Abstract
Proteinases of Staphylococcus aureus are emerging as potential virulence factors which may be involved in the pathogenecity of staphylococcal diseases. We describe here the structure of the gene encoding the metalloproteinase referred to as aureolysin. This gene occurs in two allelic forms and is strongly conserved among S. aureus strains, implying the possibility that the proteinase may have important housekeeping functions.
Staphylococcus aureus is a major human pathogen of increasing importance because of its rapid development of antibiotic resistance. Due to the possession of a broad array of potential virulence factors, including several toxins, adhesins, and exoenzymes, this organism is able to colonize a broad range of tissues in the host and, as a result, cause many grave infections (9). Recently, a renewed interest has been focused on proteinases from S. aureus which are under the control of agr (accessory gene regulator) and sar (staphylococcal accessory regulator), the main regulators of S. aureus virulence determinant genes (3, 4, 8). In this case, the expression of a specific serine glutamyl endopeptidase (V8 protease) was found to be important for the full display of bacterial virulence in animal models of staphylococcal infection (5).
In addition to the glutamyl endopeptidase, S. aureus secretes at least one papain-like cysteine proteinase as well as a typical metalloproteinase that is commonly referred to as aureolysin. The primary and tertiary structures of the latter enzyme have been determined (1), revealing a polypeptide chain of 301 amino acids which is folded into a β-pleated N-terminal domain and an α-helical C-terminal domain, a typical fold for the thermolysin family of metalloproteinases. This diverse family of proteinases also encompasses several enzymes which are acknowledged virulence factors, including Pseudomonas aeruginosa elastase, Legionella pneumophila and Listeria monocytogenes metalloproteinases, Vibrio cholerae hemagglutinin protease, Staphylococcus epidermidis elastase, and the lambda toxin of Clostridium perfringens. In contrast to these proteinases, however, little is known about the exact role of aureolysin in the pathogenicity of S. aureus. In vitro, aureolysin has been shown to cleave the plasma proteinase inhibitors, α1-antichymotrypsin and α1-proteinase inhibitor (13, 14), and to activate prothrombin in human plasma (20). It may also affect the stimulation of T and B lymphocytes by polyclonal activators and display inhibitory activity against immunoglobulin production by lymphocytes (15). In addition, aureolysin activates the precursor of the glutamyl endopeptidase (V8 protease) secreted by this same microorganism (6).
In order to cast more light on the potential importance of staphylococcal proteinases in pathogenicity, we have focused the present study on the genetic analysis of aureolysin, including distribution, copy number, and genetic variability of the aureolysin gene in both clinical isolates and laboratory strains of S. aureus.
The PCR technique was first applied to clone a fragment of the S. aureus aureolysin gene by using the genomic DNA of the S. aureus strain V8-BC10 and the oligodeoxynucleotides 5′-TGGATTGGTGATAAAATGAT-3′ and 5′-ACTTCATTCCATGCTTCATA-3′. These were chosen on the basis of reverse translation of the sequences WIGDKMI and YEAWNEV at positions 117 to 123 and 292 to 298, respectively, in the mature form of the V8-BC10 aureolysin (1) and taking into account the codon usage bias of S. aureus (11). PCR was performed for a total of 30 cycles of 94°C for 1 min, 37°C for 1 min, and 72°C for 3 min by using the Expand High Fidelity System (Boehringer Mannheim, Mannheim, Germany). The resulting 550-bp amplicon was sequenced by using a Thermo Sequenase dye terminator cycle sequencing kit (Amersham Life Science, Cleveland, Ohio) and was analyzed with an Applied Biosystem DNA sequencer. The predicted amino acid sequence encoded by the fragment was in agreement with the published amino acid sequence of aureolysin (1).
The cloning of the remaining part of the aureolysin gene and flanking regions was achieved by inverse PCR. The genomic DNA was restricted with ClaI or NsiI (Boehringer Mannheim), the fragments produced were circularized with T4 DNA ligase (Boehringer Mannheim), and PCR was performed with two outward-pointing oligonucleotide primers, 5′-GTGGTGTGCATACGAATTCT-3′ and 5′-ACTCATGTGCTACTACGTCA-3′, complementary to sequences inside of the internal 550-bp fragment by using the following cycling parameters (30 cycles): 94°C for 1 min, 60°C for 1 min, and 72°C for 6 min. The resulting PCR products of 1.3 and 1.4 kb for the NsiI and ClaI inverse PCRs, respectively, were cloned into the pUC19 vector and sequenced. Analysis revealed that the complete C-terminal region of the aureolysin gene was included in the ClaI inverse PCR product, whereas the N terminus was lacking in both inverse PCR products. In order to obtain the full-length aureolysin gene, a third inverse PCR with SfuI-digested, ligated chromosomal DNA was conducted, resulting in the amplification of a 5.5-kb fragment. A 1.3-kb BclI internal fragment of this was subcloned into pUC19 and was sequenced. The combined sequence information revealed a coding sequence (CDS) of 1,527 nucleotides starting with GTG with the potential of encoding the aureolysin preproenzyme of 509 amino acids with a molecular mass of 56,321 Da and the putative ribosomal binding site, AGGAGG, eight nucleotides upstream of the CDS. We propose to name this gene aur.
Figure 1 shows the alignment of the deduced amino acid sequences of aureolysin from six different strains of S. aureus (see below) and the sequence of metalloproteinase from S. epidermidis. By analogy to the S. epidermidis metalloproteinase (17), and in accordance with data on the mature aureolysin (1), we propose that the nascent translation product of the aur gene includes an N-terminal signal peptide of 27 amino acids with a typical signal peptidase cleavage site (19), followed by a profragment and a mature proteinase composed of 181 and 301 residues, respectively. The prometalloproteinase activation apparently occurs by cleavage of the Glu208-to-Ala209 peptide bond releasing a mature aureolysin with a calculated molecular mass of 33,306 Da. A comparison of the aureolysin sequence inferred from the gene structure to that obtained by direct sequencing on the protein level revealed five differences that are likely due to errors in the amino acid sequencing of the protein (1).
FIG. 1.
Sequence comparison of metalloproteinases from S. aureus and S. epidermidis. The preliminary sequences for strains 8325 and COL were obtained from University of Oklahoma and TIGR, respectively; S. aureus strains 022, V8-BC10, 017, and 01 were obtained from this study (EMBL accession no. AJ249167, AJ249166, AJ249169, and AJ249168, respectively; and S. epidermidis was obtained as described in reference 17. A gap, indicated by a dot, was introduced by the program PILEUP (Wisconsin Package; Genetics Computer Group, Madison, Wis.) in order to obtain maximal alignment. Black shading indicates identical amino acids. The arrows marked above and below the alignment of the deduced amino acid sequences indicate the predicted sites of cleavage by signal peptidase for S. aureus and S. epidermidis, respectively. The cleavage site in the proenzyme is shown by a double arrow. Asterisks indicate the putative zinc-binding sequence.
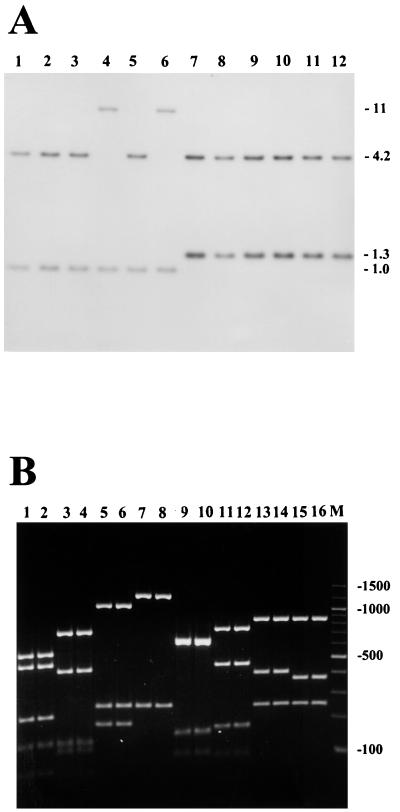
To assay for the distribution and copy number of the aur gene among S. aureus strains, 53 strains isolated from healthy as well as diseased persons in southern Poland from 1996 to 1998 were included in this study. Southern blot analysis was performed on total DNA from S. aureus strains digested with restriction enzymes SpeI, ClaI, and BclI. Endonuclease SpeI does not cleave within the S. aureus V8-BC10 aur gene sequence, whereas ClaI and BclI each recognize a single site within the gene. To detect DNA sequences homologous to the aur gene, a 1.4-kb NsiI fragment encoding amino acids 8 to 482 in the preproform of aureolysin was used as a DNA probe (Fig. 1). For all strains, a single hybridizing SpeI fragment was found, whereas ClaI and BclI each resulted in two bands upon autoradiography (representative results for BclI are shown in Fig. 2A). Thus, the aur gene is a single-copy gene and, apparently, is widespread among S. aureus strains isolated from humans.
FIG. 2.
Conservation of the aur gene among S. aureus strains. (A) Southern blot of BclI-digested chromosomal DNA of 12 S. aureus strains probed with a 1.4-kb fragment of the aureolysin gene. Lanes 1 to 6, strain N75, N66, A12, A9, 017, and 01 representatives of aureolysin type II; lanes 7 to 12, strain N72, N68, A11, A3, 022, and V8-BC10 representatives of aureolysin type I. Molecular size markers (in kilobases) are indicated to the right. (B) Agarose gel electrophoresis of PCR-amplified aureolysin gene products from genomic DNA of representative S. aureus strains. Lanes 1 and 2, representatives of aureolysin-type I digested with ApoI; lanes 3 and 4, representatives of aureolysin type II digested with ApoI; lanes 5 and 6, representatives of aureolysin type I digested with HincII; lanes 7 and 8, representatives of aureolysin type II digested with HincII; lanes 9 and 10, representatives of aureolysin type I digested with HinfI; lanes 11 and 12, representatives of aureolysin type II digested with HinfI; lanes 13 and 14, representatives of aureolysin type I digested with NdeI; lanes 15 and 16, representatives of aureolysin type II digested with NdeI; M, 100-bp ladder.
To further investigate the degree of conservation of the aureolysin gene, PCR-restriction fragment length polymorphism (RFLP) was applied. Primers for PCR amplification were designed by selecting sequences conserved between the published metalloproteinase structures as determined from S. epidermidis (17) and the aureolysin gene cloned in this work. The nucleotide sequences of primers were forward primer 5′-AATGTGAGGAAATTTTCAAGAT-3′ and reverse primer 5′-CCATGCTTCATATACTTGTTC-3′, with PCR consisting of 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 3 min, resulting in amplicons of 1.5 kb. Based on digestion of the amplified fragments with four different restriction enzymes, ApoI, HincII, HinfI, and NdeI, followed by agarose gel electrophoresis, we detected only two distinct restriction patterns, designated I and II, respectively (Fig. 2B). The presence of only two types of the aur gene is not caused merely by a very limited number of clonal types in the collection of strains analyzed, since we noted that the fingerprints of the strains differed considerably in the genomic DNA digests for Southern blot analysis (data not shown). The PCR-RFLP pattern I contained 16 isolates, including 7 from the nasal passages of healthy individuals, 4 from patients with furuncles, and 5 from patients with atopic dermatitis, while pattern II contained 37 isolates, including 7 from healthy individuals, 13 from patients with furuncles, and 17 from patients with atopic dermatitis. This may suggest that type II is more often isolated from skin diseases than type I, although the differences observed are not statistically significant (Fisher exact test, P = 0.09).
In order to compare the aureolysin encoded by the two PCR-RFLP types of the aur gene, strain 022 of type I and strains 01 and 017 of type II were selected for sequencing of the aur gene. During this work, preliminary results from two S. aureus genome sequencing projects were released, strain COL (isolated in England in 1965) from The Institute for Genomic Research (TIGR) and strain 8325 (isolated in England in the 1950s) from the University of Oklahoma. Each of these genome teams has released sequences which are homologous to the aur gene, and these data together with those obtained for the strain V8 were used to design primers for PCR amplification of the aureolysin gene from the additional three strains selected for sequencing (sequence data for COL and NCTC 8325 were obtained from the TIGR website [http://www.tigr.org] and from the University of Oklahoma website [http://www.genome.ou.edu], respectively). Similar to that of the V8 strain, all five aur genes contained a CDS starting with GTG and with the potential of encoding an aureolysin preproenzyme of 509 amino acids (Fig. 1). The deduced amino acid sequences of the open reading frames were almost identical within each of the two aur PCR-RFLP types, whereas homology at the gene level between the two types was only 89% (Fig. 3). The CDS of the two strains COL and 8325 were identical and were closely related to that of the type II strains. It remains to be examined whether these differences are reflected in differences in biochemical properties such as enzymatic specificity and activity.
FIG. 3.
Sequences of polymorphic sites within and 100 nucleotides up- and downstream of the four S. aureus aur genes analyzed in this study. The strains included were V8-BC10 (aureusV8) and 022 (aureus022) of PCR-RFLP type I and 01 (aureus01) and 017 (aureus017) of PCR-RFLP type II. The first G in the GTG start codon is assigned number 1 and the TTA stop codon ending the CDS is at positions 1528 to 1530.
Taken together, the data presented in this report indicate that the structure of the aureolysin gene is strongly conserved among S. aureus strains. This argues in favor of the likelihood that the enzyme may have an important housekeeping function in the proteolytic processing of other staphylococcal proteins, including the glutamyl endopeptidase (6), lipase (16), and autolysin (7). In addition, aureolysin may participate in modification of bacterial surface proteins, including clamping factor B (12), as has been found with a streptococcal cysteine proteinase (streptopain) (2) and S. aureus serine proteinase (10). It may also exert a direct effect on receptor function on the surface of host cells, as staphylococcal metalloproteinase activity has previously been implicated in shedding of the interleukin-6 receptor from human cells (18). In this respect, knowledge of the structure of the aureolysin gene may be valuable for future experiments aimed at delineating the pathophysiological function of the expressed enzyme, as well as mechanisms regulating the proteinase production.
Nucleotide sequence accession number.
The sequence of aur has been deposited in the EMBL Data Library under accession no. AJ249166.
Acknowledgments
We thank Antoine Danchin (Institut Pasteur, Paris, France) for helpful discussion and kind support and Renata Filipek and Wojciech Strzalka (Institute of Molecular Biology, Jagiellonian University, Krakow, Poland) for assistance during the course of this study. Sequence data for COL and NCTC 8325 were obtained from TIGR and the University of Oklahoma, respectively.
This work was supported by grant 6 PO4A 051 16 from the Committee of Scientific Research (KBN, Warsaw, Poland) (to K.K.) and by grants from the National Institutes of Health (to J.T.).
REFERENCES
- 1.Banbula A, Potempa J, Travis J, Fernandez-Catalan C, Mann K, Huber R, Bode W, Medrano F J. Amino-acid sequence and three-dimensional structure of the Staphylococcus aureus metalloproteinase at 1.72 Å resolution. Structure. 1998;6:1185–1193. doi: 10.1016/s0969-2126(98)00118-x. [DOI] [PubMed] [Google Scholar]
- 2.Berge A, Bjorck L. Streptococcal cysteine proteinase releases biologically active fragments of streptococcal surface proteins. J Biol Chem. 1995;270:9862–9867. doi: 10.1074/jbc.270.17.9862. [DOI] [PubMed] [Google Scholar]
- 3.Chan P F, Foster S J. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J Bacteriol. 1998;180:6232–6241. doi: 10.1128/jb.180.23.6232-6241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chien Y, Cheung A L. Molecular interactions between two global regulators, sar and agr, in Staphylococcus aureus. J Biol Chem. 1998;273:2645–2652. doi: 10.1074/jbc.273.5.2645. [DOI] [PubMed] [Google Scholar]
- 5.Coulter S N, Schwan W R, Ng E Y, Langhorne M H, Ritchie H D, Westbrock-Wadman S, Hufnagle W O, Folger K R, Bayer A S, Stover C K. Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol Microbiol. 1998;30:393–404. doi: 10.1046/j.1365-2958.1998.01075.x. [DOI] [PubMed] [Google Scholar]
- 6.Drapeau G R. Role of a metalloprotease in activation of the precursor of the staphylococcal protease. J Bacteriol. 1978;136:607–613. doi: 10.1128/jb.136.2.607-613.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foster S J. Molecular characterization and functional analysis of the major autolysin of Staphylococcus aureus 8325/4. J Bacteriol. 1995;177:5723–5725. doi: 10.1128/jb.177.19.5723-5725.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janzon L, Arvidson S. The role of the delta-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 1990;9:1391–1399. doi: 10.1002/j.1460-2075.1990.tb08254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lowy F D. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 10.McGavin M J, Zahradka C, Rice K, Scott J E. Modification of the Staphylococcus aureus fibronectin binding phenotype by V8 protease. Infect Immun. 1997;65:2621–2628. doi: 10.1128/iai.65.7.2621-2628.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from the international DNA sequence databases. Nucleic Acids Res. 1998;26:334. doi: 10.1093/nar/26.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ni Eidhin D, Perkins S, Francois P, Vaudaux P, Hook M, Foster T J. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol Microbiol. 1998;30:245–257. doi: 10.1046/j.1365-2958.1998.01050.x. [DOI] [PubMed] [Google Scholar]
- 13.Potempa J, Fedak D, Dubin A, Mast A, Travis J. Proteolytic inactivation of α1-anti-chymotrypsin. Sites of cleavage and generation of chemotactic activity. J Biol Chem. 1991;266:21482–21487. [PubMed] [Google Scholar]
- 14.Potempa J, Watorek W, Travis J. The inactivation of human α1-proteinase inhibitor by proteinases from Staphylococcus aureus. J Biol Chem. 1986;261:14330–14334. [PubMed] [Google Scholar]
- 15.Prokesova L, Porwit-Bóbr Z, Baran K, Potempa J, Pospisil M, John C. Effect of metalloproteinase from Staphylococcus aureus on in vitro stimulation of human lymphocytes. Immunol Lett. 1991;27:225–230. doi: 10.1016/0165-2478(91)90156-5. [DOI] [PubMed] [Google Scholar]
- 16.Rollof J, Normark S. In vivo processing of Staphylococcus aureus lipase. J Bacteriol. 1992;174:1844–1847. doi: 10.1128/jb.174.6.1844-1847.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teufel P, Gotz F. Characterization of an extracellular metalloprotease with elastase activity from Staphylococcus epidermidis. J Bacteriol. 1993;175:4218–4224. doi: 10.1128/jb.175.13.4218-4224.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vollmer P, Walev I, Rose-John S, Bhakdi S. Novel pathogenic mechanism of microbial metalloproteinases: liberation of membrane-anchored molecules in biologically active form exemplified by studies with the human interleukin-6 receptor. Infect Immun. 1996;64:3646–3651. doi: 10.1128/iai.64.9.3646-3651.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wegrzynowicz Z, Heczko P B, Drapeau R G, Jeljaszewicz J, Pulverer G. Prothrombin activation by a metalloprotease from Staphylococcus aureus. J Clin Microbiol. 1990;12:138–139. doi: 10.1128/jcm.12.2.138-139.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]