Abstract
Plesiomonas shigelloides, a member of the family Vibrionaceae, is a gram-negative, rod-shaped, facultative anaerobic bacterium with flagella. P. shigelloides has been isolated from such sources as freshwater, surface water, and many wild and domestic animals. P. shigelloides contains 102 Oantigens and 51 H-antigens. The diversity of O-antigen gene clusters is relatively poorly understood. In addition to O1 and O17 reported by other laboratories, and the 12 O serogroups (O2, O10, O12, O23, O25, O26, O32, O33, O34, O66, O75, and O76) reported previously by us, in the present study, nine new P. shigelloides serogroups (O8, O17, O18, O37, O38, O39, O44, O45, and O61) were sequenced and annotated. The genes for the O-antigens of these nine groups are clustered together in the chromosome between rep and aqpZ. Only O38 possesses the wzm and wzt genes for the synthesis and translocation of O-antigens via the ATP-binding cassette (ABC) transporter pathway; the other eight use the Wzx/Wzy pathway. Phylogenetic analysis using wzx and wzy showed that both genes are diversified. Among the nine new P. shigelloides serogroups, eight use wzx/wzy genes as targets. In addition, we developed an O-antigen-specific PCR assay to detect these nine distinct serogroups with no cross reactions among them.
Keywords: Plesiomonas shigelloides, O-antigen gene clusters, serogroups, PCR assay
Introduction
Plesiomonas shigelloides, a member of the family Vibrionaceae, is a rod-shaped, gram-negative facultative anaerobic bacterium with flagella. P. shigelloides has been isolated from such sources as freshwater, surface water, and many wild and domestic animals [1]. Infection with P. shigelloides has been associated with travel to or residence in tropical and subtropical countries and with the ingestion of raw seafood, particularly oysters. Despite the epidemiological association, the causal relationship between P. shigelloides and diarrhea remains to be confirmed [2].
Aldová et al. recognized 102 O-antigens and 51 H-antigens of P. shigelloides [3]. P. shigelloides strain 302-73, which was originally isolated in Japan [4], was determined and titled as the ‘O1 representative strain.’ In 2010, the complete chemical structures of its O-antigen and lipopolysaccharide (LPS) core were characterized [5]. The whole genome was sequenced in 2013 and deposited in GenBank (AQQO01000000) [6]. P. shigelloides O17, which carries the same O-antigen as Shigella sonnei, was used in vaccine production against shigellosis [7]. Our previous study reported the diversity of P. shigelloides O-antigen gene clusters, and also established an identification and detection method for molecular serotyping [8]. The variability of the O-antigen provides the primary basis for serotyping schemes for P. shigelloides [9]. Based on the 12 O-antigen clusters of P. shigelloides, serogroups O2, O10, O12, O23, O25, O26, O32, O33, O34, O66, O75, and O76, it became clear that the O-antigen gene clusters group together and are located on the chromosome between the rep and aqpZ genes. Among these 12 serogroups, nine use the Wzx/Wzy pathway to synthesize and translocate O chains, and the other three use the wzm/wzt-encoded ATP-binding cassette (ABC) transporter pathway [10]. Furthermore, a suspension array that detected 12 distinct O-antigens was developed [8]. In the present study, nine new O-antigen gene clusters of P. shigelloides (O8, O17, O18, O37, O38, O39, O44, O45, and O61) were sequenced and annotated. Like the 12 previously identified serogroups, the genes for the O-antigens group together in chromosomes between rep and aqpZ, which encode a helicase and an aquaporin, respectively. Only O38 possesses the wzm and wzt genes for the synthesis and translocation of O chains via the ABC transporter pathway, and the other eight use the Wzx/Wzy pathway.
The identity of Wzx/Wzy and Wzm/Wzt is low (30-50%); therefore, they are used as determinants for each serovar and have the potential to be DNA molecular markers for serotyping. In our previous work, wzx and wzy were used to serotype Escherichia coli, Shigella spp. [11], Salmonella spp. [12], and Pseudomonas aeruginosa [13]; wzm and wzt were used to serotype Legionella pneumophila; and wzx, wzy, and wzm were used to serotype P. shigelloides [8]. The phylogenetic analysis of wzx and wzy showed that both of these genes are diversified and are potential candidates for molecular serotyping. Thus, out of the nine P. shigelloides serogroups, eight employed wzx/wzy/wzm genes as targets.
In the present study we used the PCR assay as the detection technique. The PCR has several advantages in terms of safety, saving on cost/time, high specificity, sensitivity, and reproducibility. We show that all nine new P. shigelloides serogroups could be detected without any cross reactions among them.
Materials and Methods
Strains and Genomic DNA Extraction
The P. shigelloides reference strains used in this study are shown in Table 1. All the strains were cultured in Tryptone Soya Broth (TSB) (QingDao ShuiRi Bio-Technologies Co., Ltd., China) at 37°C with aeration at 200 rpm. Genomic DNA was extracted from 1.5 ml of the overnight bacterial cultures (approximately 108 CFU/ml) using a DNA extraction kit according to the manufacturer's instructions (CWBiotech, China).
Table 1.
Plesiomonas shigelloides strains used in this study.
| Plesiomonas shigelloides | Lab number | Original number | Original source | Reference/Clinical Sample |
|---|---|---|---|---|
| O8H22 | G5267 | E7 | Italya | Salerno et al., 2010 |
| O17H11 | G5878 | CNCTC Aer 42/89 | CNCTCb | Maciejewska et al., 2013 |
| O18H2 | G6000 | CNCTC Aer7007 54/89 | CNCTCb | |
| O37 | G5883 | CNCTC Aer 39/89 | CNCTCb | Kaszowska et al., 2013 |
| O38H4 | G6003 | CNCTC Aer7009 59/89 | CNCTCb | |
| O39H11 | G6004 | CNCTC Aer6410 41/89 | CNCTCb | Maciejewska et al., 2013 |
| O44H8 | G6005 | CNCTC Aer6416 52/89 | CNCTCb | |
| O45H2 | G5884 | CNCTC Aer 44/89 | CNCTCb | Maciejewska et al., 2013 |
| O61H2 | G5998 | CNCTC Aer6999 31/89 | CNCTCb | |
| Clinical isolates | ||||
| 1 | G6370 | ps-1 | CDCc | diarrhea |
| 2 | G6371 | ps-4 | CDCc | diarrhea |
| 3 | G6372 | ps-5 | CDCc | watery diarrhea |
| 4 | G6373 | ps-6 | CDCc | diarrhea |
| 5 | G6374 | ps-7 | CDCc | diarrhea |
| 6 | G6375 | ps-8 | CDCc | diarrhea |
| 7 | G6376 | ps-9 | CDCc | diarrhea |
| 8 | G6377 | ps-16 | CDCc | mucinous feces |
| 9 | G6378 | ps-17 | CDCc | watery diarrhea |
| 10 | G6379 | ps-18 | CDCc | diarrhea |
| 11 | G6380 | ps-19 | CDCc | diarrhea |
| 12 | G6381 | ps-22 | CDCc | diarrhea |
| 13 | G6382 | ps-23 | CDCc | watery diarrhea |
| 14 | G6383 | V14-017 | CDCd | diarrhea |
| 15 | G6384 | V14-009 | CDCd | diarrhea |
| 16 | G6385 | V14-017 | CDCd | diarrhea |
aDepartment of Environmental Sciences, Parthenope University of Naples, Naples, Italy.
bCzech National Collection of Type Cultures (CNCTC), Czech Republic.
cChinese Center for Disease Control and Prevention, Beijing, China.
dGuangzhou Center for Disease Control and Prevention, Guangdong, China.
Library Construction
A gene sequencing library was constructed as previously described [8]. Briefly, DNA was fragmented by nebulization, end-repaired, and phosphorylated. An A-tail and adaptors were ligated to the end-repaired DNA fragments according to the manufacturer’s instructions (Illumina Inc., United Kingdom).
Genome Sequencing, Assembly, and Annotation
According to a previous report [14], the whole genomes of nine P. shigelloides reference strains were sequenced using an Illumina/Solexa Genome Analyzer IIx (Illumina, USA), assembled using VelvetOptimiser v2.2, and identified by BLAST search against the NCBI Prokaryotic Genome Annotation Pipeline. For genes involved in sugar synthesis, the UniProt/SwissProt database was used. For genes involved in O-antigen processing and translocation, the program TMHMM Server v2.0 was used to confirm the characteristics of wzx/wzy or wzm/wzt. For genes involved in glycosyl transferase (GT), the Pfam protein motif database (http://pfam.xfam.org/) was used.
Target Gene Selection and Phylogenetic Analysis
All the wzx/wzy sequences in this study were extracted from the O-antigen gene clusters, aligned using the ClustalW program [15], and the identity of the wzx/wzy gene was acquired using BioEdit software [16]. The phylogenetic trees based on the wzx/wzy sequences were constructed using MEGA 6.0 [17].
Primers for the PCR Assay
The primers were designed using Primer Premier 5.0 software (Premier Biosoft International, USA) based on the sequences of the processing genes (Table 2). The formation of ring hairpin structures, self-dimers, and cross-dimers was avoided. The specificity of primers was evaluated by BLAST alignments against the GenBank database [18]. The length of the primers was about 25 nt, and Tm was between 55 and 60°C. The length of the generated PCR product was 50–200 bp. The primers were synthesized by Invitrogen (China). The numbers and sequence information of each primer are listed in Table 2.
Table 2.
Primers used in this study.
| Serogroup | Target gene | Primer name | Sequence(5’-3’) | Length (nt) | Tm (℃) |
|---|---|---|---|---|---|
| O8 | wzy | wln-15653 | CCACTTTATGGTTTTATCTCATCTGATT | 28 | 58 |
| wln-15654 | CCACTCTTGCTTTGCAACAACT | 22 | 58 | ||
| O17 | wzx | wln-15209 | AAATGACCACACCAATACAATACGA | 25 | 58 |
| wln-15210 | AATGAACTTTATCTTGTGTGTAGTGGAAA | 29 | 59 | ||
| O18 | wzx | wln-15211 | GGCACCTAACACTCGAGTCAAAT | 23 | 58 |
| wln-15212 | GGTGATGCAAGGCGTTAACTATT | 23 | 58 | ||
| O37 | wzx | wln-15213 | ATGGCTAGTGACACCTCCTGAA | 22 | 58 |
| wln-15214 | ACCTGGACAACACCCAACTTTA | 22 | 58 | ||
| O38 | wzt | wln-13770 | CAATTTCTTTTGCCGCTCCTA | 21 | 58 |
| wln-13771 | TTTTGTCAGAGAGCTGTTTTGTTGA | 25 | 59 | ||
| O39 | wzy | wln-15659 | GATGCTGGACTTTTTCTGCATAGTT | 25 | 58 |
| wln-15660 | AATACCAGTGTACACCACAAACCAA | 25 | 59 | ||
| O44 | wzx | wln-15663 | TTTACAGTAGTAGCAAGTCTGCGATATG | 28 | 58 |
| wln-15664 | TGGCCGCTTCAGTATTGCTT | 20 | 59 | ||
| O45 | wzx | wln-15217 | GATCTTGAAGTTCTCCACTATGGCTATA | 28 | 58 |
| wln-15218 | GCACTCTCAACTCCATCATATGAAA | 25 | 58 | ||
| O61 | fcl | wln-15720 | CCAATCTGTATGGCGAAAACG | 21 | 59 |
| wln-15721 | CAGGGCCGGAATCACATG | 18 | 59 |
Development of a Serotyping PCR Assay
The amplification reactions were carried out in a total volume of 25 μl, including 12.5 μl of 2× Premix Ex Taq (Takara, China), 1 μl of 10 μΜ forward primer, 1 μl of 10 μΜ reverse primer, 1 μl of DNA, and 10.5 μl of ddH2O. The reaction parameters were as follows: 95°C for 3 min; 35 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min; followed by a final extension at 72°C for 5 min. A 2 μl aliquot of the PCR product was visualized by 1.5% agarose gel electrophoresis, and the images were captured digitally using the Gel Image System (Tanon 3500, China).
Sensitivity with Genomic DNA
To analyze the sensitivity of the PCR assay for genomic DNA, serial ten-fold gradient dilutions of the genomic DNA of P. shigelloides O61 genomic DNA (100 ng, 10 ng, 1 ng, 0.1 ng, 0.01 ng, 1 pg, 0.1 pg, and 0.01 pg) were used as templates for PCR amplification to determine the minimum detection level.
GenBank Accession Numbers
The nine O-antigen gene clusters of P. shigelloides serogroups O8, O17, O18, O37, O38, O39, O44, O45, and O61 have been submitted to GenBank with the accession numbers MN115821–MN115829, respectively.
Results
Identification of O-Antigen Gene Clusters in Nine P. shigelloides Genome Sequences
The O-antigen gene clusters of nine P. shigelloides serogroups were sequenced and localized in the type strains O8, O17, O18, O37, O38, O39, O44, O45, and O61 (Table 1). All nine clusters were located between rep and aqpZ in the chromosome and comprised 24,307 bp, 16,842 bp, 16,858 bp, 23,694 bp, 20,811 bp, 27,237 bp, 25,003 bp, 18,828 bp, and 21,560 bp, respectively. Twenty-one, 14, 16, 19, 15, 21, 19, 17 and 22 open reading frames (ORFs), respectively, were identified in the nine serotypes (Tables S 1-9). The functions of the genes were assigned according to their similarities with the genes with known functions in the sugar pathway and others from available databases (Figs. 1 and 2).
Fig. 1.
The O-antigen gene clusters of nine P. shigelloides serogroups O8, O17, O18, O37, O38, O39, O44, O45 and O61.
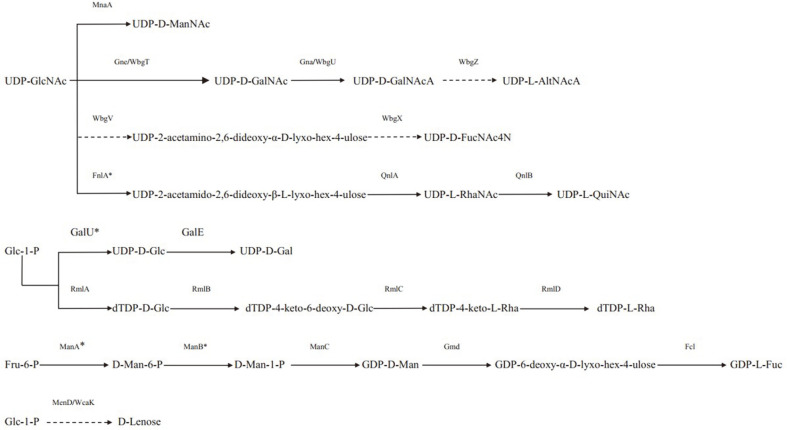
Fig. 2. Proposed biosynthesis pathways for sugars in the nine P. shigelloides serogroups clusters.
*Indicates the genes that were found outside the OPS clusters. Putative pathways are denoted by a broken line. MnaA, UDP-Nacetylglucosamine- 2-epimerase; Gne, UDP-N-acetylglucosamine-4-epimerase; Gna, UDP-GalNAcA synthetase; WbgZ, C-5 epimerase; FnlA*, 4,6-dehydratase, 3- and 5-epimerase; QnlA, dTDP-4-dehydrorhamnose reductase; QnlB, C-2 epimerase; GalU*, UTP-glucose-1-phosphate uridylyltransferase; GalE, UDP-glucose-4-epimerase; RmlA, glucose-1-phosphate thymidylyltransferase; RmlB, dTDP-D-glucose 4,6-dehydratase; RmlC, dTDP-4-keto-6-deoxy-Dglucose3,5-epimerase; RmlD, dTDP-6-deoxy-L-mannose-dehydrogenase. ManA*, phosphomannose isomerase; ManB*, phosphomannomutase; ManC*, mannose-1-phosphate guanylyltransferase; Gmd, GDP-mannose 4,6-dehydratase; Fcl, GDP-L-fucose synthetase. DManNAc, 2-acetamido-2-deoxy-D-mannose; D-GalNAc, 2-acetamido-2-deoxy-D-galactose; D-GalNAcA, 2-Acetamido-2- deoxy-D-galacturonic acid; L-AltNAcA,2-Acetamido-2-deoxy-L-altruronic acid; D- FucNAc4N, 2-Acetamido-4-amino-2, 4- dideoxy-D-fucose; L-RhaNAc, 2-acetamido-2,6-dideoxy-L-mannose (N-acetyl-L-rhamnosamine); D-GlcNAc, 2-acetamido- 2-deoxy-D-glucose; L-QuiNAc, 2-Acetamido-2-deoxy-L-quinovose (2-acetamido-2,6-dideoxy-L-glucose); D-Glc, Dglucose; D-Gla, D-galactose; L-Rha, L-rhamnose (6-deoxy-L-mannose); L-Fuc, L-fucose; D-Man, D-mannose.
O-Antigen Cluster of P. shigelloides O8
In the P. shigelloides O8 cluster, orf16 shares 81% identity with MnaA from Vibrio cholerae, indicating there is a putative rare sugar, ManNAc, in the structure. Orf12 shares 53% identity with Wzx from Aeromonas hydrophila, and orf14 shares 34% identity with Wzy in Aminobacterium colombiense. The presence of wzx and wzy genes suggested the Wzx/Wzy pathway for O-antigen processing in O8. O8 has four GTs, encoded by orf10, orf15, orf18, and orf19, which share 50% identity with YifM from Marinobacter antarcticus, 83% identity with a GT from Vibrio ordalii, 58% identity with WbuB from Photorhabdus luminescens, and 97% identity with a GT from S. sonnei, respectively.
O-Antigen Cluster of P. shigelloides O17
For P. shigelloides O17, orf5 and orf6 share 100% identity with WbgT and WbgU from P. shigelloides and were renamed as Gne and Gna by Liu et al. [19]. Working together with WbgZ, a UDP-GlcNAc can be changed to a putative rare sugar, UDP-L-AltNAcA, in this serotype. Orf9 and orf11 share 99% identity with WbgV and WbgX from S. sonnei, indicating a putative rare sugar, UDP-D-FucNAcA, might be present in the structure. Orf7 shares 100% identity with Wzy from P. shigelloides and orf8 shares 99% identity with Wzx from S. sonnei. The set of wzx and wzy genes indicated the presence of Wzx/Wzy pathway-related O-antigen processing in O17. There is one GT encoded by the orf10, which shares 99% identity with WbgW from P. shigelloides.
O-Antigen Cluster of P. shigelloides O18
In the P. shigelloides O18 cluster, orf8 shares 63% identity with GalE from Providencia stuartii, indicating a putative UDP-Gal in the structure in O18. Orf12 shares 45% identity with Wzx in Proteus vulgaris and orf14 shares 45% identity with Wzy in Edwardsiella ictaluri. The presence of wzx and wzy genes indicated the presence of Wzx/Wzy pathway involvement in O-antigen processing. There are three GTs encoded by orf9, orf13, and orf15, which share 99%, 97%, and 99% identity with GTs from P. shigelloides.
O-Antigen Cluster of P. shigelloides O37
In the P. shigelloides O37 cluster, orf06 and orf07 share 96% and 100% identity with RmlB and RmlA from V. cholerae, respectively, and orf08 and orf09 share 86% and 85% identity with RmlD and RmlC in Vibrio anguillarum, respectively, suggesting a putative rhamnose in the structure. Furthermore, the chemical structure of P. shigelloides O37 reported in 2013 showed that there is a rare sugar, α-D-Lenose ((2S)-O-(4-oxopentanoic acid)-a-D-Glcp), which has not been found in nature [20]. In the cluster, orf12 shares 60% identity with MenD from Vibrio mimicus, and orf13 shares 84% identity with WcaK from Photobacterium sp. These two proteins work together to catalyze D-Glc to the rare sugar D-lenose. Orf10 and orf16 share 30% identity with Wzy and Wzx from E. coli. The presence of the wzx and wzy genes suggests that the Wzx/Wzy pathway is involved in O-antigen processing in O37. There are four GTs encoded by orf10, orf14, orf15, and orf17, which share 65% identity with a GT from E. coli, 48% identity with a GT from Shewanella loihica, 53% identity with WbsX from Flexilinea flocculi, and 35% identity with a GT from Cytophaga hutchinsonii, respectively.
O-Antigen Cluster of P. shigelloides O38
In the P. shigelloides O38 cluster, orf04 shares 87% identity with RmlD from A. hydrophila; however, whether rhamnose is present in the structure cannot be predicted using the presence of the rmlD gene only. Orf5 shares 62% identity with Wzm from Aeromonas veronii, and orf6 shares 42% identity with Wzm from Legionella santicrucis; therefore, the set of wzm and wzt genes suggests the presence of the transporter pathway in O38. There are three GTs encoded by the orf8, orf11, and orf12, which share 89% identity with a GT from Aeromonas salmonicida, 36% identity with a GT from V. cholerae, and 73% identity with a GT from A. hydrophila, respectively.
O-Antigen Cluster of P. shigelloides O39
In the P. shigelloides O39 cluster, orf19 and orf15 share 75% identity with QnlA from Vibrio sp. and 98% identity with QnlB from V. cholerae, respectively, suggesting a putative rare sugar, UDP-L-QuiNAc, in the structure. Orf8 shares 33% identity with Wzx from E. coli, and orf10 shares 55% identity with Wzy from V. cholera. Therefore, the set of wzx and wzy genes suggests that the Wzx/Wzy pathway is involved in O-antigen processing. There are three GTs encoded by orf11, orf14, and orf18, which share 61% identity with a GT from Pseudomonas saudiphocaensis, 82% identity with a GT from A. veronii, and 89% identity with WbuB from V. cholerae.
O-Antigen Cluster of P. shigelloides O44
In the P. shigelloides O44 cluster, no rare sugar could be predicted. Orf11 and orf12 share 35% and 80% identity with Wzx from Moorella sp. and Wzy from V. cholerae, respectively; therefore, the set of wzx and wzy genes suggests the presence of the Wzx/Wzy pathway in O-antigen processing. There are two GTs: Orf13 shares 55%identity with a GT from Pseudomonas mosselii, and orf15 shares 50% identity with a GT from V. cholerae.
O-Antigen Cluster of P. shigelloides O45
In the P. shigelloides O45 cluster, orf6, orf8, and orf9 share 89% identity with RlmB from P. shigelloides, 92%identity with RlmA from Aeromonas piscicola, and 66% identity with RlmB from A. hydrophila, respectively, which indicates a putative rhamnose in the structure. Orf11 and orf13 share 38% and 38% identity with Wzx from Pseudomonas sp. 286 and Wzy from E. coli, respectively; thus, the set of wzx, wzy, and wzz genes suggests the presence of the Wzx/Wzy pathway in O-antigen processing. There are three GTs encoded by orf12, orf14, and orf15, which share 44% identity with WbiP from Salmonella enterica, 44% identity with a GT from Vibrio ishigakensis, and 48% identity with a GT from Citrobacter freundii, respectively.
O-Antigen Cluster of P. shigelloides O61
In the P. shigelloides O61 cluster, orf14, orf15, and orf17 share 94% identity with Gmd from Aeromonas caviae, 87% identity with Fcl from A. hydrophila, and 75% identity with ManC from Providencia alcalifaciens, respectively, which indicates a putative rhamnose in the structure. Orf10 shares 35% identity with Wzx from E. coli, and orf11 shares 32% identity with Wzy from P. vulgaris; thus, the set of wzx and wzy genes indicates the presence of the Wzx/Wzy pathway in O-antigen processing. There are three GTs: orf12 and orf13 share 58% and 64% identity with GTs from E. coli, and orf18 shares 61% identity with WfgS from Vibrio vulnificus.
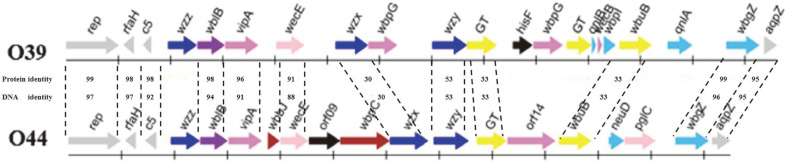
Comparative Analysis of the O-Antigen Gene Clusters of O39 and O44
The comparison of the O-antigen gene clusters of O39 and O44 showed that except for the conservative and anchor genes of rep, rfaH, c5 and aqpZ, there are some other genes shared by both clusters (Fig. 3). For example, both occupy wzx, wzy and wzz, and Wzx from O39 shares 30% identity with that from O44; Wzy, 53%; and Wzz, 42%. Also, the three GTs encoded by orf11, orf14, and orf18 in O39 cluster were blasted with the two GTs encoded by orf13 and orf15 in O44. We found that the GT of orf11 of O39 shares 33% identity with that of orf13 of O44, while the GT of orf18 of O39 shares 33% identity with that of orf15 of O44, which goes by the name of WbuB. By the number of GTs, we speculate that there are at least four sugars in the structure of O39, including the putative rare sugar UDP-L-QuiNAc represented by QnlA and QnlB; while O44 has three sugars. Moreover, WblB of O39 shares 98% identity with that of O44; VipA, 96%; WecE, 91%; and WbgZ, 99%.
Fig. 3.
Comparison of the O-antigen gene clusters of P. shigelloides O39 and O44.
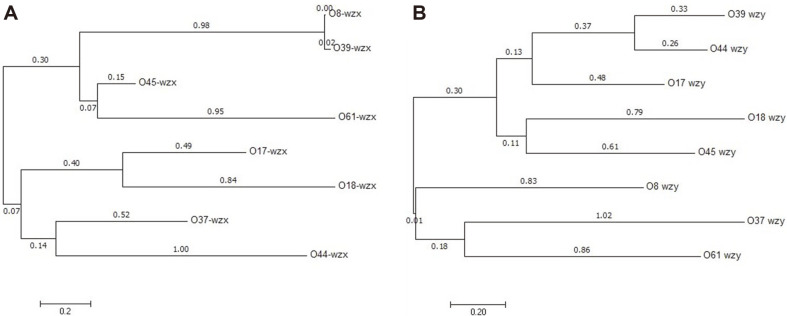
Phylogenetic Analysis
Dendrogram analysis of wzx was used for eight P. shigelloides serogroups (O8, O17, O18, O37, O39, O44, O45, and O61 (O38 does not have wzx) (Fig. 4). The phylogenetic tree showed two major clades, the upper one includes four serogroups of O8, O39, O45, and O61, and the lower one includes the other four serogroups of O17, O18, O37, and O44. In the upper clade, the distance of O8 is the same as that of O39, and both are close to the two-member group of O45 and O61. In the lower clade, there are two parallel groups; O17 is grouped together with O18, and O37 is grouped together with O44 (Fig. 4A).
Fig. 4. Phylogenetic analysis.
An unrooted phylogenetic tree constructed by the neighbor-joining method based on the wzx (A) and wzy (B) genes is shown. Bootstrap values were based on 1000 replications and only values greater than 50% are shown.
The wzy gene-based phylogenetic tree showed two major clades, the bigger one includes the five serogroups of O39, O44, O17, O18, and O45, and the smaller one includes the three serogroups of O8, O37, and O61. In the bigger clade, there are two subclades. The upper subclade consists of O17, O39, and O44, in which O39 is close to O44 and is grouped together with O17; the lower subclade consists of O18 and O45. In the smaller clade, O37 is close to O61 and is grouped together with O8 (Fig. 4B).
Target Genes and PCR Serotyping with Specific Primers
In our previous studies we suggested that the genes involved in O-antigen synthesis, either wzx and wzy, or wzm and wzt, are highly specific to individual O serotypes [8]. Therefore, they were selected as the target genes for the development of a molecular serotyping method. Among the nine O-antigens, O38 possesses wzm and wzt, and the other eight possess wzx and wzy. Accordingly, specific primers were designed based on the wzx genes of O17, O18, O37, O44, and O45; the wzy genes of O8 and O39; the wzt gene of O38; and the fcl gene of O61.
After optimization, a nine singleplex PCR assay with 9 primer pairs specific to the nine P. shigelloides serogroups was established (Table 2). The interpretation is based on the presence or absence of the characteristic PCR amplicons (~100 - 150 bp in length) for the serotype; for example, O61 can only generate PCR product with its own specific primer pair with 100% specificity and accuracy (Figs. 5A and S1). Samples isolated from the environment usually comprise more than one serotype. The mock samples containing two (O37 and O61) and three serotypes (O37, O38 and O61) were also tested with 100% specificity and accuracy (Fig. 5A). The PCR approach is useful for the simultaneous detection of multiple serotypes.
Fig. 5. PCR detection and sensitivity.
A. Line 1: Layout of the nine single-plex PCR with specific primers for each serotype. Line 2: Negative control. Line 3: Specificity of the PCR for P. shigelloides O61 without cross reaction with the other eight serogroups. Line 4: Mock sample with two serotypes of P. shigelloides 37 and O61. Line 5: Mock sample with three serotypes of P. shigelloides O37, O38 and O61. B. Sensitivity of detection with the genomic DNA of P. shigelloides O61. M, DL 500 bp Marker; Lane 1: 100 ng; Lane 2: 10 ng; Lane 3: 1 ng; Lane 4: 0.1 ng; Lane 5: 0.01 ng; Lane 6: 1 pg; Lane 7: 0.1 pg; Lane 8: 0.01 pg; and Lane 9: negative control.
DNA Sensitivity for PCR Assay
For genomic DNA of P. shigelloides O61, serial ten-fold dilutions ranging from 100 ng to 0.01 pg were used to test the sensitivity of the PCR assay. The minimum detection level was determined as 0.1 pg with the genomic DNA of O61 shown in Fig. 5B.
Detection of Clinical Isolates
Out of the total 16 clinical isolates collected, 13 were from the Chinese Center for Disease Control and Prevention Beijing, China, and the other 3 from Guangzhou Center for Disease Control and Prevention, Guangdong, China. The established serotyping PCR assay was applied to all the 16 samples and the data showed that three of them, clinical samples ps-1, ps-9, and ps-16, generated positive results as serotype O37 (Fig. S2). DNA sequencing of the PCR fragments confirmed the results (Fig. S3).
Discussion
Of the nine novel O-antigens, only the O37 chemical structure, which possesses the rare sugar D-Lenose that has not been found in the nature, has been reported [20]. In addition, several other rare sugars can be deduced based on the highly homologous sugar synthetic genes; for example, dTDP-L-Rha in O37 and O45; UDP-ManNAc in O8; UDP-L-AltNAcA and UDP-D-FucNAcA in O17; UDP-Gal in O18, and UDP-L-QuiNAc in O39. No rare sugars were predicted in O38 and O44, however.
In the O-antigen clusters of the nine P. shigelloides serogroups, transporter pathways encoded by wzm/wzt are employed for O-antigen processing by O38 and the wzt gene was selected as its target; Wzx/Wzy pathways encoded by wzx/wzy are employed for the other eight serogroups (O8, O17, O18, O37, O39, O44, O45, and O61). Among them, wzx genes were selected as the targets for five serogroups (O17, O18, O37, O44, and O45). In the wzx dendrogram tree, O8 and O39 are almost identical; however, in the wzy dendrogram tree, they are far from each other and therefore the wzy genes were selected as targets for both of them.
Interestingly, the eight O-antigens with Wzx/Wzy pathways possess not only wzx and wzy, but also wzz, which is the gene downstream of c5 in the cluster. Wzz is a well-known regulator of O-antigen chain-length [21]. Generally wzz is located within the chromosome but outside of the O-antigen clusters, as in the case of Shigella [19], Salmonella [9], Citrobacter [22], Providencia [23], and A. hydrophila [24]. However, together with the previous publication of 12 P. shigelloides, 17 of the 21 O-antigens employ the Wzx/Wzy pathway, and all 17 clusters have wzz genes. Similar to P. shigelloides, in P. aeruginosa, wzz is the next gene downstream of himD in the O-antigen clusters of all 20 identified serotypes [25].
In conclusion, O-antigen variability has been used as the basis for serotyping schemes established for many gram-negative bacteria, and serotyping plays a key role in detection and epidemiological surveillance. The frequent intra-species recombination events of P. shigelloides might lead to the expansion and diversity of serotypes which are important for understanding the evolution of this pathogen and other bacterial strains. In the present study, we described a traditional PCR procedure to serotype nine P. shigelloides serogroups based on their wzx/wzy/wzt/fcl gene sequences. Our method provides a rapid, reliable and reproducible tool for the detection, serotyping and epidemiological surveillance of P. shigelloides.
Supplemental Materials
Supplementary data for this paper are available on-line only at http://jmb.or.kr.
Acknowledgments
X. W. performed the sequence analyses and developed the molecular serotyping system. D. X. and X. G. purchased the strains and prepared the sample DNA. Y. L. and J. Y. conducted the bioinformatics analyses. J. Z. provided the clinical isolates. B. C. conceived the project and prepared the manuscript. All authors read and approved the final manuscript. This work was supported by the National Key Programs for Infectious Diseases of China (2017ZX10303405-001, 2017ZX10104002-001-006, and 2018ZX10712001-017).
Footnotes
Conflict of Interest
The authors have no financial conflicts of interest to declare.
REFERENCES
- 1.Niedziela T, Lukasiewicz J, Jachymek W, Dzieciatkowska M, Lugowski C, Kenne L. Core oligosaccharides of Plesiomonas shigelloides O54:H2 (strain CNCTC 113/92): structural and serological analysis of the lipopolysaccharide core region, the O-antigen biological repeating unit, and the linkage between them. J. Biol. Chem. 2002;277:11653–11663. doi: 10.1074/jbc.M111885200. [DOI] [PubMed] [Google Scholar]
- 2.Herrington DA, Tzipori S, Robins-Browne RM, Tall BD, Levine MM. In vitro and in vivo pathogenicity of Plesiomonas shigelloides. Infect. Immun. 1987;55:979–985. doi: 10.1128/IAI.55.4.979-985.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aldová E, Shimada T. New O and H antigens of the international antigenic scheme for Plesiomonas shigelloides. Folia Microbiol (Praha). 2000;45:301–304. doi: 10.1007/BF02817550. [DOI] [PubMed] [Google Scholar]
- 4.Shimada T, Sakazaki R. New O and H antigens and additional serovars of Plesiomonas shigelloides. Jpn J. Med. Sci. Biol. 1985;38:73–76. doi: 10.7883/yoken1952.38.73. [DOI] [PubMed] [Google Scholar]
- 5.Pieretti G, Carillo S, Lindner B, Lanzetta R, Parrilli M, Jimenez N, et al. The complete structure of the core of the LPS from Plesiomonas shigelloides 302-73 and the identification of its O-antigen biological repeating unit. Carbohydr. Res. 2010;345:2523–2528. doi: 10.1016/j.carres.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 6.Piqué N, Aquilini E, Alioto T, Miñana-Galbis D, Tomás JM. Genome Sequence of Plesiomonas shigelloides Strain 302-73 (Serotype O1) Genome Announc. 2013;1:e00404–13. doi: 10.1128/genomeA.00404-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sayeed S, Sack DA, Qadri F. Protection from Shigella sonnei infection by immunisation of rabbits with Plesiomonas shigelloides (SVC 01) J. Med. Microbiol. 1992;37:382–384. doi: 10.1099/00222615-37-6-382. [DOI] [PubMed] [Google Scholar]
- 8.Xi D, Wang X, Ning K, Liu Q, Jing F, Guo X, Cao B. O-Antigen gene clusters of Plesiomonas shigelloides serogroups and its application in development of a molecular serotyping scheme. Front. Microbiol. 2019;10:741. doi: 10.3389/fmicb.2019.00741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu B, Knirel YA, Feng L, Perepelov AV, Senchenkova SN, Reeves PR, Wang L. Structural diversity in Salmonella O antigens and its genetic basis. FEMS Microbiol. Rev. 2014;38:56–89. doi: 10.1111/1574-6976.12034. [DOI] [PubMed] [Google Scholar]
- 10.Samuel G, Reeves P. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and Oantigen assembly. Carbohydr. Res. 2003;338:2503–2519. doi: 10.1016/j.carres.2003.07.009. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, Liu D, Cao B, Han W, Liu Y, Liu F, et al. Development of a serotype-specific DNA microarray for identification of some Shigella and pathogenic Escherichia coli strains. J. Clin. Microbiol. 2006;44:4376–4383. doi: 10.1128/JCM.01389-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo D, Liu B, Liu F, Cao B, Chen M, Hao X, et al. Development of a DNA microarray for molecular identification of all 46 Salmonella O serogroups. Appl. Environ. Microbiol. 2013;79:3392–3399. doi: 10.1128/AEM.00225-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Du Y, Qian C, Li L, Jiang L, Jiang X, et al. Establishment of a suspension array for Pseudomonas aeruginosa O-antigen serotyping. J. Microbiol. Methods. 2018;155:59–64. doi: 10.1016/j.mimet.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Hu D, Liu B, Dijkshoorn L, Wang L, Reeves PR. Diversity in the major polysaccharide antigen of Acinetobacter baumannii assessed by DNA sequencing, and development of a molecular serotyping scheme. PLoS One. 2013;8:e70329. doi: 10.1371/journal.pone.0070329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benvenuto D, Cella E, Fogolari M, De Florio L, Borsetti A, Donati D, et al. The transmission dynamic of Madariaga Virus by bayesian phylogenetic analysis: Molecular surveillance of an emergent pathogen. Microb. Pathog. 2019;132:80–86. doi: 10.1016/j.micpath.2019.04.039. [DOI] [PubMed] [Google Scholar]
- 17.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6. 0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu B, Knirel YA, Feng L, Perepelov AV, Senchenkova SN, Wang Q, et al. Structure and genetics of Shigella O antigens. FEMS Microbiol. Rev. 2008;32:627–653. doi: 10.1111/j.1574-6976.2008.00114.x. [DOI] [PubMed] [Google Scholar]
- 20.Kaszowska M, Jachymek W, Lukasiewicz J, Niedziela T, Kenne L, Lugowski C. The unique structure of complete lipopolysaccharide isolated from semi-rough Plesiomonas shigelloides O37 (strain CNCTC 39/89) containing (2S)-O-(4-oxopentanoic acid)-α-D-Glcp (α-D-Lenose) Carbohydr. Res. 2013;378:98–107. doi: 10.1016/j.carres.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 21.Islam ST, Lam JS. Synthesis of bacterial polysaccharides via the Wzx/Wzy-dependent pathway. Can. J. Microbiol. 2014;60:697–716. doi: 10.1139/cjm-2014-0595. [DOI] [PubMed] [Google Scholar]
- 22.Qian C, Du Y, Li H, Wu P, Wang L, Wei Y, et al. Development of rapid and simple experimental and in silico serotyping systems for Citrobacter. Future Microbiol. 2018;13:1511–1522. doi: 10.2217/fmb-2018-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du Y, Li H, Yin Z, Rozalski A, Torzewska A, Yang P, et al. Development of a molecular serotyping scheme and a multiplexed luminex-based array for Providencia. J. Microbiol. Methods. 2018;153:14–23. doi: 10.1016/j.mimet.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 24.Cao H, Wang M, Wang Q, Xu T, Du Y, Li H, et al. Identifying genetic diversity of O antigens in Aeromonas hydrophila for molecular serotype detection. PLoS One. 2018;13:e0203445. doi: 10.1371/journal.pone.0203445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raymond CK, Sims EH, Kas A, Spencer DH, Kutyavin TV, Ivey RG, et al. Genetic variation at the O-antigen biosynthetic locus in Pseudomonas aeruginosa. J. Bacteriol. 2002;184:3614–3622. doi: 10.1128/JB.184.13.3614-3622.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.