host combination were obtained by plotting OD600 after blank correction (baseline adjustment) against time. Each growth curve was transformed into a single numerical value by calculating the area under the curve (AUC) using the Trapezoid method. Then, AUCs were normalized as a percentage of the AUC of their corresponding uninfected control following the equation,
The resulting value, defined as the “liquid assay score”, represents how well the phage strain can repress the growth of a bacterial population over the course of the 8-hour experiment. No inhibition of bacterial growth would result in a liquid assay score of 0, and complete suppression would translate into a score of 100. Liquid assay scores were averaged using data from three biological replicates.
One-step growth curve
Phage burst size was determined by one-step growth curve experiments. The host PAO1 strain was grown in LBM (LB supplemented with 10 mM MgSO4) media to OD600 ~0.4 at 37°C with aeration at 250 rpm. 1 ml of the cell culture was then mixed with the phage lysate to achieve an MOI of 0.01. The mixture was incubated in a 37°C heat block for 10 min for infection. 100 μl of the infection mixture was then added to 4.9 ml of fresh, pre-warmed LBM and incubated at 37°C with shaking at 250 rpm. The initial phage titer was determined by phage titration. At 1 min from this point, 950 μl of the culture was transferred into an Eppendorf tube supplied with 50 μl of chloroform, followed by phage titration to calculate the number of free phages. Samples were collected at 15-min or 10-min intervals for ~120 min, and phage titers were determined immediately. Phage titers at different time points were plotted against time to determine the latent period and burst size of each phage strain. Phage burst sizes were calculated by dividing the average phage titers at the plateau phase by the initial phage titers after subtracting free phages. Assays were performed at least three biological replicates for each phage strain and burst sizes are the mean of three or four measurements.
Single-cell infection assay
A single colony was inoculated in 1 ml of LBM supplied with 50 μg/ml gentamicin (if necessary), and grown at 37°C with 250 rpm shaking for overnight. The overnight culture was diluted 100-fold in 5 ml of LBM and grown at 37°C with 250 rpm shaking to an OD600 of ~0.4. Next, 1 ml of the cell culture was collected by centrifugation at 3,000 × g for 2 min at room temperature and concentrated by 25-fold in fresh LBM. 10 μl of cells were then mixed with 10 μl of appropriate phage strains to reach an appropriate MOI, followed by incubation at 30°C for 10 min to allow for phage infection. The infection mixture was further diluted by 10-fold into 50 μl of fresh LBM at room temperature. 1 μl of the diluted culture was gently placed onto a piece of agarose pad (~1 mm thick) with 1:5 diluted LBM, arabinose (0.8%), and DAPI (5 μg/ml; Invitrogen™, No. D1306). A coverslip (No.1.5, Fisher Scientific) was gently laid over the agarose pad and the sample was imaged under the fluorescence microscope at 30°C within a cage incubator to maintain temperature and humidity.
Fluorescence microscopy and imaging
Microscopy was performed on an inverted epifluorescence (Ti2-E, Nikon, Tokyo, Japan) equipped with the Perfect Focus System (PFS) and a Photometrics Prime 95B 25-mm camera. Image acquisition and processing were performed using Nikon Elements AR software (5.02.00 64-bit). During a time-lapse movie, the specimen was typically imaged at a time interval of 5 min at the focal plane for 2.5~3 h, through channels of phase contrast (200 ms exposure, for cell recognition), blue (DAPI, 200 ms exposure, for phage DNA), and green (GFP, 300 ms exposure, for Gp93-mNeonGreen).
Next-generation sequencing (NGS)
To isolate phage genomic DNA, purified high titer lysates were treated with Benzonase Nuclease (Sigma) for 30 min at 37°C. Phage genomic DNA was extracted following a modified Wizard DNA Clean-Up kit (Promega) protocol. For binding DNA to the column, we kept a ratio of 1:2 lysate:Wizard® DNA Clean-Up Resin. In the elution step,100 μl of pre-warmed H2O was added to the column and immediately centrifuged at 13,000 × g for 1 min to elute DNA. DNA samples were quantified with AccuGreen Broad Range dsDNA quantification kit (Biotium, USA) in a Qubit Fluorometer 2.0.
Purified phage genomic DNA was processed following Illumina DNA Preparation Protocol. Samples were sequenced on a MiSeq system (Illumina) with 300 cycles of paired-end sequencing, and loading concentration of 12 pM. Illumina short reads were downsampled to ~50–100× coverage and de novo assembled using SPAdes. The sequences of mutant phage strains were aligned to the reference genome in Geneious with the Mauve alignment algorithm to confirm the intended genomic edits.
The isolated orf54 “pseudo knock-out” phage strain (“Δorf54”) was sequenced using long-read sequencing. DNA samples were processed using SQK-LSK109 kit (Oxford Nanopore Technologies, UK). Libraries were sequenced using an R10.3 flow cell until the desired number of reads was achieved. Oxford Nanopore long reads were filtered for the longest high quality reads using Nanofilt and de novo assembled using Flye.
Extended Data
Extended Data Fig. 1. Plaque efficiency assays of distinct crRNAs of CRISPR-Cas13a targeting transcripts of diverse ΦKZ genes.
Different crRNAs show varied degrees of Cas13a interference efficiency against ΦKZ. The crRNA targeting orf120 highlighted in the red frame has been used for ΦKZ genome engineering. NT, non-targeting.
Extended Data Fig. 2. One-step growth curves of engineered ФKZ variants.
One-step growth curve experiment was performed to determine the latent time period and burst size of engineered phages. Representative plots are shown for each phage strain. The burst sizes are shown in brackets after of each ФKZ variant, representing the mean ± standard error of three or four biologically independent replicates.
Extended Data Fig. 3. Sequence alignment of wild type ΦKZ and three escape mutants at the engineered genomic site.
Escape mutants were isolated and verified by PCR and sequencing. The WT orf120 sequence is highlighted in blue and the downstream region is highlighted in grey. The stop codon (TGA) of orf120 is highlighted in green and Escape mutant #3 reconstitutes it to TAG. The sequence in the red frame matches the spacer sequence of the crRNA that was used to target and eliminate WT phages. Deletions were indicated by dashed lines and their corresponding numbers of absent base pairs.
Extended Data Fig. 4. Determination of host range of ΦKZ ΔphuZ and Δorf93 mutants by plaque assay on P. aeruginosa clinical strains.
Spot-titration of the indicated ΦKZ phages on lawns of clinical isolates of P. aeruginosa (FB-XX).
Extended Data Fig. 5. Failure of genetic editing the shell gene (orf54) in ΦKZ.
(A) Schematic of genomes of WT ΦKZ and three mutated orf54 variants, “Δorf54”, “FP-orf54”, and “orf54-FP”, at the editing site. orf54, acrVIA1, and fluorescent protein (FP) are shown as blue, red, and green rectangles, respectively. F and R indicate forward and reverse primers, respectively, for PCR confirmation of orf54 engineering. (B) PCR confirmation of the indicated orf54 mutants using their corresponding pair of primers. All three mutants generated multiple bands, including a band in the same size as the single band produced by WT. PCR-based screening for engineered ΦKZ orf54 variants have been independently repeated at least three times yielding similar results. (C) Genome alignment of WT phage with the isolated orf54 “pseudo knock-out” mutant (“Δorf54”). A gene cluster of ~ 7 kbp (orf206 - orf216) was missing in the mutant, likely as a result of phage packaging capacity. The majority of the editing plasmid used to generate recombinants was at the editing site, leaving orf54 intact.
Extended Data Fig. 6. Host range assay of engineered OMKO1 variants.
Host ranges were determined by microplate liquid assay at MOI of 0.01 and 1 on 22 P. aeruginosa clinical strains. The values are presented as the mean liquid assay scores across three independent experiments. Asterisks (*) indicate significant difference between WT and engineered strains as determined by two-sided Students’ T-tests (p < 0.05). The color intensity of each phage-host combination reflects the liquid assay score, which represents how well the phage strain can repress the growth of a given bacterial host. No inhibition of bacterial growth is reflected by a liquid assay score of 0, and complete suppression would result in a score of 100.
Supplementary Material
Acknowledgements
J.B.-D. was supported by the National Institutes of Health (R01GM127489), the Vallee Foundation, the Searle Scholarship, the Innovative Genomics Institute, and the UCSF Program for Breakthrough Biomedical Research funded in part by the Sandler Foundation. This work was also supported by research funds from Felix Biotechnology.
We are grateful to Luciano Marraffini (The Rockefeller University) for providing the plasmid pAM383. We thank Paul Turner and Ben Chan (Yale University) for providing phage OMKO1 and P. aeruginosa clinical isolates. We thank Gabriel Guarneros Peña at Centro de Investigación y de Estudios Avanzados for providing phage PaMx41. We thank Tomer Rotstein for his generous assistance with NGS data processing and interpretation. We thank members of the Bondy-Denomy laboratory for productive conversations and generous suggestions for our work.
Footnotes
Competing Interests
J.B.-D. is a scientific advisory board member of SNIPR Biome, Excision Biotherapeutics, and Leapfrog Bio, and a scientific advisory board member and co-founder of Acrigen Biosciences. The Bondy-Denomy lab receives research support from Felix Biotechnology. The remaining authors declare no competing interests.
Reviewer recognition statement:
Nature Microbiology thanks Anne Chevallereau and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
All relevant data are included in the paper and/or its supplementary information files. The complete genome sequence of OMKO1 was deposited in GenBank under accession number ON631220. All strains and plasmids are available from the corresponding author upon request. Source data are provided with this paper.
Code availability
Data analysis code is available from public repositories at https://zenodo.org/record/6324407#.YiAjrejMI2w
REFERENCES
- 1.Kortright KE, Chan BK, Koff JL, and Turner PE (2019). Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe 25, 219–232. [DOI] [PubMed] [Google Scholar]
- 2.Pires DP, Cleto S, Sillankorva S, Azeredo J, and Lu TK (2016). Genetically Engineered Phages: a Review of Advances over the Last Decade. Microbiol Mol Biol Rev 80, 523–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doss J, Culbertson K, Hahn D, Camacho J, and Barekzi N (2017). A Review of Phage Therapy against Bacterial Pathogens of Aquatic and Terrestrial Organisms. Viruses 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nobrega FL, Costa AR, Kluskens LD, and Azeredo J (2015). Revisiting phage therapy: new applications for old resources. Trends Microbiol 23, 185–191. [DOI] [PubMed] [Google Scholar]
- 5.Lusiak-Szelachowska M, Zaczek M, Weber-Dabrowska B, Miedzybrodzki R, Klak M, Fortuna W, Letkiewicz S, Rogoz P, Szufnarowski K, Jonczyk-Matysiak E, et al. (2014). Phage Neutralization by Sera of Patients Receiving Phage Therapy. Viral Immunol 27, 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weber-Dabrowska B, Jonczyk-Matysiak E, Zaczek M, Lobocka M, Lusiak-Szelachowska M, and Gorski A (2016). Bacteriophage Procurement for Therapeutic Purposes. Front Microbiol 7, 1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu TK, and Koeris MS (2011). The next generation of bacteriophage therapy. Curr Opin Microbiol 14, 524–531. [DOI] [PubMed] [Google Scholar]
- 8.Lenneman BR, Fernbach J, Loessner MJ, Lu TK, and Kilcher S (2020). Enhancing phage therapy through synthetic biology and genome engineering. Curr Opin Biotechnol 68, 151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ando H, Lemire S, Pires DP, and Lu TK (2015). Engineering Modular Viral Scaffolds for Targeted Bacterial Population Editing. Cell Syst 1, 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahichi F, Synnott AJ, Yamamichi K, Osada T, and Tanji Y (2009). Site-specific recombination of T2 phage using IP008 long tail fiber genes provides a targeted method for expanding host range while retaining lytic activity. FEMS Microbiol Lett 295, 211–217. [DOI] [PubMed] [Google Scholar]
- 11.Matsuda T, Freeman TA, Hilbert DW, Duff M, Fuortes M, Stapleton PP, and Daly JM (2005). Lysis-deficient bacteriophage therapy decreases endotoxin and inflammatory mediator release and improves survival in a murine peritonitis model. Surgery 137, 639–646. [DOI] [PubMed] [Google Scholar]
- 12.Monteiro R, Pires DP, Costa AR, and Azeredo J (2019). Phage Therapy: Going Temperate? Trends Microbiol 27, 368–378. [DOI] [PubMed] [Google Scholar]
- 13.Kilcher S, and Loessner MJ (2019). Engineering Bacteriophages as Versatile Biologics. Trends Microbiol 27, 355–367. [DOI] [PubMed] [Google Scholar]
- 14.Marinelli LJ, Hatfull GF, and Piuri M (2012). Recombineering: A powerful tool for modification of bacteriophage genomes. Bacteriophage 2, 5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deveau H, Garneau JE, and Moineau S (2010). CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol 64, 475–493. [DOI] [PubMed] [Google Scholar]
- 16.Hille F, Richter H, Wong SP, Bratovic M, Ressel S, and Charpentier E (2018). The Biology of CRISPR-Cas: Backward and Forward. Cell 172, 1239–1259. [DOI] [PubMed] [Google Scholar]
- 17.Mayo-Munoz D, He F, Jorgensen JB, Madsen PK, Bhoobalan-Chitty Y, and Peng X (2018). Anti-CRISPR-Based and CRISPR-Based Genome Editing of Sulfolobus islandicus Rod-Shaped Virus 2. Viruses 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samson JE, Magadan AH, Sabri M, and Moineau S (2013). Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol 11, 675–687. [DOI] [PubMed] [Google Scholar]
- 19.Malone LM, Birkholz N, and Fineran PC (2020). Conquering CRISPR: how phages overcome bacterial adaptive immunity. Curr Opin Biotechnol 68, 30–36. [DOI] [PubMed] [Google Scholar]
- 20.Mendoza SD, Nieweglowska ES, Govindarajan S, Leon LM, Berry JD, Tiwari A, Chaikeeratisak V, Pogliano J, Agard DA, and Bondy-Denomy J (2020). A bacteriophage nucleus-like compartment shields DNA from CRISPR nucleases. Nature 577, 244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malone LM, Warring SL, Jackson SA, Warnecke C, Gardner PP, Gumy LF, and Fineran PC (2020). A jumbo phage that forms a nucleus-like structure evades CRISPR-Cas DNA targeting but is vulnerable to type III RNA-based immunity. Nat Microbiol 5, 48–55. [DOI] [PubMed] [Google Scholar]
- 22.Guan J, and Bondy-Denomy J (2020). Intracellular Organization by Jumbo Bacteriophages. J Bacteriol 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E, Minakhin L, et al. (2016). C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meeske AJ, Nakandakari-Higa S, and Marraffini LA (2019). Cas13-induced cellular dormancy prevents the rise of CRISPR-resistant bacteriophage. Nature 570, 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meeske AJ, Jia N, Cassel AK, Kozlova A, Liao J, Wiedmann M, Patel DJ, and Marraffini LA (2020). A phage-encoded anti-CRISPR enables complete evasion of type VI-A CRISPR-Cas immunity. Science 369, 54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.East-Seletsky A, O’Connell MR, Knight SC, Burstein D, Cate JH, Tjian R, and Doudna JA (2016). Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meeske AJ, and Marraffini LA (2018). RNA Guide Complementarity Prevents Self-Targeting in Type VI CRISPR Systems. Mol Cell 71, 791–801 e793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.L MI, Anantharaman V, Krishnan A, Burroughs AM, and Aravind L (2021). Jumbo Phages: A Comparative Genomic Overview of Core Functions and Adaptions for Biological Conflicts. Viruses 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Al-Shayeb B, Sachdeva R, Chen LX, Ward F, Munk P, Devoto A, Castelle CJ, Olm MR, Bouma-Gregson K, Amano Y, et al. (2020). Clades of huge phages from across Earth’s ecosystems. Nature 578, 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aylett CH, Izore T, Amos LA, and Lowe J (2013). Structure of the tubulin/FtsZ-like protein TubZ from Pseudomonas bacteriophage PhiKZ. J Mol Biol 425, 2164–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaikeeratisak V, Nguyen K, Egan ME, Erb ML, Vavilina A, and Pogliano J (2017). The Phage Nucleus and Tubulin Spindle Are Conserved among Large Pseudomonas Phages. Cell Rep 20, 1563–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaikeeratisak V, Khanna K, Nguyen KT, Sugie J, Egan ME, Erb ML, Vavilina A, Nonejuie P, Nieweglowska E, Pogliano K, et al. (2019). Viral Capsid Trafficking along Treadmilling Tubulin Filaments in Bacteria. Cell 177, 1771–1780 e1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kraemer JA, Erb ML, Waddling CA, Montabana EA, Zehr EA, Wang H, Nguyen K, Pham DS, Agard DA, and Pogliano J (2012). A phage tubulin assembles dynamic filaments by an atypical mechanism to center viral DNA within the host cell. Cell 149, 1488–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaikeeratisak V, Nguyen K, Khanna K, Brilot AF, Erb ML, Coker JK, Vavilina A, Newton GL, Buschauer R, Pogliano K, et al. (2017). Assembly of a nucleus-like structure during viral replication in bacteria. Science 355, 194–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu W, Thomas JA, Cheng N, Black LW, and Steven AC (2012). Bubblegrams reveal the inner body of bacteriophage phiKZ. Science 335, 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas JA, Weintraub ST, Wu W, Winkler DC, Cheng N, Steven AC, and Black LW (2012). Extensive proteolysis of head and inner body proteins by a morphogenetic protease in the giant Pseudomonas aeruginosa phage phiKZ. Mol Microbiol 84, 324–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan BK, Sistrom M, Wertz JE, Kortright KE, Narayan D, and Turner PE (2016). Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci Rep 6, 26717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan BK, Turner PE, Kim S, Mojibian HR, Elefteriades JA, and Narayan D (2018). Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol Med Public Health 2018, 60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sepulveda-Robles O, Kameyama L, and Guarneros G (2012). High diversity and novel species of Pseudomonas aeruginosa bacteriophages. Appl Environ Microbiol 78, 4510–4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cruz-Plancarte I, Cazares A, and Guarneros G (2016). Genomic and Transcriptional Mapping of PaMx41, Archetype of a New Lineage of Bacteriophages Infecting Pseudomonas aeruginosa. Appl Environ Microbiol 82, 6541–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huiting E, Athukoralage J, Guan J, Silas S, Carion H, and Bondy-Denomy J (2022). Bacteriophages antagonize cGAS-like immunity in bacteria. bioRxiv doi: 10.1101/2022.03.30.486325. [DOI] [Google Scholar]
- 42.Skennerton CT, Angly FE, Breitbart M, Bragg L, He S, McMahon KD, Hugenholtz P, and Tyson GW (2011). Phage encoded H-NS: a potential achilles heel in the bacterial defence system. PLoS One 6, e20095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pul U, Wurm R, Arslan Z, Geissen R, Hofmann N, and Wagner R (2010). Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol Microbiol 75, 1495–1512. [DOI] [PubMed] [Google Scholar]
- 44.Hampton HG, Watson BNJ, and Fineran PC (2020). The arms race between bacteria and their phage foes. Nature 577, 327–336. [DOI] [PubMed] [Google Scholar]
- 45.Vlot M, Houkes J, Lochs SJA, Swarts DC, Zheng P, Kunne T, Mohanraju P, Anders C, Jinek M, van der Oost J, et al. (2018). Bacteriophage DNA glucosylation impairs target DNA binding by type I and II but not by type V CRISPR-Cas effector complexes. Nucleic Acids Res 46, 873–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bryson AL, Hwang Y, Sherrill-Mix S, Wu GD, Lewis JD, Black L, Clark TA, and Bushman FD (2015). Covalent Modification of Bacteriophage T4 DNA Inhibits CRISPR-Cas9. mBio 6, e00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y, Dai L, Dong J, Chen C, Zhu J, Rao VB, and Tao P (2020). Covalent Modifications of the Bacteriophage Genome Confer a Degree of Resistance to Bacterial CRISPR Systems. J Virol 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davidson AR, Lu WT, Stanley SY, Wang J, Mejdani M, Trost CN, Hicks BT, Lee J, and Sontheimer EJ (2020). Anti-CRISPRs: Protein Inhibitors of CRISPR-Cas Systems. Annu Rev Biochem 89, 309–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, Charpentier E, Cheng D, Haft DH, Horvath P, et al. (2020). Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol 18, 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adler B, Hessler T, Cress B, Mutalik V, Barrangou R, Banfield J, and Doudna JA (2022). RNA-targeting CRISPR-Cas13 Provides Broad-spectrum Phage Immunity. bioRxiv doi: 10.1101/2022.03.25.485874. [DOI] [Google Scholar]
- 51.Krylov VN, Smirnova TA, Minenkova IB, Plotnikova TG, Zhazikov IZ, and Khrenova EA (1984). Pseudomonas bacteriophage phi KZ contains an inner body in its capsid. Can J Microbiol 30, 758–762. [DOI] [PubMed] [Google Scholar]
- 52.van Beljouw SPB, Haagsma AC, Rodriguez-Molina A, van den Berg DF, Vink JNA, and Brouns SJJ (2021). The gRAMP CRISPR-Cas effector is an RNA endonuclease complexed with a caspase-like peptidase. Science 373, 1349–1353. [DOI] [PubMed] [Google Scholar]
- 53.Ozcan A, Krajeski R, Ioannidi E, Lee B, Gardner A, Makarova KS, Koonin EV, Abudayyeh OO, and Gootenberg JS (2021). Programmable RNA targeting with the single-protein CRISPR effector Cas7–11. Nature 597, 720–725. [DOI] [PubMed] [Google Scholar]
- 54.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, et al. (2000). Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406, 959–964. [DOI] [PubMed] [Google Scholar]
- 55.Qiu D, Damron FH, Mima T, Schweizer HP, and Yu HD (2008). PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl Environ Microbiol 74, 7422–7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.