Abstract
We have previously demonstrated the existence of Klebsiella pneumoniae clinical isolates deficient in the lipopolysaccharide O side chain, the major factor for resistance to complement-mediated killing in this bacterial species. These isolates are complement resistant, and their mechanisms to resist complement were investigated by selecting transposon-generated complement-sensitive mutants. One mutant with a drastically reduced capacity to grow in nonimmune human serum carried the transposon inserted in an open reading frame of a gene cluster involved in capsule synthesis. This mutant produced less capsule, bound more molecules of the complement component C3, and was more sensitive to complement-mediated and opsonophagocytic killings than was the parent strain. Four additional clinical isolates representing four different K serotypes were studied, and results showed that capsular polysaccharide is a major complement resistance factor in these O side chain-deficient isolates.
Klebsiella pneumoniae is a common pathogen of the urinary tract that occasionally invades the bloodstream and the lungs, causing more-threatening infections. K. pneumoniae strains constitutively express a polysaccharide capsule that is critical for the organism's ability to resist complement-mediated opsonophagocytic killing (10). However, contribution of the capsule to complement resistance is not completely clear. On the other hand, it is well known that lipopolysaccharide (LPS) is critical for the protection of K. pneumoniae against complement-mediated killing. Our studies have shown that the LPS core impedes the binding of C1q to porins, thus restricting the activation of the complement classical pathway (CP) (3). In addition, the alternative pathway (AP) of the complement system is only poorly activated by the O side chain, avoiding formation of the membrane attack complex and lysis of the microorganism (1). In consequence, laboratory mutant strains lacking the LPS O side chain are sensitive to complement-mediated killing (12).
Recently, we investigated the prevalence of LPS O types among 638 K. pneumoniae clinical isolates. In that study, 17.4% of the strains were nontypeable (8). When their LPSs were studied by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining (13), almost half of the nontypeable clinical isolates (8.3% of the total) lacked the O side chain of the LPS (O− strains) independently of their country of origin (Denmark, Spain, or the United States) and isolation source (urine, blood, or other sites) (8). Since the LPS O side chain is the major complement resistance factor described for K. pneumoniae, we decided to characterize these O− clinical isolates and their mechanisms to resist complement-mediated killing.
The ability of the O− clinical strains to resist the bactericidal activity of the serum (serum resistance) was demonstrated by bacterial survival experiments, performed as we previously described (3). In these experiments, after 1 h of incubation at 37°C in 25% nonimmune human serum (NHS), O− clinical isolates maintained their viability (Tables 1 and 2). We purified the LPS by the method of Westphal and Jann (14) and verified by SDS-PAGE analysis and silver staining (13) that the strains remained O− even after overnight incubation in 25% NHS (Fig. 1). To investigate the mechanisms adopted by the O− clinical isolates to resist complement-mediated killing, we generated a set of mutants by conjugation of the representative O− clinical isolate USA0352/78 (O−:K47) with Escherichia coli strain S17-1 λpir carrying plasmid pUTmini-Tn5 Km1 (6). Among 900 mutants, 11 were selected because of their reduced growth in serum. One of these mutants, USA0352/78-3, showed a drastic reduction (approximately 2 logs [Table 1]) in its viability in serum compared to that of the parental strain USA0352/78, for which no reduction (Table 1) was observed. Southern blot analysis using a specific probe of the transposon demonstrated that the serum-sensitive mutant had a single copy of the minitransposon in its genome (data not shown). Cloning of the minitransposon-containing fragment from the genomic DNA of mutant USA0352/78-3 in pBluescript (Stratagene) and sequencing of the minitransposon flanking regions showed 95% identity with ORF3 of a capsule synthesis cluster of K. pneumoniae strain Chedid (4). Capsular polysaccharide isolation from the parent and mutant strains by the method of Wilkinson and Sutherland (15) and quantitation using an inhibition enzyme-linked immunosorbent assay (ELISA) (2) demonstrated that the mutant strain USA0352/78-3 produced fourfold-less capsule than did the parental strain USA0352/78 (Table 1). Furthermore, SDS-PAGE analysis of the outer membrane proteins and LPS demonstrated no differences between parent and mutant strains (data not shown). The amount of complement component C3 bound to the bacterial cells was measured as described previously (1). Mutant strain USA0352/78-3 incubated in NHS bound threefold-more C3 molecules than did the parent strain (Table 1). In addition, using sera with only the CP or the AP functional (1), we determined that the binding of C3 to the cells was due to activation of both complement pathways (Table 1). CP activation was independent of the presence of specific antibodies, since the serum used as a complement source in the assays was a nonimmune serum preabsorbed with organisms used in the study to remove specific antibodies. These results suggest that the complement activators described for this species, porins and LPS, are more exposed in the capsule mutant than in the parent strain. This was further supported by experiments using bacteriophages FC3-10 (5) and FC3-11 (9), specific for the LPS core and for porin OmpK36, respectively. These phages infected the capsule mutant but not the parent strain. Susceptibility to opsonophagocytic killing was also studied. In these experiments, bacterial cells previously opsonized with NHS at 4°C for 15 min were incubated with freshly isolated polymorphonuclear cells (7) and plated to count viable cells. Opsonized cells of the capsule mutant were completely cleared by phagocytes, whereas a reduction of 3 logs was observed for the parent strain USA0352/78 (Table 1).
TABLE 1.
Phenotypic characteristics of K. pneumoniae clinical isolate USA0352/78 (O−:K47) and its derived mutant USA0352/78-3
| Characteristic | Value for strain:
|
|
|---|---|---|
| USA0352/78 | USA03552/78-3 | |
| Mini-Tn5 insertions (no.) | 0 | 1 |
| Capsule productiona | 87 ± 3 | 21 ± 2 |
| C3 binding (NHS)b | 0.3 ± 0.07 | 0.97 ± 0.08 |
| C3 binding (CPS)b | 0.15 ± 0.05 | 0.535 ± 0.05 |
| C3 binding (APS)b | 0.15 ± 0.05 | 0.950 ± 0.015 |
| Phage FC3-10 infection | − | + |
| Phage FC3-11 infection | − | + |
| Complement killingc | 3 × 106/3 × 106 | 3.3 × 106/5 × 104 |
| Opsonophagocytic killingc | 9.3 × 106/5 × 103 | 3 × 106/0 |
Cell-associated capsule polysaccharide (femtograms per CFU) quantified by a competitive ELISA.
Data are means (in arbitrary A405 units) ± standard deviations from ELISAs done in triplicate at least twice. The results from control wells coated with nonopsonized cells were always less than 0.1 U. CPS, CP serum; APS, AP serum.
CFU per milliliter at time (0 h/1 h).
TABLE 2.
Capsule production and complement and opsonophagocytic killing of K. pneumoniae O− strains and their derived capsule mutants
| Strain | Capsule productiona | Complement killing (CFU/ml) at time (h):
|
Opsonophagocytic killing (CFU/ml) at time (h):
|
||
|---|---|---|---|---|---|
| 0 | 1 | 0 | 1 | ||
| M9844 (O−:K47) | 80 ± 2 | 30 × 106 | 1 × 106 | 0.5 × 106 | 2.6 × 102 |
| M9844-2 (O−:K−) | 15 ± 1 | 57 × 106 | 8 × 104 | 2.8 × 106 | 86 |
| USA2073/79 (O1:K47) | 75 ± 4 | 1.9 × 106 | 5.3 × 106 | 1.9 × 106 | 1.3 × 106 |
| KD341 (O−:K3) | 84 ± 3 | 30 × 106 | 10 × 106 | 1.5 × 106 | 8.5 × 103 |
| KD341-7 (O−:K−) | 9.6 ± 0.5 | 10 × 106 | 20 × 103 | 0.6 × 106 | 0 |
| USA1555 (O−:K35) | 78 ± 2 | 23 × 106 | 10 × 106 | 1.3 × 106 | 1.6 × 104 |
| USA1555-8 (O−:K−) | 1.2 ± 0.2 | 120 × 106 | 1.1 × 103 | 3.2 × 106 | 0 |
| KD57 (O−:K2) | 425 ± 16 | 80 × 106 | 30 × 106 | 1.8 × 106 | 75 |
| KD57-14 (O−:K−) | 15 ± 2 | 20 × 106 | 10 × 104 | 0.8 × 106 | 0 |
| 52145 (O1:K2) | 400 ± 24 | 0.6 × 106 | 3.2 × 106 | 0.6 × 106 | 0.9 × 106 |
Cell-associated capsule polysaccharide (femtograms per CFU) quantified by a competitive ELISA.
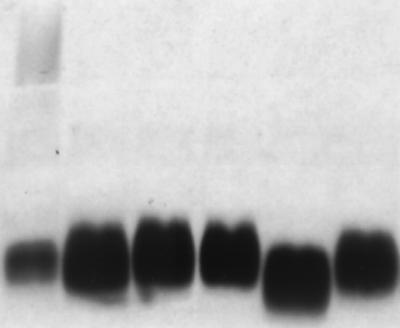
FIG. 1.
SDS-PAGE analysis and silver staining of purified LPS (50 ng) from K. pneumoniae strains grown overnight at 37°C in 25% NHS. Lanes correspond, from left to right, to LPS extracted from strains C3 (O1:K66), KD57 (O−:K2), KD341 (O−:K3), M9844/93 (O−:K47), USA0352/78 (O−:K47), and USA1555 (O−:K35).
In summary, a reduction in the amount of capsule produced by the mutant strain USA0352/78-3 caused an exposition of the complement activators, increased deposition of C3, and a subsequent increase in sensitivity to complement-mediated and opsonophagocytic killing. To extend these results to other K. pneumoniae O− clinical isolates with different K serotypes, we created capsule mutants with the mini-Tn5 Km1 transposon in four additional K. pneumoniae O− blood isolates: KD57 (O−:K2), KD341 (O−:K3), M9844 (O−:K47), and USA1555 (O−:K35). All capsule mutants, compared to their parent strains, produced between three- and fivefold-less capsular polysaccharide and had increased sensitivities to both complement-mediated and opsonophagocytic killing (Table 2).
Two major mechanisms of resistance to the bactericidal activity of complement have been identified for K. pneumoniae. The most frequently identified mechanism is expression of an LPS O side chain, which prevents access of complement components to activators (porins and rough LPS). In a minority of strains, resistance is due to coverage of the activators by the polysaccharide capsule. While some particular K serotypes have been involved in complement resistance in K. pneumoniae (K1, K10, and K16) (11), most K serotypes studied, including K2, K7, K19, K21, K22, and K66 (11), do not impede complement activation, and resistance is due to expression of LPS O side chain. Moreover, K. pneumoniae O+:K− strains are resistant to complement-mediated killing (11). Our results indicate that in K. pneumoniae O− clinical isolates the capsular polysaccharide protects the microorganism against complement-mediated killing independently of the K serotype, since in all strains studied, representing four different K serotypes, capsule expression was required to resist complement activity. These data suggest that the amount of capsular polysaccharide produced by the cells is more important for resistance against complement than is the chemical composition (or K type) of the capsule polysaccharide.
At the opsonophagocytosis level, except for some particular K types that interact with the human macrophage mannose receptor and are eliminated by phagocytosis (10), the capsular polysaccharide provides the organisms with a physical antiphagocytic barrier impeding the interaction between complement opsonins and complement receptors on the phagocytic cells. The presence of the LPS O side chain, which reduces the binding of opsonic complement components to the outer surface of the microorganism (1), together with the physical barrier provided by the capsule, makes the pathogen less susceptible to opsonophagocytic killing. Probably for this reason, we found that the K. pneumoniae strains expressing O antigen and serotypes K47 and K2 were more resistant to complement and opsonophagocytosis than were O− strains with the same K serotypes, independently of the amount of capsule produced (Table 2).
In summary, we have identified the capsular polysaccharide as the major factor for resistance to complement in K. pneumoniae clinical isolates deficient in the O side chain of LPS.
Acknowledgments
This work was supported by grants from the Comisión Interministerial de Ciencia y Tecnología (CICYT) and project FEDER 2FD97-0287. D.A. was supported by a predoctoral fellowship from CICYT.
We thank Dennis Hansen (Statens Seruminstitut, Copenhagen, Denmark) for providing antisera against different K types.
REFERENCES
- 1.Albertí S, Alvarez D, Merino S, Casado M T, Vivanco F, Tomás J M, Benedí V J. Analysis of complement C3 deposition and degradation on Klebsiella pneumoniae. Infect Immun. 1996;64:4726–4732. doi: 10.1128/iai.64.11.4726-4732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albertí S, Imperial J, Tomás J M, Benedí V J. Bacterial lipopolysaccharide extraction in silicagel containing tubes. J Microbiol Methods. 1991;14:63–69. [Google Scholar]
- 3.Albertí S, Marqués G, Camprubí S, Merino S, Tomás J M, Vivanco F, Benedí V J. C1q binding and activation of the complement classical pathway by Klebsiella pneumoniae outer membrane proteins. Infect Immun. 1993;61:852–860. doi: 10.1128/iai.61.3.852-860.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arakawa Y, Wacharotayankun R, Nagatsuka T, Ito H, Kato N, Ohta M. Genomic organization of the Klebsiella pneumoniae cps region responsible for serotype K2 capsular polysaccharide synthesis in the virulent strain Chedid. J Bacteriol. 1995;177:1788–1796. doi: 10.1128/jb.177.7.1788-1796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camprubí S, Merino S, Benedí V J, Tomás J M. Isolation and characterisation of bacteriophage FC3-10 from Klebsiella spp. FEMS Microbiol Lett. 1991;83:291–298. doi: 10.1016/0378-1097(91)90491-r. [DOI] [PubMed] [Google Scholar]
- 6.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fotino M, Merson E J, Allen F H. Micromethod for rapid separation of lymphocytes from peripheral blood. Ann Clin Sci. 1971;1:131–133. [PubMed] [Google Scholar]
- 8.Hansen D S, Mestre F, Albertí S, Hernández-Allés S, Alvarez D, Doménech A, Gil J, Merino S, Tomás J M, Benedí V J. Klebsiella pneumoniae lipopolysaccharide O-typing: revision of prototype strains and O-type distribution among clinical isolates of different sources and countries. J Clin Microbiol. 1999;37:56–62. doi: 10.1128/jcm.37.1.56-62.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hernández-Allés S, Albertí S, Rubirés X, Merino S, Tomás J M, Benedí V J. Isolation of FC3-11, a bacteriophage specific for the Klebsiella pneumoniae porin OmpK36, and its use for the isolation of porin-deficient mutants. Can J Microbiol. 1996;41:399–406. [Google Scholar]
- 10.Kabha K, Nissimov L, Athamna A, Keisari Y, Parolis H, Parolis L A S, Grue R M, Schlepper-Schafer J, Ezekowitz A R B, Ohman D E, Ofek I. Relationships among capsular structure, phagocytosis, and mouse virulence in Klebsiella pneumoniae. Infect Immun. 1995;63:847–852. doi: 10.1128/iai.63.3.847-852.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merino S, Camprubí S, Albertí S, Benedí V J, Tomás J M. Mechanisms of Klebsiella pneumoniae resistance to complement-mediated killing. Infect Immun. 1992;60:2529–2535. doi: 10.1128/iai.60.6.2529-2535.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomás J M, Benedí V J, Ciurana B, Jofre J. Role of capsule and O antigen in resistance of Klebsiella pneumoniae to serum bactericidal activity. Infect Immun. 1986;54:85–89. doi: 10.1128/iai.54.1.85-89.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai C M, Frasch C E. A sensitive silver strain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 14.Westphal O, Jann K. Bacterial lipopolysaccharides: extraction with phenol-water and further applications of the procedure. Methods Carbohydr Chem. 1965;5:83–91. [Google Scholar]
- 15.Wilkinson J F, Sutherland I W. Chemical extraction methods of microbial cells. Methods Microbiol. 1971;5B:345–383. [Google Scholar]