Summary
The classical Cre-LoxP system is time consuming. Here we detail a protocol that leverages Rosa26-LSL-Cas9;Adiponectin-Cre mice to restrict Cas9 expression in adipocytes. This enables specific deletion of target genes in brown adipocytes within 6 weeks by local injection of AAV-sgRNA into interscapular brown adipose tissue. We also describe an adiponectin-promoter-driven AAV vector to express sgRNA-resistant cDNA-encoded protein for subsequent rescue. This protocol thus provides an efficient means to specifically knockout and overexpress genes in brown adipocytes in vivo.
For complete details on the use and execution of this protocol, please refer to Xue et al. (2022).1
Subject areas: Metabolism, Model Organisms, Molecular Biology, CRISPR
Graphical abstract

Highlights
-
•
Generation of adipocyte-specific Cas9 transgenic mice for gene editing
-
•
Brown-adipocyte-specific knockout by local injection of AAV-sgRNA within 6 weeks
-
•
Adiponectin-promoter-driven expression of sgRNA-resistant protein for rescue
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
The classical Cre-LoxP system is time consuming. Here we detail a protocol that leverages Rosa26-LSL-Cas9;Adiponectin-Cre mice to restrict Cas9 expression in adipocytes. This enables specific deletion of target genes in brown adipocytes within 6 weeks by local injection of AAV-sgRNA into interscapular brown adipose tissue. We also describe an adiponectin-promoter-driven AAV vector to express sgRNA-resistant cDNA-encoded protein for subsequent rescue. This protocol thus provides an efficient means to specifically knockout and overexpress genes in brown adipocytes in vivo.
Before you begin
This protocol describes how to specifically and efficiently knockout genes of interest and perform their rescues in brown adipocytes in vivo. This protocol leverages key technique advances towards the uses of AAVs to target adipose tissues and the methods to inject interscapular brown adipose tissue (iBAT).2,3,4,5,6,7,8
Ucp1Cre transgenic mice are utilized in the Cre-LoxP system to allow “BAT-specific” gene knockout. However, Kristin E. Claflin et al. recently reported that the Ucp1Cre expression is not restricted to BAT but throughout the brain.9 Thus, the interpretation of many findings obtained by using Ucp1Cre-mediated knockout mouse models should consider their potential non-specific effects through the central nervous system. In this protocol, we employ Rosa26-LSL-Cas9;AdiponectinCre mice to restrict Cas9 expression in adipocytes and further combine with iBAT local injection of AAV-sgRNA, which provides a dual guarantee of the specificity of gene knockout in brown adipocytes.
To overcome potential off-target mutations by CRISPR/Cas9-mediated gene knockout, we suggest using at least two different sgRNA to independently verify the phenotypes. In addition, we develop an adiponectin promoter-driven AAV vector to express sgRNA-resistant cDNA encoded wild-type protein as a rescue, which serves as an on-target control for the sgRNA. Moreover, one also can express sgRNA-resistant cDNA encoded functional mutant protein such as kinase dead one.
Institutional permissions
The experiments should be approved by the Institutional Animal Care and Use Committee (IACUC) of laboratory animals to protect the welfare of animals. All mouse experiments were performed according to PKU IACUC guidelines.
Preparation of Rosa26-LSL-Cas9;AdipoqCre mice
Timing: n/a
The Rosa26-LSL(loxP-stop-loxP)-Cas9 knock-in mice10 were crossed with AdiponectinCre mice to generate Rosa26-LSL-Cas9;AdipoqCre mice that restrict Cas9 expression in adipocytes. Local injection of AAV-sgRNA into iBAT of Rosa26-LSL-Cas9;AdipoqCre mice allows a specific deletion of the target gene in brown adipocytes (Figure 1). In this protocol, AAV injections were performed with sex-matched mice at the age of 6–8 weeks.
Figure 1.
Breeding strategy for generating Rosa26-LSL-Cas9;AdipoqCre mice
sgRNA design and synthesis
Timing: 1 day
sgRNAs can be designed using online CRISPR design tools such as Benchling, Broad Institute GPP, CRISPOR, etc. In this protocol, we use CRISPOR to design sgRNAs targeting mouse gene Smdt1 (encoding essential MCU regulator, EMRE).11
-
1.
Obtain the genomic sequence of the target gene from the NCBI-Genome Data Viewer by searching the gene name.
-
2.
Select the first exon containing the coding sequence preferentially, and input the sequence (starting from ATG) on CRISPOR. Select Mus musculus genome and spCas9 PAM motif.
-
3.
Choose higher-scoring sgRNAs from the predicted guide sequences, which may have higher editing efficiency and lower off-target effects. Design at least three sgRNAs and screen for efficient ones (as described later in the protocol).
-
4.
Synthesize two sgRNA oligonucleotides as follows:
sgRNA-F: 5′-CACCGNNNNNNNNNNNNNNNNNNNN- 3′.
sgRNA-R: 5′-AAACNNNNNNNNNNNNNNNNNNNNC-3′.
Note: The 20 bp sgRNA sequence is shown as polyN. The sgRNA-F (forward) contains the sgRNA sequence upstream of the PAM motif.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-EMRE (dilutions: 1:500) | Santa Cruz Biotech | Cat. #sc-86337; RRID: AB_2250685 |
| Mouse polyclonal anti-NDUFS1 (dilutions: 1:2000) | Abcam | Cat. #ab22094; RRID: AB_2151098 |
| Bacterial and virus strains | ||
| Adeno-Associated Virus Serotype 8 (AAV8) | In-house purified | N/A |
| T1 Phage Resistant Chemically Competent cells | In-house preparetion | N/A |
| Stbl3 Chemically Competent Cells (Stbl3) | In-house preparetion | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| NaCl | Sigma-Aldrich | Cat. #S7653 |
| Yeast extract | Sigma-Aldrich | Cat. #Y1625 |
| Tryptone | Thermo Scientific | Cat. # LP0042B |
| Agar | Sigma-Aldrich | Cat. # W201201 |
| BsmBI | NEB | Cat. #R0580 |
| NEbuffer 3.1 | NEB | Cat. #B7203S |
| T4 ligase | NEB | Cat. #M0202 |
| T4 PNK | NEB | Cat. # M0201S |
| Polyethylenimine (PEI) | Polysciences | Cat. #23966-1 |
| Puromycin dihydrochloride | BioVision | Cat. #295160 |
| AgeI-HF | NEB | Cat. #R3552 |
| Sall-HF | NEB | Cat. #R3138 |
| DpnI | NEB | Cat. #R0176 |
| BspQI | NEB | Cat. #T0712 |
| Phanta Max Super-Fidelity DNA Polymerase | Vazyme | Cat. # P505-d1 |
| Exonuclease III | NEB | Cat. #M0206 |
| NEbuffer 1 | NEB | Cat. #B7001 |
| 2,2,2-Tribromoethanol | Sigma-Aldrich | Cat. #T48402 |
| 2-Methyl-2-butanol | Sigma-Aldrich | Cat. #19954 |
| Tolfedine | Vetoquinol | N/A |
| Bovine Serum Albumin (fatty-acid free) | Yuanye Biotech | Cat. #S25762 |
| Digitonin | Biosynth | Cat. #D3200 |
| Sucrose | Sigma-Aldrich | Cat. #V900116 |
| EGTA | Sigma-Aldrich | Cat. #E3889 |
| Complete protease inhibitor cocktail | Roche | Cat. #4693116001 |
| HEPES | Sigma-Aldrich | Cat. #V900477 |
| Experimental models: Cell lines | ||
| Mouse melanoma cell B16F10 | ATCC | CRL-6475 |
| Experimental models: Organisms/strains | ||
| Mouse: Rosa26-LSL-Cas9 knock-in | The Jackson Laboratory | JAX: 026175 |
| Mouse: AdiponectinCre | The Jackson Laboratory | JAX: 010803 |
| Oligonucleotides | ||
| sgRNA forward: 5′-CACCGNNNNNN NNNNNNNNNNNNNN-3′ |
This paper | N/A |
| sgRNA reverse: 5′-AAACNNNNNNNN NNNNNNNNNNNNC-3′ |
This paper | N/A |
| sgRNA forward: 5′-ACCGNNNNNN NNNNNNNNNNNNNN-3′ |
This paper | N/A |
| sgRNA reverse: 5′-AACANNNNNN NNNNNNNNNNNNNNC-3′ |
This paper | N/A |
| sgRNA targeting sequence against Emre #1: CTGGGTTGCAGTTCGACCCG |
This paper | N/A |
| sgRNA targeting sequence against Emre #2: GGCGATGTCTACACCGTACC |
This paper | N/A |
| sgRNA targeting sequence against Emre #3: GTCTCAGCCAGGTACCGTCG |
This paper | N/A |
| sgRNA targeting sequence against Emre #4: TGGCGATGTCTACACCGTAC |
This paper | N/A |
| sgRNA targeting sequence against Emre #5: GCCTGGGTTGCAGTTCGACC |
This paper | N/A |
| sgRNA targeting sequence against LacZ: CCCGAATCTCTATCGTGCGG |
Addgene | Cat. #74179 |
| Seq-F: CTTCCCGTGTGTGTCACGAG | This paper | N/A |
| Seq-R: TGAGTCCTATGTCCGGTCCC | This paper | N/A |
| SLIC-EMRE-F: GGTTGGGGCAACCGGTA TGGCGTCCACGGCGGCTCG |
This paper | N/A |
| SLIC-EMRE-R: AGGCCCGGGCGTCGACC ATCGTCGTCGTCGTCATCCT |
This paper | N/A |
| sgRNA-resis-mutation-F: GGGGAGGAGAGGTGGAGACGTG TATACGGTTCCCTCCAGCTCAGG |
This paper | N/A |
| sgRNA-resis-mutation-R: CTGAGACCTGAGCTGGAGGGAACC GTATACACGTCTCCACCTCTC |
This paper | N/A |
| Recombinant DNA | ||
| lentiCRISPRv2 | Addgene | Cat. #52961 |
| pAAV-U6-gRNA-CBh-mCherry | Addgene | Cat. #91947 |
| pAAV-ADP-MCS-FLAG | Addgene | Cat. #192360 |
| pRC2/8 | Addgene | Cat. #112864 |
| pHelper | Addgene | Cat. #112867 |
| Software and algorithms | ||
| CRISPOR | TEFOR Infrastructure | Version 5.01 |
| ICE | Synthego | Version 3.0 |
| Other | ||
| Surgical swabs | N/A | N/A |
| Animal hair clipper | N/A | N/A |
| Heating pad | N/A | N/A |
| Surgical scissors | Fine Science Tools | Cat. #14002-12 |
| Curved forceps | Fine Science Tools | Cat. #11052-10 |
| Surgical suture clips | Fine Science Tools | Cat. #12022-09 |
| Clip applicator | Fine Science Tools | Cat. #12018-12 |
| Microsyringe (50 μL) | Hamilton | Cat. #80500 |
| Polyethylene tubing | Smiths Medical | Cat. #10793527 |
| Nanopass33 (33G needle for pen injectors) | Terumo Corp. | N/A |
Materials and equipment
LB (Lysogeny Broth) Medium
| Reagent | Final concentration | Amount |
|---|---|---|
| NaCl | 1% | 10 g |
| Yeast extract | 0.5% | 5 g |
| Trypton | 1% | 10 g |
| H2O | N/A | Bring up to 1 L |
| Total | N/A | 1 L |
The LB medium should be sterilized by autoclaving.
2,2,2-Tribromoethanol stock solution
| Reagent | Final concentration | Amount |
|---|---|---|
| 2,2,2-Tribromoethanol | N/A | 5 g |
| 2-Methyl-2-butanol | N/A | 10 mL |
| Total | N/A | N/A |
The solution should be freshly made and fully dissolved with vortex mixer.
2,2,2-Tribromoethanol working solution
| Reagent | Final concentration | Amount |
|---|---|---|
| 2,2,2-Tribromoethanol stock solution | N/A | 1.25 mL |
| 0.9% NaCl | N/A | Bring up to 50 mL |
| Total | N/A | 50 mL |
The working solution should be sterilized using a 0.22 μm filter and stored at 4°C away from light for a couple of weeks.
BAT mitochondrion isolation buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Sucrose | 250 mM | 8.56 g |
| 1 M Hepes | 10 mM | 1 mL |
| EGTA | 1 mM | 0.038 g |
| BSA (fatty-acid free) | 0.3% | 0.3 g |
| 3 M KOH | N/A | Adjust pH to 7.0 |
| ddH2O | N/A | Bring up to 1 L |
| Total | N/A | 100 mL |
The buffer should be sterilized using a 0.22 μm filter and stored at 4°C for a couple of weeks.
Step-by-step method details
Screen efficient sgRNAs in mouse melanoma cells using pAAV-sgRNA vector
Timing: 10 days
The sgRNAs targeting mouse genes with editing efficiency are screened using lentiCRISPRv2 vector transfected in mouse melanoma cells.
-
1.Prepare lentiCRISPRv2-sgRNA vector.
-
a.Anneal the sgRNA oligonucleotides as inserts.
-
i.Anneal each pair of sgRNA oligonucleotides as the following reaction.
Reagent Amount Oligo 1 (100 μM) 1 μL Oligo 2 (100 μM) 1 μL 10× T4 ligation buffer (NEB) 1 μL T4 PNK (NEB) 0.5 μL ddH2O 6.5 μL -
ii.Anneal in a thermocycler at 37°C for 30 min, then 95°C for 5 min and ramp down to 25°C (5°C/min).
-
i.
-
b.Digest 2 μg of lentiCRISPRv2 plasmid at 55°C for 40 min and purify the digested plasmids from the 0.8% agarose gel using Thermo Scientific GeneJET Gel Extraction Kit (K0691) following the manufacturer’s instructions.(https://www.thermofisher.cn/document-connect/document-connect.html?url=https://assets.thermofisher.cn/TFS-Assets%2FLSG%2Fmanuals%2FMAN0012661_GeneJET_Gel_Extraction_UG.pdf).
Reagent Amount lentiCRISPRv2 2 μg BsmBI 1 μL NEbuffer3.1 (10×) 2 μL ddH2O 15 μL -
c.Clone the annealed oligos into the digested lentiCRISPRv2 vector.
-
i.Dilute the annealed oligos at 1:10.
-
ii.Set up the ligation reaction as follows and incubate the reaction at 25°C for 2 h.
Reagent Amount Digested and purified lentiCRISPRv2 50 ng Diluted annealed oligos 1 μL 10× T4 ligation buffer (NEB) 1 μL T4 ligase (NEB) 1 μL ddH2O Bing up to 10 μL -
iii.Transform the ligation mixture into Stbl3 competent cells. Transform 2 μL of each ligation mixture into 50 μL of Stbl3 competent cells and incubate the mixture for 20 min on ice. After heat shock at 42°C for 90 s, the transformation mixture is immediately incubated on ice for 2 min. Then, add 200 μL of LB medium to the mixture and allow the transformed cells to recover at 37°C for 1 h with 220 rpm shaking.
-
iv.Add the transformed cells onto the LB plate with 100 mg/mL ampicillin and incubate the LB plate at 37°C for 14 h to allowing colonies to grow.
-
v.Pick 2 colonies from each lentiCRISPRv2-sgRNA plate into 5 mL LB medium with 100 mg/mL ampicillin to be incubated at 37°C with 220 rpm shaking for 8–10 h and perform plasmid miniprep using Thermo Scientific GeneJET Plasmid Miniprep Kit (K0502) following the manufacturer’s instructions (https://www.thermofisher.cn/document-connect/document-connect.html?url=https://assets.thermofisher.cn/TFS-Assets%2FLSG%2Fmanuals%2FMAN0012655_GeneJET_Plasmid_Miniprep_UG.pdf).
-
vi.Sequence each colony to confirm the sgRNA insertion into lentiCRISPRv2 vector.
-
i.
-
a.
-
2.Screen sgRNAs with editing efficiency in mouse melanoma cells (troubleshooting 1) (troubleshooting 2).
-
a.Mouse melanoma cells (B16F10) are cultured in DMEM supplemented with 10% FBS at 37°C in a 5% CO2 incubator.
-
b.Plate B16F10 cells onto 12-well plate in DMEM supplemented with 10% FBS.
-
c.When cells reach 80%–90% confluent, transfect 1 μg of lentiCRISPRv2-sgRNA plasmid into each well using PEI as follows. Preheat PEI solution and DMEM at 37°C and prepare plasmid/DMEM mixture for each well of cells. Add 3 μL of PEI into the plasmid/DMEM mixture (the ratio of PEI (μL): plasmid (μg) is 3:1), mix well, and place at room temperature for 10 min. Add the transfection mixture into each well of B16F10 cells.
Reagent Amount lentiCRISPRv2-sgRNA 1 μg DMEM N/A PEI 3 μL Total 100 μL Note: Each lentiCRISPRv2-sgRNA plasmid is transfected in 2 wells, one for genomic DNA sequencing and another one for western blot analysis. CRITICAL: The control sgRNA targets a gene not existing in the mouse genome (LacZ here).
CRITICAL: The control sgRNA targets a gene not existing in the mouse genome (LacZ here). -
d.24 h after transfection, pass the B16F10 cells from the 12-well plate onto the 6-well plate. Culture one well of cells in 2 mL of DMEM supplemented with 10% FBS and 10 μM puromycin for 48–60 h to remove non-infected dead cells.
-
e.Wash cells with PBS buffer to remove the dead cells and collect the remaining cells for genomic DNA extraction.
-
f.Genomic DNA is extracted using Invitrogen Purelink Genomic DNA Kit (K182001) following the manufacturer’s instructions (https://www.thermofisher.cn/document-connect/document-connect.html?url=https://assets.thermofisher.cn/TFS-Assets%2FLSG%2Fmanuals%2Fpurelink_genomic_man.pdf).
-
g.Amplify the sgRNA target region on genomic DNA via PCR using a pair of PCR primers (Seq-F/R primers) to produce 400–600 bp amplicons with the sgRNA target sequence in the center. Set up the PCR reaction as follows.
Reagent Amount 2× Phanta Max Buffer 25 μL dNTP Mix (10 mM each) 1 μL 500 ng/μL Genomic DNA 3 μL 10 μM Seq-F primer 1 μL 10 μM Seq-R primer 1 μL Phanta Max Super-Fidelity DNA Polymerase (Vazyme) 1 μL ddH2O Up to 50 μL Then run PCR using the following cycling condition.Steps Temperature Time Cycles Initial Denaturation 95°C 3 min 1 Denaturation 95°C 15 s 29 cycles Annealing 58°C 15 s Extension 72°C 30 s/kb Final extension 72°C 5 min 1 Hold 4°C forever -
h.Purify the PCR products from the 2% agarose gel using Thermo Scientific GeneJET Gel Extraction Kit (K0691).
-
i.Sequence the amplicons of control sgRNA and target gene sgRNA samples using Seq-F primer.
-
j.Analyze the editing efficiency through uploading sgRNA sequence and sequencing results of control and experimental amplicons onto the software ICE (https://ice.synthego.com/#/).
-
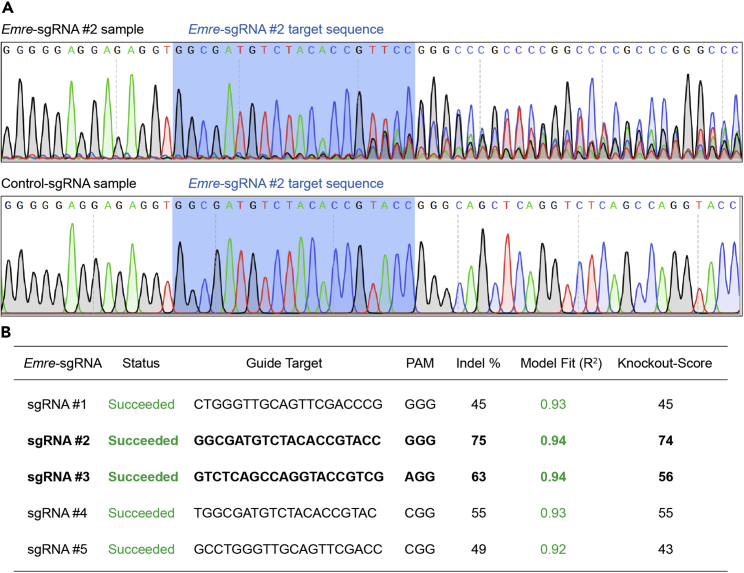
k.Select at least two sgRNAs with the highest editing activity based on the Indel %, Model Fit and Knockout-Score got from ICE analyses (Figure 2).
-
l.Another well of transfected cells is used for protein extraction to confirm the knock-out efficiency of the target gene on protein level by western blot.
-
a.
-
3.pAAV-sgRNA-mCherry vector construction.
-
a.sgRNA synthesis. Select two sgRNAs with highest editing efficiency verified in step 2 and synthesize the oligonucleotides as follows.sgRNA-F: 5′-ACCGNNNNNNNNNNNNNNNNNNNN-3′.sgRNA-R: 5′-AACANNNNNNNNNNNNNNNNNNNNC-3′.
-
b.Anneal the sgRNA oligos as inserts.
-
c.Digest 2 μg of pAAV-U6-gRNA-CBh-mCherry plasmid with BspQI at 50°C for 40 min.
Reagent Amount pAAV-U6-gRNA-CBh-mCherry 2 μg BsmBI 1 μL NEbuffer3.1 (10×) 2 μL ddH2O 15 μL -
d.Purify the digested plasmids from the 0.8% agarose gel using Thermo Scientific GeneJET Gel Extraction Kit (K0691).
-
e.Ligate the annealed oligos into the digested pAAV-U6-gRNA-CBh-mCherry plasmid.
-
i.Dilute the annealed oligos at 1:10.
-
ii.Set up the ligation reaction as follows and incubate the reaction at 25°C for 2 h.
Reagent Amount Digested and purified pAAV-U6-gRNA-CBh-mCherry 50 ng Diluted annealed oligos 1 μL 10× T4 ligation buffer (NEB) 1 μL T4 ligase (NEB) 1 μL ddH2O Up to 10 μL -
iii.Transform the ligation mixture into T1 competent cells. Transform 2 μL of the ligation mixture into 50 μL of T1 competent cells and incubate the mixture for 20 min on ice. After heat shock at 42°C for 90 s, the transformation mixture is immediately incubated on ice for 2 min.
-
iv.Plate transformed cells onto the LB plate with 100 mg/mL ampicillin and incubate the LB plate at 37°C for 14 h allowing colonies to grow.
-
v.Pick 2 colonies from each pAAV-U6-gRNA-CBh-mCherry plate into 5 mL LB medium with 100 mg/mL ampicillin to be incubated at 37°C with 220 rpm shaking for 8–10 h and perform plasmid miniprep.
-
vi.Sequence each colony to confirm the sgRNA insertions into the pAAV-U6-gRNA-mCherry vector.
-
i.
-
a.
Figure 2.
Analysis of the editing efficiency of the sgRNAs by sequencing the amplicons of sgRNA-targeted region on genomic DNA
(A) The disorderly sequencing peaks suggest potential editing activity of Emre-sgRNAs compared to control-sgRNA.
(B) The editing efficiency analyzed by the ICE software.
Design sgRNA-resistant cDNA of target gene and construct AAV vector
Timing: 6 days
To avoid recognition by sgRNA-CRISPR, sgRNA-resistant cDNA of the target gene is designed by the introduction of synonymous mutations on the critical “GG” in the PAM motif (NGG) and on the sgRNA-targeted sequence region. Using sgRNA-resistant cDNA, we can rescue with tagged wild-type or mutant protein according to experimental purpose in the knockout context of the endogenous gene.
-
4.Construct pAAV-ADP (adiponectin promoter)-EMRE-FLAG (troubleshooting 4).
-
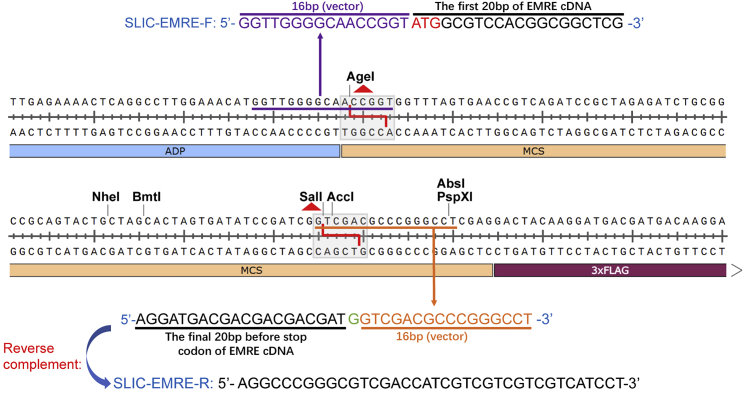
a.Use SLIC (sequence and ligation-independent cloning) to insert EMRE-cDNA into the MCS (multiclonal site) of pAAV-ADP-MCS-FLAG. Design SLIC PCR primers according to the selected dual enzyme cleavage sites in the MCS of pAAV-ADP-MCS-FLAG.Note: In this case we select AgeΙ and SalI as dual enzyme cleavage sites, and synthesize the SLIC PCR primers as shown in Figure 3.
-
b.Use the above SLIC PCR primers to amplify the EMRE insert. Perform PCR using Vazyme Phanta Max Super-Fidelity DNA Polymerase kit (P505d1) and set up PCR reaction as below.
Reagent Amount 2× Phanta Max Buffer 25 μL dNTP Mix (10 mM each) 1 μL 100 ng/μL template plasmid containing EMRE cDNA 1 μL 10 μM SLIC-EMRE-F 1 μL 10 μM SLIC-EMRE-R 1 μL Phanta Max Super-Fidelity DNA Polymerase (Vazyme) 1 μL ddH2O Up to 50 μL -
c.Run PCR using the following cycling condition.
Steps Temperature Time Cycles Initial Denaturation 95°C 3 min 1 Denaturation 95°C 15 s 29 cycles Annealing 58°C 15 s Extension 72°C 30 s/kb Final extension 72°C 5 min 1 Hold 4°C forever -
d.Purify the PCR amplicons as SLIC inserts using Thermo Scientific GeneJET Gel Extraction Kit (K0691).
-
e.Digest 2 μg of pAAV-ADP-MCS-FLAG plasmid with AgeI and SalI at 37°C for 60 min as below.
Reagent Amount pAAV-ADP-MCS-3×FLAG 2 μg AgeI-HF (NEB) 1 μL SalI-HF (NEB) 1 μL 10× Cutsmart buffer (NEB) 2 μL ddH2O 14 μL -
f.Purify the digested plasmid as the SLIC vector using Thermo Scientific GeneJET Gel Extraction Kit (K0691).
-
g.Perform SLIC reaction to construct pAAV-ADP-EMRE-3×FLAG.
-
i.Set up SLIC reaction as below.
Reagent Amount SLIC insert 50 ng SLIC vector 25 ng 10× NEbuffer 1 0.5 μL ddH2O Up to 5 μL -
ii.Place the reaction tube on ice for 5 min and the following steps should be performed on ice.
-
iii.Add 0.25 μL Exonuclease III (40 units) to the tube and mix well by pipetting for several times, and then place on ice for 30 min.
-
iv.Add 0.5 μL 0.5 M EDTA (pH 8.0) into the tube and mix by pipetting for several times, and then incubate the mixture at 65°C for 5 min to stop the reaction.
-
v.Centrifuge the tube to concentrate the mixture.
-
vi.Replace the reaction tube on ice to cool down for 5 min.
-
vii.Transform the total reaction mixture into 50 μL T1 competent cells and incubate the mixture for 20 min on ice. After heat shock at 42°C for 90 s, the mixture is immediately incubated on ice for 2 min.
-
viii.Plate transformed cells onto the LB plate with 100 mg/mL ampicillin and incubate the LB plate at 37°C for 14 h allowing colonies to grow.
-
ix.Pick 2 colonies from the plate into 5 mL LB medium with 100 mg/mL ampicillin to be incubated at 37°C with 220 rpm shaking for 8–10 h and perform plasmid miniprep.
-
x.Sequence each colony to confirm the correct construction of pAAV-ADP-EMRE-3×FLAG.
-
i.
-
a.
-
5.Design synonymously mutagenic primers for sgRNA-resistant cDNA of target gene (troubleshooting 2).
-
a.Synonymously mutate the “GG” in the PAM motif (NGG) which is critical for the recognition by sgRNA-spCas9.Note: In this case, since synonymous mutations cannot be performed on the “GG”, we mutate the glycine containing “GG” to serine in smdt1 of homo sapiens, as illustrated in Figure 4.
-
b.Synonymously mutate the sgRNA-targeting sequence as illustrated in Figure 4.
-
c.Design and synthesize the synonymously mutagenic primers as follows.sgRNA-resis-mutation-F:5′-GGGGAGGAGAGGTGGAGACGTGTATACGGTTCCCTCCAGCTCAGG-3′.sgRNA-resis-mutation-R:5′-CTGAGACCTGAGCTGGAGGGAACCGTATACACGTCTCCACCTCTC-3′.
-
a.
-
6.Take pAAV-ADP-EMRE-3×FLAG plasmid as PCR template and use synonymously mutagenic primers to produce sgRNA-resistant pAAV-ADP-EMRE-resis-3×FLAG.
-
a.Set up PCR reaction as follows to perform site-directed mutagenesis using Vazyme Phanta Max Super-Fidelity DNA Polymerase kit.
Reagent Amount 2× Phanta Max Buffer 5 μL dNTP Mix (10 mM each) 0.2 μL Template plasmid 20 ng 10 μM sgRNA-resis-mutation-F 0.25 μL Phanta Max Super-Fidelity DNA Polymerase (Vazyme) 0.2 μL ddH2O Up to 9.5 μL -
b.Run PCR using the following cycling condition.
Steps Temperature Time Cycles Initial Denaturation 95°C 3 min 1 Denaturation 95°C 15 s 9 cycles Annealing 58°C 15 s Extension 72°C 30 s/kb Final extension 72°C 5 min 1 Hold 4°C forever -
c.Add 0.5 μL of 10 μM sgRNA-resis-mutant-R to the PCR reaction mixture and run PCR using the following cycling condition.
Steps Temperature Time Cycles Initial Denaturation 95°C 3 min 1 Denaturation 95°C 15 s 19 cycles Annealing 58°C 15 s Extension 72°C 30 s/kb Final extension 72°C 5 min 1 Hold 4°C forever -
d.Digest the PCR amplicons with DpnI at 37°C for 2 h to remove the template plasmid as follows.
Reagent Amount PCR amplicons 10 μL DpnI (NEB) 0.2 μL 10× Cutsmart buffer (NEB) 1 μL -
e.Add 5 μL of the digestion mixture into 50 μL T1 competent cells and incubate the mixture on ice for 20 min. After heat shock at 42°C for 90 s, the mixture is immediately incubated on ice for 2 min.
-
f.Plate the transformed cells onto LB plate with 100 mg/mL ampicillin and incubate the LB plate at 37°C for 14 h allowing colonies to grow.
-
g.Pick 2 colonies from the plate into 5 mL LB medium with 100 mg/mL ampicillin to be incubated at 37°C with 220 rpm shaking for 8–10 h and perform plasmid miniprep.
-
h.Sequence each colony to confirm the correct mutation of pAAV-ADP-EMRE-resis-3×FLAG.
-
i.Use SLIC to insert EMRE-resis-cDNA into the MCS of pAAV-ADP-MCS-3×FLAG via SLIC-EMRE-F/R primers to avoid vector mutation, following step 4.
-
a.
Figure 3.
Design SLIC PCR primers to clone EMRE-cDNA into pAAV-ADP-MCS-3×FLAG
Figure 4.
Design synonymously mutagenic primers for sgRNA-resistant cDNA of target gene
AAV production
Timing: 1 week
In this protocol, we use AAV8 serotype that can effectively infect BAT.12 AAV-U6-Emre-sgRNA-mCherry, AAV-U6-Ctrl-sgRNA-mCherry and AAV-ADP-EMRE-resis-3×FLAG are produced following the previous step-by-step instructions.13 The AAV titer for the following AAV injection experiments should be above 1 × 1013 vg/mL (vector genomes per mL).
Local injection of AAV into intrascapular BAT (iBAT)
Timing: 3 weeks
-
7.Preparation for local injection of AAV.
-
a.Prepare surgical equipment as in key resources table.Note: all the surgical equipment should be sterilized with 75% ethanol before surgical operation.
-
b.Set up injection device.
-
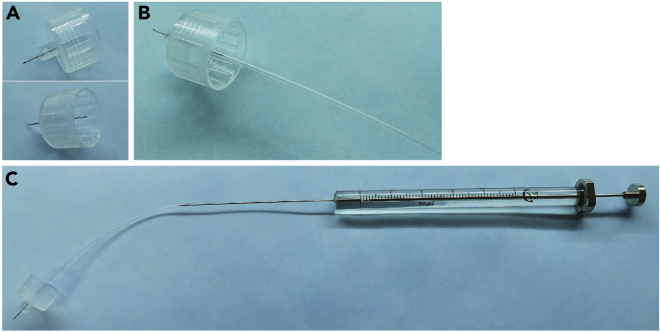
i.Connect the tail end of a pen needle to a polyethylene tubing (5 cm in length), as illustrated in Figures 5A and 5B.
-
ii.Connect a needle of microsyringe (50 μL) to the other side of the polyethylene tubing, as illustrated in Figure 5C.
-
iii.Test the assembled injection device and make sure all the connections are sealed well without leakage.
-
i.
-
c.Prepare virus solution (troubleshooting 5).
-
i.Dilute AAVs with PBS as follows to achieve the volume for injection (50 μL for each lobe of iBAT, 100 μL for each mouse).
Reagent Amount AAV-U6-Emre/Ctrl-sgRNA-mCherry 1×1012 vg AAV-ADP-EMRE-resis-3×FLAG 5×1011 vg PBS Up to 100 μL -
ii.Draw 50 μL of virus solution with microsyringe and connect the injection device.
-
i.
-
a.
-
8.Surgical operation for local injection of AAV into iBAT (troubleshooting 3) (Methods video S1).
-
a.Anaesthetize mice with 2,2,2-Tribromoethanol.Inject mice with 2,2,2-Tribromoethanol working solution (20 μL/g body weight) through intraperitoneal injection. Two min after injection, the mice will be anaesthetized deeply enough to perform surgical operation.Note: The anaesthetized mice should be on a heating pad during all the surgical process.
-
b.Remove the hair above iBAT using an animal hair clipper and wipe the exposed skin with ethanol (Figure 6A).
-
c.Cut a 0.5–0.8 cm incision in the skin above iBAT with a surgical scissor to expose the two lobes of iBAT (Figures 6B–6D).
-
d.Hold the injection device and carefully insert the pen needle into the fat pad of one iBAT lobe, as illustrated in Figure 6E. When the needle is inserted well, inject 5 μL virus for one point and inject 10 points (50 μL virus) to distribute evenly throughout one lobe of iBAT.
 CRITICAL: The depth of needle insertion should be 1–2 mm avoiding too deep or too superficial.
CRITICAL: The depth of needle insertion should be 1–2 mm avoiding too deep or too superficial. -
e.Draw another 50 μL of virus solution rapidly for another lobe of iBAT.
-
f.After injection, close the incision with two surgical suture clips (9 mm) (Figure 6F).Note: The suture clips should be clamped tightly using a clip applicator avoiding shedding.
-
g.Inject mice with analgesic (Tolfedine, 2 mg/kg body weight) by a single intramuscular injection.
-
h.Keep the mice on a heating pad until revivtication.
-
i.Place one mouse per cage to avoid fighting. Monitor the health status of mice daily and inject the mice with analgesic (Tolfedine, 2 mg/kg body weight) at 24 h and 48 h after the surgery.Note: Recovery time after surgery is about 7 days. It needs 3 weeks after AAV injection to allow sufficient CRISPR editing in vivo and enough overexpression of protein driven by adiponectin promoter.
Methods video S1. Surgical operation for local injection of AAV into iBAT, related to step 8Download video file (15.2MB, mp4) -
a.
Figure 5.
Set up injection device
(A) 33G needle for pen injectors.
(B) Connect the tail end of a pen needle to a polyethylene tubing.
(C) The injection device assembled by connecting a needle of microsyringe (50 μL) to the other side of the polyethylene tubing.
Figure 6.
Surgical operation for local injection of AAV into iBAT
(A) Remove the hair above iBAT to exposure the skin.
(B) Cut a 0.5–0.8 cm incision in the skin above iBAT as indicated by the red dotted line.
(C) The left lobe of iBAT illustrated as the red dotted circle.
(D) The right lobe of iBAT illustrated as the red dotted circle.
(E) Insert the pen needle into the fat pad of iBAT, and the injection point is indicated by the red arrow.
(F) Close the incision with two surgical suture clips (9 mm) as indicated by the red arrows.
Knockout and rescue efficiency detection
Timing: 2 days
-
9.Isolation of BAT mitochondria.
-
a.Isolate the iBAT from mice injected with AAV for 3 weeks and immediately transfer the iBAT into 2 mL tube with 0.5 mL BAT mitochondrion isolation buffer.Note: The whole isolation process should be performed on ice. The buffer and equipment used for mitochondrion isolation should be precooled on ice.
-
b.Mince the iBAT into small pieces (1 mm3 each) with a scissor in 2-mL tube and transfer the content into a Teflon-glass Dounce homogenizer (Wheaton, 2 mL). Wash the tube with 0.5 mL isolation buffer and collect it into the homogenizer (total 1 mL content in the homogenizer).
-
c.Homogenize the iBAT pieces with a teflon pestle for 10 strokes. Transfer the homogenate into a new 2 mL tube and wash the homogenizer with 0.5 mL isolation buffer and collect it into the 2 mL tube (total 1.5 mL homogenate in the tube).
-
d.Centrifuge at 8,500 g for 10 min to separate the lipids from the cell lysate in brown adipocytes.
-
e.After centrifugation, discard the upper lipid layer and the supernatant, and keep the pellet.
-
f.Resuspend the pellet with 0.5 mL isolation buffer and transfer it into homogenizer. Wash the tube with 0.5 mL isolation buffer and collect it into the homogenizer (total 1 mL content in the homogenizer).
-
g.Homogenize the resuspended pellet with a teflon pestle for 5–8 strokes and transfer the homogenate into a new 2 mL tube. Wash the homogenizer with 0.5 mL isolation buffer and collect it into the 2 mL tube (total 1.5 mL homogenate in the tube).
-
h.Centrifuge at 700 g for 10 min to separate mitochondria from other cell components.
-
i.Transfer the supernatant containing mitochondria to a new 2 mL tube and centrifuge at 8,500 g for 10 min to precipitate the mitochondria.
-
j.Discard the supernatant and dissolve the mitochondrion pellet with 1% digitonin buffer containing 150 mM NaCl, 50 mM Tris-HCl (pH 7.5), 1 mM EDTA and 1× complete protease inhibitor.
-
k.After dissolution on ice for 30 min, centrifuge at 20,000 g for 10 min at 4°C, and the supernatant (mitochondrial protein lysate) can be used to detect the knockout and rescue efficiency by western blot.
-
a.
Expected outcomes
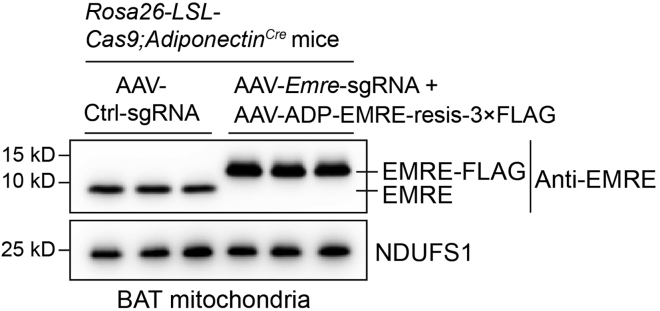
Through local injection of AAV-sgRNA into the iBAT of Rosa26-LSL-Cas9;AdipoqCre mice with adipocyte-specific expression of Cas9, we can efficiently and specifically knock out Emre in brown adipocytes of iBAT (Figure 7). Meanwhile, we can specifically rescue with tagged sgRNA-resistant protein in brown adipocytes of iBAT using AAV-ADP (Figure 7).
Figure 7.
Knockout and rescue efficiency analysis of EMRE protein in BAT mitochondria via Western blot
Limitations
One limitation of this protocol is that the CRISPR-knockout system requires Rosa26-LSL-Cas9;AdipoqCre mice generated by crossing Rosa26-LSL (loxP-stop-loxP)-Cas9 with AdiponectinCre mice. Another limitation is that the pAAV-ADP vector can only express proteins encoded cDNA no longer than 1.9 kb due to AAV package limitation.
Troubleshooting
Problem 1
Low knockout efficiency (in step 2 “Screen sgRNAs with editing efficiency in mouse melanoma cells”).
Potential solution
The knockout efficiency in this protocol mainly relies on the editing efficiency of sgRNA for the target gene. Thus, more than three sgRNAs should be designed and screened in mouse melanoma cells so that at least two sgRNAs have relatively high editing activity.
Problem 2
Off-target effects of CRISPR-Cas9 system (in step 2 “Screen sgRNAs with editing efficiency in mouse melanoma cells” and step 5 “Design synonymously mutagenic primers for sgRNA-resistant cDNA of target gene”).
Potential solution
Use at least two sgRNAs targeting different coding regions and use rescue experiments via expressing sgRNA-resistant cDNA of the target gene to validate the on-target phenotypes.
Problem 3
Low efficiency of gene knockout or overexpression due to nonstandard operation of AAV local injection, i.e., without widespread infection of AAV in iBAT (in step 8 “Surgical operation for local injection of AAV into iBAT”).
Potential solution
The virus solution should be injected into each lobe of iBAT by 10 evenly dispersed points to allow thorough infection. The injection depth of the needle should be in the middle of the fat pad. One can practice injection operation using trypan blue solution.
Problem 4
Low success rate of vector construction by SLIC (in step 4 “Construct pAAV-ADP (adiponectin promoter)-EMRE-3×FLAG”).
Potential solution
SLIC PCR primers for amplifying SLIC insert should be designed strictly according to the rules in Figure 3. Insert and vector used for SLIC must be obtained from gel purification. When setting up SLIC reaction, amount of insert should be 2 folds more than that of vector.
Problem 5
Poor control of the expression level of wild-type and mutant protein to a comparable level in the rescue experiments (in step 7 “Prepare virus solution”).
Potential solution
The titers of AAVs expressing wild-type or mutant protein should be measured at the same time to avoid titration errors among batches. For proteins with the same molecular weight, an equal amount of AAVs should be injected. Usually, AAVs carrying cDNA encoding smaller protein will express more, such that one should inject mice with less amount of AAVs to achieve comparable protein expression.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yifu Qiu (yifu.qiu@pku.edu.cn).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
This work was supported by grants from the National Key R&D Program of China (2018YFA0800702 and 2021YFA0804801), National Natural Science Foundation of China (31671227 and 91642113), and the Thousand Young Talents Program of The Chinese Government (to Y.Q.).
Author contributions
K.X., D.W., and Y.Q. conceptualized the study and designed experiments. K.X. performed experiments and analyzed data. K.X., D.W., and Y.Q. wrote the paper.
Declaration of interests
A Chinese patent application (No. 202210961425.3) related to this work was filed (Y.Q., K.X., D.W., and H.S.).
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2022.101895.
Contributor Information
Kaili Xue, Email: kaili.xue@pku.edu.cn.
Yifu Qiu, Email: yifu.qiu@pku.edu.cn.
Data and code availability
This study did not generate any unique datasets or code.
References
- 1.Xue K., Wu D., Wang Y., Zhao Y., Shen H., Yao J., Huang X., Li X., Zhou Z., Wang Z., Qiu Y. The mitochondrial calcium uniporter engages UCP1 to form a thermoporter that promotes thermogenesis. Cell Metab. 2022;34:1325–1341.e6. doi: 10.1016/j.cmet.2022.07.011. [DOI] [PubMed] [Google Scholar]
- 2.Romanelli S.M., Lewis K.T., Nishii A., Rupp A.C., Li Z., Mori H., Schill R.L., Learman B.S., Rhodes C.J., MacDougald O.A. BAd-CRISPR: inducible gene knockout in interscapular brown adipose tissue of adult mice. J. Biol. Chem. 2021;297:101402. doi: 10.1016/j.jbc.2021.101402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jimenez V., Muñoz S., Casana E., Mallol C., Elias I., Jambrina C., Ribera A., Ferre T., Franckhauser S., Bosch F. In vivo adeno-associated viral vector-mediated genetic engineering of white and brown adipose tissue in adult mice. Diabetes. 2013;62:4012–4022. doi: 10.2337/db13-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Neill S.M., Hinkle C., Chen S.J., Sandhu A., Hovhannisyan R., Stephan S., Lagor W.R., Ahima R.S., Johnston J.C., Reilly M.P. Targeting adipose tissue via systemic gene therapy. Gene Ther. 2014;21:653–661. doi: 10.1038/gt.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang W., Queen N.J., Cao L. rAAV-mediated gene delivery to adipose tissue. Methods Mol. Biol. 2019;1950:389–405. doi: 10.1007/978-1-4939-9139-6_23. [DOI] [PubMed] [Google Scholar]
- 6.Gómez-Banoy N., Lo J.C. Genetic manipulation with viral vectors to assess metabolism and adipose tissue function. Methods Mol. Biol. 2017;1566:109–124. doi: 10.1007/978-1-4939-6820-6_11. [DOI] [PubMed] [Google Scholar]
- 7.Balkow A., Hoffmann L.S., Klepac K., Glöde A., Gnad T., Zimmermann K., Pfeifer A. Direct lentivirus injection for fast and efficient gene transfer into brown and beige adipose tissue. J. Biol. Methods. 2016;3:e48. doi: 10.14440/jbm.2016.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bagchi D.P., Forss I., Mandrup S., MacDougald O.A. SnapShot: niche determines adipocyte character I. Cell Metab. 2018;27:264–264.e1. doi: 10.1016/j.cmet.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claflin K.E., Flippo K.H., Sullivan A.I., Naber M.C., Zhou B., Neff T.J., Jensen-Cody S.O., Potthoff M.J. Conditional gene targeting using UCP1-Cre mice directly targets the central nervous system beyond thermogenic adipose tissues. Mol. Metab. 2022;55:101405. doi: 10.1016/j.molmet.2021.101405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Platt R.J., Chen S., Zhou Y., Yim M.J., Swiech L., Kempton H.R., Dahlman J.E., Parnas O., Eisenhaure T.M., Jovanovic M., et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Concordet J.P., Haeussler M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018;46:W242–W245. doi: 10.1093/nar/gky354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y., Zeng X., Huang X., Serag S., Woolf C.J., Spiegelman B.M. Crosstalk between KCNK3-mediated ion current and adrenergic signaling regulates adipose thermogenesis and obesity. Cell. 2017;171:836–848.e13. doi: 10.1016/j.cell.2017.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fripont S., Marneffe C., Marino M., Rincon M.Y., Holt M.G. Production, purification, and quality control for adeno-associated virus-based vectors. J. Vis. Exp. 2019 doi: 10.3791/58960. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code.