Highlights
-
•
The Brain Imaging Data Structure (BIDS) eases neuroimaging data sharing and analysis.
-
•
The conversion of DICOM data into BIDS standard is cumbersome and time-consuming.
-
•
BIDS Managing and Analysis Tool (BMAT) is a complete open-source software.
-
•
BMAT provides the possibility to automatically convert DICOM into BIDS datasets.
-
•
BMAT provides the possibility to run analysis for neuroimaging studies.
Keywords: Neuroimaging, Software, BIDS, Multiple Sclerosis, MRI
Abstract
Magnetic Resonance Imaging (MRI) is an established technique to study in vivo neurological disorders such as Multiple Sclerosis (MS). To avoid errors on MRI data organization and automated processing, a standard called Brain Imaging Data Structure (BIDS) has been recently proposed. The BIDS standard eases data sharing and processing within or between centers by providing guidelines for their description and organization. However, the transformation from the complex unstructured non-open file data formats coming directly from the MRI scanner to a correct BIDS structure can be cumbersome and time consuming. This hinders a wider adoption of the BIDS format across different study centers. To solve this problem and ease the day-to-day use of BIDS for the neuroimaging scientific community, we present the BIDS Managing and Analysis Tool (BMAT). The BMAT software is a complete and easy-to-use local open-source neuroimaging analysis tool with a graphical user interface (GUI) that uses the BIDS format to organize and process brain MRI data for MS imaging research studies. BMAT provides the possibility to translate data from MRI scanners to the BIDS structure, create and manage BIDS datasets as well as develop and run automated processing pipelines, and is faster than its competitor. BMAT software propose the possibility to download useful analysis apps, especially applied to MS research with lesion segmentation and processing of imaging contrasts for novel disease biomarkers such as the central vein sign and the paramagnetic rim lesions.
1. Introduction
Magnetic Resonance Imaging (MRI) offers the unique opportunity to safely and non-invasively investigate brain pathology. The study of neurological disorders, such as Multiple Sclerosis (MS), often requires the acquisition of multiple MRI sequences, producing complex data in different formats (Marek et al., 2022, Smith, 2012). Furthermore, MRI data sharing is common in large-scale multicenter neuroimaging studies, as it allows increasing the sample size and produces generalizable results (Cheng et al., 2017, Dashevsky et al., 2018). In this framework, a well-structured organization of neuroimaging data is required in order to facilitate data sharing, ensure generalizability and ease the adoption of automated image-processing pipelines.
The Brain Imaging Data Structure (BIDS) format (Gorgolewski et al., 2016) proposes a universal standard for naming and organizing neuroimaging data. Indeed, adopting the BIDS format ensures an organized and homogeneous structuration of neuroimaging data, eases data sharing in the context of multicenter studies and facilitates the development of automated neuroimaging tools (Gorgolewski et al., 2017).
In a nutshell, the root directory of a BIDS dataset contains all raw data converted in an open file format such as Neuroimaging Informatics Technology Initiative (NIfTI) for images. These raw data are stored in specific subfolders according to the subject, session, and class of data. Metadata are stored in dictionary JavaScript Object Notation (JSON) files or Tabular Separated Value (TSV) files. Each file is named following a strict but explicit standardized convention according to a <key-value> scheme (e.g. sub-001_ses-01_part-mag_T2starw.nii.gz) (BIDS-Contributors 2022). The root directory is also composed of two other folders, sourcedata and derivatives, containing the original and the processed data, respectively. Finally, the dataset_description.json is used to store information about the study dataset (e.g. dataset name, authors, version, etc.) while the participants.tsv contains information about the study subjects (e.g. ID, age, sex, etc.) (BIDS-Contributors., 2022, Gorgolewski et al., 2016).
Although the BIDS format is a promising solution to organize neuroimaging datasets, its diffusion and applicability are limited by the high number of restrictive principles required to manage different types of neuroimaging data. Applying BIDS rules by hand is tricky, time-consuming and potentially associated with intra and inter-user errors. On the other hand, the use of shell scripts to automatically apply BIDS principles prevents non-informatics experts (such as medical neuroimaging scientists) to apply these principles by themselves. Therefore, software able to automatically implement the BIDS format are needed to broadcast its utilization. Furthermore, since several funding agencies, including the European Research Council (https://erc.europa.eu), now require a detailed Data Management Plan as part of new research proposals, we envision an increasing need and a more straightforward application of the BIDS format to neuroimaging projects in the near future.
Several solutions have already been proposed. The “BIDS manager” is a software designed to create and manage BIDS datasets (Roehri et al., 2021). However, the import of new MRI data within the dataset is performed one sequence at a time and requires the user to provide the name of the MRI sequences to the software every time, which make the software less user-friendly. “BIDS manager” is more restrictive than the BIDS specification and does not permit the importation or naming of every possible BIDS compliant sequences, potentially by lack of update. It does not allow either to work with research sequence not included in the BIDS specification yet. This ensures that the BIDS principles are well implemented but makes the software more restrictive and with limited flexibility.
“BrainLife” is another existing solution, a free online platform to perform neuroscience data analysis based on the BIDS format (https://brainlife.io/). BrainLife is an efficient data organizer and analysis tool, allowing to create and manage BIDS datasets as well as to run automated tasks (https://brainlife.io/docs/). BrainLife works exclusively online and offers cloud-computing resources to run extensive analysis. However, uploading MRI images online can raise relevant ethical questions for neuroimaging scientists, especially when working with sensitive patients’ data.
We propose the BIDS Managing and Analysis Tool (BMAT), a local offline neuroimaging analysis tool based on the BIDS format. BMAT desktop application is a Graphical User Interface (GUI) wrapper, working with existing command-line neuroimaging tools. BMAT can create and manage BIDS datasets, providing a user-friendly interface for running automated tasks on the dataset and for viewing analysis results. This software has been specifically developed and tested in the framework of multiple Sclerosis (MS) MRI research studies with multicentric data and dedicated analysis tools.
2. Materials and methods
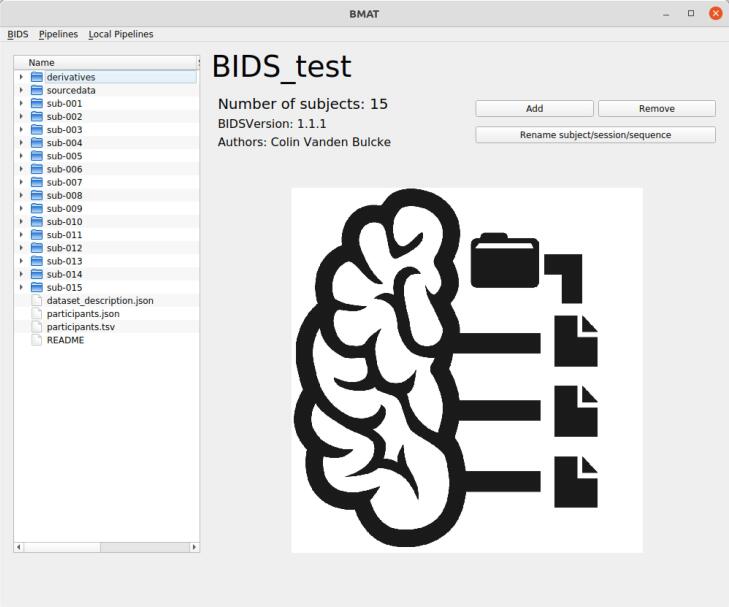
This section describes the proposed BMAT software in detail, starting with the Main Window structure, and then presenting more in depth some MS relevant BMAT analysis pipelines. Fig. 1 shows a view of the software’s Main Window.
Fig. 1.
A view of the software’s Main Window. The top central widget displays metainformation about the dataset. The left widget allows to safely navigate the BIDS root directory. The bottom central widget is used to view files and the right widget are buttons to apply actions on the dataset. The BIDS and Pipelines drop-down menu are respectively used to apply less often used actions and run preprogrammed automated tasks on the dataset.
2.1. Download and installation
BMAT can be downloaded for Linux, Windows and MacOSX operating system (OS) directly from its GitHub repository (https://github.com/ColinVDB/BMAT), which contains the source code of the software as well as a detailed documentation describing how to install and use the software. As will be described hereafter, BMAT requires a number of additional dependencies and the overall installation process takes one to two hours, depending on the OS and the informatic background of the user. Here is a list of the required dependencies:
-
•
Python (3.8): to run the software
-
•
dcm2niix: used for the conversion of the DICOM files to NifTI
-
•
ITK-SNAP: 3D viewer with annotation tools
-
•
Docker: used to run several tools
2.2. Graphical user interface (GUI) and functions
After the software has been downloaded and installed, once launched, BMAT asks to select a folder corresponding to a BIDS directory. If there is no BIDS directory available, the user can select an empty folder to create a new dataset. The Main Window is composed of several widgets, including:
-
•Dataset Information: the top central widget of the Main Window displays a short description of the dataset with useful information:
-
•Name of the BIDS directory
-
•Number of subjects in the dataset
-
•The BIDS version of the directory
-
•The Authors of the dataset
-
•Most of these pieces of information come from the previously described dataset_description.json file present in the directory.
-
•
-
•
Directory Tree Viewer: placed on the left of the Main Window, this widget allows to view and navigate the BIDS directory in a safe manner. Indeed, its use is similar to a classic File Explorer, without the risk of modifying, moving or deleting files in the dataset.
-
•
Quick Viewer: the central bottom widget allows the user to observe the files in the dataset. By default, it shows the icon of the software (like in Fig. 1). When opening any non-image file in the Directory Tree viewer by double-clicking on it, the software will open a dedicated viewer in this widget to observe the file. For image files, simple-clicking opens one slice as preview in the BMAT widget while double-clicking allows users to completely open the image in a separated ITK-SNAP window, an efficient advanced viewer for 3D medical volumes (Yushkevich et al., 2006; https://www.itksnap.org/pmwiki/pmwiki.php).
-
•
BIDS actions: this corresponds to the right widget and contains buttons to trigger specific actions on the dataset such as importing data, removing subjects and sessions, and renaming subjects, sessions, or sequences. These actions automatically edit all the files that needs to be updated. BMAT offers the possibility to automatically rename a sequence across the whole dataset. This is particularly useful if the user made a mistake in following the BIDS naming convention.
-
•
BIDS drop-down menu: it provides the user with other, less often used, actions such as creating or selecting another BIDS directory, updating the dataset_description.json file, or checking the BIDS validity of a dataset. The software uses the BIDS Validator tool to provide useful information to the user about the quality of its BIDS dataset. BIDS Validator is a proven tool highlighted in the BIDS specification (BIDS-Contributors 2022). This feature also provides the user with useful links towards the BIDS specification.
-
•
Pipelines drop-down menu: this menu allows the user to run specific tasks on the dataset in a fully automatic way. The pipelines are coming from the BMAT community and can be downloaded directly through the GUI. Some already available pipelines will be described below.
-
•
Local Pipelines drop-down menu: it allows the user to add his own pipeline locally. For that, the user needs to already have sufficient coding knowledge in the use of python and PyQt5. How to to add locally a pipeline is detailed in the supplementary materials.
2.3. Dataset management
The first objective of the software is to provide researchers with a user-friendly interface to create and manage BIDS datasets. This section focuses on the software’s usability and convenience.
The first step after creating a BIDS dataset is to import new subjects. Neuroimaging data are nowadays exported from MRI devices in a DICOM format (Graham, Perriss, and Scarsbrook 2005). This format is universal and contains relevant information about the sequence but is dependent on the service provider and usually heavy and not convenient to manipulate (Mildenberger, Eichelberg, and Martin 2002). Older formats, such as PAR/REC format from Philips, can also be used. In order to follow the BIDS guidelines, the images need to be converted into NIfTI format, which is an open-source format accessible by all and easy to manipulate (https://NIfTI.nimh.nih.gov/). In parallel, a JSON file containing the metadata is always associated with the image, and for some sequences such as diffusion MRI, additional specific files are created. BMAT uses dicom2niix to convert the DICOM files into NIfTI format (Li et al., 2016). The NIfTI file’s name is mainly based on the ProtocolName and SeriesDescription attributes in the respective DICOM. Although these attributes are free-form and non-guaranteed to be unique, as a common practice, the same SeriesDescription attribute is generally not used for more than one sequence within the same study MRI protocol. Also, the choice of DICOM attributes for the naming of the NIfTI files is limited by dcm2niix. After this step, the NIfTI files are renamed following the BIDS specification and organized into the right folder of the BIDS dataset. The renaming strategy is user-dependent and based on a mapping between the NIfTI file name and the corresponding BIDS modality, described in the sequences.csv file in the source code of the software. The sequences.csv file should follow the BIDS specification (BIDS-Contributors 2022) and needs to be completed only once at the beginning of the study; the software will be then able to recognize and work with these specific MRI sequences. The renaming strategy is further detailed in the supplementary materials.
Information about the subjects of the dataset is essential for neuroimaging studies and is contained in the participants.tsv file, completed automatically by BMAT. This file is composed of one row per subject and contains by default its BIDS ID, age, sex, and the dates of its different MRI sessions. As an additional feature, BMAT proposes the option for the user to automatically retrieve any information from the DICOM files of the subjects and store it in the participants.tsv file.
2.4. MS image analysis tools
As previously mentioned, BMAT can also incorporate different image analysis tools. Here we present some image analysis pipelines which are of specific interest for MS MRI studies, but also for neuroimaging studies in general.
A viewer is essential when carrying out neuroimaging studies as it allows to observe the images, and the results of computations performed on the images. Another essential feature is the possibility to manually segment and annotate regions of interest. This is particularly true in the case of MS MRI studies, where the segmentation of White Matter (WM) lesions is a fundamental starting point in many research studies, including MRI-based clinical trials. Lesion segmentation can be performed automatically (see below “Brain lesion segmentation” pipeline) or manually. To perform this task manually, our tool works with ITK-SNAP, an open-source easy-to-use viewer that allows to annotate 3D images (Yushkevich et al., 2006; https://www.itksnap.org/pmwiki/pmwiki.php). It also allows to open multiple images at the same time and provides a synchronization option when several windows are open to concurrently observe and annotate pathological details on different MRI images. Files edited using ITK-SNAP needs to be manually saved by the user in the right BIDS folder via the save button in ITK-SNAP.
Neuroimaging studies often use automatic computational tools, called pipelines, to produce specific resources and results. Thanks to the BIDS structure, the pipeline can be scripted to perform the same automatic task on all subjects’ images. For BMAT, the implementation of pipelines is community-driven and pipelines can be downloaded directly through the GUI from a dedicated GitHub organization (https://github.com/BMAT-Apps). Depending on the ongoing study, pipelines may differ between research groups. Here, we propose 4 representative BMAT pipelines, which can be used in the context of MS MRI studies:
-
•
Image registration: this pipeline provides a tool to perform a rigid registration of two images from a single subject using ANTs Registration (Avants et al., 2008, Avants et al., 2011) and to script this operation for any subject of the dataset. It also allows the possibility to apply the same transformation to another image by using the transformation matrix computed during the original registration. An image registration takes about 2 two three minutes to be computed.
-
•
Brain lesion segmentation: Lesion segmentation is a fundamental task in MS MRI studies. The brain segmentation pipeline uses SAMSEG (Cerri et al., 2021) to automatically segment WM lesions based on the Fluid Attenuated Inversion Recovery (FLAIR) and, if available, the Magnetization Prepared - RApid Gradient Echo (MPRAGE) contrasts. SAMSEG can produce an output segmentation in around 15 to 20 min.
-
•
FLAIR* computation: The FLAIR* contrast allows the simultaneous visualization of WM lesions and parenchymal veins in MS, and is a widely accepted technique to detect the central vein sign (CVS) diagnostic biomarker (Maggi et al., 2018, Ontaneda et al., 2021, Sati et al., 2016, Sinnecker et al., 2019). The FLAIR* computation pipeline allows to compute a FLAIR* image by voxel-wise multiplication of the registered FLAIR and T2* images (Sati et al., 2012), using a previously proposed docker container (Blakedewey/Flairstar - Docker Image | Docker Hub, n.d.). FLAIR* computation can take up to around 20 to 25 min.
-
•
Phase Unwrapping: this pipeline allows to unwrap and filter phase images obtained from susceptibility-based MRI using a previously proposed docker container (Dewey, 2022, Blakedewey/Phase_unwrap - Docker Image | Docker Hub, n.d.). The analysis of unwrapped phase images (in combination with other MRI contrasts) (Marcille et al., 2022) allows to detect Paramagnetic Rim Lesions (PRL), an emerging neuroimaging prognostic biomarker of chronic active inflammation in MS (Absinta et al., 2021, Absinta et al., 2019, Maggi et al., 2020). Phase unwrapping is computed in around 3 to 5 min.
Besides, the conception of the Local Pipelines drop-down menu allows the implementation of new pipelines to run specific tasks related to the study being conducted. To add a new pipeline locally to the software, the user must add its source code in python containing a dedicated graphical interface using PyQt5 and the computation code, as well as an associated JSON file containing metainformation about the pipeline, in the ‘LocalPipelines’ folder of the software source code. The computation part of the pipeline can use docker containers or can be implemented locally, allowing adaptability of the software to a wide range of neuroimaging applications. The steps for adding new local pipelines are explained in detail in the supplementary materials. To use this feature, the user needs good programming skills, especially in python and PyQt5. Afterwards, the new pipeline can be shared with the community through the GitHub organization (https://github.com/BMAT-Apps).
2.5. BMAT performance and differences with existing BIDS managing solutions
BMAT performance against “BIDS manager” and “BrainLife” (presented in the Introduction) for the conversion of a representative MRI session containing 15 sequences (14210 DICOM files) was tested by a researcher with good experience in data processing and BIDS conversion (CVB).
Additionally, five independent researchers from four separate universities (Université Catholique de Louvain, Ecole Polytechnique Fédérale de Lausanne, Icahn School of Medicine at Mount Sinai, University of Berne) have tested BMAT and filled out a survey on the installation and utilization of the program. The full survey can be accessed in the supplemental materials.
3. Results
This section presents the results of the utilization of BMAT, first through an illustrative example of a typical workflow when using the software for MS MRI studies and then by comparing the performance of BMAT with respect to “BIDS manager” and “BrainLife”, to convert source DICOM data into the BIDS format.
The example consists of 1) managing MS patients MRI DICOM data following the BIDS principles, 2) performing image registration, 3) WM lesion segmentation as well as 4) FLAIR* and 5) phase unwrapping for the CVS and PRL diagnostic and prognostic work-up.
3.1. Dataset management
The first task BMAT proposes is to import the data to create a new BIDS dataset for a new study. The MRI data presented here were acquired on different MRI machines from 3 different vendors: General Electrics (GE), Siemens and Philips. For the import to work properly, the user must first edit the sequences.csv file with the names of the imaging sequences that were used in the study. For example, “_3DT2_0.65mm_iso_20180824081221_11_.nii.gz”, and “_3DT2_0.65mm_iso_20180824081221_12__ph.nii.gz” are the names of the magnitude and phase images respectively obtained from a 3D segmented T2*-weighted echo-plan-imaging (EPI) sequence acquired on a Siemens Prisma scanner after conversion to NIfTI format (Sati et al., 2014). For the renaming scheme, the first step is to select a specific keyword that captures the modality of the sequence (‘3DT2’), search in the BIDS specification for the corresponding BIDS modality (‘T2starw’) and MRI type (‘anat’). Now, the BIDS fields remain to be completed; the phase and the magnitude part needs to be differentiated thanks to the “part” field. The “_ph” keyword can differentiate the phase from the magnitude. Similarly, the names of the same 3D-EPI sequence acquired on a GE and on a Philips scanner are “_p_Sag_3D_EPI_20220628104007”/ ”SWIphase__p_Sag_3D_EPI_20220628104007” and “_SWI_NEW_S2_20201229103505_”/“_SWI_NEW_S2_20201229103505__ph” respectively. Following the same process as explained above, the sequences.csv file can be completed accordingly (Table 1).
Table 1.
Representative section of a sequences.csv file to homogenize the BIDS-renaming of a 3D-EPI MRI sequence acquired on different scanners from different vendors. Abbreviations: modality, keyword from the sequence name identifying the type of MRI sequence; modality_bids, corresponding BIDS MRI modality; part, keyword from the sequence name identifying a specific part or a specific image derived from the MRI sequence (optional); part_bids, corresponding BIDS part (optional); MRI_type, BIDS MRI type value corresponding to the modality.
| modality | modality_bids | part | part_bids | MRI_type |
|---|---|---|---|---|
| 3DT2 | T2starw | _ph | phase | anat |
| 3DT2 | T2starw | _ | mag | anat |
| 3D_EPI | T2starw | SWIphase | phase | anat |
| 3D_EPI | T2starw | _ | mag | anat |
| SWI_NEW_S2 | T2starw | _ph | phase | anat |
| SWI_NEW_S2 | T2starw | _ | mag | anat |
At this point, the software can recognize three different names of the same 3D-EPI MRI sequence implemented on 3 different scanners (Siemens, GE, Philips) and rename them according to the BIDS specification (sub-xxx_ses-xx_part-mag_T2starw.nii.gz and sub-xxx_ses-xx_part-phase_T2starw.nii.gz).
Once the renaming file has been updated, the process of importing new data can begin. Compared to other existing BIDS managing software which import MRI sequences one by one (BIDS Manager), BMAT can convert at once the whole MRI session (containing several sequences acquired in series). This reduces the time spent on importing new data and increases the comfort of use. Thanks to the user interface, one can select any DICOM folder, compressed (ZIP file) or uncompressed (folder), that correspond to an MRI session of the subject. The user must then tell the software which patient this session corresponds to, by providing the corresponding BIDS_ID (e.g. sub-013) and session (e.g. ses-02). If this information is not provided, BMAT will consider that this session corresponds to the first session of a new subject and select a new available BIDS_ID (i.e. sub-001 and ses-01 for the first MRI session). The software will then 1) convert all the files contained in the DICOM folder to the NIfTI format, 2) rename the files according to the renaming strategy provided (e.g. sub-013_ses-01_part-mag_T2starw.nii.gz) and 3) create the directories to store these files. The DICOM folder is copied in the BIDS dataset following the corresponding subject and session folders within the sourcedata directory. This ensures that the original DICOM are stored unmodified in the dataset, so that the user can always retrieve this information and have a backup of the images in case an error is made. Finally, it will also modify the participants.tsv file to add the information relative to this new session.
When the import is complete, the user may assess the BIDS validity of its dataset by running the BIDS quality control tool and manually edit the dataset based on the feedback provided and on the BIDS specification.
3.2. MS image analysis tools
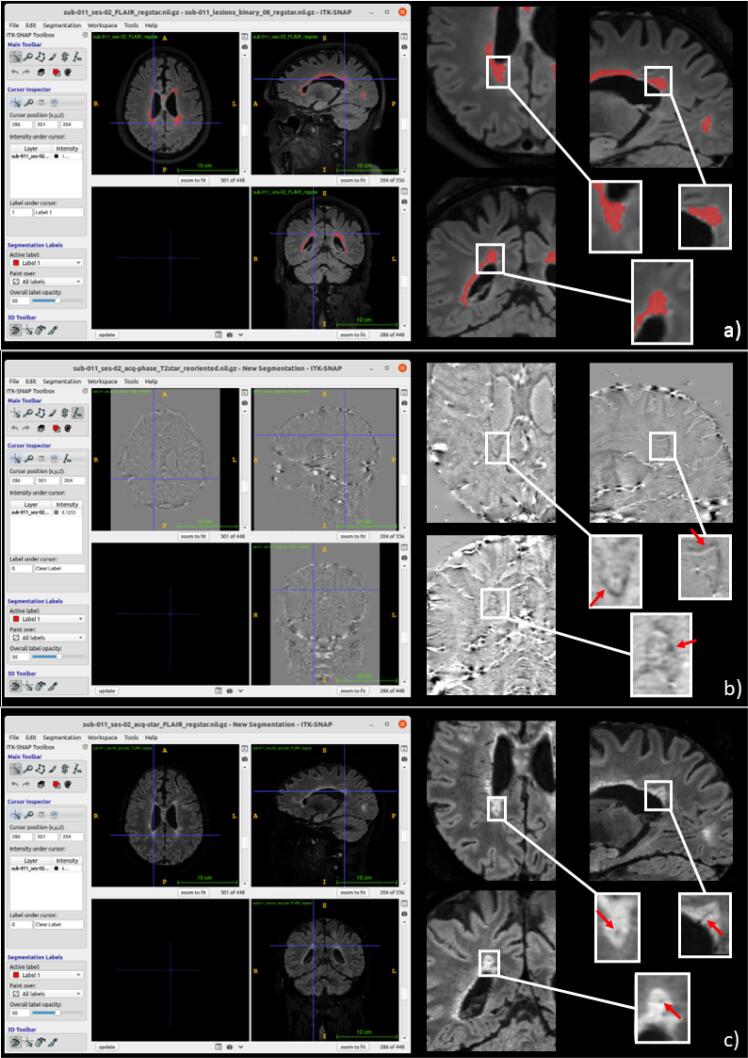
The next step corresponds to the image analysis part of the study performed using one or more of the different image analysis tools provided by the software (cf. Materials and methods – Image analysis tools). Here we present a representative BMAT MS MRI study analysis. The results of pipelines computations are shown in Fig. 2.
Fig. 2.
Results of the processing pipelines for the detection of the PRL and the CVS imaging biomarkers in MS viewed in ITK-SNAP (left) and in details (right). Lesion segmentation is performed automatically with the SAMSEG pipeline (a). A PRL (arrow) can be observed on unwrapped phase images (b), computed using the Phase Unwrapping pipeline; while the CVS work-up is performed on FLAIR* images, obtained with the FLAIR* computation pipeline (c). Abbreviations: PRL, paramagnetic rim lesions; CVS, central vein sign; MS, multiple sclerosis.
The first step is to segment WM lesions which can be automatically performed by the SAMSEG pipeline, using as input 3D FLAIR alone or in combination with 3D-MPRAGE images (see Fig. 2.a). Then, to perform the CVS and PRL biomarkers work-up, a 3D-EPI MRI sequence, giving magnitude and phase images, is required. In order to have all the different MRI contrast images in the same space, images are registered together using the registration pipeline (and the image transformation matrix can be easily applied to lesion masks if needed). FLAIR* and unwrapped phase images can be obtained using the corresponding BMAT pipelines and the segmented lesions can be evaluated for the presence of the PRL (Fig. 2b) and CVS (Fig. 2c) imaging biomarkers. Thanks to use of ITK-SNAP, the user is able to manually edit an existing lesion mask or to create new lesion masks.
3.3. BMAT performance and differences with existing BIDS managing solutions
The main differences and the performance of BMAT with respect to “BIDS manager” and “BrainLife” (presented in the Introduction) are summarized in Table 2. Both BIDS Manager and BMAT require the user to manually provide the name of the MRI sequences to the software (by filling in the requirements.json and sequences.csv file, respectively), see “Preprocessing before conversion Time” in Table 2. While this process is inevitably time consuming, it needs to be done only once at the beginning of each new imaging study (assuming that the MRI protocol does not change during the study) and makes the overall renaming step accurate using BMAT. As a drawback, BIDS Manager requires the user to import the whole session sequence by sequence and to rename by hand each sequence once imported, considerably slowing down the overall BIDS conversion process. By contrast, BrainLife automatically recognize and rename MRI sequences. However, the different MRI sequences within a session can be wrongly renamed (especially research MRI sequences), requiring the researcher to check and eventually fix the sequence name every time a new session is imported. Moreover, BrainLife requires to upload online the entire session before BIDS conversion, considerably slowing down the overall process. We believe that the major difference between BMAT and previous BIDS managing solutions lies in the ability of BMAT to convert DICOM to BIDS independently. Indeed, once the sequences.csv file is completed, the BMAT user can launch the conversion of several MRI sessions at a time, while both BIDS Manager and BrainLife requires the user intervention at some stage of the conversion.
Table 2.
Comparison of BMAT against “BIDS manager” and “BrainLife” for the conversion of an MRI session containing 15 sequences (14210 DICOM files). Abbreviations: Platform, platform on which operating system the software is available; Analysis possibility, possibility to run additional post-processing analysis on a BIDS dataset; Image viewer, presence/absence of an integrated complete 3D viewer; MRI import, possibility to importa new MRI session sequence by sequence or entirely; Preprocessing before converversion time, time needed to complete a possible preprocessing step before conversion; Conversion time; time needed to perform the complete conversion of an MRI session; Total time (first conversion), total time for the first conversion of the study; Total time: total time of conversion for a conversion; Scriptable conversion, does the software allow the conversion of several MRI session in one go; Flexibility, is the software more restrictive (<), equivalent (=) or less restrictive (>) than the BIDS specification.
| BIDS Manager | BrainLife | BMAT | |
|---|---|---|---|
| Platform | Windows, Linux | Online | Linux, Windows, MacOSX |
| Analysis possibility | No | Yes | Yes |
| Image viewer | No | Yes | Yes |
| MRI import | Sequence by sequence | Entire Session | Entire Session |
| Preprocessing before conversion time | requirements.json: 1′39″38 (once) | Upload: 38′30″ |
sequences.csv: 4′40″26 (once) |
| Conversion time | 6′26″09 | 3′57″02 | 1′06″39 |
| Total time (first conversion) |
8′05″47 | 44′27″02 | 5′46″65 |
| Total time | 6′26″09 | 44′27″02 | 1′06″39 |
| Scriptable conversion | No | No | Yes |
| Flexibility | < BIDS | =BIDS | > BIDS |
Finally, five independent researchers from four separate universities (Université Catholique de Louvain, Université de Lausanne, Icahn School of Medicine at Mount Sinai, University of Berne) have tested BMAT and gave their feedback on the installation and utilization of the program. Based on the BMAT survey, the installation process was rated 4.4/5. Indeed, although some issues were encountered during this step, all users had a medium-good level of informatics expertise that allowed them to solve basic problems and successfully complete the installation. The overall utilization of BMAT was rated at 4.8/5 with 5/5 for the GUI design and usability, but 4.2/5 for the format conversion with difficulties experienced in completing the sequences.csv file. Finally, all users were favorable to recommend BMAT to fellow researchers, describing it as having an intuitive GUI and being at least equivalent to other existing tools for data conversion into the BIDS format (see supplementary material for the full survey results).
4. Discussion and conclusion
BMAT is a complete local and open-source neuroimaging analysis tool that uses the BIDS format to organize and process brain MRI data for MS imaging research studies. The software enables the creation of well-structured datasets and the import of new data from different MRI scanners and vendors, including GE, Philips and Siemens MRI scanners. The quality of the BIDS dataset is ensured by using a BIDS validator tool, giving instantaneous feedback to the user about the validity of its dataset. BMAT also enables the use of pipelines, downloadable as BMAT-Apps, to automatically perform specific post-processing and image analysis tasks on the entire dataset. Here we present four relevant pipelines for MS imaging studies: image registration, image segmentation, FLAIR*computation, and phase-unwrapping. Many other MS and non-MS relevant image analysis pipelines can be implemented and shared open-source via the BMAT-Apps GitHub organization with the scientific community. Being based on the BIDS format, BMAT eases the sharing of datasets and automated MRI analysis tools between multiple institutions.
When compared to other available solutions, BMAT stands out by being at least equivalent or better than the competition, faster for conversion and more intuitive and complete. Part of its intuitiveness is due to the freedom it gives to its users to manage their dataset, by being less restrictive than the BIDS specification. A significant advantage of this feature is to allow the use of research imaging sequences not present in the BIDS specification. However, the software does not entirely enforce BIDS principles and lets the responsibility to the user, while providing the tools, to follow the specification.
The renaming scheme offers the possibility to use the software with any MRI sequences. BMAT allow users to add new analysis pipelines implemented by the community as well as adding personalized scripts to compute ad-hoc analysis on the dataset, thanks to the implementation of the local pipelines feature. These three options make the software generalizable to any kind of neuroimaging study.
As it has been implemented as an intuitive GUI, BMAT would especially be useful for non-computer experts, such as medical neuroimaging researchers, to implement the BIDS format and have a complete analysis tool for their studies. It also allows to run image processing pipelines through a simple GUI. This software can also be very useful for graduate students to take in hand the BIDS format that could be otherwise overwhelming, especially for early-stage researchers.
The principal limitations of this software are its installation and setting up. Since this software is integrated with several other tools (e.g. ITK-SNAP, Docker, etc.), these dependencies make the installation tedious. The use of Docker makes the software available on every OS. However, Docker performance is influenced by the OS, specifically Mac with an M1 chip can considerably slow Docker compared to Linux. As another limitation, the renaming scheme requires the user to complete the sequences.csv file following the BIDS specification before starting a new MRI study. However, this is only needed once at the beginning. Finally, as the conversion from DICOM to NIfTI is performed using dicom2niix, possible new issues can arise from the conversion of new specific research sequences that have not yet been implemented in this tool.
In the future, we aim to integrate the software with DataLad. DataLad is a free and open-source distributed data management system that keeps track of your data, creates structure, ensures reproducibility, supports collaboration, and integrates with widely used data infrastructure (Halchenko et al., 2021). This will allow the versioning and sharing of datasets with other researchers. This last point raises some ethical question that needs to be addressed when implementing this feature.
The BMAT software is a research software completely open-source and will be maintained through a GitHub forum to report issues. Other contributors are welcomed to join the project or give feedback and ideas for improvement.
Overall, BMAT is a complete and local software based on the BIDS format to carry out neuroimaging studies. It provides a user-friendly framework to create and manage as well as run automated tasks on BIDS dataset. BMAT is convenient to use and customizable by the user to adapt to any kind of neuroimaging study.
Funding
Colin Vanden Bulcke has the financial support of the Fédération Wallonie Bruxelles - Fonds Spéciaux de Recherche (F.S.R.).
Maxence Wynen is supported by Swiss government excellence scholarship (#2021.0087).
Francesco La Rosa is supported by the Swiss National Foundation (SNF) Postdoc Mobility Fellowship (P500PB_206833), the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie project TRABIT (agreement No 765148), and the Novartis Foundation for Medical-Biological Research (#21A032).
Martina Absinta is supported by the Conrad N. Hilton Foundation (Marilyn Hilton Bridging Award for Physician-Scientists, grant #17313), the Roche Foundation for Independent Research, the Cariplo Foundation (grant #1677), the FRRB Early Career Award (grant#1750327), and the International Progressive MS alliance (PA-2107-38081).
Pietro Maggi is supported by the Fund for Scientific Research (F.R.S.–FNRS, grant #40008331) and by the Fonds de Recherche Clinique (FRC), Université catholique de Louvain.
CRediT authorship contribution statement
Colin Vanden Bulcke: Conceptualization, Methodology, Software, Visualization, Investigation, Data curation, Writing – original draft, Writing – review & editing. Maxence Wynen: Conceptualization, Methodology, Software, Visualization, Investigation, Data curation, Writing – original draft, Writing – review & editing. Jules Detobel: Data curation, Writing – original draft, Writing – review & editing. Francesco La Rosa: Visualization, Investigation, Data curation, Writing – original draft, Writing – review & editing, Writing – review & editing. Martina Absinta: Data curation, Writing – original draft, Writing – review & editing. Laurence Dricot: Data curation, Writing – original draft, Writing – review & editing. Benoît Macq: Data curation, Writing – original draft, Writing – review & editing. Meritxell Bach Cuadra: Data curation, Writing – original draft, Writing – review & editing, Supervision. Pietro Maggi: Conceptualization, Methodology, Software, Data curation, Writing – original draft, Writing – review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank the Radiology departments of the Cliniques universitaires Saint-Luc (UCLouvain, Brussels, Belgium), of the Hôpital Erasme, (Université Libre de Bruxelles, Brussels, Belgium) and of the Lausanne University Hospital (Lausanne, Switzerland) for helping with MRI data acquisition; Oscar Esteban for his time and precious feedback on BMAT; Sebastien Jodogne for his for his time and help in the deployment of biomedical software.
Contributor Information
Colin Vanden Bulcke, Email: colin.vandenbulcke@uclouvain.be.
Pietro Maggi, Email: pietro.maggi@saintluc.uclouvain.be.
Data availability
Data will be made available on request.
References
- Absinta M., Sati P., Masuzzo F., Nair G., Sethi V., Kolb H., Ohayon J., Tianxia W.u., Cortese I.C.M., Reich D.S. Association of chronic active multiple sclerosis lesions with disability in vivo. JAMA Neurol. 2019;76(12):1474–1483. doi: 10.1001/jamaneurol.2019.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Absinta M., Maric D., Gharagozloo M., Garton T., Smith M.D., Jin J., Fitzgerald K.C., et al. A Lymphocyte–microglia–astrocyte axis in chronic active multiple sclerosis. Nature. 2021;597(7878):709–714. doi: 10.1038/s41586-021-03892-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants, Brian, Nick Tustison, and Gang Song. 2008. ‘Advanced Normalization Tools (ANTS)’. Insight J 1–35 (November). https://doi.org/10.54294/uvnhin.
- Avants B.B., Tustison N.J., Song G., Cook P.A., Klein A., Gee J.C. A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage. 2011;54(3):2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BIDS-Contributors. 2022. ‘The Brain Imaging Data Structure (BIDS) Specification’, February. https://doi.org/10.5281/ZENODO.3686061.
- ‘Blakedewey/Flairstar - Docker Image | Docker Hub’. n.d. Accessed 5 March 2022. https://hub.docker.com/r/blakedewey/flairstar.
- ‘Blakedewey/Phase_unwrap - Docker Image | Docker Hub’. n.d. Accessed 5 March 2022. https://hub.docker.com/r/blakedewey/phase_unwrap.
- Cerri S., Puonti O., Meier D.S., Wuerfel J., Mühlau M., Siebner H.R., Van Leemput K. A contrast-adaptive method for simultaneous whole-brain and lesion segmentation in multiple sclerosis. NeuroImage. 2021;225(January) doi: 10.1016/j.neuroimage.2020.117471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A., Kessler D., Mackinnon R., Chang T.P., Nadkarni V.M., Hunt E.A., Duval-Arnould J., Lin Y., Pusic M., Auerbach M. Conducting multicenter research in healthcare simulation: lessons learned from the INSPIRE network. Adv. Simulat. 2017;2(1):6. doi: 10.1186/s41077-017-0039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashevsky B.Z., Bercu Z.L., Bhosale P.R., Burton K.R., Chatterjee A.R., Alexandre L., Frigini R., Heacock L., et al. Multicenter research studies in radiology. Acad. Radiol. 2018;25(1):18–25. doi: 10.1016/j.acra.2017.05.019. [DOI] [PubMed] [Google Scholar]
- Dewey, Blake E. 2022. ‘Laplacian-Based Phase Unwrapping in Python (v1.0)’. Zenodo. https://doi.org/10.5281/zenodo.7198991.
- Gorgolewski K.J., Auer T., Calhoun V.D., Cameron Craddock R., Das S., Duff E.P., Flandin G., et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci. Data. 2016;3(1) doi: 10.1038/sdata.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgolewski K.J., Alfaro-Almagro F., Auer T., Bellec P., Mihai Capotă M., Chakravarty M., Churchill N.W., et al. BIDS apps: improving ease of use, accessibility, and reproducibility of neuroimaging data analysis methods. PLoS Comput. Biol. 2017;13(3):e1005209. doi: 10.1371/journal.pcbi.1005209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.N.J., Perriss R.W., Scarsbrook A.F. DICOM demystified: A review of digital file formats and their use in radiological practice. Clin. Radiol. 2005;60(11):1133–1140. doi: 10.1016/j.crad.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Halchenko Y.O., Meyer K., Poldrack B., Solanky D.S., Wagner A.S., Gors J., MacFarlane D., et al. DataLad: distributed system for joint management of code, data, and their relationship. J. Open Source Software. 2021;6(63):3262. doi: 10.21105/joss.03262. [DOI] [Google Scholar]
- Li X., Morgan P.S., Ashburner J., Smith J., Rorden C. The first step for neuroimaging data analysis: DICOM to NIfTI conversion. J. Neurosci. Methods. 2016;264(May):47–56. doi: 10.1016/j.jneumeth.2016.03.001. [DOI] [PubMed] [Google Scholar]
- Maggi P., Absinta M., Grammatico M., Vuolo L., Emmi G., Carlucci G., Spagni G., et al. Central vein sign differentiates multiple sclerosis from central nervous system inflammatory vasculopathies. Ann. Neurol. 2018;83(2):283–294. doi: 10.1002/ana.25146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi P., Sati P., Nair G., Cortese I.C.M., Jacobson S., Smith B.R., Nath A., et al. Paramagnetic rim lesions are specific to multiple sclerosis: an international multicenter 3T MRI study. Ann. Neurol. 2020;88(5):1034–1042. doi: 10.1002/ana.25877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcille M., Rúa S.H., Tyshkov C., Jaywant A., Comunale J., Kaunzner U.W., Nealon N., et al. Disease correlates of rim lesions on quantitative susceptibility mapping in multiple sclerosis. Sci. Rep. 2022;12(1):4411. doi: 10.1038/s41598-022-08477-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek S., Tervo-Clemmens B., Calabro F.J., Montez D.F., Kay B.P., Hatoum A.S., Donohue M.R., et al. Reproducible brain-wide association studies require thousands of individuals. Nature. 2022;603(7902):654–660. doi: 10.1038/s41586-022-04492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mildenberger P., Eichelberg M., Martin E. Introduction to the DICOM Standard. Eur. Radiol. 2002;12(4):920–997. doi: 10.1007/s003300101100. [DOI] [PubMed] [Google Scholar]
- Ontaneda, D., P. Sati, P. Raza, M. Kilbane, E. Gombos, E. Alvarez, C. Azevedo, et al. 2021. ‘Central Vein Sign: A Diagnostic Biomarker in Multiple Sclerosis (CAVS-MS) Study Protocol for a Prospective Multicenter Trial’. NeuroImage: Clinical 32 (January): 102834. https://doi.org/10.1016/j.nicl.2021.102834. [DOI] [PMC free article] [PubMed]
- Roehri N., Villalon S.M., Jegou A., Colombet B., Giusiano B., Ponz A., Bartolomei F., Bénar C.-G. Transfer, collection and organisation of electrophysiological and imaging data for multicentre studies. Neuroinformatics. 2021;19(4):639–647. doi: 10.1007/s12021-020-09503-6. [DOI] [PubMed] [Google Scholar]
- Sati P., Dm Thomasson, Li N., Dl Pham, Nm Biassou, Ds Reich, Ja Butman. Rapid, high-resolution, whole-brain, susceptibility-based MRI of multiple sclerosis. Multiple Sclerosis (Houndmills, Basingstoke, England) 2014;20(11) doi: 10.1177/1352458514525868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sati P., George I.C., Shea C.D., Gaitán M.I., Reich D.S. FLAIR*: A Combined MR Contrast Technique for Visualizing White Matter Lesions and Parenchymal Veins. Radiology. 2012;265(3):926–932. doi: 10.1148/radiol.12120208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sati P., Jiwon O.h., Todd Constable R., Evangelou N., Guttmann C.R.G., Henry R.G., Klawiter E.C., et al. The central vein sign and its clinical evaluation for the diagnosis of multiple sclerosis: A consensus statement from the North American imaging in multiple sclerosis cooperative. Nature Rev. Neurol. 2016;12(12):714–722. doi: 10.1038/nrneurol.2016.166. [DOI] [PubMed] [Google Scholar]
- Sinnecker T., Clarke M.A., Meier D., Enzinger C., Calabrese M., De Stefano N., Pitiot A., et al. Evaluation of the central vein sign as a diagnostic imaging biomarker in multiple sclerosis. JAMA Neurol. 2019;76(12):1446–1456. doi: 10.1001/jamaneurol.2019.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. Brain imaging: FMRI 2.0. Nature. 2012;484(7392):24–26. doi: 10.1038/484024a. [DOI] [PubMed] [Google Scholar]
- Yushkevich P.A., Piven J., Hazlett H.C., Smith R.G., Ho S., Gee J.C., Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. NeuroImage. 2006;31(3):1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.