Abstract
Background and Objectives:
Identification of pnemococcal serotypes and antimicrobial resistance provides helpful information for the use of suitable vaccines and antibiotics; however, very limited data is available on these issues in Vietnam. The present study aimed to find the serotype distribution and drug resistance patterns of Streptococcus pneumoniae isolated from unvaccinated children less than 5 years of age with pneumonia at a province in centre Vietnam.
Materials and Methods:
A total of 126 clinical pnemococcal strains isolated from unvaccinated children less than 5 years of age with pneumonia at the Nghe An province, Vietnam between Nov 2019 and Mar 2021. All strains were identified using conventional microbiological method, VITEK® 2 Compact system, specific PCR and sequencing. The serotypes and antimicrobial resistance patterns of pnemococcal strains were determined using the multiplex PCR assays and VITEK® 2 Compact system.
Results:
The results showed that, eight different pneumococcal serotypes were identified. The most common serotypes were 19F (67.46%), followed by 23F (10.32%), 19A (9.52%), 6A/B (3.17%), 15A (2.38%), 9V (3.17%), 11A (1.59%) and 14 (0.80%), respectively. More than half of the pneumococcal strains were non-susceptible to penicillin. The resistance rate to ceftriaxone and cefotaxime were 41.3% and 50.8%. The percentage of pneumococci strains resistant to clarithromycin, azithromycin, erythromycin, cotrimoxazole, tetracyclin, and clindamycin were more than 93% of all strains. All pneumococcal serotypes were highly resistant to clarithromycin, azithromycin, erythromycin, cotrimoxazole, and clindamycin.
Conclusion:
Our findings showed high antibiotic resistance rates of the strains causing pneumococcal pneumonia, mostly macrolide resistance, among unvaccinated children.
Keywords: Streptococcus pneumoniae, Serotypes, Antibiotic resistance, Children, Pneumonia
INTRODUCTION
The bacterium Streptococcus pneumoniae (S. pneumoniae, pneumococcus) causes pneumococcal disease. This is a Gram-positive, facultative anaerobic bacterium that is an important pathogen causing community pneumonia, sinusitis, otitis media, and invasive infections such as bacteremia and bacterial meningitis, which are the leading causes of morbidity and mortality among children less than 5 years of age (1–3). The occurrence of diseases caused by this etiological agent are a prominent global public health issues (4, 5). According to the study reported by Wahl et al. (2018), diseases caused by S. pneumoniae kill 317,300 children under five years of age every year, mostly in lower income countries (6). S. pneumoniae also occurs as a cause of invasive infections in elderly persons (7).
Pneumococcus is currently divided into more than ninety serotypes based on the antigenic capsular polysaccharide (CPS) (8). The CPS is an important virulence factor of S. pneumoniae and of the pneumococcal serotypes, serotypes 6, containing four confirmed serotypes (6A–6D), are reported to account for the most common serotypes of pneumococcal disease worldwide (5, 9, 10). Epidemiological studies around the world have showed that distribution of pneumococcal serotypes varies by age and geographical area (11–13). According to previous studies, although there are many pneumococcal serotypes, only certain types lead to invasive diseases worldwide such as serotypes 1, 4, 6A/6B, 7F, 9V, 14, 15B/15C, 18C, 19F, 19A, and 23F, which were found to account for about 80–90% of invasive pneumococcal disease (IPD) in children, especially in unvaccinated areas (2, 14, 15). Therefore, the type of pneumococcal vaccines should be selected in accordance with the circulating serotypes in each country (5, 11).
The antibiotic treatment for pneumococcal infections seems to be the primary choice. However, increasing antibiotic drug-resistance of pneumococcus, mainly to β-lactams, has been noted worldwide, especially in Asia, that has made the role of antibiotics is limited (11, 13). This showed that the importance of disease prevention (11). Vaccines have demonstrated to be an effective means of preventing pneumococcal disease worldwide (13). In the US, after the introduction of seven-valent pneumococcal conjugate vaccines (PCV7) to prevent pneumococcal diseases, the rate of IPD has significantly reduced from >200 cases/100 000 persons to >50 cases/100 000 persons (7). To date, 146 member countries out of WHO members have added pneumococcal conjugate vaccine (PCV) into their National Immunization Program (16). Nevertheless, only about 55% (approximately 74 million) of the global infant population are receiving PCV (7).
In addition, antibiotic resistance in S. pneumoniae strains is rising in all parts of the world, including Vietnam (17). Thus, an understanding of the serotype distribution and antibiotic resistance patterns of pneumococcus is necessary to guidance for the use of suitable vaccines and antibiotics (3). The aim of this study was to determine the serotypes and patterns of antibiotic resistance of S. pneumoniae isolated from unvaccinated children less than 5 years of age with pneumonia at a province in centre Vietnam.
MATERIALS AND METHODS
Bacterial isolates and identification of S. pneumoniae. In current study, a total of 126 S. pneumoniae clinical isolates were isolated from sputum samples of pneumonia children, aged between 2 and 59 months, at the Nghe An Obstetrics and Pediatrics Hospital (500 beds), Nghe An province, Vietnam, during the period between November 2019 and March 2021. Sputum specimens for each patient were taken by trained nurses using a clean suction and were then transported to the clinical microbiology laboratory within 2 h for isolation of S. pneumoniae. All samples were inoculated onto agar plates containing 5% sheep blood (Himedia, India) at 37°C in 5% CO2 atmosphere for 18–24 h. The samples that no growth on the agar after 24 h were followed up for a further 24 h before being pronounced as negative. Colonies of suspected isolates was taken to identify as S. pneumonia using conventional microbiological method (Gram staining, the alpha hemolysis test, the optochin sensitivity test) in combination with the VITEK® 2 Compact system (bioMérieux, North Carolina 27712, USA) according to the manufacturer’s instructions and PCR analyses using species-specific primers as described previously (1). All isolates were stored at −80°C in cryotubes containing trypticase soy broth (Merck, Germany), 20% glycerol (Merck, Germany) and 10% horse serum for further analysis.
Determination of pneumococcal serotypes. Genomic DNA of S. pneumoniae was extracted from the bacterial cultures using G-spin™ Genomic DNA Extraction Kit (iNtRON Biotechnology, Korea), following the manufacturer’s protocol. First of all, the pneumococcal isolates were confirmed by molecular method using the specific primer pair of cpsA-F (5′-GCA GTA CAG CAG TTT GTT GGA CTG ACC-3′) and cpsA-R (5′-GAA TAT TTT CAT TAT CAG TCC CAG TC-3′) (Integrated DNA Technologies, USA) for amplification of cpsA gene (8). And then, the most common serotypes of S. pneumoniae isolates were identified by multiplex PCR (mPCR) using twenty-one capsular specific primer pairs (Table 1) as described in previous reports (1, 8). The capsular types were collected on five groups as follows: Types 14, 19A, 19F and 23F; 6A/B, 9V, 15A and 15B/C; 1, 3, 10A and 11A; 4, 5, 7C and 17F; 7F, 8, 12A, 20 and 23B (Table 1). The mPCR reactions were carried out in 25 μl volumes containing 2 μl of DNA solution, 12.5 μl 2× Master mix (Cat.# M7505, Promega, USA), 0.5 μl of each primer (0.2 μM) and distilled water up to 25 μl. Thermal cycling was performed in Thermo Mastercycler Gradient system (Thermo Fisher Scientific, USA) under the following conditions: 94°C for 5 minutes; followed by 35 cycles at 94°C for 45 seconds, 54°C for 45 seconds and 65°C for 150 seconds; and a final extension of 72°C for 10 minutes. The mPCR products were analyzed on a 2% agarose gel containing 0.5 μg/ml ethidium bromide at 100V for 60 minutes and visualized with UV transillumination (UVP, Canada). The sizes of the mPCR products were determined by 100bp DNA Ladders (Cleaver, UK). Pneumococcal isolates that could not be serotyped by mPCR were classified as non-typeable. Total DNA isolated from S. pneumoniae strain ATCC 49619 were used for quality control.
Table 1.
Primers used to confirm and identify serotypes of S. pneumoniae
| Reaction | Serotype/Primer | Primer sequence (5′-3′) | Product size (bp) |
|---|---|---|---|
| 1 | 14-F | CTT GGC GCA GGT GTC AGA ATT CCC TCT AC | 208 |
| 14-R | GCC AAA ATA CTG ACA AAG CTA GAA TAT AGC C | ||
| 19A-F | GTT AGT CCT GTT TTA GAT TTA TTT GGT GAT GT | 478 | |
| 19A-R | GAG CAG TCA ATA AGA TGA GAC GAT AGT TAG | ||
| 19F-F | GTT AAG ATT GCT GAT CGA TTA ATT GAT ATC C | 304 | |
| 19F-R | GTA ATA TGT CTT TAG GGC GTT TAT GGC GAT AG | ||
| 23F-F | GTA ACA GTT GCT GTA GAG GGA ATT GGC TTT TC | 384 | |
| 23F-R | CAC AAC ACC TAA CAC ACG ATG GCT ATA TGA TTC | ||
| 2 | 6A/B-F | AAT TTG TAT TTT ATT CAT GCC TAT ATC TGG | 250 |
| 6A/B-R | TTA GCG GAG ATA ATT TAA AAT GAT GAC TA | ||
| 9V-F | CTT CGT TAG TTA AAA TTC TAA ATT TTT CTA A | 753 | |
| 9V-R | GTC CCA ATA CCA GTC CTT GCA ACA CAA G | ||
| 15A-F | ATT AGT ACA GCT GCT GGA ATA TCT CTT C | 436 | |
| 15A-R | GAT CTA GTG AAC GTA CTA TTC CAA AC | ||
| 15B/C-F | TTG GAA TTT TTT AAT TAG TGG CTT ACC TA | 496 | |
| 15B/C-R | CAT CCG CTT ATT AAT TGA AGT AAT CTG AAC C | ||
| 3 | 1-F | CTC TAT AGA ATG GAG TAT ATA AAC TAT GGT TA | 280 |
| 1-R | CCAAAGAAAATACTAACATTA TCA CAA TAT TGG C | ||
| 3-F | ATG GTG TGA TTT CTC CTA GAT TGG AAA GTA G | 371 | |
| 3-R | CTT CTC CAA TTG CTT ACC AAG TGC AAT AAC G | ||
| 10A-F | GGT GTA GAT TTA CCA TTA GTG TCG GCA GAC | 628 | |
| 10A-R | GAA TTT CTT CTT TAA GAT TCG GAT ATT TCT C | ||
| 11A-F | GGA CAT GTT CAG GTG ATT TCC CAA TAT AGT G | 463 | |
| 11A-R | GAT TAT GAG TGT AAT TTA TTC CAA CTT CTC CC | ||
| 4 | 4-F | CTG TTA CTT GTT CTG GAC TCT CGA TAA TTG G | 430 |
| 4-R | GCC CAC TCC TGT TAA AAT CCT ACC CGC ATT G | ||
| 5-F | ATA CCT ACA CAA CTT CTG ATT ATG CCT TTG TG | 362 | |
| 5-R | GCTCGATAAACATAATCAATATTTGAAAAA GTA TG | ||
| 7C-R | CTATCTCAGTCATCTATTGTTAAAGTTTACGACGGGA | 260 | |
| 7C-R | GAA CAT AGA TGT TGA GAC ATC TTT TGT AAT TTC | ||
| 17F-F | TTC GTG ATG ATA ATT CCA ATG ATC AAA CAA GAG | 693 | |
| 17F-R | GAT GTA ACA AAT TTG TAG CGA CTA AGG TCT GC | ||
| 5 | 7F-F | CCT ACG GGA GGA TAT AAA ATT ATT TTT GAG | 826 |
| 7F-R | CAA ATA CAC CAC TAT AGG CTG TTG AGA CTA AC | ||
| 8-F | GAT GCC ATG AAT CAA GCA GTG GCT ATA AAT C | 294 | |
| 8-R | ATC CTC GTG TAT AAT TTC AGG TAT GCC ACC | ||
| 12A-F | ACT CTT CCA AAT TCT TAT GCT TTT ATT GAT TC | 656 | |
| 12A-R | ATG AAT GAG AAA AGG AAC TTA AAA TTC ATA GC | ||
| 20-F | GAG CAA GAG TTT TTC ACC TGA CAG CGA GAA G | 514 | |
| 20-R | CTA AAT TCC TGT AAT TTA GCT AAA ACT CTT ATC | ||
| 23B-F | TTG TTA GTG GTA TTA AAT TGG GGA CTA CTA GG | 216 | |
| 23B-R | ATA CCT ATC TGA AGT GTT ATT AAC CCA CCA AC | ||
| Positive control | cpsA-F | GCA GTA CAG CAG TTT GTT GGA CTG ACC | 160 |
| cpsA-R | GAA TAT TTT CAT TAT CAG TCC CAG TC |
16S rRNA gene sequencing. Two PCR primers, namely 27F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492R (5′-GGT TAC CTT GTT ACG ACT T-3′), were chosen to amplify the 16S rDNA gene (18). PCR products of 16S ribosomal RNA genes from twenty-two isolates were randomly selected and were sent to Apical Scientific Sdn. Bhd (Selangor, Malaysia) for purification and automatic DNA sequencing, using the same primer pair. Species confirmed S. pneumoniae isolates were accurately examined by two-directional sequencing. The 16S rRNA gene sequences of these strains were deposited in the DDBJ/EMBL/GenBank databases under accession number MW672550-MW672562 and MZ007491-MZ007499, respectively.
Antimicrobial susceptibility testing. The antimicrobial susceptibility tests of each isolate with penicillin (PEN, 0.0625–8.0 μg/mL), cefotaxim (CXM, 0.125–8.0 μg/mL), ceftriaxone (CEF, 0.125–8 μg/mL), chloramphenicol (CLP, 1.0–16.0 μg/mL), azithromycin (AZM, 0.125–8.0 μg/mL), clarithromycin (CLA, 0.25–16.0 μg/mL), erythromycin (ERY, 0.125–8.0 μg/mL), clindamycin (CLI, 0.25–1.0 μg/mL), levofloxacin (LEV, 0.25–16.0 μg/mL), linezolid (LIN, 2.0–8.0 μg/mL), moxifloxacin (MXF, 0.0625–4.0 μg/mL), rifampicin (RIF, 0.0625–4.0 μg/mL), tetracyclin (TET, 0.25–16.0 μg/mL), vancomycin (VAN, 0.125–8.0 μg/mL) and trimethoprim-sulfamethoxazole (SXT, 10.0–320.0 μg/mL) were performed for each strain using VITEK® 2 Compact system according the manufacturer’s instructions. The breakpoints used for S. pneumoniae were classified in accordance with the Clinical and Laboratory Standards Institute (CLSI) 2020 criteria. For quality control of the susceptibility tests, S. pneumoniae ATCC 49619 was chosen as the reference strain.
Statistical analysis. The statistical analysis was carried out by IBM SPSS Statistics software, version 20.0 developed by IBM Corp. (Armonk, NY, USA). Chi-squared and Fisher’s exact tests were performed to check the significance of the data. P values less than 0.05 were considered significant statistically. The 16S gene sequences of pneumococcal isolates were compared to the publicly available DNA sequences in the Genbank databases, using BLAST programs (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Ethics approval and consent to participate. The purpose and benefits of the current study were informed to parents/legal guardians of each participant who also signed a written informed consent before the study procedure was performed. The study protocols was accepted by the Ethical Committee of the National Institute of Malariology, Parasitology and Entomology (Ha Noi, Vietnam) in March 2018 (ethics code: 225/QĐ-VSR). Furthermore, this study is based on the Declaration of Helsinki Principles.
RESULTS
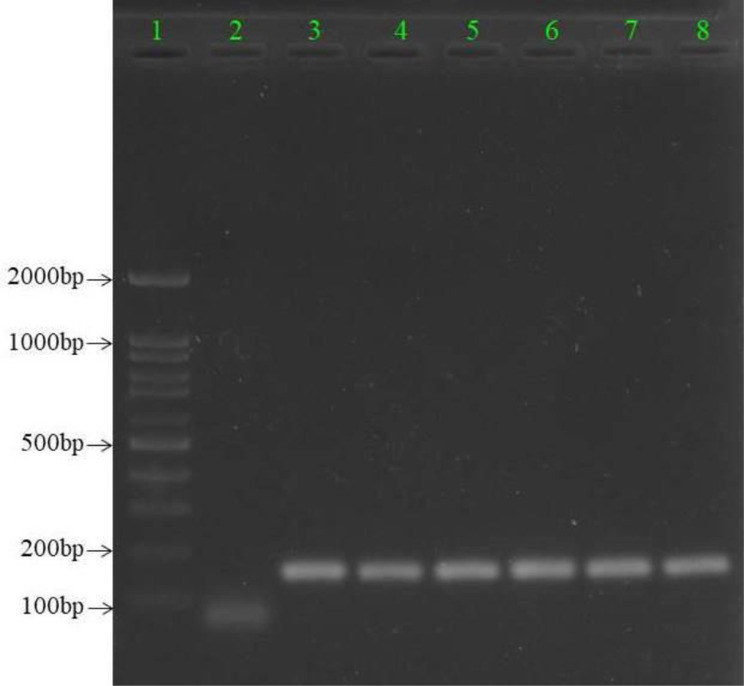
All of the 126 individual isolates which were S. pneumoniae culture-positive also indicated a positive PCR result for the cpsA gene (Fig. 1). 22 sequences of the 16S rDNA regions of different pneumococcal isolates were also deposited in the NCBI database under accession number MW672550-MW672562 and MZ007491-MZ007499, respectively.
Fig. 1.
Gel electrophoresis of S. pneumoniae-specific PCR products targeting the 160 bp cpsA gene
Lane 1: DNA Ladder 100 bp Standard; lane 2: negative control; lanes 3–7 (strain Sp8107, Sp8279, Sp8281, Sp8294, and Sp8298): clinical samples; lane 8: positive control
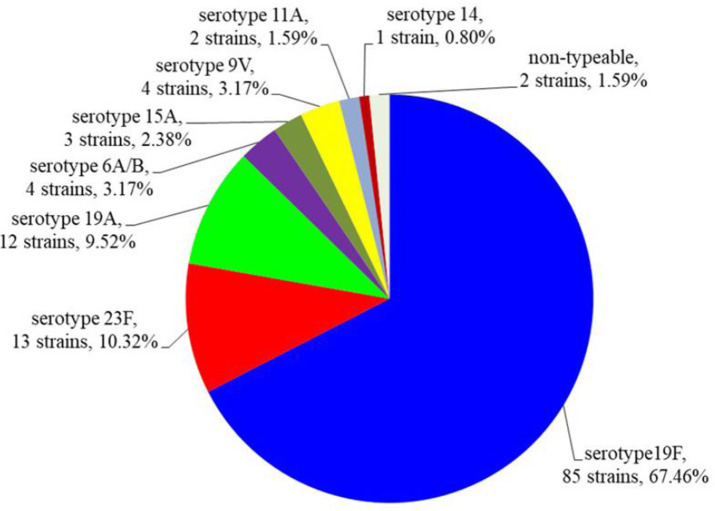
By multiplex PCR assays, of the 126 S. pneumoniae isolates analyzed, 124 strains (98.41%) could be serotyped, of which the eight different pneumococcal serotypes were classified. Only one serotype per patient was detected. The remaining 2 isolates (1.59%) could not be serotyped. The serotype distribution is shown in Figs 2 and 3.
Fig. 2.
The distribution of the pneumococcal serotypes
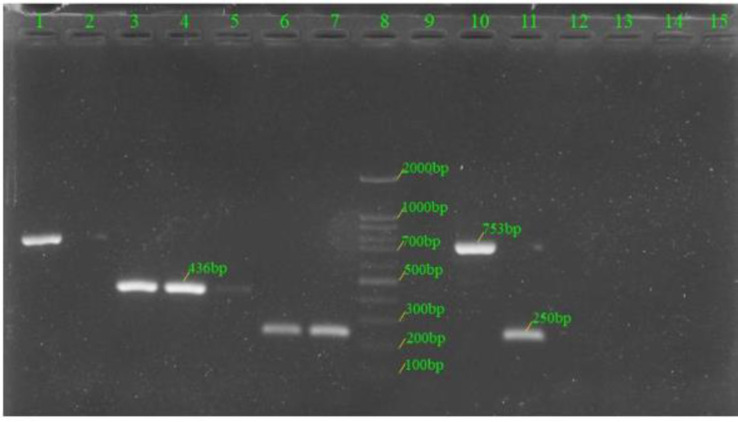
Fig. 3.
The multiplex PCR patterns of serotypes 6A/B, 9V, 15A and 15B/C (reaction 2)
Lanes 1 and 10 denoted to those of serotype 9V; lanes 3–5 denoted to those of serotype 15A; lanes 6, 7 and 11 denoted to those of serotype 6A/B; lanes 2, 9, 12–14 denoted to those of non-typeable; lane 8: DNA Ladder 100bp Standard; lane 15: negative control.
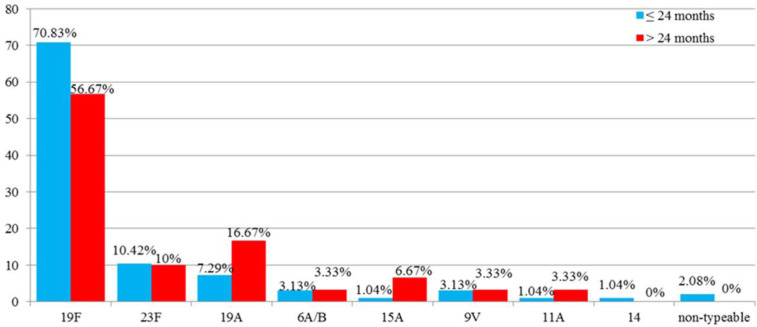
Pneumococcal serotype distribution varied between age groups (Fig. 4), but the difference was not statistically significant between the two groups (p > 0.05).
Fig. 4.
Serotype distribution according to age groups
Table 2 showed the trends of antimicrobial resistance patterns of S. pneumoniae strains. Accordingly, the observed resistance rates of 126 S. pneumoniae isolates to CLA, AZM, SXT, TET, CLI, ERY, CEF, and CXM were high, i.e. 100% (126), 100% (126), 93.7% (118), 96% (121), 96% (121, 99.2% (125), 41.3% (52), and 50,8 (64), respectively. This bacteria showed 100% susceptibility to the RIF, CLP, VAN, LIN and MXF. The susceptibility rates of pneumococcus to levofloxacin were 97.6%.
Table 2.
Antimicrobial resistance rates of 126 pneumococcal isolates of different serotypes against 16 antimicrobial agents
| Antibiotic | % of isolates | Resistance of serotype s (%) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||
| S | I | R |
19F
No. 85 |
23F
No. 13 |
19A
No. 12 |
6A/B
No. 4 |
15A
No. 3 |
9V
No. 4 |
11A
No. 2 |
14
No. 1 |
NT
No. 2 |
||||
| PEN | 46.8 | 48.4 | 4.8 | 4.7 | 0 | 8.33 | 0 | 33.33 | 0 | 0 | 0 | 0 | |||
| CXM | 22.2 | 27.0 | 50.8 | 64.7 | 7,7 | 25 | 50 | 66.7 | 25 | 0 | 0 | 0 | |||
| CEF | 25.4 | 33.3 | 41.3 | 55.3 | 7.7 | 0 | 50 | 0 | 25 | 0 | 100 | 0 | |||
| LEV | 97.6 | 0 | 2.4 | 3.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| MXF | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| ERY | 0.8 | 0 | 99.2 | 100 | 100 | 100 | 100 | 100 | 100 | 50 | 100 | 100 | |||
| CLI | 4.0 | 0 | 96.0 | 95.3 | 100 | 100 | 100 | 100 | 100 | 50 | 100 | 100 | |||
| LIN | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| VAN | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| TET | 4.0 | 0 | 96.0 | 96.5 | 100 | 83.3 | 100 | 100 | 100 | 100 | 100 | 100 | |||
| CLP | 100 | 0 | 0 | 1.2 | 0 | 0 | 25 | 0 | 0 | 0 | 0 | 0 | |||
| RIF | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| SXT | 5.5 | 0.8 | 93.7 | 98.8 | 92.3 | 100 | 100 | 0 | 75 | 50 | 100 | 50 | |||
| AZM | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | |||
| CLA | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | |||
Abbreviation: S susceptible, I Intermediate, R Resistant; NT non-typeable
Antimicrobial resistance of pneumococcus among different serotypes is shown in the Table 2. These results indicated that the observed resistance rates of serotype 19F were highest. All of 8 serotypes exhibited high rates of resistance to macrolides, tetracyclin and clindamycin.
DISCUSSION
Invasive pneumococcal disease is known to be a major cause of morbidity and mortality among children under five years of age, especially those under 2 years of age, although preventive actions have been implemented in many countries (1, 19). According to the previous studies, the distribution of pneumococcal serotypes, which plays an important role in the cause of invasive infections, varies in different geographical area (11–13). Thus, routine screening for local serotype distribution of S. pneumoniae was necessary to inform the developing a safe and effective vaccine and guidance for the use of appropriate antibiotics (3). According to the WHO, universal vaccination is the best way against pneumococcal disease (2).
The epidemiological data from around the world have indicated that serotypes 1, 4, 6A/B, 7F, 9V, 14, 15B/C, 18C, 19F, 19A, and 23F represent around 80–90% of IPD in children, especially in unvaccinated areas (2, 14, 15). The current study detected pneumococcal serotypes 6A/B, 9V, 11A, 14, 15A, 19F, 19A, and 23F among unvaccinated children under five years of age in Nghe An province. The three major serotypes 19F, 19A, and 23, were found to account for about 90% of S. pneumoniae isolates, while serotypes 6A/B, 9V, 11A, 14, and 15A were represented in lower percentages, ranging from 0.80 to 3.17%. Notably, serotype 19A has a rather high prevalence. The serotype distribution in this study was quite similar to those found previous studies in southern Vietnam, ASEAN countries, and Taiwan (20–24). Serotypes 6B, 23F and 19F are the most prevalent among children in Japan, while serotypes 1, 5, 6ABC, and 19F predominate in Egypt (25, 26). Serotype 14 was most common among strains from Paulo, Brazil and Casablance, Morocco (27, 28). Serotypes 23F, 14 and 3 are the most common in Tehran, Iran (11). In China, several studies have demonstrated the distribution of serotypes of S. pneumoniae varies between cities and different years (3, 29, 30). The results from different studies indicate that the prevalence of pneumococcal serotypes varies different depending on the population, region, and change over time (29, 31, 32). Thus, additional investigations should be carry out to identify the pneumococcal serotypes in different regions of Vietnam to provide information for the development of appropriate vaccines.
The emergence of resistance to antibiotics in pneumococci is increasing and it is becoming increasingly important predictive factor since it is directly related to persistent disease or disease mortality (3, 33, 34). The results of this study indicated that antimicrobial resistance patterns of S. pneumoniae is a matter of great concern. The resistance rates of S. pneumoniae isolates to AZM, CLA, CLI, ERY, SXT, and TET were higher than 96%. The previous studies in China, Taiwan and other Asian countries, including Vietnam also indicated bad in vitro activity of macrolides (ERY, CLA and AZM), lincosamide (CLI), tetracyclines (TET), and SXT against S. pneumoniae isolates (3, 23, 24). Our results suggested that these antibiotics are not appropriate for the treatment of pneumococcal disease in Vietnam. Besides, the rates of decreased susceptibility to penicillins and cephalosporins showed a rising trend. This result is in agreement with previous studies conducted in China, Taiwan and Vietnam, where more than 50% of patients infected with non-susceptible to penicillins and cephalosporins (3, 24, 29, 30, 35). In our study, all the S. pneumoniae strains were susceptible to RIF, CLP, VAN, MXF and LIN. Our findings also indicated the prevalence of LEV resistance in S. pneumoniae isolates were low (2,4%). The results of current study have shown that RIF, CLP, VAN, MXF, LIN and LEV may provide an opportunity for treating β-lactam, macrolides, lincosamide, tetracyclines, and cotrimoxazole-resistant pneumococcal disease in Vietnam.
In the present study, the prevalence of multidrug resistance of all eight serotypes were 100%. This rate was higher in the current research than previously findings in southern Vietnam (20–22). Notably, the high rates of antimicrobial resistance of serotype 19A were observed. This serotype is not covered by the PCV-7, thus the use of these vaccines may not be effective in preventing pneumococcal disease (3, 11). Therefore, PCV-13 should be recommended for future vaccination in Nghe An because of its broader serotype coverage.
CONCLUSION
In the current study, eight different pneumococcal serotypes were identified in Nghe An, Vietnam. Among that, 19F, 23F and 19A were the most prevalent serotypes. The high frequency of serotype 19A was a notable characteristic. In addition, the rate of antibiotic resistance of S. pneumoniae is considerable. Cautious use of antibiotics is extremely important and necessary to prevent the appearance of resistant pneumococci.
ACKNOWLEDGEMENTS
This work was partially supported by the Department of Science and Technology of Nghe An province, Vietnam, (grant no. 901/HĐ-SKHCN).
REFERENCES
- 1.Ahn JG, Choi SY, Kim DS, Kim KH. Enhanced detection and serotyping of Streptococcus pneumoniae using multiplex polymerase chain reaction. Korean J Pediatr 2012; 55: 424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Houri H, Tabatabaei SR, Saee Y, Fallah F, Rahbar M, Karimi A. Distribution of capsular types and drug resistance patterns of invasive pediatric Streptococcus pneumoniae isolates in Teheran, Iran. Int J Infect Dis 2017; 57: 21–26. [DOI] [PubMed] [Google Scholar]
- 3.Liu C, Xiong X, Xu W, Sun J, Wang L, Li J. Serotypes and patterns of antibiotic resistance in strains causing invasive pneumococcal disease in children less than 5 years of age. PLoS One 2013; 8(1): e54254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van de Vooren K, Duranti S, Curto A, Garattini L. Cost effectiveness of the new Pneumococcal vaccines: A systematic review of European studies. Pharmacoeconomics 2014; 32: 29–45. [DOI] [PubMed] [Google Scholar]
- 5.Shi W, Zhou K, Yuan L, Meng Q, Dong F, Gao W, et al. Serotype distribution, antibiotic resistance patterns and molecular characteristics of serogroup 6 Streptococcus pneumoniae isolates collected from Chinese children before the introduction of PCV13. J Glob Antimicrob Resist 2018; 14: 23–28. [DOI] [PubMed] [Google Scholar]
- 6.Wahl B, O’Brien KL, Greenbaum A, Majumder A, Liu L, Chu Y, et al. Burden of Streptococcus pneumoniae and Haemophilus influenzae type b disease in children in the era of conjugate vaccines: global, regional, and national estimates for 2000–15. Lancet Glob Health 2018; 6(7): e744–e757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.International Vaccine Access Center . Johns Hopkins Bloomberg School of Public Health. VIEW-hub report: Global vaccine introduction and Implementation, March 2019. [Google Scholar]
- 8.Beheshti M, Jabalameli F, Feizabadi MM, Hahsemi FB, Beigverdi R, Emaneini M. Molecular characterization, antibiotic resistance pattern and capsular types of invasive Streptococcus pneumoniae isolated from clinical samples in Tehran, Iran. BMC Microbiol 2020; 20: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Liu F, Ao P, Li X, Zheng H, Wu D, et al. Detection of serotype distribution and drug resistance of Streptococcus Pneumoniae isolated from pediatric patients. Lab Med 2017; 48: 39–45. [DOI] [PubMed] [Google Scholar]
- 10.Huang S, Liu X, Lao W, Zeng S, Liang H, Zhong R, et al. Serotype distribution and antibiotic resistance of Streptococcus pneumoniae isolates collected at a Chinese hospital from 2011 to 2013. BMC Infect Dis 2015; 15: 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Habibi Ghahfarokhi S, Mosadegh M, Ahmadi A, Pourmand MR, Azarsa M, Rahbar M, et al. Serotype distribution and antibiotic susceptibility of Streptococcus pneumoniae isolates in Tehran, Iran: A surveillance study. Infect Drug Resist 2020; 13: 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hausdorff WP, Feikin DR, Klugman KP. Epidemiological differences among pneumococcal serotypes. Lancet Infect Dis 2005; 5: 83–93. [DOI] [PubMed] [Google Scholar]
- 13.Zhao C, Li Z, Zhang F, Zhang X, Ji P, Zeng J, et al. Serotype distribution and antibiotic resistance of Streptococcus pneumoniae isolates from 17 Chinese cities from 2011 to 2016. BMC Infect Dis 2017; 17: 804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song JY, Nahm MH, Moseley MA. Clinical implications of pneumococcal serotypes: invasive disease potential, clinical presentations, and antibiotic resistance. J Korean Med Sci 2013; 28: 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nuorti JP, Whitney CG. Prevention of pneumococcal disease among infants and children - use of 13-valent pneumococcal conjugate vaccine and 23-valent pneumococcal polysaccharide vaccine - recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm Rep 2010; 59: 1–18. [PubMed] [Google Scholar]
- 16.International Vaccine Access Center . Johns Hopkins Bloomberg School of Public Health. VIEW-hub Report: Global Vaccine Introduction and Implementation, March 2020. [Google Scholar]
- 17.Reinert RR. The antimicrobial resistance profile of Streptococcus pneumoniae. Clin Microbiol Infect 2009; 15 Suppl 3: 7–11. [DOI] [PubMed] [Google Scholar]
- 18.Miller CS, Handley KM, Wrighton KC, Frischkorn KR, Thomas BC, Banfield JF. Short-read assembly of full-length 16S amplicons reveals bacterial diversity in subsurface sediments. PLoS One 2013; 8(2): e56018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan TQ. Pediatric invasive pneumococcal disease in the United States in the era of pneumococcal conjugate vaccines. Clin Microbiol Rev 2012; 25: 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vo TT, Phan T, Ngo HTM, Pham H, Ho T. Antibiotic susceptibility of invasive Streptococcus pneumoniae isolates in southern Vietnam. Int J Infect Dis 2020; 101: 53–54. [Google Scholar]
- 21.Parry CM, Diep TS, Wain J, Hoa NT, Gainsborough M, Nga D, et al. Nasal carriage in Vietnamese children of Streptococcus pneumoniae resistant to multiple antimicrobial agents. Antimicrob Agents Chemother 2000; 44: 484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parry CM, Duong NM, Zhou J, Mai NTH, Diep TS, Thinh LQ, et al. Emergence in Vietnam of Streptococcus pneumoniae resistant to multiple antimicrobial agents as a result of dissemination of the multiresistant Spain(23F)-1 clone. Antimicrob Agents Chemother 2002; 46: 3512–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jauneikaite E, Jefferies JM, Hibberd ML, Clarke SC. Prevalence of Streptococcus pneumoniae serotypes causing invasive and non-invasive disease in South East Asia: A review. Vaccine 2012; 30: 3503–3514. [DOI] [PubMed] [Google Scholar]
- 24.Wu CJ, Lai JF, Huang IW, Shiau YR, Wang HY, Lauderdale TL. Serotype distribution and antimicrobial susceptibility of Streptococcus pneumoniae in pre- and post- PCV7/13 Eras, Taiwan, 2002–2018. Front Microbiol 2020; 11: 557404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakata H. Invasive pneumococcal diseases in children in Hokkaido, Japan from April 2000, to March 2015. J Infect Chemother 2016; 22: 24–26. [DOI] [PubMed] [Google Scholar]
- 26.El-Kholy A, Badawy M, Gad M, Soliman M. Serotypes and antimicrobial susceptibility of nasopharyngeal isolates of Streptococcus pneumoniae from children less than 5 years old in Egypt. Infect Drug Resist 2020; 13: 3669–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Medeiros MIC, Almeida SCG, Guerra MLLS, Da Silva P, Carneiro AMM, De Andrade D. Distribution of Streptococcus pneumoniae serotypes in the northeast macro-region of São Paulo state/Brazil after the introduction of conjugate vaccine. BMC Infect Dis 2017; 17: 590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diawara I, Zerouali K, Katfy K, Zaki B, Belabbes H, Najib J, et al. Invasive pneumococcal disease among children younger than 5 years of age before and after introduction of pneumococcal conjugate vaccine in Casablanca, Morocco. Int J Infect Dis 2015; 40: 95–101. [DOI] [PubMed] [Google Scholar]
- 29.Pan F, Han L, Huang W, Tang J, Xiao S, Wang C, et al. Serotype distribution, antimicrobial susceptibility, and molecular epidemiology of Streptococcus pneumoniae isolated from children in Shanghai, China. PLoS One 2015; 10(11): e0142892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu Y-Y, Xie X-H, Ren L, Deng Y, Gao Y, Zhang Y, et al. Epidemiological characteristics of nasopharyngeal Streptococcus pneumoniae strains among children with pneumonia in Chongqing, China. Sci Rep 2019; 9: 3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson HL, Deloria-Knoll M, Levine OS, Stoszek SK, Freimanis Hance L, Reithinger R, et al. Systematic evaluation of serotypes causing invasive Pneumococcal disease among children under five: the Pneumococcal global serotype project. PLoS Med 2010; 7(10): e1000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ktari S, Jmal I, Mroua M, Maalej S, Ben Ayed NE, Mnif B, et al. Serotype distribution and antibiotic susceptibility of Streptococcus pneumoniae strains in the south of Tunisia: A five-year study (2012–2016) of pediatric and adult populations. Int J Infect Dis 2017; 65: 110–115. [DOI] [PubMed] [Google Scholar]
- 33.McGee L, McDougal L, Zhou J, Spratt BG, Tenover FC, George R, et al. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J Clin Microbiol 2001; 39: 2565–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lonks JR, Garau J, Gomez L, Xercavins M, De Echagüen AO, Gareen IF, et al. Failure of macrolide antibiotic treatment in patients with bacteremia due to erythromycin-resistant Streptococcus pneumoniae. Clin Infect Dis 2002; 35: 556–564. [DOI] [PubMed] [Google Scholar]
- 35.Kim SH, Song J-H, Chung DR, Thamlikitkul V, Yang Y, Wang H, et al. Changing trends in antimicrobial resistance and serotypes of Streptococcus pneumoniae isolates in Asian countries: an Asian Network for Surveillance of Resistant Pathogens (ANSORP) study. Antimicrob Agents Chemother 2012; 56: 1418–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]