Abstract
Purpose
To identify candidate variants in genes possibly associated with premature ovarian insufficiency (POI).
Methods
Fourteen women, from 7 families, affected by idiopathic POI were included. Additionally, 98 oocyte donors of the same ethnicity were enrolled as a control group. Whole-exome sequencing (WES) was performed in 14 women with POI to identify possibly pathogenic variants in genes potentially associated with the ovarian function. The candidate genes selected in POI patients were analysed within the exome results of oocyte donors.
Results
After the variant filtering in the WES analysis of 7 POI families, 23 possibly damaging genetic variants were identified in 22 genes related to POI or linked to ovarian physiology. All variants were heterozygous and five of the seven families carried two or more variants in different genes. We have described genes that have never been associated to POI pathology; however, they are involved in important biological processes for ovarian function. In the 98 oocyte donors of the control group, we found no potentially pathogenic variants among the 22 candidate genes.
Conclusion
WES has previously shown as an efficient tool to identify causative genes for ovarian failure. Although some studies have focused on it, and many genes are identified, this study proposes new candidate genes and variants, having potentially moderate/strong functional effects, associated with POI, and argues for a polygenic etiology of POI in some cases.
Keywords: Premature ovarian insufficiency, Whole-exome sequencing, Candidate variants, Ovarian function, Polygenic origin
Introduction
Premature ovarian insufficiency (POI) is defined as the loss of ovarian function before the age of 40 and it is considered as a common cause of female infertility affecting 2–4% of women [1]. The main characteristics of POI are absence of regular menstrual cycles, increased follicle-stimulating hormone (FSH) levels and low serum oestrogen levels. Anti-Müllerian hormone (AMH) is another biochemical marker that can be used for POI diagnosis since it is correlated with the number of developing follicles in the ovary and declines to very low values at menopause independently of hypothalamic-gonadal axis. The POI condition has a heterogeneous etiology, including iatrogenic, autoimmune or genetic causes. Chromosomal and genetic alterations could explain approximately 20% of cases. Total or mosaic Turner syndrome, X chromosome deletions or X,autosomal translocations are the main causes of POI, followed by premutations in fragile X mental retardation I (FMR1) gene [2]. In the past decades, many genes associated with ovarian function have been discovered, especially since the incorporation of next-generation sequencing (NGS) [3–5]. Moreover, through exome sequencing studies, new variants in several candidate genes related with premature ovarian insufficiency (POI) are being identified, not only in genes associated with folliculogenesis, but also involved in processes such as metabolism, DNA repair, apoptosis, meiosis and others [6–8]. Therefore, genetic causes of POI seem to be more common than previously thought. Initially, premature ovarian insufficiency was considered as a monogenic condition; however, recent studies have showed a digenic or polygenic origin [9–11]. In the study performed by Bouilly et al. [9], 36% of patients carried mutations in two different genes, while Patiño et al. [10] and Tang et al. [11] found at least two possible candidate variants in distinct genes in a 42% and 85% of the POI patients, respectively, suggesting a synergistic effect of different mutations in several genes. Despite intense research in this field, many genes involved in this pathology remain to be discovered and more studies are necessary. The aim of this study was to identify candidate variants in genes possibly associated with the ovarian failure in a cohort of 7 families affected by POI.
Materials and methods
Study design and population
Fourteen women, from 7 families, affected by idiopathic POI were included in the study from October 2019 to September 2020. Seven POI patients were recruited when they came to our clinic to undergo assisted reproductive treatment. In the anamnesis, it was found that they had at least one first-degree relative with a diagnosis of POI, who were also recruited for the study (with the exception of one family, where the patient’s maternal aunt was included). All of them were of Caucasian ethnicity. The inclusion criteria were amenorrhea before 38 years old and ultrasound/analytical signs of ovarian insufficiency (FSH ≥ 25 IU/L and/or AMH ≤ 0.1 ng/ml). Additionally, 46,XX peripheral blood karyotype and normal FMR1 premutation study. The exclusion criteria were iatrogenic or autoimmune causes of POI, endometriosis and endocrine pathologies. The clinical parameters recorded were menarche age, age at POI diagnosis and serum FSH and/or AMH levels (the latter were only available in index cases, not in relatives).
Moreover, we selected 100 women from our oocyte donation program as an initial control group of the same ethnicity. All women with a 46,XX karyotype and no FMR1 premutations had an apparently normal ovarian reserve by ultrasound and no family history of POI. The oocyte donors had performed at least one controlled ovarian stimulation (COS) following the standard protocol in our clinic, varying the dose of gonadotropins according to their characteristics (antral follicle count and weight). Ninety-eight of these donors had an adequate response to ovarian stimulation treatments and two of them had a suboptimal response, so they were excluded from the control group. Therefore, in the end, the control group consisted of 98 donors with a normal ovarian reserve and no history of POI.
All work was conducted with the formal approval of the Instituto Bernabeu Institutional Review Board and it follows the principles of the Declaration of Helsinki. Informed consent was obtained from all participants prior to the study.
Whole-exome sequencing
Peripheral blood sample was obtained from all participants. Genomic DNA was extracted using the MagMAX DNA Multi-Sample Ultra 2.0 kit (Thermo Fisher Scientific, Colchester, UK) on a KingFisher™ Duo Prime system (Thermo Fisher Scientific, Colchester, UK), following the manufacturer’s instructions. DNA was quantified using Qubit™ dsDNA HS Assay Kit with the Qubit Fluorometer (Thermo Fisher Scientific, Colchester, UK). Whole-exome sequencing (WES) of the genomic DNA was performed using Trusight One Sequencing Panel (Illumina®) with 150 paired-end reads on NextSeq 550 (Illumina®), according to the manufacturer’s protocol. Sequenced data were aligned to the human genome 19 (hg19) through Burrows-Wheeler Aligment tool (BWA) and GATK algorithm was used for single nucleotide variations (SNVs)/InDel identification. Variant Call Format files (VCF) were annotated using Variant Interpreter software. In the POI families, only the variants shared by each family were extracted for analysis, and the following criteria were used for filtering and annotation of candidate variants: (1) minor allele frequency (MAF) < 0.05 in the gnomAD and 1000 genomes project for European population, (2) exonic/splicing variants in genes previously associated with POI, genes involved in biological ovarian functions or involved in DNA repair, transcription, meiosis and cell division processes, (3) variants having potentially strong/moderate functional effects on the protein (nonsense, frameshift, inframe deletion, splice region and missense variants). Within the variants with a potentially moderate effect, the missense variants were evaluated using three in silico prediction algorithms (SIFT, PolyPhen-2 and MutationTaster). Other moderate variants, as inframe deletions or variants in splice regions were considered separately. Moreover, the variants were assessed with the guidelines from the American College of Medical Genetics and Genomics (ACMG) [12], and only pathogenic/likely pathogenic variants, or variants of uncertain significance (VUS) were included. According to ACMG score: extremely low frequency in gnomAD population databases was recognized moderate evidence of pathogenicity (PM2). Null variant in a gene (as stop gain or frameshift) where loss of function is a known mechanism of disease was considered as pathogenic (PVS1). Protein length changes resulting from in-frame deletions/insertions in a non-repeat region or a stop-loss variant was recognized pathogenic moderate (PM4). Missense or splicing region variant was assigned as pathogenic, if computational prediction tools unanimously support a deleterious effect on the gene (PP3). Missense variant in a gene with low rate of benign missense mutations and for which missense mutation is a common mechanism of a disease was included within the possibly pathogenic variants (PP2). Allele frequency is greater than expected for disorder (BS1). Variants observed in a homozygous state in population databases more than expected for disease (BS2). For a missense or a splice region variant, computational prediction tools unanimously support a benign effect on the gene (BP4).
Whole-exome sequencing was performed in oocyte donors using the same protocol. However, in donors, only the candidate genes possibly related with the POI phenotype found in POI families were analysed within the exome sequencing data. The filtering process for variant calling and annotation to identify possibly pathogenic variants was the same as the one detailed above for POI families. The candidate variants identified by WES, in the POI patients/relatives and oocyte donors, were confirmed by Sanger sequencing using BigDye™ Terminator v3.1 Cycle Sequencing Kit and SeqStudio Genetic Analyzer (Applied Biosystems).
Results
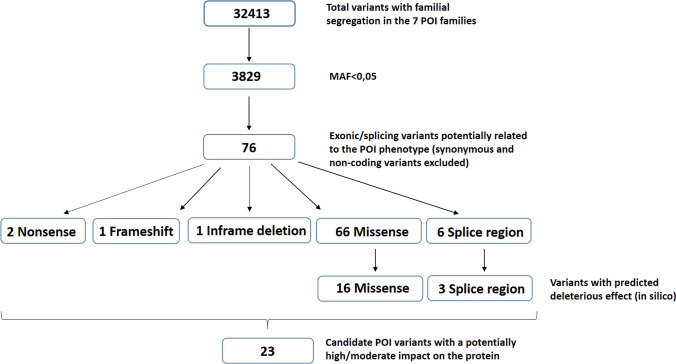
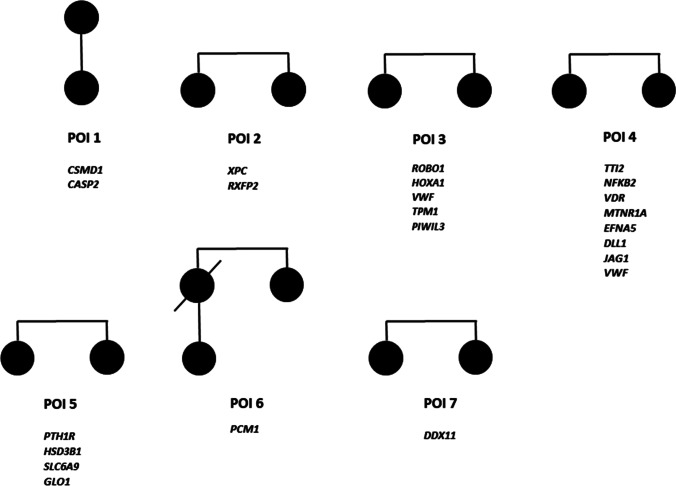
A total of 14 women, from 7 families (POI 1–7), affected by idiopathic POI were included, all of them with secondary amenorrhea. The mean menarche age was 13.1 ± 1.0 years and the mean age at POI diagnosis was 34.2 ± 2.3 years, being similar within each family. AMH and FSH levels were available only in the index cases and the mean was 0.04 ± 0.03 ng/ml for AMH and 33.8 ± 6.7 IU/L for FSH (Table1). After samples sequencing, the mean read depths were 100–180 × (varying for each sample), and over 98% of bases were covered at a minimum of 10 × depth in all samples. 32,413 variants with a familial segregation were identified in the WES results of the 14 women, and 3,829 of these, with a minor allele frequency (MAF) < 0.05. Exonic/splicing variants in genes potentially related to the POI phenotype were annotated manually obtaining 76 variants in the seven families (synonymous and non-coding variant were excluded). Among these candidate variants, 2 were nonsense, 1 frameshift, 1 inframe deletion, 66 missense and 6 splice region. Within the missense variants 16 were predicted to have deleterious effects by minimum two of the three in silico algorithms used (SIFT, PolyPhen-2 and MutationTaster), and according to the ACMG classification, we discarded 3 splice region with likely benign annotation based on computational prediction tools. After all filtering process, the final number of candidate variants were 23 (in 22 genes) (Fig. 1), within them, three variants were likely pathogenic and 20 of unknown significance (ACMG criteria). All variants were heterozygous, and five families carried two or more candidate variants. Figure 2 shows the family pedigrees with the genes identified in each family. The variants details are shown in Table 2.
Table 1.
Clinical characteristics of POI patients and their relatives
| Family ID | Patient-relative ID | Menarche age (y) | Age at POI diagnosis (y) | FSH (IU/L) | AMH (ng/mL) |
|---|---|---|---|---|---|
| POI 1 | P1 | 12 | 30 | 35 | 0.11 |
| R1 (mother) | 11 | 33 | - | - | |
| POI 2 | P2 | 14 | 37 | 40 | 0.02 |
| R2 (sister) | 14 | 35 | - | - | |
| POI 3 | P3 | 13 | 30 | 31 | 0.01 |
| R3 (sister) | 14 | 34 | - | - | |
| POI 4 | P4 | 13 | 34 | 30 | 0.05 |
| R4 (sister) | 13 | 33 | - | ||
| POI 5 | P5 | 12 | 37 | 38 | 0.02 |
| R5 (sister) | 13 | 36 | - | - | |
| POI 6 | P6 | 15 | 35 | - | 0.02 |
| R6 (maternal aunt) | 14 | 37 | - | - | |
| POI 7 | P7 | 13 | 35 | 39 | 0.07 |
| R7 (sister) | 13 | 33 | - | - |
Menarche age and age at POI diagnosis of all POI patients/relatives are specified. FSH (follicle-stimulating hormone) and AMH (Anti-Mullerian Hormone) were available only in index cases
Fig. 1.
Flowchart describing variant filtering process. Only the variants with familial segregation were selected (32,413), and filtered according to their minor allele frequency (MAF) < 0.05 (3,829 variants). Exonic/splicing variants potentially related to the POI phenotype were annotated manually (excluding synonymous and non-coding variants), resulting in 76 variants. Finally, we retained 23 variants with a potentially deleterious effect on the protein
Fig. 2.
Pedigrees in POI families (1–7). Filled shapes represent affected individuals. Candidate genes identified in each family are listed
Table 2.
Candidate variants identified in POI families (POI 1–7) and oocyte donors (D1 and D2)
| Family/Donor ID | Gene | Transcript | dsSNP | Nucleotide variation | Amino acid change | Effect | GnomAD | S/P/M | ACMG score | ACMG classification |
|---|---|---|---|---|---|---|---|---|---|---|
| POI 1 | CSMD1 | NM_033225.5 | rs1435513451 | c.5844 + 3A > G | - | Splice region | 0.000072 | - | PM2/PP3 | VUS |
| CASP2 | NM_032982.3 | rs1026228070 | c.1144C > T | p.Arg382Ter | Stop gain | 0.000029 | D/-/D | PM2/PVS1 | Likely pathogenic | |
| POI 2 | XPC | NM_004628.4 | rs1346001152 | c.2306 T > C | p.Ile769Thr | Missense | 0 | D/B/D | PM2 | VUS |
| RXFP2 | NM_130806.3 | rs150652160 | c.1826A > G | p.Tyr609Cys | Missense | 0.001 | D/D/D | PM2/BS2 | VUS | |
| POI 3 | ROBO1 | NM_002941.3 | rs375897144 | c.703G > A | p.Ala235Thr | Missense | 0.0001 | T/D/D | PM2 | VUS |
| HOXA1 | NM_005522.4 | - | c.62G > A | p.Gly21Glu | Missense | 0 | D/D/D | PM2 | VUS | |
| VWF | NM_000552.4 | rs376285757 | c.8324C > T | p.Ser2775Phe | Missense | 0.00002 | D/B/D | PM2/PP2 | VUS | |
| TPM1 | NM_001018005.1 | rs397516376 | c.548C > T | p.Ala183Val | Missense | 0.00004 | D/D/D | PM2/PM1/PP2/PP3/PP5 | Likely pathogenic | |
| PIWIL3 | NM_001008496.3 | rs142590557 | c.727C > T | p.Arg243Cys | Missense | 0.00072 | D/D/- | PM2 | VUS | |
| POI 4 | TTI2 | NM_025115.3 | rs140977711 | c.1135C > T | p.Arg379Trp | Missense | 0.0001 | D/D/D | PM2 | VUS |
| NFKB2 | NM_001077494.3 | rs200139098 | c.1993A > T | p.Thr665Ser | Missense | 0.00023 | D/D/D | PM2/PP2 | VUS | |
| VDR | NM_001017536.1 | rs756858031 | c.479G > A | p.Arg160Gln | Missense | 0.00011 | T/D/D | PM2 | VUS | |
| MTNR1A | NM_005958.4 | rs138837121 | c.1002A > T | p.Lys334Asn | Missense | 0.0022 | D/D/D | PM2/BS2 | VUS | |
| EFNA5 | NM_001962.2 | - | c.168C > A | p.Asp56Glu | Missense | 0 | D/D/D | PM2/PP3 | VUS | |
| DLL1 | NM_005618.3 | rs776343632 | c.2167-3del | - | Splice region | 0.0018 | - | BS1 | VUS | |
| JAG1 | NM_000214.2 | rs375017114 | c.1627C > T | p.Arg543Cys | Missense | 0.00007 | D/D/D | PM2/PP2 | VUS | |
| VWF | NM_000552.4 | rs368366214 | c.3101_3103del | p.Thr1034del | Inframe deletion | 0.0002 | - | PM4/BS1 | VUS | |
| POI 5 | PTH1R | NM_000316.2 | - | c.822_831del | p.Ala275ThrfsTer8 | Frameshift | 0 | - | PVS1/PM2 | Likely pathogenic |
| HSD3B1 | NM_000862.2 | rs775120619 | c.258 T > G | p.Ile86Met | Missense | 0.00008 | D/D/D | PM2 | VUS | |
| SLC6A9 | NM_201649.3 | rs757420753 | c.577G > A | p.(Gly193Ser) | Missense | 0.00002 | T/D/D | PM2 | VUS | |
| GLO1 | NM_006708.2 | rs141465532 | c.383 T > C | p.(Ile128Thr) | Missense | 0.0003 | D/D/D | PM2 | VUS | |
| POI 6 | PCM1 | NM_006197.3 | rs148806955 | c.4057G > T | p.Glu1353Ter | Stop gain | 0.003 | - | PM2 | VUS |
| POI 7 | DDX11 | NM_152438.1 | rs2111769 | c.1242 + 4 T > C | - | Splice region | 0.000008 | - | BP4 | VUS |
| D1 | PIWIL3 | NM_001008496.3 | rs747731185 | c.106del | p.Gln36ArgfsTer41 | Frameshift | 0.0016 | - | PM2 | VUS |
| D2 | VWF | NM_000552.4 | rs1800386 | c.4751A > G | p.Tyr1584Cys | Missense | 0.004 | D/D/D | PM2/PP2/PP5 | Likely pathogenic |
| CSMD1 | NM_033225.5 | rs763163101 | c.8926G > A | p.Val2976Met | Missense | 0.00002 | D/D/D | PM2 | VUS |
GnomAD, the variant frequency in the genomAD database for non-Finish European population; S/P/M, The prediction of SIFT/Poyphen2/MutationTaster bioinformatics tools (SIFT D, deleterious; T, tolerated/Polyphen2; D, probably damaging; B, benign/MutationTaster; D, disease causing). ACMG, American College of Medical Genetics
In 2 families (POI 1 and 3), we found variants in genes previously associated with POI phenotype (CSMD1 and PIWIL3) [13, 14]. The remaining candidate genes (CASP2, XPC, RXFP2, ROBO1, HOXA1, VWF, TPM1, TTI2, NFKB2, VDR, MTNR1A, EFNA5, DLL1, JAG1, PTH1R, HSD3B1, SLC6A9, GLO1, PCM1 and DDX11) had never been reported in POI patients; however, they are involved in biological ovarian functions or in cell division, apoptosis, DNA repair or transcription processes, and most of them are expressed in ovary, oocytes or granulosa cells. In the case of VWF gene, different variants were observed in two families. The gene information is specified in Table 3.
Table 3.
Candidate genes identified in 14 women from 7 families (POI 1–7) with premature ovarian insufficiency
| Family ID | Gene | Full name (HGNC) | OMIM | Biological process related to ovarian function |
|---|---|---|---|---|
| POI 1 | CSMD1 | CUB and Sushi multiple domains 1 | 608397 | Reproductive structures development |
| CASP2 | Caspase 2 | 600639 | Cellular apoptosis | |
| POI 2 | XPC | XPC complex subunit, DNA damage recognition and repair factor | 613208 | DNA damage repair |
| RXFP2 | Relaxin family peptide receptor 2 | 606655 | Follicle formation and development | |
| POI 3 | ROBO1 | Roundabout guidance receptor 1 | 602430 | Follicle formation and development |
| HOXA1 | Homeobox A1 | 142955 | Transcription | |
| VWF | Von Willebrand factor | 613160 | Follicle atresia | |
| TPM1 | Tropomyosin 1 | 191010 | Follicle formation and development | |
| PIWIL3 | Piwi like RNA-mediated gene silencing 3 | 610314 | Maintaining genome integrity in mammalian oocytes | |
| POI 4 | TTI2 | TELO2 interacting protein 2 | 614426 | DNA damage repair |
| NFKB2 | Nuclear factor kappa B subunit 2 | 164012 | Transcription | |
| VDR | Vitamin D receptor | 601769 | Follicle formation and development | |
| MTNR1A | Melatonin receptor 1A | 600665 | Follicle formation and development | |
| EFNA5 | Ephrin A5 | 601535 | Follicle formation and development | |
| DLL1 | Delta like canonical Notch ligand 1 | 606582 | Follicle formation and development | |
| JAG1 | Jagged canonical Notch ligand 1 | 601920 | Follicle formation and development | |
| VWF | Von Willebrand factor | 613160 | Follicle atresia | |
| POI 5 | PTH1R | Parathyroid hormone 1 receptor | 168468 | Follicle formation and development |
| HSD3B1 | 3 beta- and steroid delta-isomerase 1 | 109715 | Follicle formation and development | |
| SLC6A9 | Solute carrier family 6 member 9 | 601019 | Oocyte growing | |
| GLO1 | Glyoxalase I | 138750 | Follicle atresia | |
| POI 6 | PCM1 | Pericentriolar material 1 | 600299 | Cell division |
| POI 7 | DDX11 | DEAD/H-box helicase 11 | 601150 | Cell division and DNA repair |
HGNC, HUGO Gene Nomenclature Committee
When we analysed the 22 candidate genes in the 98 oocyte donors who formed the control group, we did not find potentially pathogenic variants. The ovarian stimulation of these donors was optimal according to our results within the oocyte donation program with an average of 14.96 oocytes and 11.57 MII retrieved in the oocyte pick-up. However, in the two donors with suboptimal results in the COS cycles who were excluded from the control group, we found one candidate variant in PIWIL3 gene in the first donor (D1) and two variants in VWF and CSMD1 genes in the second donor (D2) (Table 2). In the first donor, carrying a variant in PIWIL3 gene, the COS was cancelled on day 7 due to low follicular development after daily administration of 300 IU of rFSH. In the second one (with variants in VWF and CSMD1 genes), only 5 MII were retrieved, after 8 days of stimulation with 300 IU of rFSH.
Discussion
In the current study, we report 23 probably damaging genetic variants in 22 genes expressed, mostly in ovary, oocyte or granulosa cells and related to POI or possibly linked to ovarian physiology. The variants are not yet classified as pathogenic, but as likely pathogenic or VUS. However, as in previous studies [11, 15], they have been selected because they are very rare in the general population and in silico predictions confer a deleterious effect on protein function. Functional studies are required to confirm the expected pathogenic effect of the variants. In five of the seven POI families (71.4%), we identified two or more variants in distinct genes, suggesting a polygenic aetiology of POI in some cases, according to previous studies [9–11]. For each family, we will discuss the function of the different genes and their possible relationship to the POI phenotype or the ovarian function.
Family POI 1: CSMD1 and CASP2
CSMD1 is a complement regulatory protein that is enriched at the germ-cell/somatic-cell interface in both male and female gonads. Knockout females show significant reduction in ovarian quality and breeding success [16]. In humans, Jaillard et al. [13] identified a copy number variation (CNV) including CSMD1 gene in a patient younger than 40 years with ovarian failure, and propose this gene as candidate potentially implicated in reproductive function. Caspase-2 (CASP2) plays a central role in the execution-phase of cell apoptosis. Several caspases, including caspase-2, have been implicated in granulosa cell death during atresia of maturing antral follicles in the rat [17], and in oocyte apoptosis occurring developmentally [18, 19].
Family POI 2: XPC and RXFP2
The protein encoded by XPC gene plays an important role in the early steps of genome nucleotide excision repair (NER). It is expressed in the ovary and oocytes where it performs a tissue-specific DNA repair activity [20, 21]. On the other hand, RXFP2 gene encodes a member of the GPCR (G protein-coupled, 7-transmembrane receptor) family. The receptors for glycoprotein hormones such as follicle-stimulating hormone (FSH) and thyroid-stimulating hormone (TSH) are also GPCRs. Kawamura et al. [22] reported paracrine regulation of mammalian oocyte maturation, demonstrating the importance of the RXFP2 receptor in mediating gonadotropin actions. Moreover, several studies have reported the presence of RXFP2 receptor and is ligand INSL3 in the ovary of different animals during the development of antral follicles, which implies a potential role of the ligand-receptor pair in female reproduction in mammals [23, 24].
Family POI 3: ROBO1, HOXA1, VWF, TPM1 and PIWIL3
Previous studies have showed that the SLIT/Roundabout (ROBO) pathway is involved in follicle formation during foetal ovary development [25, 26]. HOXA1 is a transcription factor that is preferentially expressed in the oocytes and is essential for folliculogenesis and the regulation of oocyte-specific gene expression in the mouse [27]. VWF has been suggested to be associated with follicular atresia in mammals [28] and, recently, Wang et al. [29] have proposed VWF (an ovarian endothelial cell marker) as a new diagnostic biomarker in age-related human ovarian disorders. TPM1 is a protein located in the cytoskeleton of granulosa cells and is involved in the regulation of follicular growth [30, 31]. Finally, PIWIL3 has an important role in maintaining genome integrity in mammalian oocytes [32], and in a recent study, PIWIL3 has been proposed as causal and candidate gene for primary ovarian insufficiency [14].
Family POI 4: TTI2, NFKB2, VDR, MTNR1A, EFNA5, JAG1, DL11 and VWF
The TTI2 protein is a component of the Triple T complex (TTT) which also includes telomere length regulation protein and TELO2 interacting protein 1. The TTT complex is involved in cellular resistance to DNA damage. On the other hand, NFKB2 gene encodes a subunit of the transcription factor complex, and may have a role in ovarian function [33]. As for the VDR gene, Vitamin D receptor-knockout mice fail to produce mature oocytes, indicating that vitamin D is crucial for folliculogenesis in mice. The presence of vitamin D receptor (VDR) in gonadal tissues in humans, as well as the existence of several animal models are showing adverse reproductive outcomes related to the lack of VDR signalling. Different studies have demonstrated that vitamin D and its receptor play important roles in female reproduction [34, 35]. MTNR1A gene encodes one of two high affinity forms of a receptor for melatonin, the primary hormone secreted by the pineal gland. Talpur et al. [36] showed that MTNR1 suppression interferes in the normal physiological function of the ovary by enhancing follicular apoptosis, inhibiting proliferation, and influencing hormonal signalling, suggesting that Melatonin receptor 1 performs a critical role in the regulation of the animal reproductive system, particularly in follicular growth. EphrinA5 (EFNA5) is a neurogenic factor; however, recent findings suggest that it has a novel role in female mouse fertility. Worku et al. [37] demonstrated in an in vitro study with mouse granulose cells that EFNA5 is a pro-apoptotic agent and plays an important role in folliculogenesis by mediating apoptosis, proliferation, and steroidogenesis in female mouse. Human jagged1 (JAG1) is the ligand for the receptor notch 1, which is involved in signalling processes. In the mammalian ovary, Notch signalling pathway plays an important role in the mammalian folliculogenesis, regulating primordial follicle formation and coordinating follicular growth [38, 39]. In a mouse model with conditional deletion of Jagged1 within germ cells (J1KO), the histological study showed changes in follicle dynamics, including perturbations in the primordial follicle pool and antral follicle development, and consequently, J1KO female mice were subfertile [40]. On the other hand, Hubbard et al. [41] generated a transgenic Notch Reporter (TNR) mice, displaying that JAG1 is necessary for activation of Notch signalling in the developing ovary. Related to this gene, DLL1 is a human homolog of the Notch Delta ligand and is a member of the delta/serrate/jagged family. It plays a role in cell-to-cell communication. Li et al. [42] demonstrated, using in vitro ovary culture system, that Notch signalling–related genes including DLL1 were involved in early ovarian follicle development. Finally, VWF gene has been suggested to be associated with follicular atresia in a previous study [28].
Family POI 5: PTH1R, HSD3B1, SLC6A9, GLO1
Guo et al. [43] in 2019 showed that parathyroid hormone-like hormone (PTHLH) and its receptor (PTH1R) play an important role in chicken follicle selection by stimulating cell proliferation and steroidogenesis. In the same way, it could have a similar function in humans. HSD3B1 enzyme plays a crucial role in the biosynthesis of all classes of hormonal steroids and steroid signalling is required for ovarian primordial follicle formation. Fowler et al. [44] studied the steroid signalling pathways during primordial follicle formation in the human foetal ovary, showing that it is required for this process. GLYT1-mediated glycine transport (SLC6A9) is the main cell volume-homeostatic mechanism in mouse eggs and early preimplantation embryos. The SLC6A9 protein that is responsible for GLYT1 activity and SLC6A9 transcripts are present in growing oocytes and increased over the course of oogenesis. Furthermore, SLC6A9 becomes localized to the oocyte plasma membrane as the oocyte grows. Thus, oocytes acquire the ability to regulate their cell volume by releasing adhesion to the ZP and activating GLYT1 as they approach the end of oogenesis [45, 46]. On the other hand, Zhang et al. [47] investigated proteins differentially expressed in the ovaries of menopausal women in comparison to childbearing women, showing that glyoxalase I (GLO1) displayed an altered expression pattern, with higher expression in the atresic follicles of menopausal women. These data suggest that this protein may play a role in the regulation of follicle atresia in menopausal women.
Family POI 6: PCM1
PCM1 is a component of centriolar satellites, essential for the correct localization of different centrosomal proteins, and for joining microtubules to the centrosome. It shows high level of expression in testis and ovaries.
Family POI 7. DDX11
DDX11 protein is a RNA helicase implicated in a number of cellular processes involving alteration of RNA secondary structure and may function at the interface of replication-coupled DNA repair and sister chromatid cohesion. Based on their distribution patterns, some members of this family are believed to be involved in embryogenesis, spermatogenesis, and cellular growth and division.
The results of this study support the hypothesis that POI is a genetically heterogeneous condition, with a polygenic origin in many cases and still poorly studied. Candidate variants were found in known POI-associated genes (CSMD1 and PIWIL3). In addition, we have identified variants in 20 genes that have never been described in POI patients, but which are expressed in the ovary, and are involved in processes previously associated with POI. Most of these novel genes can be classified into the following categories: apoptosis (CASP2 and EFNA5), DNA damage repair (XPC, TTI2 and DDX11), transcription (NFKB2 and HOXA1), cell division (PCM1 and DDX11), follicle formation and development (RXFP2, ROBO1, TPM1, VDR, MTNR1A, EFNA5, JAG1, DL11, PTH1R and HSD3B1) and follicle atresia (VWF and GLO1).
Initially, premature ovarian insufficiency has been considered as a monogenic condition and most papers describe homozygous or compound heterozygous mutations in a gene. The fact that only one rare heterozygous variant has been detected in families POI 6 and 7 could mean that in these families, PCM1 and DDX11 genes, both of which are involved in cell division, may be causing POI in the autosomal dominant mode of inheritance. There are a few descriptions of POI associated with monoallelic pathogenic variants in normally autosomal recessively inherited genes, also with an important role in cell division processes, e.g. FANCA, FANCL and MCM9. Heterozygous variants of these genes cause haploinsufficiency and contribute to the pathogenesis of POI, especially secondary amenorrhea [5, 48–51].
Moreover, the presence of two or more heterozygous possibly damaging variants in different genes, in a high percentage of families, means that in some patients ovarian failure could be caused by mutations in genes involved in different processes, as previous studies have shown [9–11]. The phenomenon, in which different genetic variants influence a biological function, as could be ovarian function, is known as synergistic heterozygosity. This is where heterozygous variants of recessive inheritance in different genes partially affect the function of the respective genes and their combination affects intersecting biological routes leading to the phenotype. Synergistic heterozygosity is a special case of multigenic condition where variations in two or more genes are necessary for phenotypic manifestation [52]. However, to test the contribution of these multiple heterozygous variants to ovarian function, a functional validation with RNA and protein expression assays are required.
The finding that no potentially pathogenic variants in the POI candidate genes in this study were found in the control group donors, and that candidate variants were found in two donors who had poor COS cycle outcomes, argues in favour that these genes could be important for ovarian function. Although the phenotype of these two donors is completely different from the POI phenotype of the patients, the low response to ovarian stimulation in these young women could mean a diminished ovarian reserve (DOR) and could be a precondition for a future ovarian failure. In fact, previous studies suggest that POI and DOR are different stages of the same process that may have a common genetic cause [11, 53].
This study has identified potentially pathogenic variants in candidate genes associated to the ovarian function in 14 women affected by POI, from seven families, and two young women with a very low response to the ovarian stimulation cycle. These findings may help future research in this complex pathology. However, a limitation of this study is that in addition to the proband and one POI-affected relative in each family, there were no other affected or non-affected relatives available for analysis. Further studies identifying these genes in other women or families with ovarian failure are necessary to establish a definitive correlation between them and the POI phenotype, especially in case of the polygenic origin.
In summary, it is well demonstrated that the WES is a very useful tool to identify candidate causative mutations in POI patients. This is not only important at the level of diagnosis, but also at the level of prevention in risk patients who can be counselled for oocyte cryopreservation before the ovarian reserve decreases. Moreover, the discovering of new genes potentially involved in ovarian decline will supply better understanding of the ovarian function and the physiopathological mechanisms of POI.
Acknowledgements
The authors would like to thank Ania Pitas for the revision and correction of the language.
Declarations
Conflict of interest
None.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Webber L, Davies M, Anderson R, Bartlett J, Braat D, Cartwright B, et al. ESHRE Guideline: management of women with premature ovarian insufficiency. Hum Reprod. 2016;31(5):926–937. doi: 10.1093/humrep/dew027. [DOI] [PubMed] [Google Scholar]
- 2.Man L, Lekovich J, Rosenwaks Z, Gerhardt J. Fragile X-associated diminished ovarian reserve and primary ovarian insufficiency from molecular mechanisms to clinical manifestations. Front Mol Neurosci. 2017;10:290. doi: 10.3389/fnmol.2017.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tenenbaum-Rakover Y, Weinberg-Shukron A, Renbaum P, Lobel O, Eideh H, Gulsuner S, et al. Minichromosome maintenance complex component 8 (MCM8) gene mutations result in primary gonadal failure. J Med Genet. 2015;52(6):391–399. doi: 10.1136/jmedgenet-2014-102921. [DOI] [PubMed] [Google Scholar]
- 4.Li L, Wang B, Zhang W, Chen B, Luo M, Wang J, et al. A homozygous NOBOX truncating variant causes defective transcriptional activation and leads to primary ovarian insufficiency. Hum Reprod. 2017;32(1):248–255. doi: 10.1093/humrep/dew271. [DOI] [PubMed] [Google Scholar]
- 5.Yang X, Zhang X, Jiao J, Zhang F, Pan Y, Wang Q, et al. Rare variants in FANCA induce premature ovarian insufficiency. Hum Genet. 2019;138(11–12):1227–1236. doi: 10.1007/s00439-019-02059-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaillard S, Bell K, Akloul L, Walton K, McElreavy K, Stocker WA, et al. New insights into the genetic basis of premature ovarian insufficiency: novel causative variants and candidate genes revealed by genomic sequencing. Maturitas. 2020;141:9–19. doi: 10.1016/j.maturitas.2020.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Jiao X, Ke H, Qin Y, Chen ZJ. Molecular genetics of premature ovarian insufficiency. Trends Endocrinol Metab. 2018;29(11):795–807. doi: 10.1016/j.tem.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Tucker EJ, Grover SR, Bachelot A, Touraine P, Sinclair AH. Premature ovarian insufficiency: new perspectives on genetic cause and phenotypic spectrum. Endocr Rev. 2016;37(6):609–635. doi: 10.1210/er.2016-1047. [DOI] [PubMed] [Google Scholar]
- 9.Bouilly J, Beau I, Barraud S, Bernard V, Azibi K, Fagart J, et al. Identification of multiple gene mutations accounts for a new genetic architecture of primary ovarian insufficiency. J Clin Endocrinol Metab. 2016;101(12):4541–4550. doi: 10.1210/jc.2016-2152. [DOI] [PubMed] [Google Scholar]
- 10.Patiño LC, Beau I, Carlosama C, Buitrago JC, González R, Suárez CF, et al. New mutations in non-syndromic primary ovarian insufficiency patients identified via whole-exome sequencing. Hum Reprod. 2017;32(7):1512–1520. doi: 10.1093/humrep/dex089. [DOI] [PubMed] [Google Scholar]
- 11.Tang R, Yu Q. Novel variants in women with premature ovarian function decline identified via whole-exome sequencing. J Assist Reprod Genet. 2020;37(10):2487–2502. doi: 10.1007/s10815-020-01919-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaillard S, Akloul L, Beaumont M, Hamdi-Roze H, Dubourg C, Odent S, et al. Array-CGH diagnosis in ovarian failure: identification of new molecular actors for ovarian physiology. J Ovarian Res. 2016;9(1):63. doi: 10.1186/s13048-016-0272-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorsi B, Hernandez E, Moore MB, Moriwaki M, Chow CY, Coelho E, et al. Causal and candidate gene variants in a large cohort of women with primary ovarian insufficiency. J Clin Endocrinol Metab. 2022;107(3):685–714. doi: 10.1210/clinem/dgab775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turkyilmaz A, Alavanda C, Ates EA, Geckinli BB, Polat H, Gokcu M, Karakaya T, Cebi AH, Soylemez MA, Guney Aİ, Ata P, Arman A. Whole-exome sequencing reveals new potential genes and variants in patients with premature ovarian insufficiency. J Assist Reprod Genet. 2022;39(3):695–710. doi: 10.1007/s10815-022-02408-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee AS, Rusch J, Lima AC, Usmani A, Huang N, Lepamets M, et al. Rare mutations in the complement regulatory gene CSMD1 are associated with male and female infertility. Nat Commun. 2019;10(1):4626. doi: 10.1038/s41467-019-12522-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flaws JA, Kugu K, Trbovich AM, DeSanti A, Tilly KI, Hirshfield AN, et al. Interleukin-1 beta-converting enzyme-related proteases (IRPs) and mammalian cell death: dissociation of IRP-induced oligonucleosomal endonuclease activity from morphological apoptosis in granulosa cells of the ovarian follicle. Endocrinology. 1995;136(11):5042–5053. doi: 10.1210/endo.136.11.7588240. [DOI] [PubMed] [Google Scholar]
- 18.Bergeron L, Perez GI, Macdonald G, Shi L, Sun Y, Jurisicova A, et al. Defects in regulation of apoptosis in caspase-2-deficient mice. Genes Dev. 1998;12(9):1304–1314. doi: 10.1101/gad.12.9.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanoux V, Pairault C, Bakalska M, Habert R, Livera G. Caspase-2 involvement during ionizing radiation-induced oocyte death in the mouse ovary. Cell Death Differ. 2007;14(4):671–681. doi: 10.1038/sj.cdd.4402052. [DOI] [PubMed] [Google Scholar]
- 20.Men NT, Kikuchi K, Furusawa T, Dang-Nguyen TQ, Nakai M, Fukuda A, et al. Expression of DNA repair genes in porcine oocytes before and after fertilization by ICSI using freeze-dried sperm. Anim Sci J. 2016;87(11):1325–1333. doi: 10.1111/asj.12554. [DOI] [PubMed] [Google Scholar]
- 21.Sabatella M, Thijssen KL, Davó-Martínez C, Vermeulen W, Lans H. Tissue-specific DNA repair activity of ERCC-1/XPF-1. Cell Rep. 2021;34(2):108608. doi: 10.1016/j.celrep.2020.108608. [DOI] [PubMed] [Google Scholar]
- 22.Kawamura K, Kumagai J, Sudo S, Chun SY, Pisarska M, Morita H, et al. Paracrine regulation of mammalian oocyte maturation and male germ cell survival. Proc Natl Acad Sci U S A. 2004;101(19):7323–7328. doi: 10.1073/pnas.0307061101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanna CB, Yao S, Patta MC, Jensen JT, Wu X. Expression of insulin-like 3 (INSL3) and differential splicing of its receptor in the ovary of rhesus macaques. Reprod Biol Endocrinol. 2010;8:150. doi: 10.1186/1477-7827-8-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Satchell L, Glister C, Bleach EC, Glencross RG, Bicknell AB, Dai Y, et al. Ovarian expression of insulin-like peptide 3 (INSL3) and its receptor (RXFP2) during development of bovine antral follicles and corpora lutea and measurement of circulating INSL3 levels during synchronized estrous cycles. Endocrinology. 2013;154(5):1897–1906. doi: 10.1210/en.2012-2232. [DOI] [PubMed] [Google Scholar]
- 25.Dickinson RE, Hryhorskyj L, Tremewan H, Hogg K, Thomson AA, McNeilly AS, et al. Involvement of the SLIT/ROBO pathway in follicle development in the fetal ovary. Reproduction. 2010;139(2):395–407. doi: 10.1530/REP-09-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin N, Fan XC, Zhang YY, Xu XX, Tyasi TL, Jing Y, et al. New insights into implication of the SLIT/ROBO pathway in the prehierarchical follicle development of hen ovary. Poult Sci. 2015;94(9):2235–2246. doi: 10.3382/ps/pev185. [DOI] [PubMed] [Google Scholar]
- 27.Huntriss J, Hinkins M, Picton HM. cDNA cloning and expression of the human NOBOX gene in oocytes and ovarian follicles. Mol Hum Reprod. 2006;12(5):283–289. doi: 10.1093/molehr/gal035. [DOI] [PubMed] [Google Scholar]
- 28.Feranil JB, Isobe N, Nakao T. Immunolocalization of von Willebrand factor and vascular endothelial growth factor during follicular atresia in the swamp buffalo ovary. J Reprod Dev. 2005;51(4):419–426. doi: 10.1262/jrd.17011. [DOI] [PubMed] [Google Scholar]
- 29.Wang S, Zheng Y, Li J, Yu Y, Zhang W, Song M, et al. Single-cell transcriptomic atlas of primate ovarian aging. Cell. 2020;180(3):585–600.e19. doi: 10.1016/j.cell.2020.01.009. [DOI] [PubMed] [Google Scholar]
- 30.McRae RS, Johnston HM, Mihm M, O'Shaughnessy PJ. Changes in mouse granulosa cell gene expression during early luteinization. Endocrinology. 2005;146(1):309–317. doi: 10.1210/en.2004-0999. [DOI] [PubMed] [Google Scholar]
- 31.Li J, Li C, Li Q, Li WT, Li H, Li GX, et al. Identification of the Key microRNAs and miRNA-mRNA interaction networks during the ovarian development of hens. Animals (Basel). 2020;10(9). 10.3390/ani10091680. [DOI] [PMC free article] [PubMed]
- 32.Tan M, Tol HTAV, Rosenkranz D, Roovers EF, Damen MJ, Stout TAE, et al. PIWIL3 forms a complex with TDRKH in mammalian oocytes. Cells. 2020;9(6). 10.3390/cells9061356. [DOI] [PMC free article] [PubMed]
- 33.Kurowska P, Mlyczyńska E, Dawid M, Dupont J, Rak A. Role of vaspin in porcine ovary: effect on signaling pathways and steroid synthesis via GRP78 receptor and protein kinase A†. Biol Reprod. 2020;102(6):1290–1305. doi: 10.1093/biolre/ioaa027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson LE, DeLuca HF. Vitamin D receptor null mutant mice fed high levels of calcium are fertile. J Nutr. 2001;131(6):1787–1791. doi: 10.1093/jn/131.6.1787. [DOI] [PubMed] [Google Scholar]
- 35.Panda DK, Miao D, Tremblay ML, Sirois J, Farookhi R, Hendy GN, et al. Targeted ablation of the 25-hydroxyvitamin D 1alpha -hydroxylase enzyme: evidence for skeletal, reproductive, and immune dysfunction. Proc Natl Acad Sci U S A. 2001;98(13):7498–7503. doi: 10.1073/pnas.131029498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Talpur HS, Worku T, Rehman ZU, Dad R, Bhattarai D, Bano I, et al. Knockdown of melatonin receptor 1 and induction of follicle-stimulating hormone on the regulation of mouse granulosa cell function. Reprod Biol. 2017;17(4):380–388. doi: 10.1016/j.repbio.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 37.Worku T, Wang K, Ayers D, Wu D, Ur Rehman Z, Zhou H, et al. Regulatory roles of ephrinA5 and its novel signaling pathway in mouse primary granulosa cell apoptosis and proliferation. Cell Cycle. 2018;17(7):892–902. doi: 10.1080/15384101.2018.1456297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen CL, Fu XF, Wang LQ, Wang JJ, Ma HG, Cheng SF, et al. Primordial follicle assembly was regulated by Notch signaling pathway in the mice. Mol Biol Rep. 2014;41(3):1891–1899. doi: 10.1007/s11033-014-3038-4. [DOI] [PubMed] [Google Scholar]
- 39.Prasasya RD, Mayo KE. Notch signaling regulates differentiation and steroidogenesis in female mouse ovarian granulosa cells. Endocrinology. 2018;159(1):184–198. doi: 10.1210/en.2017-00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vanorny DA, Prasasya RD, Chalpe AJ, Kilen SM, Mayo KE. Notch signaling regulates ovarian follicle formation and coordinates follicular growth. Mol Endocrinol. 2014;28(4):499–511. doi: 10.1210/me.2013-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hubbard N, Prasasya RD, Mayo KE. Activation of notch signaling by oocytes and Jag1 in mouse ovarian granulosa cells. Endocrinology. 2019;160(12):2863–2876. doi: 10.1210/en.2019-00564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, Zhao D, Guo C, Mi Y, Zhang C. Involvement of notch signaling in early chick ovarian follicle development. Cell Biol Int. 2016;40(1):65–73. doi: 10.1002/cbin.10538. [DOI] [PubMed] [Google Scholar]
- 43.Guo X, Wang Y, Chen Q, Yuan Z, Chen Y, Guo M, et al. The role of PTHLH in ovarian follicle selection, its transcriptional regulation and genetic effects on egg laying traits in hens. Front Genet. 2019;10:430. doi: 10.3389/fgene.2019.00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fowler PA, Anderson RA, Saunders PT, Kinnell H, Mason JI, Evans DB, et al. Development of steroid signaling pathways during primordial follicle formation in the human fetal ovary. J Clin Endocrinol Metab. 2011;96(6):1754–62. doi: 10.1210/jc.2010-2618. [DOI] [PubMed] [Google Scholar]
- 45.Richard S, Baltz JM. Preovulatory suppression of mouse oocyte cell volume-regulatory mechanisms is via signalling that is distinct from meiotic arrest. Sci Rep. 2017;7(1):702. doi: 10.1038/s41598-017-00771-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richard S, Tartia AP, Boison D, Baltz JM. Mouse oocytes acquire mechanisms that permit independent cell volume regulation at the end of oogenesis. J Cell Physiol. 2017;232(9):2436–2446. doi: 10.1002/jcp.25581. [DOI] [PubMed] [Google Scholar]
- 47.Zhang LQ, Zhang XN, Gao Y, Ma XB, Dai LS, Jiang H, et al. Identification of differentially expressed proteins in the ovaries of menopausal women. Arch Gynecol Obstet. 2014;290(6):1179–1186. doi: 10.1007/s00404-014-3357-7. [DOI] [PubMed] [Google Scholar]
- 48.Liu H, Wei X, Sha Y, Liu W, Gao H, Lin J, et al. Whole-exome sequencing in patients with premature ovarian insufficiency: early detection and early intervention. J Ovarian Res. 2020;13(1):114. doi: 10.1186/s13048-020-00716-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Y, Guo T, Liu R, Ke H, Xu W, Zhao S, et al. FANCL gene mutations in premature ovarian insufficiency. Hum Mutat. 2020;41(5):1033–1041. doi: 10.1002/humu.23997. [DOI] [PubMed] [Google Scholar]
- 50.Guo T, Zheng Y, Li G, Zhao S, Ma J, Qin Y. Novel pathogenic mutations in minichromosome maintenance complex component 9 (MCM9) responsible for premature ovarian insufficiency. Fertil Steril. 2020;113(4):845–852. doi: 10.1016/j.fertnstert.2019.11.015. [DOI] [PubMed] [Google Scholar]
- 51.Daum H, Zlotogora J. Fanconi anemia gene variants in patients with gonadal dysfunction. Reprod Sci. 2022;29(5):1408–1413. doi: 10.1007/s43032-021-00582-7. [DOI] [PubMed] [Google Scholar]
- 52.Chakravorty S, Hegde M. Inferring the effect of genomic variation in the new era of genomics. Hum Mutat. 2018;39(6):756–773. doi: 10.1002/humu.23427. [DOI] [PubMed] [Google Scholar]
- 53.Zhao M, Feng F, Chu C, Yue W, Li L. A novel EIF4ENIF1 mutation associated with a diminished ovarian reserve and premature ovarian insufficiency identified by whole-exome sequencing. J Ovarian Res. 2019;12(1):119. doi: 10.1186/s13048-019-0595-0. [DOI] [PMC free article] [PubMed] [Google Scholar]