Abstract
Neural circuits provide an anatomical basis for functional networks. Therefore, dissecting the structure of neural circuits is essential to understanding how the brain works. Recombinant neurotropic viruses are important tools for neural circuit tracing with many advantages over non-viral tracers: they allow for anterograde, retrograde, and trans-synaptic delivery of tracers in a cell type-specific, circuit-selective manner. In this review, we summarize the recent developments in the viral tools for neural circuit tracing, discuss the key principles of using viral tools in neuroscience research, and highlight innovations for developing and optimizing viral tools for neural circuit tracing across diverse animal species, including nonhuman primates.
Keywords: Neural circuit, Viral tool, Anterograde, Retrograde, Trans-synaptic
Introduction
Since the human brain is the most complex and mysterious structure in the universe, neuroscience research is regarded as the “ultimate frontier of science” in understanding nature. The human brain is a complicated multiscale structure. In the past several decades, neuroscience research has focused on the molecules, genes, and cells at the microscale level, or structure/function networks based on magnetic resonance imaging and behaviors at the macroscale level. However, the human brain contains about one hundred billion neurons, which connect through about one quadrillion synapses, and form an almost infinite number of neural circuits at the mesoscale level. These circuits serve as the structural basis for the coordination and execution of various basic and higher brain functions. Elucidating neural circuits is a prerequisite for understanding the principles of brain function and the pathogenesis of nervous system diseases.
The gene expression profile characterizes a cell, while its synaptic connection defines it in another dimension. For example, two neurons with similar gene expression profiles have different functions and characteristics because of their distinct synaptic connections (involving diverse neural circuits and networks). Therefore, there are no two identical individual neurons in the brain. Neurons integrate information received through dendritic spines (postsynaptic structures) from upstream neurons through axon terminals (presynaptic structures), and send signals to downstream neurons. The number of direct upstream and downstream partners for each neuron varies greatly from a few hundred to one hundred thousand, and neuronal information can be transmitted via convergent or divergent neural pathways through monosynaptic or multisynaptic connections.
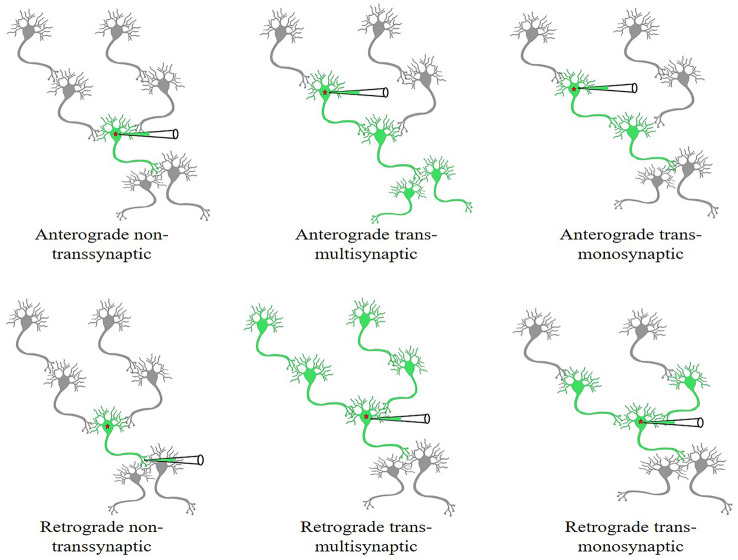
Over the past decades, many powerful labeling and imaging techniques have been developed to explore neural circuits. Currently, viral tools are widely used for neural circuit research (Table 1); they have been modified from the natural strains of neurotropic viruses, such as pseudorabies virus (PRV), herpes simplex virus (HSV), rabies virus (RV), vesicular stomatitis virus (VSV), and sindbis virus (SINV) [1–4]. In addition, some non-transsynaptic recombinant viral vectors can serve as helpers for the expression of exogenous genes, such as the adeno-associated virus (AAV) [5, 6] and lentivirus (LV) [7, 8], or efficiently label the fine structure of neurons, such as semliki forest virus (SFV) [9, 10], or demonstrate upstream projections of local brain regions, such as canine adenovirus type 2 (CAV-2) [11] and AAV2-retro [12], or efficiently cross the mouse blood-brain barrier (BBB), such as AAV9-PHP.eB [13], and retrogradely infect projection neurons after crossing the BBB in the whole brain, such as AAV9-retro [14]. A schematic of the anterograde or retrograde transmission of viral tools is shown in Fig. 1. In this review, we summarize the recent progress in the viral tools for neural circuit tracing.
Table 1.
Viral vectors commonly used for neural circuit research.
| Type | Virus (abbreviation) |
Family, genus | Genomic type | ||
|---|---|---|---|---|---|
| Non-transsynaptic (static) | Adeno-associated virus (AAV) | Parvoviridae, Dependovirus | Single-stranded DNA | ||
| Lentivirus (LV) |
Retroviridae, Lentivirus |
Single-stranded RNA | |||
| Canine adenovirus (CAV) | Adenoviridae, Mastadenovirus | Double-stranded DNA | |||
| Semliki Forest virus (SFV) | Togaviridae, Alphavirus | Single-stranded RNA | |||
| Rabies virus with the envelope glycoprotein gene deleted (RV-ΔG) |
Rhabdoviridae, Lyssavirus |
Negative-sense single-stranded RNA | |||
| Herpes simplex virus amplicon (HSV amplicon) |
Herpesviridae, α herpesvirinae |
Double-stranded DNA | |||
| Transsynaptic | Trans-multisynaptic | Anterograde, multisynaptic | Herpes simplex virus (HSV) H129 |
Herpesviridae, α herpesvirinae |
Double-stranded DNA |
| Vesicular stomatitis virus (VSV) |
Rhabdoviridae, Vesiculovirus |
Negative-sense single-stranded RNA | |||
| Sindbis virus (SINV) |
Togaviridae, Alphavirus |
Single-stranded RNA | |||
| Retrograde, multisynaptic | Pseudorabies virus, PRV Bartha |
Herpesviridae, α herpesvirinae |
Double-stranded DNA | ||
| Wild-type Rabies virus (WT RV) |
Rhabdoviridae, Lyssavirus |
Negative-sense single-stranded RNA | |||
| Trans-monosynaptic | Anterograde, monosynaptic | Herpes simplex virus with TK-deleted (HSV-ΔTK) |
Herpesviridae, α herpesvirinae |
Double-stranded DNA | |
| Adeno-associated virus, serotype 1 (AAV-1) |
Parvoviridae, Dependovirus |
Single-stranded DNA | |||
| Retrograde, monosynaptic | RVΔG-EnvA |
Rhabdoviridae, Lyssavirus |
Negative-sense single-stranded RNA | ||
| Pseudorabies virus with TK-deleted (PRV-ΔTK) |
Herpesviridae, α herpesvirinae |
Double-stranded DNA | |||
Fig. 1.
Schematics of the anterograde or retrograde transmission of non-transsynaptic (static) and transsynaptic (trans-multisynaptic or monosynaptic) viral tools. This figure is adapted from Fig. 1 in [86].
Recombinant AAV (rAAV)
The rAAV vector is the most commonly-used viral tool in biomedicine [5]. rAAV was developed as a gene transfer vector from the wild-type AAV, which belongs to the Parvoviridae family and the genus Dependovirus. It is the simplest type of replication-defective virus found so far, and is dependent on the presence of adenovirus, HSV, and other auxiliary viruses to produce progeny. The genome of AAV is a single-stranded DNA molecule ~4.7 kb long. The AAV replicase protein (Rep) and capsid protein (Cap) genes are flanked by inverted terminal repeat (ITR) sequences. The virus particle does not have an envelope and is an icosahedron with a diameter of ~20 nm. The rAAV genome only conserves the AAV-derived ITRs, and all viral DNA between the ITRs is replaced by the gene of interest. The AAV ITRs serve as cis-elements, while the AAV Rep, Cap, and helper genes are provided in trans for viral replication and packaging [15]. Therefore, rAAV not only has an excellent safety profile, but also inherits many advantages of wild-type AAV, such as low immunogenicity, multiple serotypes, a wide range of infected host cells, and the ability to mediate the long-term stable expression of foreign genes in vivo.
AAV capsid engineering and scaling-up production have attracted much attention in academic and industrial circles. To date, 13 serotypes (AAV1–13) and >100 variants have been identified from hosts. The serotypes differ in their capsid structure and display variable transduction efficiency, tissue and cell tropism, and immunological profile [16]. In AAV capsid engineering, the traditional methods for AAV mutant library construction are mainly based on DNA shuffling, the peptide display, and random point mutations [17], and now computer-aided rational design combined with directed evolution has become a hot topic for optimizing the capsid of AAV vectors [18]. Recently, a machine learning strategy has also been applied for the generation of improved AAV vectors [19, 20]. Compared to the library constructed by traditional methods, the percentage of invalid mutations in the new library is significantly lower with a reduction of redundancy. Dilution of input library DNA significantly increases packaging fidelity [21]. For library screening, new strategies are needed to focus on increasing the throughput, shortening the screening cycle, and improving efficacy. To this end, the multiplexed Cre recombination-based AAV targeted evolution (M-CREATE) platform has been developed for screening AAV variants that can efficiently transduce specific types of cells in the adult mouse brain [22], and the tropism redirection of AAV by cell type-specific expression of RNA (TRACER) platform has been established for rapid selection of AAV capsids by introducing cell type-specific promoters into the core components of the library [23]. The screening methods used in most laboratories are based on the next-generation sequencing (NGS) platform. For rAAV production, the simple, rapid, and flexible triple-plasmid transfection of HEK293 cells is the most commonly used method in the laboratory at present. The three plasmids are the pAAV-(ITR-GOI) core plasmid carrying foreign genes, the pAAV-RC plasmid containing the Rep and Cap genes, and the pAd-helper plasmid encoding the adenovirus virus-associated RNA (VA RNA), E2, and E4 genes. The HEK293 cell line containing the adenovirus E1 gene is usually used for plasmid transfection [24]. However, this method is not suitable for large-scale production due to technical and economic issues. Baculovirus-infected insect Sf9 cells with high security have been developed continuously from the early multi-baculovirus system to the one-baculovirus system [25, 26] (Table 2). Recently, a single baculovirus expression vector (BEV)-derived OneBac system suitable for scaling up the production of different serotypes of rAAV has been reported. About 1 × 1015 vector genomes of purified rAAV can be obtained from 5 L of suspension-cultured Sf9 cells flexibly, conveniently, and efficiently, based on this new OneBac system. Consequently, various serotypes of rAAV can be prepared on a large scale to meet the needs of primate research and clinical therapy [27].
Table 2.
Comparison of the characteristics of Bac systems for rAAV production.
| Bac system Taxonomy | Type and name of BEVs | Name of Cell line | Infection efficiency of BEVs | Yield of rAAV (vg/mL) | Versatility and flexibility | Large scale production | System stability |
|---|---|---|---|---|---|---|---|
| ThreeBac [25] | BEV/Rep, BEV/Cap, BEV/(ITR-GOI) | Sf9 | ~6% | 1–5 × 104 | High | Not suitable | Low |
| TwoBac [87] | BEV/Rep-Cap, BEV/(ITR-GOI) | Sf9 | ~27% | 3–9 × 104 | High | Suitable | Medium |
| OneBac [27, 88, 89] | BEV/(ITR-GOI) | Sf9/Rep-Cap | 100% | 1–5 × 105 | Low | Very suitable | Medium to High |
| BEV/Cap-(ITR-GOI) | Sf9-GFP/Rep | 100% | 0.8–2 × 105 | Medium | Very suitable | Medium | |
| BEV/Cap-(ITR-GOI)-Rep | Sf9 | 100% | 1–5 × 105 | High | Very suitable | Medium |
In the wide application in neuroscience, rAAVs are increasingly useful tools. Different AAV serotypes exhibit different tropisms and can transduce neural cells in vivo and in vitro [28, 29]. Because host cells infected with rAAVs are unable to produce new viral particles, rAAVs cannot efficiently transmit trans-synaptically between neurons. The anterograde, retrograde, and bidirectional transport properties of AAVs are serotype- and concentration-dependent [30]. Therefore, it is very important for researchers to select the appropriate serotype of AAV. In general, most AAV serotypes prefer to enter the cell bodies of neurons at the injection site, and subsequent cellular trafficking may result in the spread of transgenic products throughout the whole neuron. This property may be useful for labeling neuronal projections and manipulating projection targets. Some AAV serotypes (AAV1 and AAV9) have been shown to exhibit anterograde trans-synaptic spread [31]. High titer AAV1-Cre can infect presynaptic neurons and specifically drive Cre-dependent expression in postsynaptic neurons, thus allowing the tracing and manipulation of direct input-defined neurons. Recently, an avian AAV (A3V) has been shown to spread trans-multisynaptically in the anterograde direction and displays no extrasynaptic leakage [32]. Some engineered AAV variants (rAAV2-retro [12] and rAAV9-retro [14]) are preferentially taken up by axon terminals at the injection site, then are transported retrogradely from the axon to the cell body, which is suitable for labeling projection neurons with defined outputs. Some AAV serotypes, such as rAAV9, are able to cross the BBB to transduce brain cells, but the efficiency is limited. By using the AAV capsid engineering strategy, a series of AAV variants (AAV-PHP.B [33], AAV-PHP.eB and AAV-PHP.S [13], AAV-F [34], and AAV.Cap-B10 [35]) have been developed with enhanced BBB penetration and broader host tropism; these are ideal viral tools for brain-wide neural cell transduction after intravenous injection.
In addition, rAAVs can act as helper viruses in combination with other trans-synaptically spread viruses to visualize the structure of neural circuits (encoding fluorescent proteins), and carry optogenetic (ChR2 or eNpHR), chemogenetic (hM3Dq or hM4Di) and neurotransmitter probes (iGluSnFR or dLight), Ca2+ sensors (GCaMP-X), recombination-based gene expression systems (Cre or Flp), or genome editing systems (CRISPR-Cas9) to monitor and manipulate the function of neural circuits, as well as treating multiple neurological diseases [36].
PRV
PRV is a member of the alpha-herpesvirus subfamily, 200–250 nm in diameter, with a complicated genome of ~150 kb. It is a spherical double-stranded DNA virus with an envelope, and large exogenous genes can be inserted after molecular modification. Previously, it was reported that PRV mainly infects pigs and rodents, but not primates. In recent years, it has been reported that several naturally-occurring variants of PRV infect the human central nervous system and induce encephalitis [37], so personal protection should be emphasized during viral modification and application.
PRV strains used include Becker and Bartha strains. The Becker strain is rarely used in neural circuit research due to its significant toxicity and non-direction-specific transmission. The Bartha strain is less toxic and only transfers in a retrograde direction through neural circuits; in particular, it can transmit rapidly from the periphery across synapses to the central nervous system, which has attracted the attention of many researchers in this field. However, the weak fluorescent signal after PRV infection needs to be amplified by immunohistochemistry. To address this issue, PRV531 and PRV724 with robust EGFP and mRuby3 expression have been developed, and can be used as efficient retrograde trans-multisynaptic tracers [38]. Because these PRVs infect many different types of neurons, the lack of specificity of originally-infected neurons limits the precise elucidation of neural circuits. Therefore, PRVs that can be retrogradely transmitted from selected neurons were developed. For example, Ba2001 is a Cre-dependent retrograde trans-multisynaptic tracer encoding GFP and the thymidine kinase (TK) essential for viral replication. It only replicates in neurons expressing the Cre recombinase and in neurons synaptically connected with the originally-infected cells; the inputs of neuropeptide Y-expressing neurons in the arcuate nucleus have been traced with this method [39]. Then, Ba2017, a novel recombinant PRV derived from attenuated PRV Bartha [40], was generated with improved genetic stability and fluorescent protein expression. It has been successfully applied in identifying and characterizing a subpopulation of oxytocin neurons in the paraventricular hypothalamic nucleus that is multisynaptically connected to pancreatic islet β cells [41]. Moreover, an immediate early gene IE180-null recombinant PRV has been engineered to express site-specific Cre recombinase (PRV-hSyn-Cre). This can infect neurons efficiently and robustly in vivo and activate transgene expression from Cre-dependent vectors in local and retrogradely-projecting neurons. Therefore, it can be used for targeted gene expression in the brain, facilitating functional dissection of neural circuits in vivo [42].
RV
RV is an enveloped negative-sense single-stranded RNA virus. It is ~75 nm in diameter and 180 nm in length. It is bullet-shaped with a genome of ~12 kb and is highly neurotropic. Its replication is accomplished by a self-encoded RNA polymerase complex. Wild-type RV (WT RV) is by far the most common causative agent of rabies and is mostly transmitted by the bite of an infected dog. It infects peripheral nerve terminals at the site of the bite and proceeds through the central nervous system from neuron to neuron, exclusively retrogradely from postsynaptic to presynaptic neurons. It has been modified to be safer for applications in the laboratory by deletion of the envelope glycoprotein gene (RV-ΔG) and then used as a tracer for retrograde infection of projection neurons through their axon terminals, targeted infection of genetically specified neurons, or trans-monosynaptic tracing of direct input neurons after further modifications. For example, the recombinant RV-ΔG can express exogenous genes with a high abundance in a short time, which can be used to depict the fine structure of neurons projecting to the injection site and monitor Ca2+ activity in specific input circuits [43, 44]. The RV-ΔG pseudotyped with EnvA, the avian sarcoma and leukosis virus A protein, cannot infect mammalian neurons. It can only infect cells that are specifically engineered to express the avian cell-surface molecule tumor virus A (TVA), the receptor of EnvA. This receptor-ligand pair (TVA-EnvA) allows specific infection of target neurons. The RV-ΔG is replication-deficient, and so cannot spread beyond the initially-infected neurons unless the glycoprotein (G) is supplied. Then the virus can produce infectious progeny, which spread from these neurons to the presynaptic neurons directly connected to them. The retrograde trans-synaptic system based on the modified SAD-B19 RV strain has been widely used to map the input network of specific types of neurons [45]. However, this tracing system can label only a small fraction of the actual input neurons due to the low trans-synaptic efficiency, and its high toxicity makes it difficult to carry functional genes for monitoring and modulating neuronal activity. Therefore, there is an urgent need to improve its trans-synaptic efficiency and reduce its toxicity. Much effort has been made by researchers, but the overall effect has not been great.
Compared with the SAD-B19 strain, the retrograde trans-synaptic ability of the modified CVS-N2c strain is significantly enhanced and the toxicity to host neurons is significantly reduced, which allows functional effectors such as Ca2+ sensor and optogenetic actuators to be carried in functional circuit dissection [46, 47]. However, the difficulties in preparation and low titers limit its application. A codon-optimized chimeric glycoprotein (oG) can greatly improve the trans-synaptic efficiency of SAD-B19-ΔG, which has been applied in input network mapping [48]. However, the self-inactivating RV (SiR) based on SAD-B19-ΔG with high expectations has not been widely used [49]. Due to the low expression efficiency of the non-toxic G/L gene deletion RV (RV-ΔGL), it can only be used to monitor and manipulate neural activity by using Cre or Flp recombinase in combination with functional probes [50]. It is necessary to use appropriate viral vectors or produce transgenic animals carrying L (“large protein” gene) to achieve trans-synaptic spread, which has not been realized yet. Moreover, the trans-synaptic efficiency is significantly enhanced by a two-plasmid co-packaging system for rAAV, but the underlying mechanism is still not clear [51]. Therefore, there is still a requirement to develop a retrograde trans-synaptic tracing system with high efficiency and low toxicity. Then the oG sequence was reshuffled with a positive codon pair bias score to further optimize the glycoprotein and generate the reshuffled oG (ooG), with which the tracing efficiency increased by up to 10 times [52].
In addition, because PRV, RV-B2c, RV-N2c, RV-CVS, AAV2-retro, and HSV can infect axon terminals, they can all be used to map the input network of specific brain regions. Systematic comparisons have shown that they have different retrograde labeling efficiency and selectivity (cell type or cortical layer) [47, 53]. rAAV2-retro mainly targets cortical areas, whereas RV targets both cortical and subcortical areas, and the retrograde labeling efficiency of N2cG-enveloped RV is ten times higher than that of B19G-enveloped RV. These may provide valuable information for the selection of appropriate viral tools.
HSV
HSV1 strain H129, originally isolated from the brain of a patient suffering from acute HSV encephalitis, is widely used as an output network tracer because of its property of anterograde trans-synaptic transmission [54–56]. However, there are some shortcomings of H129 in neural circuit tracing, including low labeling brightness and non-specific retrograde labeling because of its natural property of axon terminal absorption. H129 has been reported to be capable of infecting innervating neurons through retrograde uptake and showing temporally delayed retrograde trans-neuronal spread. A comparison of retrograde labeling and local infection with H129 has not been systematically made. H306, a TK and ICP34.5 dual-deleted H129 recombinant, is replication-deficient in non-dividing postmitotic neurons. It can infect upstream neurons efficiently through axon terminal uptake. Therefore, the neural networks traced by replication-competent, trans-multisynaptic H129 might be from two types of starter cells. One is neurons local to the injection site (true starter site), and the other is the retrogradely-infected neurons (ectopic starter sites), in which H129 can replicate and spread anterogradely through their axon collaterals. As a result, when using H129 to dissect output neural networks, the axon terminal uptake phenotype should be carefully considered.
With a widely-used trans-multisynaptic H129 tracer, animals die within 3–5 days following intracranial infection, when the fluorescent protein is not strongly expressed. It is thus usually necessary to perform immunohistochemical staining to amplify the fluorescence signals for visualization of the output neural circuits. Applying a Trojan horse-like strategy, HSV integrating AAV replication elements can enhance exogenous gene expression; it has low immunogenicity and strong expression with improved fluorescent brightness. The synergism of the HSV vector and AAV replication system enables efficient exogenous gene replication and amplification both in vitro and in vivo. This approach may be useful as a general enhanced expression strategy for HSV-based tracing tools or gene-delivery vectors [57]. In combination with the strategy of the re-targeting oncolytic virus, a highly specific H129 anterograde tracing system has been developed by modifying the gD gene, an envelope glycoprotein essential for HSV to infect cells [58]. The recombinant H129 exhibits a new infection tropism by replacing the receptor recognition domain with a single-chain antibody targeting the Her2 receptor. The Her2 receptor and gD protein are pre-expressed in a specific type of neuron in a given brain region, and then the re-targeted H129 with gD gene deletion is injected to infect the cell bodies of corresponding starter neurons for rigorous anterograde monosynaptic tracing. By specifically targeting the Her2 receptors, which are not endogenously expressed in neurons, the axon terminal infection is avoided and specificity of the tracer is achieved. With helper viruses encoding truncated Her2 and wild-type gD proteins, the direct projection of a given brain region or a specific type of neuron can be elucidated.
VSV
VSV belongs to the genus Vesiculovirus in the family Rhabdoivirida. It is a bullet-shaped enveloped negative-sense RNA virus with a genome of ~11 kb, which contains genes encoding the nucleocapsid (N) protein, phosphoprotein (P), matrix (M) protein, G protein, and large polymerase (L). The G protein enables VSV to infect most mammalian cell types, and the N protein tightly encapsulates the RNA genome, forming a nuclease-resistant nucleocapsid. VSV is known for its fast replication and assembly, high efficiency of infection and exogenous gene expression, and has been shown to transmit anterogradely among neurons. These properties make it suitable for anterograde trans-multisynaptic tracing output networks efficiently. The VSV G protein can be pseudotyped with glycoproteins from other viruses to achieve different labeling characteristics and anterograde or retrograde trans-synaptic transmission [59, 60].
However, the high neurotoxicity and rapid lethality in laboratory animals hinder its application in long-term studies of the structure and function of neural circuits. To overcome these drawbacks, a recombinant VSV with mutation of the seventh amino acid of the replication-related N protein (VSV-NR7A), was developed with attenuated cytotoxicity and a more efficient anterograde trans-synaptic capability [3]. To disambiguate between monosynaptic and multisynaptic connections, the VSV that lacks the glycoprotein gene (VSV-∆G) and cannot spread on its own is complemented by providing replication-incompetent lentivirus coated with VSV glycoprotein. This thus can be used to label the direct projection targets of the initially infected neurons in a Cre-dependent manner [61].
LV
LVs are members of the Retroviridae family. They are spherical, enveloped viruses ~80–100 nm in diameter. Their genome is single-stranded RNA with gag, pol, and env genes that encode polyprotein components of the capsid, the enzymes reverse transcriptase, protease, integrase, and envelope glycoproteins, respectively. The viral RNA genome is reverse-transcribed into DNA for integration into the host genome. LV vectors based on human immunodeficiency virus 1 (HIV-1) are the most widely used in neuroscience research. They mediate stable and efficient in vivo gene transfer into differentiated neurons and infect dividing and non-dividing cells, including neurons and glia in the adult mammalian brain. They have a packaging capacity of up to 9 kb, which makes them attractive gene-delivery vehicles for gene therapy, especially for the treatment of neurological and neurodegenerative disorders [62].
The tropism of LV vectors is largely defined by the viral envelope glycoproteins which are essential for vector transduction. A variety of glycoproteins have been used to pseudotype LV vectors, typically the envelope glycoproteins from VSV (VSV-G) and RV (RV-G) [63]. The lentiviral vectors packaged with VSV-G can transduce neurons with anterograde transport of the expressed proteins throughout the cell bodies and axons, and low efficiency of retrograde gene transfer. However, the LV vectors pseudotyped with RV-G show improved retrograde axonal transport. Moreover, LV vectors pseudotyped with fusion envelope glycoprotein type B (FuG-B) show highly efficient retrograde transport (HiRet), and those pseudotyped with fusion glycoprotein type C (FuG-C) exhibit neuron-specific retrograde gene transfer (NeuRet) [64]. These viral vectors have been applied for targeting neuronal populations innervating the injection site, and thus can be used as retrograde tracers for anatomical mapping and the functional analysis of specific projection pathways.
CAV-2
Canine adenoviruses consist of 2 predominant serotypes, CAV-1 and CAV-2, which are genetically distinct from each other. CAV-1 is the causative agent of infectious canine hepatitis, while CAV-2 is a causative agent of infectious canine respiratory disease. CAV-2 is a form of adenovirus, and is a non-enveloped double-stranded DNA virus. It has been used in numerous studies in neuroscience research due to its advantages, including relatively low immunogenicity, preferential transduction of neurons, long-term transgene expression in the mammalian brain, and efficient retrograde transport from the axon terminal to the cell body. Its retrograde properties rely on the coxsackievirus and adenovirus receptor (CAR), which is a cell adhesion molecule responsible for the attachment, internalization, and axonal transport of CAV. CARs are expressed by maturing and mature neurons and are located on the cell bodies and projections [65]. Therefore, CAV-2 transduces neurons at the injection site as well as projection neurons innervating the injection site; it is transported from axon terminals via a retrograde mechanism to drive gene expression at distant cell bodies. It is thus useful for targeting neurons based on their projections.
A combination of CAV-2 expressing Cre recombinase and another vector containing Cre-inducible expression of effectors can be used to monitor or perturb neurons of one brain region that innervate a distal region. For example, the use of retrograde CAV2-Cre and anterograde AAV enables the retrograde transport of CAV-2 from distal axon terminals back to cell bodies and the Cre recombinase expression in the nuclei, and then activates the selective expression of AAV-carrying effectors in neurons projecting from the AAV injection site to the CAV-2 injection site. This dual-virus strategy can be especially useful for studying reciprocal neural pathways [66]. Furthermore, CAV has a high carrying capacity, which is suitable for the precise dissection of neural circuits. The large carrying load of CAV allows a variety of genes such as optogenetic elements, indicators of neural activity, large promoters, and recombinases, in one CAV vector. This can be used to selectively label neurons in a cell type- and/or projection-specific manner, facilitating monitoring and manipulating neural activity simultaneously [67].
SINV and SFV
SINV and SFV are small, enveloped, positive-strand RNA viruses, and closely related members of the Alphavirus genus of the Togaviridae family. Their genomes are ~12 kb, which encode four non-structural proteins (nsPs 1–4) and five structural proteins (capsid, E3, E2, 6K, and E1). The structural proteins are translated from a highly active subgenomic promoter (26S promoter). Vectors based on these two viruses have been engineered to express exogenous genes at high levels both in vitro and in vivo [68, 69].
SINV was first isolated from febrile patients and identified as a human pathogen in Uganda in 1961; it has high infection efficiency in neurons [70], and can be optimized as a tool for neuroscience research. Recent studies have found that SINV with non-structural and capsid regions from the Toto1101 strain and structural regions from the neuroadapted Sindbis virus (NSV) strain can infect neurons in the peripheral and central nervous systems of zebrafish larvae after intravenous inoculation [71] Moreover, a new anterograde trans-synaptic tracer based on SINV has been developed, which serves as a supplement to the viral tools for anterograde neural circuit tracing [4].
SFV was first isolated from mosquitoes captured in the Semliki Forest in Uganda in 1942. Wild-type SFV mainly infects neurons, and is suitable for the rapid labeling of neurons, but not suitable for long-term transgene expression, which limits its application in the functional investigation of neural circuits. Currently, a mutant virus-like particle of SFV has been developed that efficiently expresses GFP with single-round infection characteristics, which can be used for rapid and sparse labeling of neurons and combined with the fluorescence microscopic optical slice tomography system (fMOST) to delineate the detailed morphology of neurons [9, 72]. In addition, SFV strain A7 (74) infection is temperature-dependent. Contrary to conventional SFV, it exclusively transduces glial cells at 37°C, but mostly infects neurons at 31°C [73], indicating that the SFV is a potential vector to develop different tools to target specific types of cells in the nervous system.
The production of high-titer SINV and SFV is rapid, and generally requires Biosafety Level 2 facilities. Usually, SINV and SFV are produced by replicons and helper RNAs. The replicons are self-replicating with viral structural protein-encoding genes replaced by heterologous genes, and the helper RNAs provide structural proteins for the replicons [74]. The infectious particles can be packaged by co-transfection of replicons with helper RNAs into target cells, but new infectious particles cannot be produced due to the absence of structural protein genes, which can be used to decipher the fine structure of neurons [75]. Besides, the plasmid containing a promoter to initiate transcription of the viral genome can produce viral progeny, which can be used to trans-synaptically dissect neural circuits. Wild-type SINV and SFV vectors allowing fast and efficient transgene expression are cytotoxic, therefore mutant vectors with reduced cytotoxicity have been developed. For example, mutant strains of SINV, SFV-SINrep(nsP2S726), and SFV(PD) show attenuated cytotoxicity in primary cultured neurons, dissociated neurons, and brain slices, respectively [76, 77]. SINV and SFV have gradually become potent viral tools for depicting the fine structure of neurons and non-neuronal cells, and vectors for tracing neural circuits with reduced cytotoxicity in rodents and non-human primates in vivo are under development.
Viral Vectors for Neural Circuit Tracing in Non-human Primates
Recombinant viruses exhibit distinct infection tropisms across diverse animal species or neuronal types in different brain regions, and viral vectors can carry a variety of elements or probes to meet specific research needs. Selective labeling techniques and powerful microscopes can reveal the patterns of synaptic connectivity underlying the functional properties of neural circuits of diverse species, including non-human primates [78, 79]. Viral tools used in monkey brain labeling include RV, HSV1, AAV, and LV, among which AAV and LV are the most commonly used viral vectors for anterograde or retrograde targeting [6, 80]. AAV and LV differ in their transduction, specificity, and spread in the brain. AAV transduces a much larger area around the injection site, while LV is usually restricted to the injection site. AAV has a diameter of ~20 nm and LV has a larger particle size of ~100 nm, which limits LV spread through the extracellular space [8]. Moreover, LV leads to a smaller proportion of aggregates of non-native expressed proteins in cells and may thus be a better option for long-term studies. For example, LV containing ChR2 can be used to specifically activate excitatory neurons in the frontal cortex of the macaque monkey over many months [81]. Implementation of the Volumetric Imaging with Synchronized on-the-fly-scan and Readout (VISoR) and semiautomated reconstruction and tracing (SMART) techniques enables mapping long-range projections of cortical axonal fibers labeled by rAAV9 throughout the entire brain of the macaque [82]. Using a G-deleted RV vector based on a SAD-B19 strain expressing GFP, the morphology of specific projecting neurons in the primary visual cortex of the rhesus monkey has been delineated [83]. Applying the CVS-11 strain of RV, trans-multisynaptic inputs to a targeted brain region of the macaque can be dissected [84]. Without a Cre or Flp transgenic monkey line, viral tools with specific promoters can be used to drive transgene expression in specific types of neurons. The two commonly used promoters used in primates to target neurons are the human Synapsin-1 (hSyn) promoter and the mouse Ca2+ calmodulin kinase-2a (CaMKIIa) promoter. The transduction specificity is titer-dependent, but high-titer vectors may lead to off-target effects [85]. Monkeys having preexisting immunity against AAV prevent viral vectors from transducing neurons efficiently, so it is necessary to measure the immune responses before AAV vector infusion. Nevertheless, there is still a lack of useful viral tools for tracing neural circuits in the monkey brain, such as anterograde and retrograde trans-monosynaptic tracers, which we focus on in the next step.
Prospects
Structure is the basis of function, and function is the embodiment of structure. The current neural circuit tracing tools provide us with the basic means to reveal the structure and clarify the function and mechanism. The choice of viral tools in dissecting neural circuits is complex, and depends on the experimental design required to solve a specific research question or problem. It is noteworthy that many functional studies are performed on freely-moving animals over weeks or even months, so viral vectors that do not disturb neural activity are needed. Currently, replication-defective viruses such as AAV vectors which are less toxic to nerve cells are mainly used for the functional analysis of neural circuits. The viral tools that can trace neural circuits by trans-multisynaptic transmission are less virulent after modifications, but they are still unsuitable for functional research. Fortunately, the viral vectors that are transmitted monosynaptically through neural circuits both anterogradely or retrogradely are much less toxic to nerve cells and animals, and tools based on these viruses for functional dissection are under development. Moreover, the complexity of neural circuits poses an unparalleled challenge for developing techniques and tools. Viral tools need to be further improved in many ways including attenuating toxicity, increasing specificity, and enhancing sensitivity, especially in tools that combine structural and functional studies and are compatible with other approaches such as omics technologies. The China Brain Project in basic research focuses on understanding neural circuit mechanisms underlying the cognitive processes in nonhuman primates. The development of new strategies and technologies, and the endless demand of neuroscience research for new tools will promote rapid developments in this field.
Acknowledgments
This review was supported by the National Science and Technology Innovation 2030 (2021ZD0201003), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB32030200), the National Natural Science Foundation of China (31830035), and the Shenzhen Key Laboratory of Viral Vectors for Biomedicine (ZDSYS20200811142401005).
Conflict of interest
The authors claim that there are no conflicts of interest.
References
- 1.Nassi JJ, Cepko CL, Born RT, Beier KT. Neuroanatomy goes viral! Front Neuroanat. 2015;9:80. doi: 10.3389/fnana.2015.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu X, Holmes TC, Luo MH, Beier KT, Horwitz GD, Zhao F, et al. Viral vectors for neural circuit mapping and recent advances in trans-synaptic anterograde tracers. Neuron. 2020;107:1029–1047. doi: 10.1016/j.neuron.2020.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin K, Zhong X, Ying M, Li L, Tao S, Zhu X, et al. A mutant vesicular stomatitis virus with reduced cytotoxicity and enhanced anterograde trans-synaptic efficiency. Mol Brain. 2020;13:45. doi: 10.1186/s13041-020-00588-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi XW, Jia F, Lyu P, Xu FQ. A new anterograde trans-synaptic tracer based on Sindbis virus. Neural Regen Res. 2022;17:2761–2764. doi: 10.4103/1673-5374.339495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang D, Tai PWL, Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Discov. 2019;18:358–378. doi: 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kondratov O, Kondratova L, Mandel RJ, Coleman K, Savage MA, Gray-Edwards HL, et al. A comprehensive study of a 29-capsid AAV library in a non-human primate central nervous system. Mol Ther. 2021;29:2806–2820. doi: 10.1016/j.ymthe.2021.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nectow AR, Nestler EJ. Viral tools for neuroscience. Nat Rev Neurosci. 2020;21:669–681. doi: 10.1038/s41583-020-00382-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parr-Brownlie LC, Bosch-Bouju C, Schoderboeck L, Sizemore RJ, Abraham WC, Hughes SM. Lentiviral vectors as tools to understand central nervous system biology in mammalian model organisms. Front Mol Neurosci. 2015;8:14. doi: 10.3389/fnmol.2015.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jia F, Zhu X, Lv P, Hu L, Liu Q, Jin S, et al. Rapid and sparse labeling of neurons based on the mutant virus-like particle of semliki forest virus. Neurosci Bull. 2019;35:378–388. doi: 10.1007/s12264-019-00362-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia F, Miao H, Zhu X, Xu F. Pseudo-typed Semliki Forest virus delivers EGFP into neurons. J Neurovirol. 2017;23:205–215. doi: 10.1007/s13365-016-0486-8. [DOI] [PubMed] [Google Scholar]
- 11.Soudais C, Laplace-Builhe C, Kissa K, Kremer EJ. Preferential transduction of neurons by canine adenovirus vectors and their efficient retrograde transport in vivo. FASEB J. 2001;15:2283–2285. doi: 10.1096/fj.01-0321fje. [DOI] [PubMed] [Google Scholar]
- 12.Tervo DGR, Hwang BY, Viswanathan S, Gaj T, Lavzin M, Ritola KD, et al. A designer AAV variant permits efficient retrograde access to projection neurons. Neuron. 2016;92:372–382. doi: 10.1016/j.neuron.2016.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan KY, Jang MJ, Yoo BB, Greenbaum A, Ravi N, Wu WL, et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat Neurosci. 2017;20:1172–1179. doi: 10.1038/nn.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin K, Zhong X, Li L, Ying M, Yang T, Zhang Z, et al. AAV9-Retro mediates efficient transduction with axon terminal absorption and blood-brain barrier transportation. Mol Brain. 2020;13:138. doi: 10.1186/s13041-020-00679-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayuso E, Mingozzi F, Bosch F. Production, purification and characterization of adeno-associated vectors. Curr Gene Ther. 2010;10:423–436. doi: 10.2174/156652310793797685. [DOI] [PubMed] [Google Scholar]
- 16.Srivastava A. In vivo tissue-tropism of adeno-associated viral vectors. Curr Opin Virol. 2016;21:75–80. doi: 10.1016/j.coviro.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimm D, Zolotukhin S. E pluribus unum: 50 years of research, millions of viruses, and one goal—Tailored acceleration of AAV evolution. Mol Ther. 2015;23:1819–1831. doi: 10.1038/mt.2015.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li C, Samulski RJ. Engineering adeno-associated virus vectors for gene therapy. Nat Rev Genet. 2020;21:255–272. doi: 10.1038/s41576-019-0205-4. [DOI] [PubMed] [Google Scholar]
- 19.Wu Z, Kan SBJ, Lewis RD, Wittmann BJ, Arnold FH. Machine learning-assisted directed protein evolution with combinatorial libraries. Proc Natl Acad Sci U S A. 2019;116:8852–8858. doi: 10.1073/pnas.1901979116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bryant DH, Bashir A, Sinai S, Jain NK, Ogden PJ, Riley PF, et al. Deep diversification of an AAV capsid protein by machine learning. Nat Biotechnol. 2021;39:691–696. doi: 10.1038/s41587-020-00793-4. [DOI] [PubMed] [Google Scholar]
- 21.Schmit PF, Pacouret S, Zinn E, Telford E, Nicolaou F, Broucque F, et al. Cross-packaging and capsid mosaic formation in multiplexed AAV libraries. Mol Ther Methods Clin Dev. 2020;17:107–121. doi: 10.1016/j.omtm.2019.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ravindra Kumar S, Miles TF, Chen X, Brown D, Dobreva T, Huang Q, et al. Multiplexed Cre-dependent selection yields systemic AAVs for targeting distinct brain cell types. Nat Methods. 2020;17:541–550. doi: 10.1038/s41592-020-0799-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nonnenmacher M, Wang W, Child MA, Ren XQ, Huang C, Ren AZ, et al. Rapid evolution of blood-brain-barrier-penetrating AAV capsids by RNA-driven biopanning. Mol Ther Methods Clin Dev. 2021;20:366–378. doi: 10.1016/j.omtm.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao X, Li J, Samulski RJ. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J Virol. 1998;72:2224–2232. doi: 10.1128/JVI.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urabe M, Ding C, Kotin RM. Insect cells as a factory to produce adeno-associated virus type 2 vectors. Hum Gene Ther. 2002;13:1935–1943. doi: 10.1089/10430340260355347. [DOI] [PubMed] [Google Scholar]
- 26.Wu Y, Jiang L, Geng H, Yang T, Han Z, He X, et al. A recombinant baculovirus efficiently generates recombinant adeno-associated virus vectors in cultured insect cells and larvae. Mol Ther Methods Clin Dev. 2018;10:38–47. doi: 10.1016/j.omtm.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Y, Han Z, Duan M, Jiang L, Tian T, Jin D, et al. Popularizing recombinant baculovirus-derived OneBac system for laboratory production of all recombinant adeno-associated virus vector serotypes. Curr Gene Ther. 2021;21:167–176. doi: 10.2174/1566523221666210118111657. [DOI] [PubMed] [Google Scholar]
- 28.Watakabe A, Ohtsuka M, Kinoshita M, Takaji M, Isa K, Mizukami H, et al. Comparative analyses of adeno-associated viral vector serotypes 1, 2, 5, 8 and 9 in marmoset, mouse and macaque cerebral cortex. Neurosci Res. 2015;93:144–157. doi: 10.1016/j.neures.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Castle MJ, Turunen HT, Vandenberghe LH, Wolfe JH. Controlling AAV tropism in the nervous system with natural and engineered capsids. Methods Mol Biol. 2016;1382:133–149. doi: 10.1007/978-1-4939-3271-9_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salegio EA, Samaranch L, Kells AP, Mittermeyer G, San Sebastian W, Zhou S, et al. Axonal transport of adeno-associated viral vectors is serotype-dependent. Gene Ther. 2013;20:348–352. doi: 10.1038/gt.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zingg B, Chou XL, Zhang ZG, Mesik L, Liang F, Tao HW, et al. AAV-mediated anterograde transsynaptic tagging: Mapping corticocollicular input-defined neural pathways for defense behaviors. Neuron. 2017;93:33–47. doi: 10.1016/j.neuron.2016.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito T, Ono M, Matsui R, Watanabe D, Ohmori H. Avian adeno-associated virus as an anterograde transsynaptic vector. J Neurosci Methods. 2021;359:109221. doi: 10.1016/j.jneumeth.2021.109221. [DOI] [PubMed] [Google Scholar]
- 33.Deverman BE, Pravdo PL, Simpson BP, Kumar SR, Chan KY, Banerjee A, et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat Biotechnol. 2016;34:204–209. doi: 10.1038/nbt.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanlon KS, Meltzer JC, Buzhdygan T, Cheng MJ, Sena-Esteves M, Bennett RE, et al. Selection of an efficient AAV vector for robust CNS transgene expression. Mol Ther Methods Clin Dev. 2019;15:320–332. doi: 10.1016/j.omtm.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goertsen D, Flytzanis NC, Goeden N, Chuapoco MR, Cummins A, Chen Y, et al. AAV capsid variants with brain-wide transgene expression and decreased liver targeting after intravenous delivery in mouse and marmoset. Nat Neurosci. 2022;25:106–115. doi: 10.1038/s41593-021-00969-4. [DOI] [PubMed] [Google Scholar]
- 36.Haggerty DL, Grecco GG, Reeves KC, Atwood B. Adeno-associated viral vectors in neuroscience research. Mol Ther Methods Clin Dev. 2020;17:69–82. doi: 10.1016/j.omtm.2019.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Q, Wang X, Xie C, Ding S, Yang H, Guo S, et al. A novel human acute encephalitis caused by pseudorabies virus variant strain. Clin Infect Dis. 2021;73:e3690–e3700. doi: 10.1093/cid/ciaa987. [DOI] [PubMed] [Google Scholar]
- 38.Jia F, Lv P, Miao H, Shi X, Mei H, Li L, et al. Optimization of the fluorescent protein expression level based on pseudorabies virus bartha strain for neural circuit tracing. Front Neuroanat. 2019;13:63. doi: 10.3389/fnana.2019.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeFalco J, Tomishima M, Liu H, Zhao C, Cai X, Marth JD, et al. Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science. 2001;291:2608–2613. doi: 10.1126/science.1056602. [DOI] [PubMed] [Google Scholar]
- 40.Enquist LW, Card JP. Recent advances in the use of neurotropic viruses for circuit analysis. Curr Opin Neurobiol. 2003;13:603–606. doi: 10.1016/j.conb.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 41.Papazoglou I, Lee JH, Cui Z, Li C, Fulgenzi G, Bahn YJ, et al. A distinct hypothalamus-to-β cell circuit modulates insulin secretion. Cell Metab. 2022;34:285–298.e7. doi: 10.1016/j.cmet.2021.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oyibo HK, Znamenskiy P, Oviedo HV, Enquist LW, Zador AM. Long-term Cre-mediated retrograde tagging of neurons using a novel recombinant pseudorabies virus. Front Neuroanat. 2014;8:86. doi: 10.3389/fnana.2014.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wickersham IR, Finke S, Conzelmann KK, Callaway EM. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat Methods. 2007;4:47–49. doi: 10.1038/nmeth999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang Y, Li L, Sun L, Yu J, Hu Z, Lian K, et al. In vivo two-photon calcium imaging in dendrites of rabies virus-labeled V1 corticothalamic neurons. Neurosci Bull. 2020;36:545–553. doi: 10.1007/s12264-019-00452-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wickersham IR, Lyon DC, Barnard RJO, Mori T, Finke S, Conzelmann KK, et al. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reardon TR, Murray AJ, Turi GF, Wirblich C, Croce KR, Schnell MJ, et al. Rabies virus CVS-N2c(ΔG) strain enhances retrograde synaptic transfer and neuronal viability. Neuron. 2016;89:711–724. doi: 10.1016/j.neuron.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu X, Lin K, Liu Q, Yue X, Mi H, Huang X, et al. Rabies virus pseudotyped with CVS-N2C glycoprotein as a powerful tool for retrograde neuronal network tracing. Neurosci Bull. 2020;36:202–216. doi: 10.1007/s12264-019-00423-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim EJ, Jacobs MW, Ito-Cole T, Callaway EM. Improved monosynaptic neural circuit tracing using engineered rabies virus glycoproteins. Cell Rep. 2016;15:692–699. doi: 10.1016/j.celrep.2016.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ciabatti E, González-Rueda A, Mariotti L, Morgese F, Tripodi M. Life-long genetic and functional access to neural circuits using self-inactivating rabies virus. Cell. 2017;170:382–392.e14. doi: 10.1016/j.cell.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chatterjee S, Sullivan HA, MacLennan BJ, Xu R, Hou Y, Lavin TK, et al. Nontoxic, double-deletion-mutant rabies viral vectors for retrograde targeting of projection neurons. Nat Neurosci. 2018;21:638–646. doi: 10.1038/s41593-018-0091-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun P, Jin S, Tao S, Wang J, Li A, Li N, et al. Highly efficient and super-bright neurocircuit tracing using vector mixing-based virus cocktail. bioRxiv, 2020: 705772. 10.1101/705772.
- 52.Jia F, Li L, Liu H, Lv P, Shi X, Wu Y, et al. Development of a rabies virus-based retrograde tracer with high trans-monosynaptic efficiency by reshuffling glycoprotein. Mol Brain. 2021;14:109. doi: 10.1186/s13041-021-00821-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun L, Tang Y, Yan K, Yu J, Zou Y, Xu W, et al. Differences in neurotropism and neurotoxicity among retrograde viral tracers. Mol Neurodegener. 2019;14:8. doi: 10.1186/s13024-019-0308-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zemanick MC, Strick PL, Dix RD. Direction of transneuronal transport of herpes simplex virus 1 in the primate motor system is strain-dependent. Proc Natl Acad Sci U S A. 1991;88:8048–8051. doi: 10.1073/pnas.88.18.8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dum RP, Levinthal DJ, Strick PL. The spinothalamic system targets motor and sensory areas in the cerebral cortex of monkeys. J Neurosci. 2009;29:14223–14235. doi: 10.1523/JNEUROSCI.3398-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zeng WB, Jiang HF, Gang YD, Song YG, Shen ZZ, Yang H, et al. Anterograde monosynaptic transneuronal tracers derived from herpes simplex virus 1 strain H129. Mol Neurodegener. 2017;12:38. doi: 10.1186/s13024-017-0179-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Su P, Ying M, Han Z, Xia J, Jin S, Li Y, et al. High-brightness anterograde transneuronal HSV1 H129 tracer modified using a Trojan horse-like strategy. Mol Brain. 2020;13:5. doi: 10.1186/s13041-020-0544-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Su P, Ying M, Xia J, Li Y, Wu Y, Wang H, et al. Rigorous anterograde trans-monosynaptic tracing of genetic defined neurons with retargeted HSV1 H129. bioRxiv 2020, 10.1101/2020.12.01.407312.
- 59.Beier KT, Saunders A, Oldenburg IA, Miyamichi K, Akhtar N, Luo L, et al. Anterograde or retrograde transsynaptic labeling of CNS neurons with vesicular stomatitis virus vectors. Proc Natl Acad Sci U S A. 2011;108:15414–15419. doi: 10.1073/pnas.1110854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mundell NA, Beier KT, Pan YA, Lapan SW, Göz Aytürk D, Berezovskii VK, et al. Vesicular stomatitis virus enables gene transfer and transsynaptic tracing in a wide range of organisms. J Comp Neurol. 2015;523:1639–1663. doi: 10.1002/cne.23761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kler S, Ma M, Narayan S, Ahrens MB, Pan YA. Cre-dependent anterograde transsynaptic labeling and functional imaging in zebrafish using VSV with reduced cytotoxicity. Front Neuroanat. 2021;15:758350. doi: 10.3389/fnana.2021.758350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Azzouz M, Ralph GS, Storkebaum E, Walmsley LE, Mitrophanous KA, Kingsman SM, et al. VEGF delivery with retrogradely transported lentivector prolongs survival in a mouse ALS model. Nature. 2004;429:413–417. doi: 10.1038/nature02544. [DOI] [PubMed] [Google Scholar]
- 63.Mazarakis ND, Azzouz M, Rohll JB, Ellard FM, Wilkes FJ, Olsen AL, et al. Rabies virus glycoprotein pseudotyping of lentiviral vectors enables retrograde axonal transport and access to the nervous system after peripheral delivery. Hum Mol Genet. 2001;10:2109–2121. doi: 10.1093/hmg/10.19.2109. [DOI] [PubMed] [Google Scholar]
- 64.Kobayashi K, Kato S, Kobayashi K. Genetic manipulation of specific neural circuits by use of a viral vector system. J Neural Transm. 2018;125:67–75. doi: 10.1007/s00702-016-1674-7. [DOI] [PubMed] [Google Scholar]
- 65.Wehbi A, Kremer EJ, Dopeso-Reyes IG. Location of the cell adhesion molecule coxsackievirus and adenovirus receptor in the adult mouse brain. Front Neuroanat. 2020;14:28. doi: 10.3389/fnana.2020.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carter ME, Soden ME, Zweifel LS, Palmiter RD. Genetic identification of a neural circuit that suppresses appetite. Nature. 2013;503:111–114. doi: 10.1038/nature12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lavoie A, Liu BH. Canine adenovirus 2: A natural choice for brain circuit dissection. Front Mol Neurosci. 2020;13:9. doi: 10.3389/fnmol.2020.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bredenbeek PJ, Frolov I, Rice CM, Schlesinger S. Sindbis virus expression vectors: Packaging of RNA replicons by using defective helper RNAs. J Virol. 1993;67:6439–6446. doi: 10.1128/jvi.67.11.6439-6446.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kebschull JM, Garcia da Silva P, Zador AM. A New Defective Helper RNA to Produce Recombinant Sindbis Virus that Infects Neurons but does not Propagate. Front Neuroanat. 2016;10:56. doi: 10.3389/fnana.2016.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Furuta T, Tomioka R, Taki K, Nakamura K, Tamamaki N, Kaneko T. In vivo transduction of central neurons using recombinant Sindbis virus: Golgi-like labeling of dendrites and axons with membrane-targeted fluorescent proteins. J Histochem Cytochem. 2001;49:1497–1508. doi: 10.1177/002215540104901203. [DOI] [PubMed] [Google Scholar]
- 71.Passoni G, Langevin C, Palha N, Mounce BC, Briolat V, Affaticati P, et al. Imaging of viral neuroinvasion in the zebrafish reveals that Sindbis and chikungunya viruses favour different entry routes. Dis Model Mech. 2017;10:847–857. doi: 10.1242/dmm.029231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kuramoto E. Method for labeling and reconstruction of single neurons using Sindbis virus vectors. J Chem Neuroanat. 2019;100:101648. doi: 10.1016/j.jchemneu.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 73.Ehrengruber MU, Renggli M, Raineteau O, Hennou S, Vähä-Koskela MJV, Hinkkanen AE, et al. Semliki Forest virus A7(74) transduces hippocampal neurons and glial cells in a temperature-dependent dual manner. J Neurovirology. 2003;9:16–28. doi: 10.1080/13550280390173346. [DOI] [PubMed] [Google Scholar]
- 74.Ehrengruber MU, Schlesinger S, Lundstrom K. Alphaviruses: Semliki forest virus and Sindbis virus vectors for gene transfer into neurons. Curr Protoc Neurosci 2011, Chapter 4: Unit 4.22. [DOI] [PubMed]
- 75.Guo B, Chen J, Chen Q, Ren K, Feng D, Mao H, et al. Anterior cingulate cortex dysfunction underlies social deficits in Shank3 mutant mice. Nat Neurosci. 2019;22:1223–1234. doi: 10.1038/s41593-019-0445-9. [DOI] [PubMed] [Google Scholar]
- 76.Kim J, Dittgen T, Nimmerjahn A, Waters J, Pawlak V, Helmchen F, et al. Sindbis vector SINrep(nsP2S726): A tool for rapid heterologous expression with attenuated cytotoxicity in neurons. J Neurosci Methods. 2004;133:81–90. doi: 10.1016/j.jneumeth.2003.09.029. [DOI] [PubMed] [Google Scholar]
- 77.Lundstrom K, Abenavoli A, Malgaroli A, Ehrengruber MU. Novel semliki forest virus vectors with reduced cytotoxicity and temperature sensitivity for long-term enhancement of transgene expression. Mol Ther. 2003;7:202–209. doi: 10.1016/S1525-0016(02)00056-4. [DOI] [PubMed] [Google Scholar]
- 78.El-Shamayleh Y, Ni AM, Horwitz GD. Strategies for targeting primate neural circuits with viral vectors. J Neurophysiol. 2016;116:122–134. doi: 10.1152/jn.00087.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou C, Yang X, Wu S, Zhong Q, Luo T, Li A, et al. Continuous subcellular resolution three-dimensional imaging on intact macaque brain. Sci Bull. 2022;67:85–96. doi: 10.1016/j.scib.2021.08.003. [DOI] [PubMed] [Google Scholar]
- 80.An H, Cho DW, Lee SE, Yang YS, Han SC, Lee CJ. Differential cellular tropism of lentivirus and adeno-associated virus in the brain of cynomolgus monkey. Exp Neurobiol. 2016;25:48–54. doi: 10.5607/en.2016.25.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Han X, Qian X, Bernstein JG, Zhou HH, Franzesi GT, Stern P, et al. Millisecond-timescale optical control of neural dynamics in the nonhuman primate brain. Neuron. 2009;62:191–198. doi: 10.1016/j.neuron.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu F, Shen Y, Ding L, Yang CY, Tan H, Wang H, et al. High-throughput mapping of a whole rhesus monkey brain at micrometer resolution. Nat Biotechnol. 2021;39:1521–1528. doi: 10.1038/s41587-021-00986-5. [DOI] [PubMed] [Google Scholar]
- 83.Nhan HL, Callaway EM. Morphology of superior colliculus- and middle temporal area-projecting neurons in primate primary visual cortex. J Comp Neurol. 2012;520:52–80. doi: 10.1002/cne.22685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nassi JJ, Callaway EM. Multiple circuits relaying primate parallel visual pathways to the middle temporal area. J Neurosci. 2006;26:12789–12798. doi: 10.1523/JNEUROSCI.4044-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nathanson JL, Yanagawa Y, Obata K, Callaway EM. Preferential labeling of inhibitory and excitatory cortical neurons by endogenous tropism of adeno-associated virus and lentivirus vectors. Neuroscience. 2009;161:441–450. doi: 10.1016/j.neuroscience.2009.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Han Z, Shi X, Ying M, Zheng N, Li M, Zhang Z, et al. Tools for neural circuit tracing based on neurotropic viruses. Chinese J Anal Chem. 2019;47:1639–1650. [Google Scholar]
- 87.Chen H. Intron splicing-mediated expression of AAV Rep and Cap genes and production of AAV vectors in insect cells. Mol Ther. 2008;16:924–930. doi: 10.1038/mt.2008.35. [DOI] [PubMed] [Google Scholar]
- 88.Mietzsch M, Grasse S, Zurawski C, Weger S, Bennett A, Agbandje-McKenna M, et al. OneBac: platform for scalable and high-titer production of adeno-associated virus serotype 1–12 vectors for gene therapy. Hum Gene Ther. 2014;25:212–222. doi: 10.1089/hum.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu Y, Mei T, Jiang L, Han Z, Dong R, Yang T, et al. Development of versatile and flexible Sf9 packaging cell line-dependent OneBac system for large-scale recombinant adeno-associated virus production. Hum Gene Ther Methods. 2019;30:172–183. doi: 10.1089/hgtb.2019.123. [DOI] [PMC free article] [PubMed] [Google Scholar]