Abstract
Background and Objectives:
Phage therapy has gained interest as an alternative treatment for methicillin-resistant Staphylococcus aureus (MRSA) infections. The purpose of this study was to isolate and characterize an effective bacteriophage against isolates of MRSA.
Materials and Methods:
Bacteriophage was isolated from hospital sewage. Lytic activity and the titers of phage lysates were measured using spot test and double-layer plaque assay. The phage characterization was determined through transmission electron microscopy. Adsorption rate, host range and stability tests were investigated. The latent period and burst size were estimated from a one-step growth curve. The effect of bacteriophage against MRSA biofilms was determined and Real-time PCR was used to assess the effects of the bacteriophage on the expression of the biofilm-associated genes.
Results:
TEM results showed that the phage resembled the Cystoviridae family. Its latent period was 30 min, corresponding to about 71/43 phage particles per infected cell. The phage had a broad host range and it was most stable at 37°C and pH 7. It was sensitive to NaCl concentrations. The expressions of the biofilm-associated genes were significantly reduced in the presence of the phage.
Conclusion:
The isolated phage was effective against MRSA strains and it can be an optional strategy of controlling biofilm development.
Keywords: Bacteriophage, Methicillin-resistant Staphylococcus aureus, Phage therapy, Wounds
INTRODUCTION
Despite the fact that antibiotic resistance in bacteria is a natural phenomenon, the alarming increase in pathogenic bacteria refractory to various antimicrobials is attracting attention worldwide. Indeed, the World Health Organization (WHO) has recently reported a list of priority pathogens for which novel antimicrobial alternatives are urgently required. Among these pathogens, methicillin-resistant Staphylococcus aureus (MRSA) strains are perhaps the best known (1). Methicillin-resistant staphylococci were first reported in 1961 immediately after the introduction of methicillin into clinical practice. Until the late 1990s, MRSA was basically a nosocomial pathogen; however, over the last decade, methicillin-resistant staphylococci have been considered as the major cause of community-acquired infections (2). They are responsible for several diseases ranging from soft tissue and skin infections to life threatening conditions such as pneumonia, bacteremia and endocarditis especially in patients > 65 years old (3).
Methicillin resistance is mediated by mecA gene and acquired by horizontal transfer of a mobile genetic element designated staphylococcal cassette chromosome mec (SCCmec). The gene mecA encodes penicillin-binding protein 2a (PBP2a) which has a low affinity for β-lactams resulting in resistance to this entire class of antibiotics (4).
Staphylococcus aureus produces many virulence factors including toxins, immune-modulatory factors, and exoenzymes. The most important virulence factor of this bacterium is the capacity of biofilm formation. S. aureus biofilm is mediated by the icaAD-BC operon, agr locus, and other genes which can express a variety of microbial surface components. For example, polysaccharide intercellular adhesin (PIA), which is encoded by the icaADBC operon, represents more than 90% of the biomass of a biofilm (5).
Phage therapy, the application of bacteriophage viruses to treat bacterial infections, has existed for more than a hundred years (6). Due to its high specificity and effectiveness (specially against multi drug resistant bacteria), it may be a suitable alternative to antibiotic treatment (7). In the last 15 years, there has been a significant increase in the number of isolated Staphylococcus phages and many studies have indicated efficient and comprehensive antimicrobial activity of phages in vitro and in vivo (8).
Considering all the above information, this study aimed to isolate and characterize a lytic bacteriophage from hospital sewage with antibacterial activity against MRSA strains causing bedsores and diabetic wounds. Furthermore, the effects of the bacteriophage on the expression of the biofilm-associated genes were investigated.
MATERIALS AND METHODS
Bacteriophage isolation and enrichment. Bacteriophage was isolated from Mofid Children’s Hospital sewage in September 2020 in Tehran, Iran. The sample was transported to the laboratory and stored at 4°C. In order to remove or reduce bacteria and debris, 10 ml of sample was centrifuged (10, 000 × g for 10 min) and filtered through 0.22 μm pore size filters. To amplify the number of phages, clarified sewage was mixed with 10 ml of Luria–Bertani (LB) broth (Thermo Fisher Scientific, US). Then, 0.1 ml of an overnight broth culture MRSA strain ATCC 6538 was inoculated into the mixture and the flask was incubated at 37°C with gentle mixing in shaking incubator (50 rpm). After 24 h incubation, the content of the flask was centrifuged and filtered again (9, 10). Lytic activity of isolated phage was determined with a spot test, and the titers of phage lysates were measured using Double-Layer Plaque Assay (10).
Spot test assay. An overnight culture of the MRSA strain was mixed with 3 ml of LB soft agar (0.75% agar at 45°C), and the contents were poured into a petri dish containing 15 ml of 1% LB bottom agar. The plate was left to dry for a few minutes, and then 10 μl of the filtered supernatant was poured over the solidified soft agar. After it had been absorbed, the plate was incubated at 37°C overnight. The following day, the plate was checked for zones of clearing (11, 12).
Double-layer plaque assay (DLA assay). In order to measuring the titers of bacteriophage, the recovered supernatant was serially diluted in the LB broth medium. Then, 0.1 ml of the diluted phage and 0.5 ml of MRSA ATCC 6538 (adjusted to 0.5 Mc-Farland standard) were added to 3 ml soft LB agar (0.75% agar at 45°C). The mixture was then layered onto a LB agar plate and allowed to solidify before incubation at 37°C overnight. Plaque-forming units (PFU) were calculated per milliliter by determining the number of plaques×10×the inverse of the dilution factor (9, 11).
Phage characterization by transmission electron microscopy (Phage Morphology). A high titer phage preparation was deposited on a formvar-carbon coated grid Cu Mesh 300, fixed with 1% glutaraldehyde, stained with the standard negative staining using 2% uranyl acetate, and examined by an EM 208S (Zeiss, Germany) transmission electron microscope at 100 kV (8, 13).
Determination of optimal multiplicity of infection (MOI). Multiplicity of infection (MOI) is the ratio of phages added to bacteria (14). The MRSA strain was grown in LB broth at 37°C until early logarithmic growth phase (optical density at 625 nm, 0.08–0.1) corresponding to around 108 CFU. Bacterial cultures (1 ml) were inoculated with 109, 108, and 107 PFUs at the MOIs 10, 1 and 0.1, respectively. Bacterial growth was monitored by recording OD 625 nm at 30 min intervals up to 240 min (8, 15).
One-step growth curve. One-step growth curve of the isolated phage was performed using the method of Wang et al. (2016) with some modifications. Briefly, isolated phage at a MOI of 10 was added to the cells of MRSA and allowed to adsorb for 15 min at 37°C. The mixture was then centrifuged at 10,000 × g for 1 min. After the supernatants were removed, the pellets containing the phage-infected bacterial cells were suspended in fresh LB broth and incubated with shaking at 180 rpm and 37°C. Partial samples were obtained at 10 min intervals and the titrations from the aliquots were immediately done using the double-layer agar method (8). The latent period was estimated from a one-step growth curve and the burst size was measured by dividing number of phages formed during rise period with the estimated number of infected cells present at the latent period time (16).
Phage adsorption assay. The adsorption rate of the phage to the host bacterium was determined by adding 1 ml (107 PFU) of phage to 9 ml of overnight-cultured host bacterium (adjusted to 0.5 McFarland standard). The phage-host mixture was kept at 37°C, and aliquots were collected at 5 and 10 minutes. Each aliquot was centrifuged at 8,000 g for 10 minutes to sediment the phages attached to the bacteria. The titer of the unabsorbed phages in supernatant was then measured with the double-layer agar method (12, 15).
Determination of host range. The host range of the isolated bacteriophage was determined against 20 clinical MRSA isolates and Staphylococcus aureus ATCC 25923. Wound exudate samples were collected from patients hospitalized at Loghman-e Hakim Hospital, Tehran, Iran from November of 2018 to January of 2019 and transferred to Pasteur Institute of Iran. Isolates were diagnosed as S. aureus following biochemical tests and they were confirmed as MRSA based on the mecA gene PCR amplification (17). For host range determination, Spot test was employed. Bacteria were grown in LB broth for 6–8 h at 37°C to reach the turbidity of 0.5 McFarland. Bacterial lawns were formed on LB plates, and 10 μl of phage suspensions were placed on the bacterial lawns. Plates were incubated at 37°C and observed after 24 h for phage-mediated lysis (12).
Phage stability tests. Thermal stability of the isolated phage was determined by incubation of phage suspension at −20, 4, 37, 50, 60 and 70°C for 1 h, followed by determining the number of phages by the double-layer agar method.
For pH stability tests, 10 μl of phage suspension was added to 0.99 ml of LB broth at a pH range of 2–12 and incubated at 37°C for 1 h before phage titration (12).
The stability of the free phage particles in a hyper saline environment was estimated by the incubation of 0.1 ml of phage in various concentrations (5%, 10%, and 15%) of NaCl (0.9 ml) for 1 h. Then, the titer of the active phage was determined by the double-layer agar method (18).
Effect of bacteriophage in biofilm degradation. Biofilm disruption was assayed using the biofilm plate assay as described previously (19, 20). An overnight culture of MRSA was diluted 1:100 in Trypticase Soy Broth (TSB, Merck, USA) and 0.2 ml of the suspension was transferred into the 96-well flat-bottomed polystyrene tissue culture microtiter plates. After 24 h of incubation at 37°C, the wells were emptied, washed three times with phosphate-buffered saline (PBS, pH 7), and treated with 0.1 ml of the phage at different dilutions (from 10−1 to 10−10). Sterile physiological saline without phages served as a negative control. After 24 h of exposure at 37°C, the wells were washed three times with PBS and the persisting biofilm layer was stained with Triphenyl Tetrazolium Chloride (TTC, Sigma. USA) and detected with a Nano Drop device by measuring absorbance at 595 nm. This method was used to evaluate the effect of phage on 3-day and 5-day old preformed MRSA biofilms (19, 20). The biofilm inhibition percentage was calculated according to the following equation: Percentage inhibition = 100 − [{adsorption of the most effective concentration of phage / adsorption of control} × 100] (21).
Effect of bacteriophage in expression levels of biofilm-associated genes quantified by Quantitative Real-Time PCR: RNA extraction and cDNA synthesis. For the establishment of biofilm, 3 flasks containing 10 ml TSB and 0.3 g of sterile glass wool which served as a surface for attachment were used (one flask for negative control, one for positive control and one flask for phage-treated sample). In the next step, 0.1 ml of MRSA ATCC 6538 (adjusted to 0.5 McFarland standard) was added to each flask. Then, 2 of 3 flasks were incubated for 24 h at 37°C (positive control and phage-treated sample), and 1 of 3 flasks was incubated for 24 h at 37°C with mixing (100 rpm) to prevent the formation of biofilm (negative control). After 24 h, samples were washed twice by PBS. Next, 10 ml of the isolated phage was added to phage-treated sample flask and 10 ml TSB was added to each control flask. Finally, the flasks were incubated as before. After 24 h, samples were washed by PBS again and 10 ml PBS glass beads was added to each flask and shake vigorously by shaker incubator for 20 min to detach the cells attached to the glass wool. The liquid phase was removed and centrifuged at 9000 rpm for 15 min to pellet the detached cells. The pellet was resuspended in 1 ml PBS for RNA extraction (22, 23).
RNA extraction was done using a high-pure RNA isolation kit (DENAzist Asia, Iran) and the quantity and purity of the extracted RNA were measured by a Nano Drop device. Applying a cDNA synthesis kit (Takapou Zist, Iran), the extracted RNA was converted to cDNA based on the manufacturer’s instructions.
Quantitative real-time PCR. The mRNA of the studied genes was quantified with the ABI Step One Plus detection system (Applied Biosystems, USA) using the SYBR Green master mix (Amplicon Bio, Denmark). All reaction tubes contained 2 μl of the cDNA, 0.5 μl of each of forward and reverse primers, 10 μl SYBR green PCR master mix and 7 μl DEPC water. The reaction was started with an initial denaturation at 95°C for 5 min and 40 amplification cycles of 94°C for 20s, 60°C to 62°C (Annealing temperature of icaA, icaD and 16S rRNA were 62°C, 60°C and 60°C, respectively) for 20s and 72°C for 20s. All the reactions were performed in duplicate (Table 1). The formula RQ= 2−ΔΔCt was used to get relative gene expression in the comparative CT method. In this part, 16S rRNA was used as an internal control (24).
Table 1.
The sequences, length, and annealing temperature of primers used for qPCR.
| Target genes | Primer Sequence (5′ →3′) | Size (bp) | Annealing temperature (°C) | Ref |
|---|---|---|---|---|
| icaA | F: 5-GAGGTAAAGCCAACGCACTC-3 | 151 | 62 | |
| R: 5-CCTGTAACCGCACCAAGTTT-3 | ||||
| icaD | F: 5-ACCCAACGCTAAAATCATCG-3 | 211 | 60 | (24) |
| R: 5-GCGAAAATGCCCATAGTTTC-3 | ||||
| 16S rRNA | F: 5GGGACCCGCACAAGCGGTGG-3 | 191 | 60 | |
| R: 5-GGGTTGCGCTCGTTGCGGGA-3 |
Statistical analysis. GraphPad Prism 9 was used for statistical analyses. Due to the normality of the data, a one-way analysis of variance (ANOVA) test was utilized to compare means. P-value < 0.05 was considered statistically significant.
RESULTS
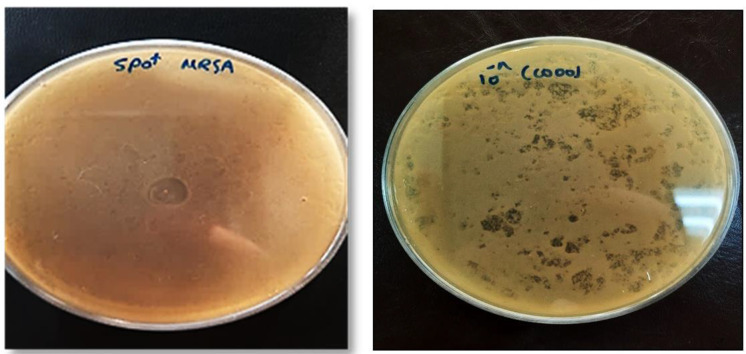
Bacteriophage isolation and enrichment. In the spot test, clear plaques were observed wherever phage lysate was spotted onto LB agar plates covered with a bacterial lawn of MRSA. Accordingly, it was revealed that the isolated phage had lytic activity (Fig. 1A). In the DLA technique, the lytic phage formed small clear plaques and the titer of the purified phage was 8 × 1010 PFU/mL−1 (Fig. 1B).
Fig. 1.
Spot test assay to check the presence of bacteriophage (A). The lytic phage formed small clear plaques (DLA assay) (B).
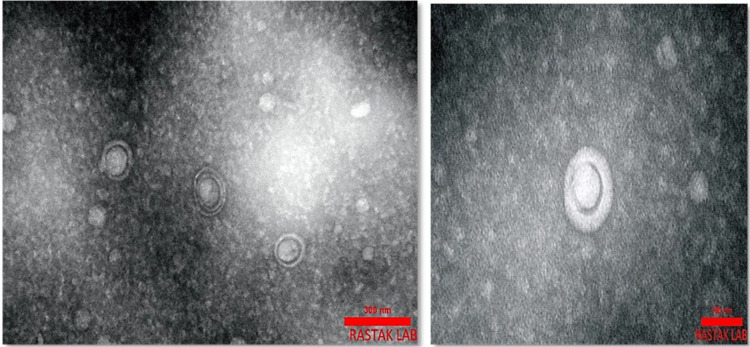
Phage characterization by transmission electron microscopy (Phage Morphology). The morphology of phage was investigated by TEM and is shown in Fig. 2. As the phage was spherical in shape with a lipid membrane around the capsomere, thus the phage can be classified into the Cystoviridae family.
Fig. 2.
Transmission Electron Microscopy of negatively-stained phage. It seemed that the isolated phage belonged to the Cystoviridae family.
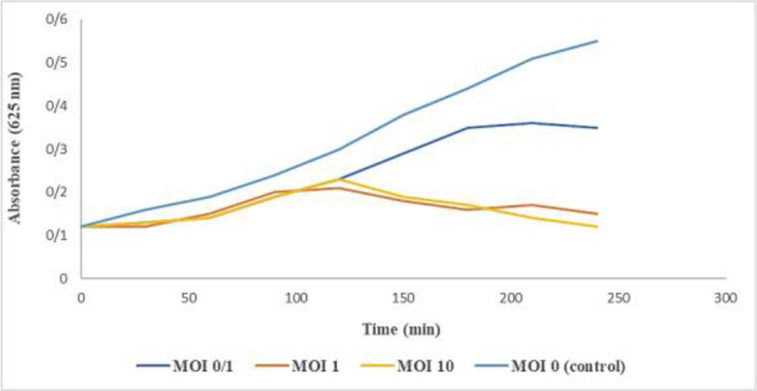
Determination of optimal multiplicity of infection (MOI). The MOI could be critical to the bacterial inactivation efficiency and it is necessary to determine the best MOI for phage therapy. As observed in Fig. 3, the reduction of pathogenic bacteria increased in parallel with MOI. all tested MOIs were effective against the host strain and the optimal MOI of phage was 10. Hence, we used the MOI 10 in further experiments.
Fig. 3.
The bacteriolytic activity of isolated phage at different MOIs (0/1, 1, and 10) against MRSA ATCC 6538 (OD 625 nm). The optimal MOI of phage was 10.
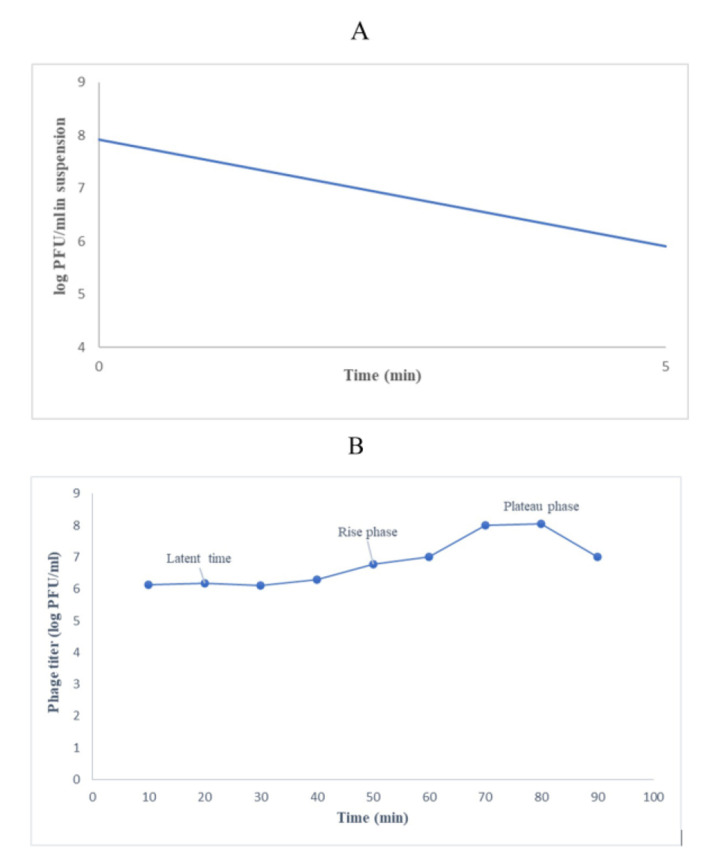
Phage adsorption assay and one-step growth curve. The phage particles were adsorbed to the bacterial host after 5 min (96%) (Fig. 4A). The one-step growth curve indicated a latent period of 30 min and a burst time of 70 min, corresponding to about 71/43 phage particles per infected cell (Fig. 4B).
Fig. 4.
Adsorption (A) and one-step growth curve of phage in MRSA ATCC 6538 (B). After 5 min, the phage exhibited rapid adsorption to the bacteria. The one-step growth curve indicated a latent period of 30 min and a burst time of 70 min, corresponding to about 71/43 phage particles per infected cell.
Determination of host range. The host range of the bacteriophage was determined against 20 clinical MRSA strains isolated from bedsore and diabetic wounds and Staphylococcus aureus ATCC 25923. This phage had a broad host range and infected 80% of clinical isolates of MRSA (16/20). It was also effective against Staphylococcus aureus ATCC 25923.
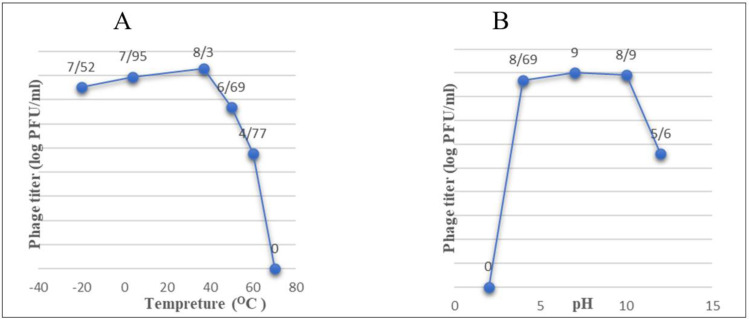
Phage stability tests. The high-level activity of the phage was observed after 1 h of incubation at 37°C. Despite remarkable reductions in the titer of the phage, full inactivation was observed only at 70°C (Fig. 5A).
Fig. 5.
The stability of the phage at different temperatures (A) and pH (B). The phage was most stable at 37°C and pH 7.
The stability of the phage at different pH values (2, 4, 7, 10, and 12) is shown in Fig. 5B. The fewest changes in the titer of the phage were observed at pH values of 4 - 10. Beyond these values, the activity decreased dramatically, and full inactivation was observed at pH 2 (Fig. 5B).
The phage’s stability at different NaCl concentrations was investigated. The titration of bacteriophage in 5%, 10%, and 15% of NaCl were 8× 106 PFU/mL−1, 13× 105 PFU/mL−1 and 104 PFU/mL−1, respectively. By increasing the NaCl concentration, the rate of phage survival decreased and the highest decrease in phage titer was observed in 15% NaCl.
Effect of bacteriophage in biofilm degradation. Isolated phage was significantly effective against all types of biofilms (i.e., 24 h, 3-day and 5-day biofilms). Different dilutions of phage were examined and the adsorption of effective concentrations and negative control are reported in Table 2. The most effective dilutions against 24h, 3-day and 5-day biofilms were 10−9 (6.5×105 PFU/ml), 10−5 (8.3×107 PFU/ml) and 10−1 (8.7×109 PFU/ml), respectively.
Table 2.
The effect of isolated phage on biofilm structures (OD 595 nm).
| 24 h biofilm | 3-day biofilm | 5-day biofilm | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Control | Phage (6.5 × 105 PFU/ml) | Control | Phage (8.3 × 107 PFU/ml) | Control | Phage (8.7 × 109 PFU/ml) | |
| Biofilm formation (OD) | 2.331 | 0.653 | 1.777 | 0.299 | 1.774 | 0.653 |
| Biofilm inhibition (%) | 0.0 | 72 | 0.0 | 84 | 0.0 | 63 |
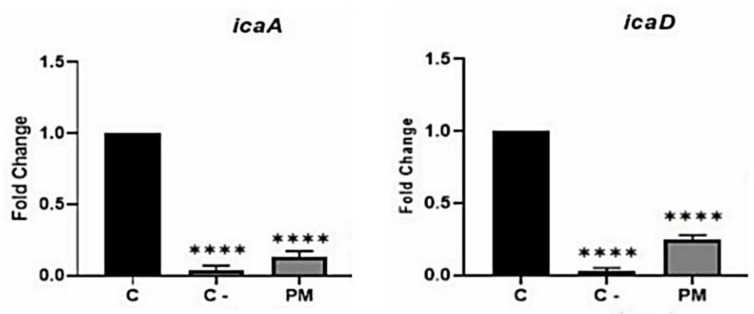
Effect of Bacteriophage in expression levels of biofilm-associated genes quantified by quantitative real-time PCR. Real-time PCR findings showed that in the presence of bacteriophage, the expressions of the biofilm-associated genes reduced remarkably. However, compared to the positive control, bacteriophage had a greater effect on the expressions of icaA. (Fig. 6, P<0.0001).
Fig. 6.
The effects of lytic bacteriophage on the expression of the genes responsible for biofilm formation by MRSA. Gene expressions before (C: positive control) and after treatment by phage (PM) have been shown ( P < 0.0001). Also, C- is a negative control in which biofilm formation was prevented.
DISCUSSION
The increasing prevalence of MRSA poses a serious threat to the world. Among the current attempts to solve this issue, phage therapy offers a promising alternative to control MRSA infections (12). In the present study, a lytic bacteriophage was isolated from the hospital sewage in Tehran which had a specific activity against MRSA. Based on TEM results and its comparison with other studies, the isolated phage can be classified into the Cystoviridae family. The genome of this family is double-stranded RNA and the virions are spherical in shape and they have a lipid membrane around the capsomere (25–27). Other studies reported different kinds of phages against MRSA based on TEM results (8, 12, 28). Rahimzadeh et al. (2021) isolated Cystoviridae phages from hospital sewage in Mazandaran, Iran, for instance (25).
Determining the optimal MOI, which is the optimal ratio of the number of phages per host cell, is necessary to produce the most phage product in the later stages of the experiment. The results showed that all tested MOIs (0/1, 1, and 10) were effective, and the optimal MOI of this phage was 10. The characteristic features of the isolated phage, i.e., the latent period of 30 min, burst time of 70 min, a burst size of 71/43 and the adsorption rate (96%) were similar to the phages reported by others (12, 28). The latent period indicated that the time needed to replicate the virus inside the host is very short and a new generation of phage will be propagated after 30 min. The amount of progeny obtained in this study was acceptable and it provided high concentrations needed for phage therapy. Moreover, the phage was adsorbed to the host bacterium very quickly, which is an advantage for antibacterial activity. All of these properties make the isolated phage a suitable candidate for treatment.
The isolated phage showed a broad range of lytic activity against MRSA and lysed 80% of MRSA clinical strains isolated from bedsore and diabetic wounds. Other studies have confirmed this result (8, 12, 28). For example, Peng et al. (2019) showed that the isolated lytic bacteriophage was able to infect 97% of healthcare- and community-associated MRSA strains (28). In general, phages with a wider host range and the capability of infecting more strains of the target bacterial species are more suitable for possible treatment. It is also best to combine phages with a narrow host range with other phages and use them as phage cocktails for treatment (12).
Due to various strains and types of structural proteins, different thermal, pH, and saline stabilities have already been reported for bacteriophages (8, 29). Therefore, it was interesting to investigate the stability of the phage under conditions mimicking wounds. In almost every case, the temperature of a wound increases by approximately 2°C when it is infected (30). According to our results, the isolated phage was stable in temperatures ranging from −20°C to 60°C (for 60 minutes). This finding indicated that low temperatures can be used for storing the phage. Furthermore, this phage can survive in temperatures ranging from 38°C to 60°C.
The results related to the effect of pH also demonstrated that the isolated phage had good lytic activity in the pH range of 4 to 10, and outside this range, the activity was severely reduced and no activity was observed at pH 2. Unlike the pH of healthy skin (which is between 4 and 6), the pH of wounds might be neutral or a little alkaline (7.15 - 8.9) (31). The optimal pH for the highest activity of the isolated phage was nearly the same as the pH of wounds.
In addition, the saline-tolerance experiment showed that the phage retained its activity in NaCl concentrations of up to 15%. As normal saline with the NaCl concentration of 0.9% is routinely used for wound cleaning, the results showed that the addition of the phage in normal saline could be a suitable way for applying the phage (32). Therefore, due to the stability of the isolated phage, it can be an appropriate candidate for the treatment of wound infections.
The isolated bacteriophage can greatly destroy the MRSA biofilm. Our results indicated that with increasing biofilm durability, low phage dilutions (in which the isolated phage concentration was higher) were more effective. Two other studies showed the effectiveness of phages on Staphylococcus aureus and MRSA biofilms (19, 20).
Normally, various enzymes and antibacterial agents can influence the expression and activity of the genes involved in biofilm formation by MRSA. For the first time, we considered the effects of the bacteriophage on the expression of the genes related to biofilm production by MRSA. Our results indicated that the lytic bacteriophage significantly reduced the expression of the assessed genes.
Generally, in this study, a lytic bacteriophage against MRSA belonging to the Cystoviridae family was isolated from hospital sewage. This phage had a broad host range and it is stable at different pH values, salt concentrations, and temperatures. Therefore, the bacteriophage can tolerate various environmental conditions and remain active. Moreover, this phage eradicated the biofilm structure and reduced the expressions of the biofilm-associated genes significantly. According to the study and its comparison with previous studies, isolated bacteriophage was effective on MRSA strains causing bedsores and diabetic wounds.
Other research is still necessary for the comprehensive molecular characterization of the bacteriophage. Also, its antibacterial activity in in vivo conditions is left to future work.
REFERENCES
- 1.Álvarez A, Fernández L, Gutiérrez D, Iglesias B, Rodríguez A, García P. Methicillin-Resistant Staphylococcus aureus in Hospitals : Latest Trends and Treatments Based on Bacteriophages. J Clin Microbiol 2019; 57(12): e01006–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borysowski J, Lobocka M, Międzybrodzki R, Weber-Dabrowska B, Górski A. Potential of bacteriophages and their lysins in the treatment of MRSA: current status and future perspectives. BioDrugs 2011; 25: 347–355. [DOI] [PubMed] [Google Scholar]
- 3.Noori Goodarzi N, Bolourchi N, Fereshteh S, Soltani Shirazi A, Pourmand MR, Badmasti F. Investigation of novel putative immunogenic targets against Staphylococcus aureus using a reverse vaccinology strategy. Infect Genet Evol 2021; 96: 105149. [DOI] [PubMed] [Google Scholar]
- 4.Turner NA, Sharma-Kuinkel BK, Maskarinec SA, Eichenberger EM, Shah PP, Carugati M, et al. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol 2019; 17: 203–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganesh PS, Veena K, Senthil R, Iswamy K, Ponmalar EM, Mariappan V, et al. Biofilm-associated Agr and Sar Quorum sensing systems of Staphylococcus aureus are inhibited by 3-Hydroxybenzoic acid derived from Illicium verum. ACS Omega 2022; 7: 14653–14665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abedon ST, Kuhl SJ, Blasdel BG, Kutter EM. Phage treatment of human infections. Bacteriophage 2011; 1: 66–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loc-Carrillo C, Abedon ST. Pros and cons of phage therapy. Bacteriophage 2011; 1: 111–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Zheng P, Ji W, Fu Q, Wang H, Yan Y, et al. SLPW: A Virulent Bacteriophage Targeting Methicillin-Resistant Staphylococcus aureus In vitro and In vivo. Front Microbiol 2016; 7: 934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nasser A, Azizian R, Tabasi M, Khezerloo JK, Heravi FS, Kalani MT, et al. Specification of bacteriophage isolated against clinical methicillin-resistant Staphylococcus aureus. Osong Public Health Res Perspect 2019; 10: 20–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Twest R, Kropinski AM. Bacteriophage enrichment from water and soil. Methods Mol Biol 2009; 501: 15–21. [DOI] [PubMed] [Google Scholar]
- 11.Sillankorva S. Isolation of bacteriophages for clinically relevant bacteria. Methods Mol Biol 2018; 1693: 23–30. [DOI] [PubMed] [Google Scholar]
- 12.Ji J, Liu Q, Wang R, Luo T, Guo X, Xu M, et al. Identification of a novel phage targeting methicillin-resistant Staphylococcus aureus In vitro and In vivo. Microb Pathog 2020; 149: 104317. [DOI] [PubMed] [Google Scholar]
- 13.Ackermann H-W. Basic phage electron microscopy. Methods Mol Biol 2009; 501: 113–126. [DOI] [PubMed] [Google Scholar]
- 14.Abedon ST. Phage therapy dosing: The problem(s) with multiplicity of infection (MOI). Bacteriophage 2016; 6(3): e1220348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drulis-Kawa Z, Mackiewicz P, Kęsik-Szeloch A, Maciaszczyk-Dziubinska E, Weber-Dąbrowska B, Dorotkiewicz-Jach A, et al. Isolation and characterisation of KP34--a novel ϕKMV-like bacteriophage for Klebsiella pneumoniae. Appl Microbiol Biotechnol 2011; 90: 1333–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nabergoj D, Modic P, Podgornik A. Effect of bacterial growth rate on bacteriophage population growth rate. Microbiologyopen 2018; 7(2): e00558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zafari M, Adibi M, Chiani M, Bolourchi N, Barzi SM, Shams Nosrati MS, et al. Effects of cefazolin-containing niosome nanoparticles against methicillin-resistant Staphylococcus aureus biofilm formed on chronic wounds. Biomed Mater 2021; 16: 035001. [DOI] [PubMed] [Google Scholar]
- 18.Seaman PF, Day MJ. Isolation and characterization of a bacteriophage with an unusually large genome from the Great Salt Plains National Wildlife Refuge, Oklahoma, USA. FEMS Microbiol Ecol 2007; 60: 1–13. [DOI] [PubMed] [Google Scholar]
- 19.Dvořáčková M, Růžička F, Benešík M, Pantůček R, Dvořáková-Heroldová M. Antimicrobial effect of commercial phage preparation Stafal® on biofilm and planktonic forms of methicillin-resistant Staphylococcus aureus. Folia Microbiol (Praha) 2019; 64: 121–126. [DOI] [PubMed] [Google Scholar]
- 20.Cha Y, Son B, Ryu S. Effective removal of staphylococcal biofilms on various food contact surfaces by Staphylococcus aureus phage endolysin LysCSA13. Food Microbiol 2019; 84: 103245. [DOI] [PubMed] [Google Scholar]
- 21.Costa GA, Rossatto FCP, Medeiros AW, Correa APF, Brandelli A, Frazzon APG, et al. Evaluation antibacterial and antibiofilm activity of the antimicrobial peptide P34 against Staphylococcus aureus and Enterococcus faecalis. An Acad Bras Cienc 2018; 90: 73–84. [DOI] [PubMed] [Google Scholar]
- 22.Benamara H, Rihouey C, Abbes I, Ben Mlouka MA, Hardouin J, Jouenne T, et al. Characterization of membrane lipidome changes in Pseudomonas aeruginosa during biofilm growth on glass wool. PLoS One 2014; 9(9): e108478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oosthuizen MC, Steyn B, Lindsay D, Brözel VS, Von Holy A. Novel method for the proteomic investigation of a dairy-associated Bacillus cereus biofilm. FEMS Microbiol Lett 2001; 194: 47–51. [DOI] [PubMed] [Google Scholar]
- 24.Atshan SS, Shamsudin MN, Karunanidhi A, Van Belkum A, Lung LT, Sekawi Z, et al. Quantitative PCR analysis of genes expressed during biofilm development of methicillin resistant Staphylococcus aureus (MRSA). Infect Genet Evol 2013; 18: 106–112. [DOI] [PubMed] [Google Scholar]
- 25.Rahimzadeh G, Saeedi M, Moosazadeh M, Hashemi SMH, Babaei A, Rezai MS, et al. Encapsulation of bacteriophage cocktail into chitosan for the treatment of bacterial diarrhea. Sci Rep 2021; 11: 15603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahimzadeh G, Saeedi M, Nokhodchi A, Moosazadeh M, Ghasemi M, Rostamkalaei SS, et al. Evaluation of in-situ gel-forming eye drop containing bacteriophage against Pseudomonas aeruginosa keratoconjunctivitis in vivo. Bioimpacts 2021; 11: 281–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ackermann HW. Phage classification and characterization. Methods Mol Biol 2009; 501: 127–140. [DOI] [PubMed] [Google Scholar]
- 28.Peng C, Hanawa T, Azam AH, LeBlanc C, Ung P, Matsuda T, et al. Silviavirus phage ϕMR003 displays a broad host range against methicillin-resistant Staphylococcus aureus of human origin. Appl Microbiol Biotechnol 2019; 103: 7751–7765. [DOI] [PubMed] [Google Scholar]
- 29.Cui Z, Feng T, Gu F, Li Q, Dong K, Zhang Y, et al. Characterization and complete genome of the virulent Myoviridae phage JD007 active against a variety of Staphylococcus aureus isolates from different hospitals in Shanghai, China. Virol J 2017; 14: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fierheller M, Sibbald RG. A clinical investigation into the relationship between increased periwound skin temperature and local wound infection in patients with chronic leg ulcers. Adv Skin Wound Care 2010; 23: 369–379. [DOI] [PubMed] [Google Scholar]
- 31.Jones EM, Cochrane CA, Percival SL. The effect of pH on the extracellular matrix and biofilms. Adv Wound Care (New Rochelle) 2015; 4: 431–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yüksel EB, Yıldırım AM, Bal A, Kuloglu T. The effect of different topical agents (silver sulfadiazine, povidone-iodine, and sodium chloride 0.9%) on burn injuries in rats. Plast Surg Int 2014; 2014: 907082. [DOI] [PMC free article] [PubMed] [Google Scholar]