Abstract
Many common pathogens are difficult or impossible to detect using conventional microbiological tests. However, the rapid and untargeted nature of metagenomic next-generation sequencing (mNGS) appears to be a promising alternative. To perform a systematic review and meta-analysis of evidence regarding the diagnostic accuracy of mNGS in patients with infectious diseases. An electronic literature search of Embase, PubMed and Scopus databases was performed. Quality was assessed using the Quality Assessment of Diagnostic Accuracy Studies-2 tool. Summary receiver operating characteristics (sROC) and the area under the curve (AUC) were calculated; A random-effects model was used in cases of heterogeneity. A total of 20 papers were eligible for inclusion and synthesis. The sensitivity and specificity of diagnostic mNGS were 75% and 68%, respectively. The AUC from the SROC was 85%, corresponding to excellent performance. mNGS demonstrated satisfactory diagnostic performance for infections and yielded an overall detection rate superior to conventional methods.
Subject terms: Microbiology, Diseases, Health care
Introduction
Infectious diseases are a leading cause of morbidity and death worldwide. However, early detection of pathogens may be challenging in many clinical scenarios. Moreover, many common pathogens are difficult or impossible to detect using conventional microbiological tests (e.g. culture, smears, immunological tests and polymerase chain reaction (PCR) assays), which makes precise diagnosis challenging. Culture methods are time-consuming and have strict limitations. Smears, immunological tests and multiplex PCR assays will only test for a specific pathogen that must be identified by the clinicians before the test is performed1. The administration of broad-spectrum antibiotics in the absence of pathogen identification, despite comprehensive testing methods, frequently confounds specific diagnoses, which could lead to more toxic and less effective antimicrobial therapy2.
Metagenomic next-generation sequencing (mNGS) is a high-throughput method that can directly detect pathogens (i.e., bacterial species) in clinical specimens and analyze functional genes without the need to pre-select target sequences3. It is especially suitable for novel, rare, and atypical etiologies of complicated infectious diseases. Due to characteristics of speed, sensitivity, culture-independent, hypothesis-free, and unbiased pathogen detection, mNGS may become a routine diagnostic tool, partly replacing more traditional detection methods4. Some investigators have even decided to upgrade their model, known as ‘Microbial Index of Pathogenic Bacteria’, by implementing whole metagenome sequencing data for species and strain- level identification of patho-genic bacteria5. To date, mNGS has been applied in the diagnosis of pathogens in bloodstream infections6,7, respiratory tract infections8,9, tuberculosis10, meningitis and encephalitis11,12. However, these studies were limited by small sample sizes. As such, we aimed to perform a systematic accuracy review of diagnostic tests and a meta-analysis to identify, quality appraise, and synthesize the available evidence to inform the implementation of mNGS in diagnosing infectious diseases.
Methods
Literature search
Results of the present systematic review and meta-analysis are reported in accordance with the Preferred Reporting Items for a Systematic Review and Meta-analysis of Diagnostic Test Accuracy studies (PRISMA-DTA)13. A comprehensive electronic literature search of Embase, PubMed and Scopus databases was performed for relevant studies published up to December 31, 2021. The medical subject heading (i.e., ‘MeSH’) search terms included 'infection' and 'Metagenomic Next-Generation Sequencing'. The reference lists of retrieved studies were also manually searched for additional, possibly eligible studies. Three reviewers independently screened the titles, and abstracts and obtained the full-text of potentially relevant studies; any disagreements were resolved by consensus discussion.
Inclusion and exclusion criteria
Cross-sectional and cohort studies including patients with clinically suspicious infection (including meningitis, bacteremia, fungemia, osteomyelitis, septic arthritis) for whom diagnostic test accuracy data for mNGS were included. Only English language articles were eligible. No restrictions were imposed on the age of the study population.
Studies reporting insufficient data to construct a 2 × 2 table (true positive, false positive, true negative, and false negative), those based on non-human samples, investigations reporting duplicate information already reported in other publications; those not reporting the reference infection diagnostic criteria; or reporting one specific pathogene and abstracts, conference presentations, case reports and letters were excluded.
Data extraction
Data were independently extracted by two reviewers using a standardized protocol and prespecified data extraction forms for diagnostic test accuracy studies14. Disagreements were resolved by a third investigator. Information regarding study characteristics (including population, period, design, country, and sample size) was extracted.
Quality assessment
The quality of the included studies was independently assessed by two reviewers, using the revised Quality Assessment of Diagnostic Accuracy Studies-2 tool15.
Statistical analysis
For each study, pooled specificity, pooled sensitivity, pooled negative predictive value (NPV), and pooled positive predictive value (PPV) were calculated based on a bivariate meta-analysis model16. They are presented as graphical representations in which the boxes mark the values and the horizontal lines represent the confidence intervals (CIs). A summary receiver operating characteristic curve (sROC) was drawn, and the area under the curve (AUC) was calculated to determine the performance of a diagnostic test17. The criteria for AUC classification were as follows: 0.50 (failure), 0.60–0.70 (poor), 0.70–0.80 (fair), 0.80–0.90 (good) and 0.90–1 (excellent). The Q* index and corresponding standard error (SE), is an additional measure which is the point on the sROC curve closest to the ideal left top-left corner (where summary sensitivities (SN) and summary specificities (SP) meet).
Heterogeneity was evaluated by calculating the I218 statistic. DerSimonian and Laird random effects models19, which include both between and within study heterogeneity, were used to generate summary SP, SN, negative likelihood ratios (− LR), positive likelihood ratios (+ LR) and diagnostic odds ratio (DOR). Heterogeneity was also assessed using forest plots of sensitivity and specificity across studies for variability of study estimates in the hierarchical sROC model (meta-regression). A Cochrane’s-Q p < 0.10 and I2 > 50% indicated significant heterogeneity, of SN and SP and LRs, respectively. Furthermore, the risk of bias in the included studies was assessed by using the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS–2) tool15. Publication bias was assessed by using a funnel plot and Deeks test20. Statistical analysis was performed using MetaDisc version 1.4, Stata version 12.0 (StataCorp LLC, College Station, TX, USA), and Review Manager 5 (version 5.3) (R Foundation for Statistical Computing, Vienna, Austria)21.
Results
Characteristics of the included studies
After removing duplicate publications and references checking for additional, potentially eligible studies, a total of 891 studies were screened. Of these, 77 separate publications underwent full text review, resulting in 20 studies included in this systematic review. The study selection process is illustrated in the PRISMA flow diagram (Fig. 1). Six studies were performed in high-income countries, whereas 14 were conducted in low and middle-income countries. The study included 2716 participants. Twelve of the studies were retrospective, and eight were prospective in design. Among the enrolled studies, participants were predominantly adults. The included studies were published between 2017 and 2021. (Table 1).
Figure 1.
Flow diagram of study selection process.
Table 1.
Characteristics of Included Studies.
| Research type | Reference standard | Participants | Age | Country | Sample type | |
|---|---|---|---|---|---|---|
| Zhang32 | Retrospective | Conventional test | 135 | Paediatric | China | Blood/CSF |
| Wang30 | Retrospective | Conventional test | 55 | Adults | China | Pulmonary biopsy/BALF |
| Rossoff33 | Retrospective | Conventional test | 79 | Pediatric | USA | Plasma |
| Miller34 | Retrospective | Conventional test | 95 | Pediatric + adult | USA | CSF |
| Blauwkamp6 | Prospective | Conventional test | 348 | Adults | USA | Plasma |
| Miao24 | Retrospective | Conventional test | 511 | Adults | Chian | Clinical specimensa |
| Madi35 | Retrospective | Conventional test | 86 | Adults | Kuwait | Respiratory samplesb |
| Parize36 | Prospective | Conventional test | 101 | Adults | France | Plasma/nasopharyngeal swabs/ biological fluid |
| Xing28 | Prospective | Conventional test | 213 | Adults | Chian | CSF |
| Wang37 | Prospective | Clinical diagnosis | 63 | Adults | Chian | Joint fluid |
| Chen38 | Retrospective | Conventional test | 235 | Adults | Chian | BALF |
| Lian39 | Retrospective | Clinical diagnosis | 51 | Adults | Chian | BALF |
| Peng40 | Retrospective | Clinical diagnosis | 49 | Adults | Chian | BALF |
| Sun41 | Prospective | Conventional test | 44 | Adults | Chian | BALF |
| Zhou42 | Prospective | Conventional test | 159 | Adults | Chian | BALF |
| Chen43 | Prospective | Conventional test | 162 | Adults | Chian | BALF |
| Jing44 | Retrospective | Clinical diagnosis | 209 | pediatric + adult | Chian | Plasma |
| Ogawa45 | Retrospective | Conventional test | 23 | Adults | Japan | Tissue |
| Lee46 | Retrospective | Clinical diagnosis | 54 | Pediatric | USA | Plasma |
| Cai47 | Prospective | Clinical diagnosis | 44 | Adults | China | Periprosthetic tissues |
a: Specimens included bronchoalveolar lavage fluid (BALF), cerebrospinal fluid (CSF), sputum, pleural fluid, tissue, pus, blood, ascetic fluid, bile, secretion, urine, herpes fluid, bone marrow, throat swab, pericardial fluid and saliva; b: The respiratory samples included nasopharyngeal aspirates/wash, nasopharyngeal swab, BALF, tracheal aspirates, sputum, throat swabs, and nasal swabs.
Risk of bias
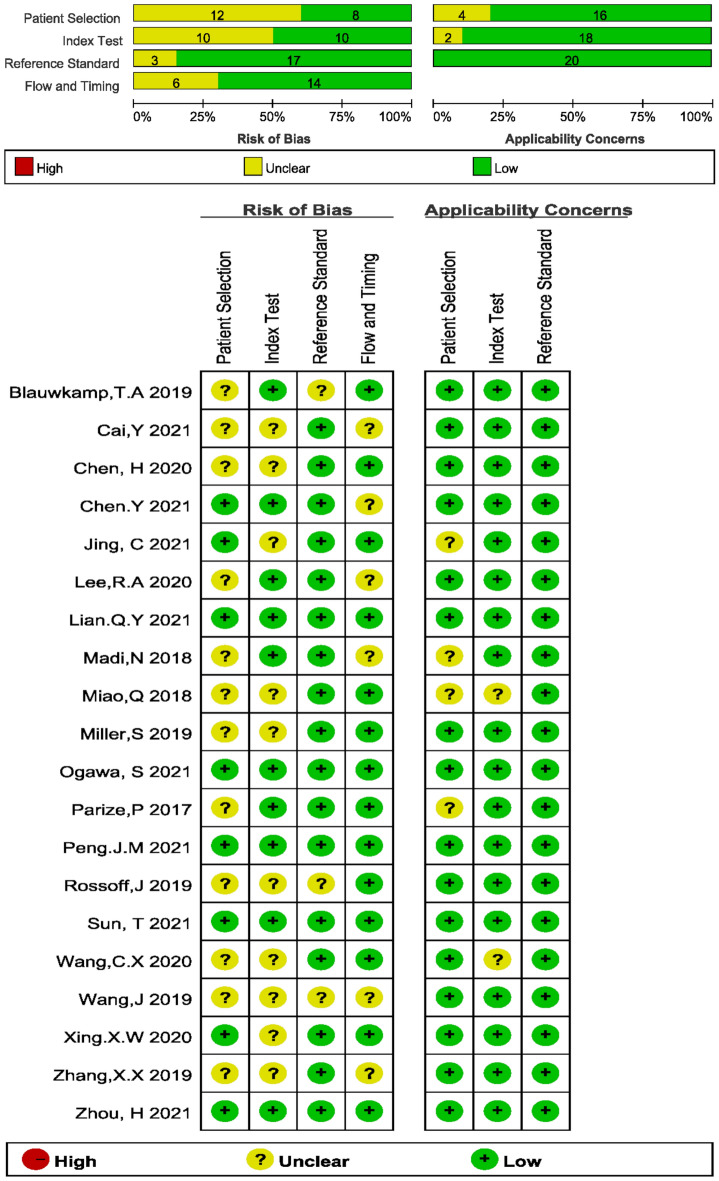
The risk of bias and applicability concerns according to the QUADAS-2 tool are shown in Fig. 2. All studies demonstrated unclear or low risks of bias.
Figure 2.
Risk of bias and applicability concerns summary.
Meta‑analysis
Heterogeneity test
No correlation was found between sensitivity logarithm and 1-specificity logarithm (Spearman correlation 0.0345 (p = 0.092)). Analysis revealed no threshold effect among the included studies. As is common with meta-analyses investigating results of diagnostic accuracy research, remarkable heterogeneity was present, with sensitivity and specificity estimates varying widely. The Cochran-Q for the pooled DOR was 207.82, (p = 0.00, I2 = 88.5%). This suggests that a non-threshold effect was the cause of heterogeneity and a random effect model was used in further analysis.
Random effect model analysis results
The reported diagnostic sensitivity of the mNGS in infectious diseases ranged between 21% and 100% (Fig. 3a), and the reported specificity ranged from 14% to 100% (Fig. 3b). The pooled summary sensitivity reached 75% (95% CI 72–77%, I2 = 93.3%) (Fig. 3a) and pooled summary specificity was computed to 68% (95% CI: 66%–70%, I2 = 97.4%) (Fig. 3b), indicating significant heterogeneity. The pooled positive LR was 2.8 (95% CI: 2.1–3.77) and the pooled negative LR was 0.32 (95% CI: 0.23–0.46) (Fig. 4a,b).
Figure 3.
Forest plot of estimates results: (a) Sensitivity; (b) Specificity.
Figure 4.
(a) Positive likelihood ratio; (b) Negative likelihood ratio.
Subgroup analysis
Subgroup analysis was also performed to explore the influence of different reference standards in the final result (Supplementary Fig. 1a–d). Two subgroups were formed based on two reference standards: conventional testing and clinical diagnosis. The results confirmed consistent performance.
Heterogeneity analysis
Four components, including “gold standard”, “experimental design”, “age” and “country income” were considered in the meta-regression analysis to explore potential risk of bias. Unfortunately, none of these components exhibited heterogeneity. Due to failure to extract more comprehensive data from the research, it was not further analyzed.
Evaluation of diagnostic accuracy
SROC curves for the mNGS in infectious diseases are presented in Fig. 5a. This figure illustrates the relationship between sensitivities and 1-specificity for the included studies in the pooled analyses. The AUC was considered excellent (AUC = 0.85 (SE = 0.03)). The point at which sensitivity and specificity were equal (Q*) was 0.78 (SE = 0.03). The pooled DOR was 11.94 (95% CI: 6.11–23.34) (Fig. 5b).
Figure 5.
Summary ROC curves.
Publication bias
Deek’s test yielded no evidence of publication bias (P = 0.795). (Supplementary Fig. 2).
Discussion
To our knowledge, the present meta-analysis was the first to systematically review the use of mNGS in diagnosing infectious diseases. Conventional techniques for the detection of pathogens are largely target-dependent tests, which detect a limited number of micro-organisms. However, NGS-based metagenome approaches are target independent and can detect unknown pathogens22. Using the pooled estimate of 75% (95% CI: 72–77%, I2 = 93.3%)) at median specificity 68% (95% CI: 66–70%, I2 = 97.4). The AUC 85%, which reflected infection using mNGS, was classified as excellent performance.
The DOR reflects the relationship between the diagnostic test and the relevant disease. The pooled DOR was 11.94, reflecting diagnostic efficacy of mNGS in infectious diseases. The pooled positive LR was 2.81 (95% CI: 2.1–3.77), which reflects that the risk of developing the disease was 2.81 times that of not having the disease when the results of next generation sequencing being positive. The pooled negative LR was 0.32 (95% CI: 0.23–0.46), which reflects that the risk of developing the disease was 0.32 times that of not having the disease when the results of NGS are negative. The sROC curve reflects merge indicators of the sensitivity and specificity. The AUC for sROC was 0.85, which reflected high diagnostic efficiency.
Some studies23–26 demonstrated that mNGS had diagnostic advantages over conventional methods for patients treated with empirical antibiotics before sample collection. The use of empirical antibiotics would significantly lower the detection rate of conventional methods by approximately 20%, while mNGS is not affected23. The reason may likely be due to the fact that culture methods require the existence of live pathogens and, therefore, are easily influenced by the administration of antimicrobials. On the other hand, high-throughput sequencing needs only to identify DNA fragments of microorganisms, which may explain its relatively higher detection rate after antimicrobial treatment. Moreover, it can shorten turnaround time and detect pathogens without bias27.
NGS also has shortcomings. First, it is not sensitive for intracellular bacteria and fungi in difficulty obtaining circulatory genome DNA23,28. RNA viruses require reverse transcription before deep sequencing and the amount of DNA segments may be reduced23. Different NGS technique may introduce bias (Supplementary Table 1). Second, mNGS is relatively expensive. Third, the criteria for diagnosing single pathogens are unclear, and are mainly based on the relative abundance of pathogens, the coverage rate or unique reads of pathogens8,29. In addition, given the untargeted nature of mNGS, background interference is a fairly common limitation.
Our study also had limitations. The first of which was considerable heterogeneity, the sources of which were extensively explored. Meta-regression results revealed that “experimental design” and “age” may have been the cause of heterogeneity. Another factor that needs to be considered is the clinical heterogeneity exhibited in the included studies such as the number of patients, antibiotic treatment, sampling methods, different reference standards and other unknown factors such as technical variations (e.g. sequencing strategies and platforms), sequence profiling software, prediction models, and batch effects. Second, the number of patients in two studies30,31 was relatively small, which may have reduced our statistical power. Third, no fourfold contingency tables were feasible for most of the studies because some of the necessary data were calculated based on reported sensitivity and specificity. Fourth, limiting the search strategy to English language publications could have potentially missed some studies. Finally, the included studies may have potentially been affected by selection bias and the use of different reference standards for infectious diseases.
Conclusions
mNGS combined with conventional microbiological testing can improve diagnostic efficiency. We believe that mNGS may be a potential step forward in diagnosing infectious diseases due to its non-invasive, rapid and untargeted characteristics.
Supplementary Information
Acknowledgements
The authors appreciate all the participants in this research.
Abbreviations
- AUC
Area under curve
- mNGS
Metagenomic next-generation sequencing
- PRISMA-DTA
Preferred reporting items for a systematic review and meta-analysis of diagnostic test accuracy
- NPV
Negative predictive value
- PPV
Positive predictive value
- CIs
Confidence intervals
- sROC
A summary receiver operating characteristic curve
- AUC
Area under the curve
- SS
Summarise sensitivities
- SP
Summarise specificities
- − LR
Negative likelihood ratios
- + LR
Positive likelihood ratios
- DOR
Diagnostic odds ratio
- HSROC
Hierarchical summary receiver operating characteristic
- PCR
Polymerase chain reaction
- ID
Infectious diseases
Author contributions
Conceived and designed the experiments: J.L. Y.Q.Q. Performed the experiments: J.L Y.Q.D J.Y. Analyzed the data: J.L J.Y. Contributed reagents/materials/analysis tools: J.L Y.Q.D. Wrote the manuscript: J.L. Revised the manuscript: Q.Z. All authors have read and approved the manuscript.
Funding
This study was supported by the Zhejiang Province Public Welfare Technology Application Research Project (CN), China (2019 LGF19H010009), National Natural Science Foundation of China (81971982) and National Natural Science Foundation of China (No. 81903849). The funders had no role in study design, data collection or analysis, decision to publish.
Data availability
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jian Liu and Qiao Zhang.
Contributor Information
Jian Liu, Email: 1516050@zju.edu.cn.
Yun-Qing Qiu, Email: qiuyq@zju.edu.cn.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-25314-y.
References
- 1.Ramanan P, Bryson AL, Binnicker MJ, Pritt BS, Patel R. Syndromic panel-based testing in clinical microbiology. Clin. Microbial. Rev. 2018 doi: 10.1128/cmr.00024-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barlam TF, et al. Implementing an antibiotic stewardship program: Guidelines by the infectious diseases society of America and the society for healthcare epidemiology of America. Clin. Infect. Dis. 2016;62:e51–77. doi: 10.1093/cid/ciw118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grumaz S, et al. Next-generation sequencing diagnostics of bacteremia in septic patients. Genome Med. 2016;8:73. doi: 10.1186/s13073-016-0326-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldberg B, Sichtig H, Geyer C, Ledeboer N, Weinstock GM. Making the leap from research laboratory to clinic: Challenges and opportunities for next-generation sequencing in infectious disease diagnostics. mBio. 2015;6:e01888–01815. doi: 10.1128/mBio.01888-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun Z, et al. Comprehensive understanding to the public health risk of environmental microbes via a microbiome-based index. J. Genet. Genomics. 2022;49:685–688. doi: 10.1016/j.jgg.2021.12.011. [DOI] [PubMed] [Google Scholar]
- 6.Blauwkamp TA, et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat. Microbiol. 2019;4:663–674. doi: 10.1038/s41564-018-0349-6. [DOI] [PubMed] [Google Scholar]
- 7.Goggin KP, et al. Evaluation of plasma microbial cell-free DNA sequencing to predict bloodstream infection in pediatric patients with relapsed or refractory cancer. JAMA Oncol. 2020;6:552–556. doi: 10.1001/jamaoncol.2019.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langelier C, et al. Metagenomic sequencing detects respiratory pathogens in hematopoietic cellular transplant patients. Am. J. Respir. Crit. Care Med. 2018;197:524–528. doi: 10.1164/rccm.201706-1097LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlaberg R, et al. Viral pathogen detection by metagenomics and pan-viral group polymerase chain reaction in children with pneumonia lacking identifiable etiology. J. Infect. Dis. 2017;215:1407–1415. doi: 10.1093/infdis/jix148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi CL, et al. Clinical metagenomic sequencing for diagnosis of pulmonary tuberculosis. J. Infect. 2020 doi: 10.1016/j.jinf.2020.08.004. [DOI] [PubMed] [Google Scholar]
- 11.Wilson MR, et al. Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N. Engl. J. Med. 2019;380:2327–2340. doi: 10.1056/NEJMoa1803396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang JZ, et al. Next-generation sequencing combined with routine methods to detect the pathogens of encephalitis/meningitis from a Chinese tertiary pediatric neurology center. J. Infect. 2019;78:409–421. doi: 10.1016/j.jinf.2019.02.011. [DOI] [PubMed] [Google Scholar]
- 13.McInnes MDF, et al. Preferred reporting items for a systematic review and meta-analysis of diagnostic test accuracy studies: The PRISMA-DTA statement. JAMA. 2018;319:388–396. doi: 10.1001/jama.2017.19163. [DOI] [PubMed] [Google Scholar]
- 14.Campbell, J. et al. The systematic review of studies of diagnostic test accuracy. Joanna Briggs Institute Reviewers’ Manual, 1–46 (2015).
- 15.Whiting PF, et al. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 16.Reitsma JB, et al. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J. Clin. Epidemiol. 2005;58:982–990. doi: 10.1016/j.jclinepi.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 17.Rutter CM, Gatsonis CA. A hierarchical regression approach to meta-analysis of diagnostic test accuracy evaluations. Stat. Med. 2001;20:2865–2884. doi: 10.1002/sim.942. [DOI] [PubMed] [Google Scholar]
- 18.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 19.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 20.Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J. Clin. Epidemiol. 2005;58:882–893. doi: 10.1016/j.jclinepi.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 21.Zamora J, Abraira V, Muriel A, Khan K, Coomarasamy A. Meta-DiSc: A software for meta-analysis of test accuracy data. BMC Med. Res. Methodol. 2006;6:31. doi: 10.1186/1471-2288-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore NE, et al. Metagenomic analysis of viruses in feces from unsolved outbreaks of gastroenteritis in humans. J. Clin. Microbiol. 2015;53:15–21. doi: 10.1128/jcm.02029-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, et al. Clinical application and evaluation of metagenomic next-generation sequencing in suspected adult central nervous system infection. J. Transl. Med. 2020;18:199. doi: 10.1186/s12967-020-02360-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miao Q, et al. Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin. Infect. Dis. 2018;67:S231–s240. doi: 10.1093/cid/ciy693. [DOI] [PubMed] [Google Scholar]
- 25.Gosiewski T, et al. Comprehensive detection and identification of bacterial DNA in the blood of patients with sepsis and healthy volunteers using next-generation sequencing method - the observation of DNAemia. J. Clin. Microbiol. Infect. Dis. 2017;36:329–336. doi: 10.1007/s10096-016-2805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhodes J, et al. Antibiotic use in Thailand: Quantifying impact on blood culture yield and estimates of pneumococcal bacteremia incidence. Am. J. Trop. Med. Hyg. 2010;83:301–306. doi: 10.4269/ajtmh.2010.09-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu W, Miller S, Chiu CY. Clinical metagenomic next-generation sequencing for pathogen detection. Annu. Rev. Pathol. 2019;14:319–338. doi: 10.1146/annurev-pathmechdis-012418-012751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xing XW, et al. metagenomic next-generation sequencing for diagnosis of infectious encephalitis and meningitis: A large, prospective case series of 213 patients. Front. Cell. Infect. Microbiol. 2020;10:88. doi: 10.3389/fcimb.2020.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, et al. Detection of pulmonary infectious pathogens from lung biopsy tissues by metagenomic next-generation sequencing. Front. Cell. Infect. Microbiol. 2018;8:205. doi: 10.3389/fcimb.2018.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, Han Y, Feng J. Metagenomic next-generation sequencing for mixed pulmonary infection diagnosis. BMC Pulm. Med. 2019;19:252. doi: 10.1186/s12890-019-1022-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boheemen SV, et al. Retrospective validation of a metagenomic sequencing protocol for combined detection of Rna and DNA viruses using respiratory samples from pediatric patients. J. Mol. Diagn. JMD. 2019 doi: 10.1016/j.jmoldx.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X-X, et al. The diagnostic value of metagenomic next-generation sequencing for identifying Streptococcus pneumoniae in paediatric bacterial meningitis. BMC Infect. Dis. 2019 doi: 10.1186/s12879-019-4132-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossoff J, et al. Noninvasive diagnosis of infection using plasma next-generation sequencing: A single-center experience. Open Forum Infect. Dis. 2019 doi: 10.1093/ofid/ofz327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller S, et al. Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal fluid. Genome Res. 2019;29:831–842. doi: 10.1101/gr.238170.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madi N, Al-Nakib W, Mustafa AS, Habibi N. Metagenomic analysis of viral diversity in respiratory samples from patients with respiratory tract infections in Kuwait. J. Med. Virol. 2018;90:412–420. doi: 10.1002/jmv.24984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parize P, et al. Untargeted next-generation sequencing-based first-line diagnosis of infection in immunocompromised adults: A multicentre, blinded, prospective study. Clin. Microbial. Infect. 2017;23:574e571–574e576. doi: 10.1016/j.cmi.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 37.Wang CX, et al. Comparison of broad-range polymerase chain reaction and metagenomic next-generation sequencing for the diagnosis of prosthetic joint infection. Int. J. Infect. Dis. 2020 doi: 10.1016/j.ijid.2020.03.055. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y, et al. Application of metagenomic next-generation sequencing in the diagnosis of pulmonary infectious pathogens from bronchoalveolar lavage samples. Front. Cell. Infect. Microbiol. 2021;11:541092. doi: 10.3389/fcimb.2021.541092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lian QY, et al. High-throughput next-generation sequencing for identifying pathogens during early-stage post-lung transplantation. BMC Pulm. Med. 2021;21:348. doi: 10.1186/s12890-021-01723-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng JM, Du B, Qin HY, Wang Q, Shi Y. Metagenomic next-generation sequencing for the diagnosis of suspected pneumonia in immunocompromised patients. J. Infect. 2021;82:22–27. doi: 10.1016/j.jinf.2021.01.029. [DOI] [PubMed] [Google Scholar]
- 41.Sun T, et al. Metagenomic next-generation sequencing for pathogenic diagnosis and antibiotic management of severe community-acquired pneumonia in immunocompromised adults. Front. Cell. Infect. Microbiol. 2021;11:661589. doi: 10.3389/fcimb.2021.661589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou H, et al. Clinical impact of metagenomic next-generation sequencing of bronchoalveolar lavage in the diagnosis and management of pneumonia: A multicenter prospective observational study. J. Mol. Diagn. 2021;23:1259–1268. doi: 10.1016/j.jmoldx.2021.06.007. [DOI] [PubMed] [Google Scholar]
- 43.Chen H, et al. Clinical utility of in-house metagenomic next-generation sequencing for the diagnosis of lower respiratory tract infections and analysis of the host immune response. Clin. Infect. Dis. 2020;71:S416–S426. doi: 10.1093/cid/ciaa1516. [DOI] [PubMed] [Google Scholar]
- 44.Jing C, et al. Clinical evaluation of an improved metagenomic next-generation sequencing test for the diagnosis of bloodstream infections. Clin. Chem. 2021;67:1133–1143. doi: 10.1093/clinchem/hvab061. [DOI] [PubMed] [Google Scholar]
- 45.Ogawa S, et al. Evaluation of infections in orthopedic patients using next-generation sequencing. J. infect. Chemother. 2021;27:1626–1633. doi: 10.1016/j.jiac.2021.07.019. [DOI] [PubMed] [Google Scholar]
- 46.Lee RA, Al Dhaheri F, Pollock NR, Sharma TS. Assessment of the clinical utility of plasma metagenomic next-generation sequencing in a pediatric hospital population. J. Clin. Microbial. 2020 doi: 10.1128/jcm.00419-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai Y, et al. Metagenomic next generation sequencing improves diagnosis of prosthetic joint infection by detecting the presence of bacteria in periprosthetic tissues. Int. J. Infect. Dis. 2020;96:573–578. doi: 10.1016/j.ijid.2020.05.125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.