Abstract
The number of thoracolumbar vertebrae in Dezhou donkeys varies from 22 to 24 and is associated with body size and carcass traits. In mammals, the latent transforming growth factor beta binding protein 2 (LTBP2) has been found to have some functions in the development of thoracolumbar vertebrae. The relationship between LTBP2 and TLN (the number of thoracolumbar vertebrae) of Dezhou donkeys is yet to be reported. The purposes of this study are as follows: 1) to quantify the effect of thoracolumbar vertebrae number variation of Dezhou donkeys on body size and carcass trait; 2) to study the distribution of single nucleotide variants (SNVs) in the LTBP2 gene of Dezhou donkeys; and 3) to explore whether these SNVs can be used as candidate sites to study the mechanism of Dezhou donkey muti-thoracolumbar vertebrae development. The TLN, body size, and carcass traits of 392 individuals from a Dezhou donkey breed were recorded. All animals were sequenced for LTBP2 using GBTS liquid chip and 16 SNVs were used for further analysis. We then analyzed the relationship between these SNVs with TLN, body size, and carcass traits. The results showed that: 1) c.5547 + 860 C > T, c.5251 + 281 A > C, c.3769 + 40 C > T, and c.2782 + 3975 A > G were complete genetic linkages and significantly associated with thoracic vertebrae number (TN) (p < 0.05) (wild-type homozygotes had more TN than heterozygotes); 2) c.1381 + 768 T > G and c.1381 + 763 G > T were significantly associated with lumber vertebrae number (LN) (p < 0.05); 3) c.1003 + 704 C > T, c.1003 + 651 C > T, c.1003 + 626 A > G, and c.812 + 22526 T > G were significantly associated with chest circumference (CHC), front carcass weight (CWF), after carcass weight (CWA), and carcass weight (CW) (p < 0.05) (wild-type homozygotes were larger than other genotypes in CHC, CWF, CWA, and CW); and 4) the effect of variation is not consistent in c.565 + 11921 A > G, c.565 + 6840 A > G, c.565 + 3453 C > T, and c.494 + 5808 C > T. These results provide useful information that the polymorphism of LTBP2 is significantly associated with TLN, body size, and carcass traits in Dezhou donkeys, which can serve as a molecule marker to improve donkey production performance.
Keywords: thoracolumbar vertebrae, Dezhou donkey, LTBP2, single nucleotide variants (SNVs), association analysis
1 Introduction
In mammals, the vertebral column consists of cervical, thoracic, lumbar, sacral, and caudal vertebrae. The number of vertebrae, especially the thoracolumbar vertebrae number, is an important economic trait in domestic animals because it is related to the length of the carcass and the size of the body (Borchers et al., 2004). The number of thoracic vertebrae (TN) is equal to the number of ribs, which are among the most valuable parts of the meat in the China market, and the number of lumbar vertebrae (LN) is bound up with abdominal muscle. The variation in the number of thoracolumbar vertebrae (TLN) has been observed in many mammalian species, such as in sheep; there are 13 thoracic vertebrae and 6 lumbar vertebrae (T13L6) for the majority of sheep, while the carcass length of T13L7 and T14L6 increases by 2.22 cm and 2.93 cm, respectively, compared with normal T13L6, and carcass weight (CW) increases about 1.68 kg and 1.90 kg (Donaldson et al., 2013; Li et al., 2017). In pig breeds, modern Western pig breeds, such as Duroc, Large White, and Landrace, have more (n = 21–23) TLN than Chinese indigenous breeds (n = 19), generally (King and Roberts, 2010; Mikawa et al., 2011; Liu et al., 2020), and one extra vertebrae expands the carcass length by 80 mm in bacon pigs (King and Roberts, 2010).
The vertebral column originates from the pre-somatic mesoderm under regulation of the notochord, which means the number of vertebrae in a region may vary connaturally, and this has been accurately determined in the embryonic stage (Greene and Copp, 2009). The variability of the vertebral column may arise from cranial-caudal border shifts, which take place when there is a somatic shift from the typical distribution of vertebral segments in a region; this may be one of the reasons for the inconsistent TLN (Thawait et al., 2012). The multi-vertebrae number trait is complex and regulated by genetics; up to now, several genes have been proved to have the function of regulating the development of vertebrae. Two quantitative trait loci (QTL) have been identified to regulate the vertebrate development in pigs: one on chromosome 1 and another one on chromosome 7; through more in-depth research, the nuclear receptor subfamily 6, group A, member 1 (NR6A1) gene (c.748 Pro 192 Leu) and vertebrae development-associated (VRTN) gene (g.20311-20312 ins291) have been proved to be candidate genes affecting spinal development (Mikawa et al., 2005; Mikawa et al., 2007; Mikawa et al., 2011; Ren et al., 2012; Fan et al., 2013; Burgos et al., 2015; Zhang et al., 2016). Association analysis has revealed significant associations between the single nucleotide variants (SNVs) (rs414302710: A > G) in the exon-8 of the NR6A1 gene with the number of lumbar vertebrae (Zhang et al., 2019). In addition to this, many genes have been found to be associated with the development of the vertebrae, such as the homeobox gene family (Rijli et al., 1995; Wellik, 2007; Gomez et al., 2008), GDF11 (Li et al., 2010), and Btg2 (Park et al., 2004), but most of these studies have focused on model animals, such as mice and fish.
The multi-vertebrae trait has also attracted donkey breeding researchers since it was discovered several decades ago owing to of multi-vertebrae advantages. Furthermore, the multi-vertebrae trait is highly heritable [heritability is 0.62 in pigs (Borchers et al., 2004)]. For the majority of Dezhou donkeys, there are 18 thoracic vertebrae and 5 lumbar vertebrae, usually labeled as T18L5. To date, we have observed five thoracic-lumbar vertebrae combination types in Dezhou donkeys (T17L5, T17L6, T18L5, T18L6, and T19L5); we have not observed the T19L6 type, nor have we seen any article reporting this. We collected skeletal specimens of three thoracic vertebrae types and two lumbar vertebrae types from the Dezhou donkey slaughterhouse (Figure 1). Many articles have reported that the LTBP2 gene may have the function of regulating the vertebrae development, but the mechanism is not clear. Zhang et al. (2016) found that growth differentiation factor 11 (Gdf11) can increase the number of ribs in mice, while some scholars have found that LTBP2 can inhibit the extracellular processing of Gdf11 through basic amino acid specific preprotein convertase 5/6 (PC5/6). In our previous studies, through the genome-wide association study (GWAS), we predicted that LTBP2 may be associated with the vertebrae number in Dezhou donkeys (Wang et al., 2020). Therefore, the current study aims to detect mutations in the LTBP2 gene and analyze their association with the TLN, body size, and carcass traits of Dezhou donkeys. This study contributes to the understanding of the mechanism based of TLN differences and provides theoretical support for multi-vertebrae molecular marker-assisted selection in donkey breeding.
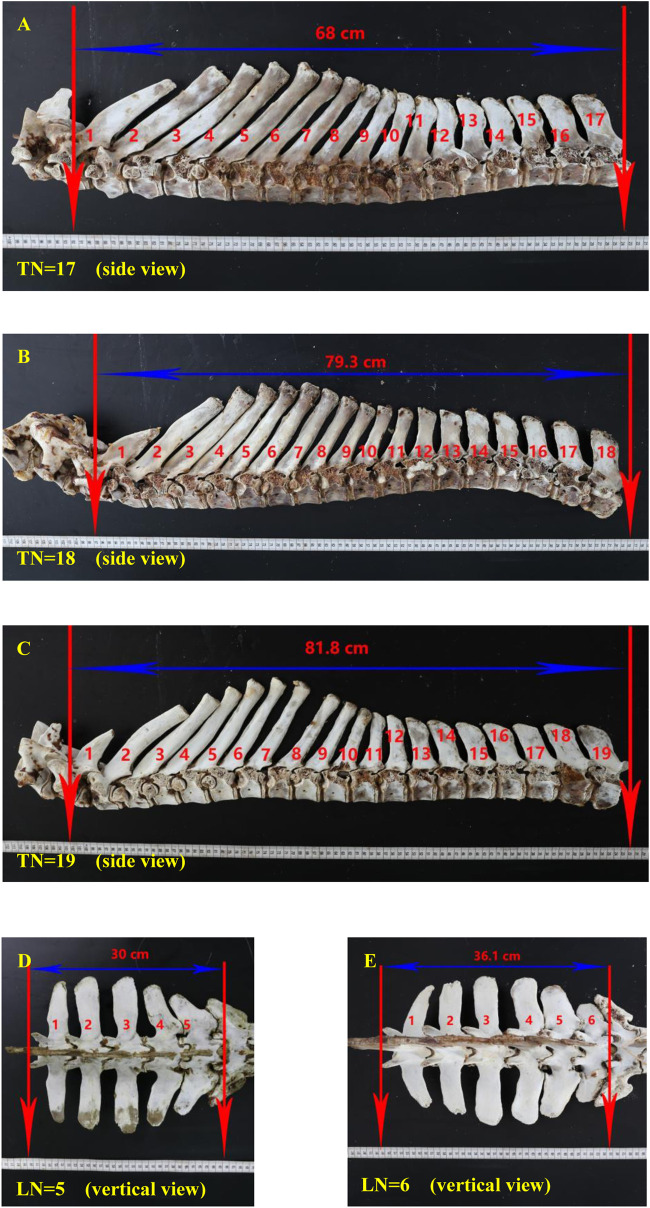
FIGURE 1.
Thoracic and lumbar vertebrae specimens of Dezhou donkeys. Notes: (A) the side view of a 17 thoracic vertebrae specimen; (B) the side view of an 18 thoracic vertebrae specimen; (C) the side view of a 19 thoracic vertebrae specimen; (D) the vertical view of 5 lumbar vertebrae specimen; (E) the vertical view of 6 lumbar vertebrae specimen. In each figure, the red arrows mark the starting and ending points of the thoracic or lumbar vertebrae, respectively. The blue arrows indicate the straight-line length of the thoracic or lumbar vertebrae. Red numbers with units indicate the straight-line length value of the thoracic or lumbar vertebrae. Red numbers without units indicate the order of the thoracic or lumbar vertebrae. “LN” means the number of lumbar vertebrae; “TN” means the number of thoracic vertebrae.
2 Materials and methods
2.1 Experimental animals and data acquisition
The study collected 392 Dezhou donkeys in Dezhou Town, Shandong Province, China, from 2019 to 2021 (the average body size data are shown in Supplementary Table S1). All donkeys were 2-year-old jackasses, fattened for meat production, with unknown pedigree information. During this study, the animal feed consisted of grass and hay ad libitum and water. The was no food intake within 12 h before slaughter, but water was unlimited. A 10 ml blood sample was collected from each Dezhou donkey using an EDTA anticoagulated blood collection tube and stored at −80°C. The body size [body height (BH), the vertical distance from the highest point of bun nail to the ground; body length (BL), the linear distance from the fore-end of humeral carina to the last internal carina of ischial tubercle; and chest circumference (CHC), make a vertical line from the back of the scapula and measure the circumference of its chest] of these 392 Dezhou donkeys was measured using Zhang et al.’s (2021) method. The measurement for collecting the thoracolumbar vertebrae number and carcass traits were as follows. The warm carcass weight [removing the head, viscus, skin, hooves, penis, testicles, and tail (CW)], the front part of the carcass weight [cut off from the boundary between the last thoracic vertebra and the first lumbar vertebra, and the carcass where the thoracic vertebra are located (CWF)], and the hind part of the carcass weight [cut off from the boundary between the thoracic vertebra and the lumbar vertebra, and the carcass where the lumbar vertebra are located (CWA)] were measured immediately after slaughter. The information regarding thoracolumbar vertebrae was measured [thoracic vertebrae number (TN), the total length of thoracic vertebrae (TL), lumbar vertebrae number (LN), and the total length of lumbar vertebrae (LL)] at the abattoir in the cold-storage room after slaughter on the left half of the carcass (Liu et al., 2022). The TIANAMP Genomic DNA Extraction Kit (DP304, TIANGEN, Beijing, China) was utilized to extract the genomic DNA from blood samples. The A260/280 ratios of all DNA samples were determined with a NanoDrop (ND 2000, NanoDrop, United States). After that, genomic DNA were diluted to a common centration 50 ng/μL and stored at −20°C.
2.2 Genotyping
All animals were sequenced for the whole LTBP2 gene with GBTS (genotyping by targeted sequencing) liquid chip (using GenoBaits and GenoPlexs technology), which was developed by our laboratory and the Shijiazhuang Breeding Biotechnology Co., Ltd. In order to confirm the accuracy of the chip sequencing results, eight pairs of specific primer sequences were designed using Primer Premier 5.0 software to amplify some polymorphic loci of the LTBP2 gene (50%). The primers are listed in Supplementary Table S2. The fragments contained 16 SNVs (c.5547 + 860 C > T, c.5251 + 281 A > C, c.1381 + 768 T > G, c.1003 + 704 C > T, c.812 + 22526 T > G, c.812 + 6591 T > C, c.565 + 3453 C > T, and c.494 + 5808 C > T) of LTBP2. The PCR system (25 μL) included: 2 μL genomic DNA (25 ng/μL); 1 μL of each primer (10 μM); 12.5 μL 2 × MasterMix (TIANGEN, Beijing, China); and 8.5 μL double-distilled H2O. The PCR protocol was as follows: 95°C for 10 min; 35 cycles of denaturing at 95°C for 30 s; annealing at Tm°C (Supplementary Table S2) for 30 s; and extension at 72°C for 1 min, with a final extension at 72°C for 10 min. After amplification, PCR products were first electrophoresed on 2% agarose gels with MF079-M5 Hipure Gelred nucleic acid strain (Mei5 Biotechnology, Co., Ltd., Beijing, China) and then sent to Sangon Biotech Biotechnology Co., Ltd. for sequencing in both directions and the sequencing results were compared using DNAMAN software (Version 5.2, Lynnon Biosoft, Vaudreuil, Canada).
2.3 Statistical analysis
The genotype frequencies of all target loci (the frequency of each genotype is greater than 5%) were calculated. Using these SNVs, haplotypes were constructed, and haplotype frequency was calculated; haplotypes with frequencies greater than 5% were used for subsequent analysis. The genotype frequencies, allelic frequencies, and Hardy–Weinberg equilibrium partial values were determined by direct counting. At the same time, the population genetic parameters were estimated using online software (http://analysis.bio-x.cn/myAnalysis.php) and (http://www.msrcall.com/Gdicall.aspx), including observed heterozygosity (Obs-Het), predicted heterozygosity (Pred-Het), homozygosity (Ho), effective number of alleles (Ne) (Nei and Roychoudhury, 1974), and polymorphism information content (PIC) (Botstein et al., 1980). The linkage relationship between D’ (LD’ coefficient) and r 2 (correlation coefficient) based on the alleles of each site was calculated using Haploview software (Version 4.2, Daly Lab at the Broad Institute Cambridge, United States) and haplotype frequency was estimated between the loci (Barrett et al., 2005). In order to explore the differences in SNVs’ genotype and haplotypes in LTBP2, which are important genetic factors for several traits, the linear model in SPSS 22.0 software (Statistical Product and Service Solutions, Version 22.0 Edition, IBM, Armonk, NY, United States) was used for correlation analysis:
where yij is the measured value of the related trait, μ is the population mean, ai is the genotype effect, and eij is the random residual.
3 Results
3.1 Variation in vertebral number in Dezhou donkeys
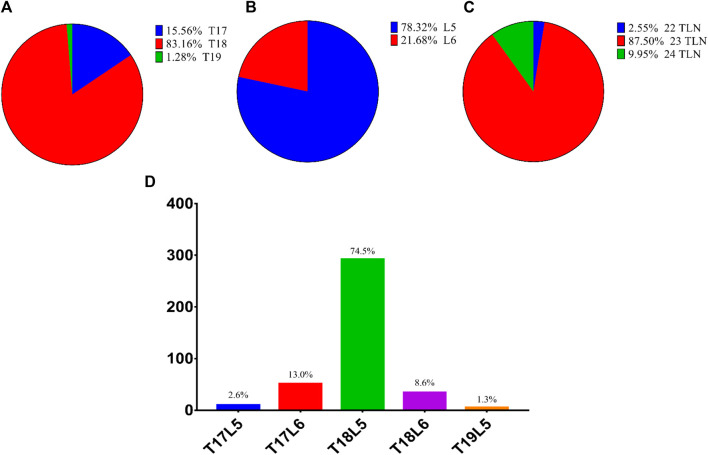
The variation in vertebral number among 392 Dezhou donkeys is shown in Figure 2. The TN, LN, and TLN ranged from 17 to 19, 5 to 6, and 22 to 24, respectively; the dominated type was 18 (83.2%), 5 (78.3%), and 23 (87.5%), respectively. The most common proportion among the experimental samples was the T18L5 thoracic-lumbar vertebrae combinational type (74.5%), and the rarest was T19L5 (1.3%).
FIGURE 2.
Variation in vertebral number of Dezhou donkeys. Notes: (A) variation in TN; (B) variation in LN; (C) variation in TLN; (D) variation in TL-type.
3.2 Effect of TLN variation on body size and carcass traits
The number of vertebrae is associated with body length, carcass weight, and other economical traits in livestock. However, for Dezhou donkeys, the effect of the vertebrae number increase on economic traits is still unclear. To evaluate their relationship, the effects of single TL, LN, TLN, and thoracic-lumbar vertebrae combinational types on carcass weight were analyzed successfully. In order to accurately calculate the effect of adding one type of vertebrae on body length and carcass weight, we studied the effect of variations in the number of thoracic vertebrae in the same population of lumbar vertebrae and vice versa (Supplementary Table S3). When LN remains unchanged, adding a thoracic vertebra means a significant increase in TL by 5 cm according to the weighted average method (TN = 17, TL = 68.73 ± 0.35; TN = 18, TL = 73.61 ± 0.18; TN = 19, TL = 75.2 ± 1.59), significant at p < 0.05. When TN remains unchanged, adding a lumbar vertebra means a significant increase in LL by 4 cm according to the weighted average method (LN = 5, LL = 23.27 ± 0.07; LN = 6, LL = 27.19 ± 0.15, p < 0.05). From the perspective of TLN, adding a thoracolumbar vertebra means the total length of thoracolumbar vertebrae (TLL) will increase significantly (TLN = 22, TLL = 91.6 ± 1.07; TLN = 23, TLL = 96.75 ± 0.22; TLN = 24, TLL = 100.59 ± 0.71; p < 0.05). Individuals with 23 or 24 TLN were about 10 kg heavier than 22 TLN individuals in carcass weight, but no significant difference was detected. Theoretically, there should be a T19L6 combinational type, but this has not been observed in our long-term observations; maybe we need a larger sample size.
3.3 Polymorphisms of 16 multi-vertebrae trait causal loci
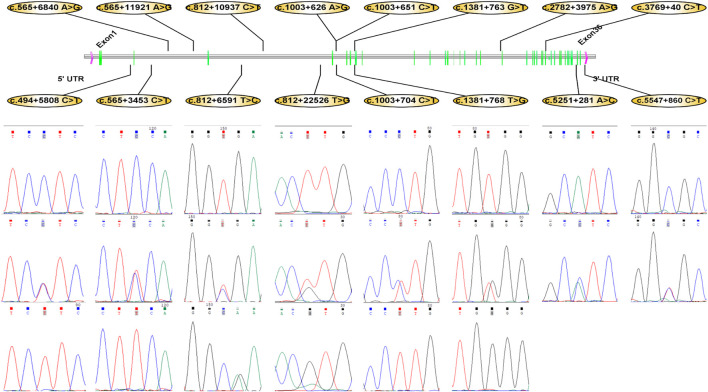
A total of 16 mutations (c.5547 + 860 C > T, c.5251 + 281 A > C, c.3769 + 40 C > T, c.2782 + 3975 A > G, c.1381 + 768 T > G, c.1381 + 763 G > T, c.1003 + 704 C > T, c.1003 + 651 C > T, c.1003 + 626 A > G, c.812 + 22526 T > G, c.812 + 10937 C > T, c.812 + 6591 T > C, c.565 + 11921 A > G, c.565 + 6840 A > G, c.565 + 3453 C > T, and c.494 + 5808 C > T) were identified as candidate sites for multi-vertebrae and carcass traits (Figure 3). In order to further confirm their association with vertebrae number, body size, and carcass traits, all samples were genotyped for these 16 alleles using GBTS liquid chip. The sequencing results of the PCR products amplified by specific primers were consistent with those of the GBTS liquid chip.
FIGURE 3.
Sixteen SNVs of the LTBP2 gene in Dezhou donkeys.
In Supplementary Table S4, three genotype frequencies (wild-type homozygote, heterozygotes, and mutant-type homozygote) and genetic indices (Obs-Het, Pred-Het, Ho, and PIC) in Dezhou donkeys’ LTBP2 gene are shown. As shown in Supplementary Table S4, we found that c.5547 + 860 C > T, c.5251 + 281 A > C, c.3769 + 40 C > T, and c.2782 + 3975 A > G did not have a mutant-type homozygote. The other 12 loci had three genotypes in total. The Hardy–Weinberg test results showed that the population was genetically balanced and belonged to the Mendelian population. The genetic diversity index showed that the PIC of Dezhou donkeys was 0.123–0.555, and the polymorphism was 1 < PIC < 0.25, which is in the moderate-high polymorphism state.
3.4 Association analysis of donkey vertebrae number, body size, and carcass traits with 16 SNV polymorphisms
The effects of the LTBP2 SNVs on the number of vertebrae, body size, and carcass traits were evaluated and are presented in Supplementary Table S5. As shown in Supplementary Table S5, individuals with the wild-type homozygote had significantly more TN than heterozygotes (p < 0.05) in c.5547 + 860 C > T, c.5251 + 281 A > C, c.3769 + 40 C > T, and c.2782 + 3975 A > G loci, which means the main effect of these four loci are significant in TN. The lumbar vertebrae number in Dezhou donkeys with wild-type homozygous and mutant-type homozygous genotypes of c.1381 + 768 T > G and c.1381 + 763 G > T was 5.05 ± 0.05 and 5.25 ± 0.03, respectively, and the difference was significant (p < 0.05). The other two mutant loci significantly related to the number of lumbar vertebrae are c.812 + 10937 C > T and c.812 + 6591 T > C. In c.812 + 10937 C > T, individuals with mutant-type homozygous T/T and heterozygous C/T genotypes had more LN than wild-type homozygotes, while in c.812 + 6591 T > C, individuals with mutant-type homozygous C/C had significantly fewer LN than wild-type homozygotes and heterozygotes. As described in Supplementary Table S5, in the c.1003 + 704 C > T, c.1003 + 651 C > T, c.1003 + 626 A > G, and c.812 + 22526 T > G mutational sites, the effect of variation on CHC, CWF, CWA, and CW was consistent. Individuals with wild-type homozygotes had significantly higher data than other genotypes among the aforementioned four traits (p < 0.05), while others (BH, BL, SW, TN, TL, STL, LN, LL, SLL, TLN, TLL, and STLL) were not significant. There are five mutational sites significantly associated with TLN (c.812 + 10937 C > T, c.565 + 11921 A > G, c.565 + 6840 A > G, c.565 + 3453 C > T, and c.494 + 5808 C > T). The effect of c.812 + 10937 C > T in TLN was consistent with that in LN. In c.565 + 11921 A > G, c.565 + 6840 A > G, c.565 + 3453 C > T, and c.494 + 5808 C > T mutational sites, individuals with the heterozygous genotype had the most TLN (23.12 ± 0.03) compared to other genotypes (significantly).
3.5 Linkage disequilibrium and haplotype analysis of LTBP2 gene mutational sites
In order to reveal the linkage relationship between the 16 mutational sites of the LTBP2 gene, the linkage disequilibrium between these loci was estimated. The analysis results showed that three blocks exist between these 16 SNVs (r 2 > 0.33) (Ardlie et al., 2002). Block1 is composed of c.5547 + 860 C > T, c.5251 + 281 A > C, c.3769 + 40 C > T, and c.2782 + 3975 A > G, block2 is composed of c.1381 + 768 T > G, c.1381 + 763 G > T, c.1003 + 704 C > T, c.1003 + 651 C > T, c.1003 + 626 A > G, and c.812 + 22526 T > G, and block3 is composed by c.565 + 11921 A > G, c.565 + 6840 A > G, c.565 + 3453 C > T, and c.494 + 5808 C > T. The linkage coefficients for other pairs of SNVs were low (r 2 < 0.33).
Haplotype analysis was performed for the three blocks, and three haplotypes with a frequency greater than 0.05 were identified and labeled as Hap1 (-CACAGTTTGGAATT-), Hap2 (-CACAGTTTGGGGCC-), and Hap3 (-CACATGCCATGGCC-). The haplotype frequencies of Hap1, Hap2, and Hap3 in Dezhou donkeys account for 37.6%, 28.3%, and 10.2%, respectively.
3.6 Association analysis between the haplotype combinations and TLN, body size, and carcass traits
In order to explore the association between SNV polymorphism and the traits of Dezhou donkeys, haplotype combination was carried out on the basis of constructed haplotypes. Six haplotype combinations (Hap1Hap1, Hap1Hap2, Hap1Hap3, Hap2Hap2, Hap2Hap3, and Hap3Hap3) were constructed, respectively. The results are shown in Supplementary Table S6; individuals with Hap2Hap3 and Hap3Hap3 made up less than 5% of the total sample size, so no follow-up analysis was conducted for these two haplotype combinations. The correlation analysis between four haplotypes and TLN, body size, and carcass traits was calculated, but no significant difference was observed.
4 Discussion
With the development of agricultural mechanization, the causative function of donkeys is gradually weakening, which is the reason why the stock of donkeys in China decreased rapidly from 1990 to 2016 (Statistics, 2021). In recent years, luckily, with the further study of the special functions of donkey meat, milk, and skin, the situation has gradually improved (D’Auria et al., 2011; Li et al., 2006; Polidori et al., 2015; Polidori et al., 2008; Yvon et al., 2018). Donkeys are raised as a small special livestock in China today; the government has enacted many policies to support the development of the donkey industry, especially in breed preservation and improvement. Vertebrae number is an important characteristic in livestock, such as pigs, cattle, sheep, and donkeys, because a higher vertebrae number means a longer body length and a heavier carcass weight (Yang et al., 2016; Zhang et al., 2017). In this study, we observed that the TN, LN, and TLN in Dezhou donkeys is 17–19, 5–6, and 22–24, respectively; no T19L6 thoracolumbar vertebrae combination type individuals were observed, which is similar to what has been reported in previous studies (Jamdar and Ema, 1982; Gao, 2020; Gao et al., 2021). The effect of increasing TN or LN is not consistent; one more TN increases TLL by about 5.14 cm, while one more LN increases TLL by about 4.05 cm (significant at p < 0.05). Interestingly, the effect of one more TN in different LN populations is unequal, such as in the 5 lumber vertebrae number population, the CW of individuals with 18 TN is 11.1 kg more than individuals with 17 TN, while in 6 lumbar vertebrae number population, the CW of individuals with 18 TN is 3 kg more than individuals with 17 TN; the disparity between the increase between the two is 8.1 kg. Similarly, the effect of one more LN in different TN populations is also unequal.
The heritability of the vertebrae is high; for example, the heritability of vertebrae in pigs is 0.62 (Borchers et al., 2004). Therefore, we believe that directional breeding of Dezhou donkeys with multiple thoracolumbar vertebrae numbers and studying the molecular mechanism of multiple thoracolumbar vertebrae numbers is worthy of attention. The development of vertebrae is a complex process, which requires the regulation of multiple signal molecules and specific components. To date, there are several regions in the genome that have been identified as having the function of regulating the development of vertebrae. Mikawa et al. (2005), Mikawa et al. (2007), and Mikawa et al. (2011) identified two QTLs’ influence on the number of vertebrae, mapped to SSC1 and SSC7. There is a 479 kb region on SSC7 including nine annotated genes, identified as a critical fragment influencing the TLN (Liu et al., 2020). The Notch receptor 1 (Notch-1) gene is a critical gene for somite development, while the activation of FOS could inhibit the expression of Notch-1 (Portanova et al., 2013; Liao and Oates, 2017).
In previous studies, many mutation sites in LTBP2 have been proved to have some functions in the development of primary congenital glaucoma (c.3028G > A, p.Asp1010Asn; c.3427delC, p.Gln1143Argfs*35) (Rauf et al., 2020), cardiomyocytes (c.2206G > A, p.Asp736Asn) (Chen et al., 2020), and lung fibroblast-to-myofibroblast differentiation (Zou et al., 2021). LTBP2 has also been found to be related to the number of teats in pigs (Martins et al., 2022). In this study, LTBP2 was used as a candidate gene associated with TLN in Dezhou donkeys based on related studies (Zhao et al., 2020; Wang et al., 2022). The liquid chip was designed based on published data from several databases and was used for genotyping all mutational loci of all samples. PCR primers were designed, based on a published donkey LTBP2 sequence (GenBank accession NC-052183.1), to amplify the eight mutational loci (Wang et al., 2020). The alignments of the genotyping result between GBTS liquid chip and PCR products with specific primers is 100%. From this, we can see that the GBTS liquid chip sequencing results are reliable.
We actually detected a total of 544 mutational sites in the whole LTBP2 gene of Dezhou donkeys; however, most of the loci cannot meet the requirements of correlation analysis, so we finally screened out 16 SNVs for subsequent analysis. Among these loci, the adjacent loci are significantly related to a certain trait (p < 0.05), centrally. For example, for c.5547 + 860 C > T, c.5251 + 281 A > C, c.3769 + 40 C > T, and c.2782 + 3975 A > G, the distance between them was 8,247 bp, 6,018 bp, and 9,253 bp, respectively, genome wide; they were all significantly associated with TN (p < 0.05). The analysis results from the Haploview software showed a strong linkage disequilibrium between them, which means when the C allele is mutated to the T allele at c.5547 + 860 C > T, the A allele at c.5251 + 281 A > C will mutate to C allele, the C allele at c.3769 + 40 C > T will mutate to T allele, and the A allele at c.2782 + 3975 A > G will mutate to G allele. In this block, individuals with wild-type homozygous had more 0.16 pieces of TN than heterozygotes, so we speculate that the mutation here has a negative effect on TN. The same situation also occurs in block2 (c.1381 + 768 T > G, c.1381 + 763 G > T, c.1003 + 704 C > T, c.1003 + 651 C > T, c.1003 + 626 A > G, and c.812 + 22526 T > G), but the function of block2 is aimed at LN, CHC, CWF, CWA, and CW. For the development of TN, mutations at c.1381 + 768 T > G and c.1381 + 763 G > T sites are beneficial, while for CHC, CWF, CWA, and CW, mutations at c.1003 + 704 C > T, c.1003 + 651 C > T, c.1003 + 626 A > G, and c.812 + 22526 T > G are negative. In block3, four SNVs (c.565 + 11921 A > G, c.565 + 6840 A > G, c.565 + 3453 C > T, and c.494 + 5808 C > T) were all significantly associated with TLN; individuals with the heterozygous genotype had higher TLN than others (p < 0.05), but the difference between wild-type homozygotes and mutant-type homozygotes was different.
In order to determine the linkage disequilibrium of 16 SNV markers located on the Dezhou donkey LTBP2 gene, a total of three haplotype blocks among 16 SNVs were identified, examining haplotypes above 5%, spanning approximately 30 kb. The most frequent haplotypes within the blocks comprised the linkage of nucleotides CACA (96.4%) for block1, GTTTGG (72.8%) for block2, and AATT (53.6%) for block3. To demonstrate the total effect of a single locus, haplotype combinations were combined for further study. The haplotype combination types with a sample size less than 5% of the total sample were removed. Our results showed no dominant type in the four haplotype combinations; the reason may be the effect of these three blocks counteracting each other. These results indicate that the LTBP2 gene is a potential functional gene for regulating the development of vertebrae in Dezhou donkeys, and this finding may provide important implications for further studies on the regulation of the TLN in Dezhou donkeys. It must be noted that our research has not explored the molecular mechanism of SNPs in LTBP2 regarding how they effect TLN development.
5 Conclusion
In summary, the effects of TLN variation on body size and carcass traits of Dezhou donkeys were quantified, and 16 trait-related SNVs have been identified in the LTBP2 gene of Dezhou donkeys. These loci have low-to-high polymorphism and there are significant differences between different genotypes among different traits. The results of this study will play an important role in the further study of the molecular mechanism of Dezhou donkey molecular breeding for muti-vertebrae breeds.
Data availability statement
The datasets presented in this study can be found in online repositories. The name of the repository and accession number can be found at: NCBI; PRJNA878942.
Ethics statement
The animal study was reviewed and approved by the Animal Welfare and Ethics Committee of Institute of Animal Sciences, Liaocheng University (No. LC2019-1). Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
CW and ZL provided the study concept and design, ZL wrote and revised the manuscript, and TW, XS, XW, WR, and BH collected and analyzed the data. All authors have read and approved the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (grant no. 31671287), the Well-bred Program of Shandong Province (grant no. 2017LZGC020), Taishan Leading Industry Talents, Agricultural Science of Shandong Province (grant no. LJNY201713), and Shandong Province Modern Agricultural Technology System Donkey Industrial Innovation Team (grant no. SDAIT-27).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.969959/full#supplementary-material
References
- Ardlie K. G., Kruglyak L., Seielstad M. (2002). Patterns of linkage disequilibrium in the human genome. Nat. Rev. Genet. 3 (4), 299–309. 10.1038/nrg777 [DOI] [PubMed] [Google Scholar]
- Barrett J. C., Fry B., Maller J., Daly M. J. (2005). Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 21 (2), 263–265. 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- Borchers N., Reinsch N., Kalm E. (2004). The number of ribs and vertebrae in a Piétrain cross: Variation, heritability and effects on performance traits. J. Anim. Breed. Genet. 121, 392–403. 10.1111/j.1439-0388.2004.00482.x [DOI] [Google Scholar]
- Botstein D., White R. L., Skolnick M., Davis R. W. (1980). Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32 (3), 314–331. [PMC free article] [PubMed] [Google Scholar]
- Burgos C., Latorre P., Altarriba J., Carrodeguas J. A., Varona L., Lopez-Buesa P. (2015). Allelic frequencies of NR6A1 and VRTN, two genes that affect vertebrae number in diverse pig breeds: A study of the effects of the VRTN insertion on phenotypic traits of a Duroc×Landrace–large white cross. Meat Sci. 100, 150–155. 10.1016/j.meatsci.2014.09.143 [DOI] [PubMed] [Google Scholar]
- Chen H. X., Yang Z. Y., Hou H. T., Wang J., Wang X. L., Yang Q., et al. (2020). Novel mutations of TCTN3/LTBP2 with cellular function changes in congenital heart disease associated with polydactyly. J. Cell. Mol. Med. 24 (23), 13751–13762. 10.1111/jcmm.15950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Auria E., Mandelli M., Ballista P., Di Dio F., Giovannini M. (2011). Growth impairment and nutritional deficiencies in a cow's milk-allergic infant fed by unmodified donkey's milk. Case Rep. Pediatr. 2011, 103825. 10.1155/2011/103825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson C. L., Lambe N. R., Maltin C. A., Knott S., Bunger L. (2013). Between- and within-breed variations of spine characteristics in sheep. J. Anim. Sci. 91 (2), 995–1004. 10.2527/jas.2012-5456 [DOI] [PubMed] [Google Scholar]
- Fan Y., Xing Y., Zhang Z., Ai H., Ouyang Z., Ouyang J., et al. (2013). A further look at porcine chromosome 7 reveals VRTN variants associated with vertebral number in Chinese and Western pigs. Plos One 8 (4), e62534. 10.1371/journal.pone.0062534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q. (2020). Study on Dezhou donkey germplasm. Shandong: Shandong Teachers' University. [Google Scholar]
- Gao Q., Wang J., Li Y., Sun Y., Yang C., Li H., et al. (2021). Preliminary study on the production performance of Dezhou donkey. J. Livest. Ecol. 42 (02), 56–61. [Google Scholar]
- Gomez C., Ozbudak E. M., Wunderlich J., Baumann D., Lewis J., Pourquie O. (2008). Control of segment number in vertebrate embryos. Nature 454 (7202), 335–339. 10.1038/nature07020 [DOI] [PubMed] [Google Scholar]
- Greene N. D., Copp A. J. (2009). Development of the vertebrate central nervous system: formation of the neural tube. Prenat. Diagn. 29 (4), 303–311. 10.1002/pd.2206 [DOI] [PubMed] [Google Scholar]
- Jamdar M. N., Ema A. N. (1982). A note on the vertebral formula of the donkey. Br. Vet. J. 138 (3), 209–211. 10.1016/s0007-1935(17)31084-9 [DOI] [PubMed] [Google Scholar]
- King J. W. B., Roberts R. C. (2010). Carcass length in the bacon pig; its association with vertebrae numbers and prediction from radiographs of the young pig. Anim. Prod. 2 (01), 59–65. 10.1017/s0003356100033493 [DOI] [Google Scholar]
- Li C., Zhang X., Cao Y., Wei J., You S., Jiang Y., et al. (2017). Multi-vertebrae variation potentially contribute to carcass length and weight of Kazakh sheep. Small Ruminant Res. 150, 8–10. 10.1016/j.smallrumres.2017.02.021 [DOI] [Google Scholar]
- Li H., Huang M. J., Zhang S. Q., Ye M. Y., Rao P. F. (2006). Major constituent proteins in donkey hide and their interaction. Zhongguo Zhong Yao Za Zhi 31 (8), 659–663. [PubMed] [Google Scholar]
- Li Z., Kawasumi M., Zhao B., Moisyadi S., Yang J. (2010). Transgenic over-expression of growth differentiation factor 11 propeptide in skeleton results in transformation of the seventh cervical vertebra into a thoracic vertebra. Mol. Reprod. Dev. 77 (11), 990–997. 10.1002/mrd.21252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B. K., Oates A. C. (2017). Delta-Notch signalling in segmentation. Arthropod Struct. Dev. 46 (3), 429–447. 10.1016/j.asd.2016.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Yue J., Niu N., Liu X., Yan H., Zhao F., et al. (2020). Genome-wide association analysis identified BMPR1A as a novel candidate gene affecting the number of thoracic vertebrae in a Large White × Minzhu intercross pig population. Animals. 10 (11), E2186. 10.3390/ani10112186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Gao Q., Wang T., Chai W., Zhan Y., Akhtar F., et al. (2022). Multi-thoracolumbar variations and NR6A1 gene polymorphisms potentially associated with body size and carcass traits of Dezhou donkey. Animals. 12 (11), 1349. 10.3390/ani12111349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins T. F., Braga Magalhães A. F., Verardo L. L., Santos G. C., Silva Fernandes A. A., Gomes Vieira J. I., et al. (2022). Functional analysis of litter size and number of teats in pigs: From GWAS to post-GWAS. Theriogenology 193, 157–166. 10.1016/j.theriogenology.2022.09.005 [DOI] [PubMed] [Google Scholar]
- Mikawa S., Hayashi T., Nii M., Shimanuki S., Morozumi T., Awata T. (2005). Two quantitative trait loci on Sus scrofa chromosomes 1 and 7 affecting the number of vertebrae. J. Anim. Sci. 83 (10), 2247–2254. 10.2527/2005.83102247x [DOI] [PubMed] [Google Scholar]
- Mikawa S., Morozumi T., Shimanuki S., Hayashi T., Uenishi H., Domukai M., et al. (2007). Fine mapping of a swine quantitative trait locus for number of vertebrae and analysis of an orphan nuclear receptor, germ cell nuclear factor (NR6A1). Genome Res. 17 (5), 586–593. 10.1101/gr.6085507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikawa S., Sato S., Nii M., Morozumi T., Yoshioka G., Imaeda N., et al. (2011). Identification of a second gene associated with variation in vertebral number in domestic pigs. BMC Genet. 12, 5. 10.1186/1471-2156-12-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Roychoudhury A. K. (1974). Sampling variances of heterozygosity and genetic distance. Genetics 76 (2), 379–390. 10.1093/genetics/76.2.379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Lee Y. J., Lee H. J., Seki T., Hong K. H., Park J., et al. (2004). B-cell translocation gene 2 (Btg2) regulates vertebral patterning by modulating bone morphogenetic protein/Smad signaling. Mol. Cell. Biol. 24 (23), 10256–10262. 10.1128/MCB.24.23.10256-10262.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polidori P., Pucciarelli S., Ariani A., Polzonetti V., Vincenzetti S. (2015). A comparison of the carcass and meat quality of Martina Franca donkey foals aged 8 or 12 months. Meat Sci. 106, 6–10. 10.1016/j.meatsci.2015.03.018 [DOI] [PubMed] [Google Scholar]
- Polidori P., Vincenzetti S., Cavallucci C., Beghelli D. (2008). Quality of donkey meat and carcass characteristics. Meat Sci. 80 (4), 1222–1224. 10.1016/j.meatsci.2008.05.027 [DOI] [PubMed] [Google Scholar]
- Portanova P., Notaro A., Pellerito O., Sabella S., Giuliano M., Calvaruso G. (2013). Notch inhibition restores TRAIL-mediated apoptosis via AP1-dependent upregulation of DR4 and DR5 TRAIL receptors in MDA-MB-231 breast cancer cells. Int. J. Oncol. 43 (1), 121–130. 10.3892/ijo.2013.1945 [DOI] [PubMed] [Google Scholar]
- Rauf B., Irum B., Khan S. Y., Kabir F., Naeem M. A., Riazuddin S., et al. (2020). Novel mutations in LTBP2 identified in familial cases of primary congenital glaucoma. Mol. Vis. 26, 14–25. [PMC free article] [PubMed] [Google Scholar]
- Ren D. R., Ren J., Ruan G. F., Guo Y. M., Wu L. H., Yang G. C., et al. (2012). Mapping and fine mapping of quantitative trait loci for the number of vertebrae in a White Duroc × Chinese Erhualian intercross resource population. Anim. Genet. 43 (5), 545–551. 10.1111/j.1365-2052.2011.02313.x [DOI] [PubMed] [Google Scholar]
- Rijli F. M., Matyas R., Pellegrini M., Dierich A., Gruss P., Dollé P., et al. (1995). Cryptorchidism and homeotic transformations of spinal nerves and vertebrae in Hoxa-10 mutant mice. Proc. Natl. Acad. Sci. U. S. A. 92 (18), 8185–8189. 10.1073/pnas.92.18.8185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statistics C. B. o. (2021). China statistical yearbook. Beijing, China: China Statistics Press. [Google Scholar]
- Thawait G. K., Chhabra A., Carrino J. A. (2012). Spine segmentation and enumeration and normal variants. Radiol. Clin. North Am. 50 (4), 587–598. 10.1016/j.rcl.2012.04.003 [DOI] [PubMed] [Google Scholar]
- Wang C., Li H., Guo Y., Huang J., Sun Y., Min J., et al. (2020). Donkey genomes provide new insights into domestication and selection for coat color. Nat. Commun. 11 (1), 6014. 10.1038/s41467-020-19813-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Ran X., Niu X., Huang S., Li S., Wang J. (2022). Whole-genome sequence analysis reveals selection signatures for important economic traits in Xiang pigs. Sci. Rep. 12 (1), 11823. 10.1038/s41598-022-14686-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellik D. M. (2007). Hox patterning of the vertebrate axial skeleton. Dev. Dyn. 236 (9), 2454–2463. 10.1002/dvdy.21286 [DOI] [PubMed] [Google Scholar]
- Yang J., Huang L., Yang M., Fan Y., Li L., Fang S., et al. (2016). Possible introgression of the VRTN mutation increasing vertebral number, carcass length and teat number from Chinese pigs into European pigs. Sci. Rep. 6, 19240. 10.1038/srep19240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvon S., Olier M., Leveque M., Jard G., Tormo H., Haimoud-Lekhal D. A., et al. (2018). Donkey milk consumption exerts anti-inflammatory properties by normalizing antimicrobial peptides levels in Paneth's cells in a model of ileitis in mice. Eur. J. Nutr. 57 (1), 155–166. 10.1007/s00394-016-1304-z [DOI] [PubMed] [Google Scholar]
- Zhang L. C., Yue J. W., Pu L., Wang L. G., Liu X., Liang J., et al. (2016). Genome-wide study refines the quantitative trait locus for number of ribs in a Large White × Minzhu intercross pig population and reveals a new candidate gene. Mol. Genet. Genomics 291 (5), 1885–1890. 10.1007/s00438-016-1220-1 [DOI] [PubMed] [Google Scholar]
- Zhang X., Li C., Li X., Liu Z., Hu S., Cao Y., et al. (2019). Association analysis of polymorphism in the NR6A1 gene with the lumbar vertebrae number traits in sheep. Genes Genomics 41 (10), 1165–1171. 10.1007/s13258-019-00843-5 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Sun Y., Du W., He S., Liu M., Tian C. (2017). Effects of vertebral number variations on carcass traits and genotyping of Vertnin candidate gene in Kazakh sheep. Asian-Australas. J. Anim. Sci. 30 (9), 1234–1238. 10.5713/ajas.16.0959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zhan Y., Han Y., Liu Z., Wang Y., Wang C. (2021). Estimation of liveweight from body measurements through best fitted regression model in Dezhou donkey breed. J. Equine Vet. Sci. 101, 103457. 10.1016/j.jevs.2021.103457 [DOI] [PubMed] [Google Scholar]
- Zhao F., Deng T., Shi L., Wang W., Zhang Q., Du L., et al. (2020). Genomic scan for selection signature reveals fat deposition in Chinese indigenous sheep with extreme tail types. Animals. 10 (5), E773. 10.3390/ani10050773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou M., Zou J., Hu X., Zheng W., Zhang M., Cheng Z. (2021). Latent transforming growth factor-β binding protein-2 regulates lung fibroblast-to-myofibroblast differentiation in pulmonary fibrosis via NF-κB signaling. Front. Pharmacol. 12, 788714. 10.3389/fphar.2021.788714 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The name of the repository and accession number can be found at: NCBI; PRJNA878942.