Abstract
Viruses contribute to food web dynamics and nutrient cycles in diverse ecosystems, yet the biogeographical patterns that underlie these viral dynamics are poorly understood, particularly in soil. Here, we identified trends in soil viral community composition in relation to habitat, moisture content, and physical distance. We generated 30 soil viromes from four distinct habitats (wetlands, grasslands, woodlands, and chaparral) by selectively capturing virus-sized particles prior to DNA extraction, and we recovered 3432 unique viral ‘species’ (dsDNA vOTUs). Viral communities differed significantly by soil moisture content, with viral richness generally higher in wet compared to dry soil habitats. However, vOTUs were rarely shared between viromes, including replicates <10 m apart, suggesting that soil viruses may not disperse well and that future soil viral community sampling strategies may need to account for extreme community differences over small spatial scales. Of the 19% of vOTUs detected in more than one virome, 93% were from the same habitat and site, suggesting greater viral community similarity in closer proximity and under similar environmental conditions. Within-habitat differences indicate that extensive sampling would be required for rigorous cross-habitat comparisons, and results highlight emerging paradigms of high viral activity in wet soils and soil viral community spatial heterogeneity.
Subject terms: Soil microbiology, Microbial ecology
Introduction
Viruses are abundant across Earth’s ecosystems, contributing to microbial dynamics and biogeochemical cycles, yet they remain understudied, particularly in terrestrial habitats [1, 2]. Soil viral abundance measurements vary substantially, ranging from nearly zero in dry deserts to over 109 virus-like particles per gram in wetlands [3]. In the better studied surface oceans, viruses lyse approximately 20% of microbial biomass daily, impacting nutrient and energy cycles [4], and recent work suggests that viruses may be similarly important in terrestrial ecosystems [3, 5–12]. For example, viruses have been suggested to affect carbon cycling in thawing permafrost peatlands by preying on methanogens and methanotrophs and by encoding glycoside hydrolases to break down complex carbon into simple sugars [2]. Soil viruses have been demonstrated to actively infect their hosts in a variety of soil environments, even in extreme environments such as arid deserts [5, 13], and soil viral communities can be spatially structured [14–16]. Despite these emerging ecological patterns, comparisons of soil viral diversity within and across habitats are limited.
Here, we compared dsDNA viromes (<0.2 µm viral size-fraction metagenomes representing the dsDNA viral community, presumably dominated by viruses of bacteria and archaea) [14] from four distinct habitats (wetlands, grasslands, chaparral shrublands, and oak woodlands) across five UC Davis Natural Reserves field sites within Northern California. We compared viral species (vOTU) richness, vOTU detection patterns, and viral community beta-diversity, according to habitat type, soil properties, and spatial distribution to better understand the fundamental relationships between soil viruses and the ecosystems that they inhabit.
Results and discussion
To compare soil viral community composition within and across terrestrial habitats on a regional scale, viromes were generated from 34 near-surface (top 15 cm) soil samples, with a total of 30 viromes included in downstream ecological analyses (see Supplementary Methods). The analyzed viromes were collected from four distinct habitats (wetlands, grasslands, chaparral shrublands, and woodlands, each with 7, 14, 4, and 5 viromes, respectively) across five field sites (Fig. S1 for sampling scheme, Table S1 for soil properties). Following quality filtering, the 30 viromes generated an average of 72,950,833 reads and 416 contigs ≥10 Kbp per virome (Table S2). Wetland viromes yielded more contigs ≥10 Kbp than viromes from other habitats, both in total and on average per virome (Table S2). We used VIBRANT to identify 3490 viral contigs in our assemblies, which were clustered into 3,432 viral operational taxonomic units (vOTUs), defined as ≥10 Kbp viral contigs sharing ≥ 95% average nucleotide identity over 85% contig length [17]. Constrained analysis of principal coordinates (CAP analysis) revealed strong clustering by habitat rather than by site, implying that, where environmental parameters are substantially different, environmental conditions are stronger drivers of viral community composition than geographic distance (Fig. S2).
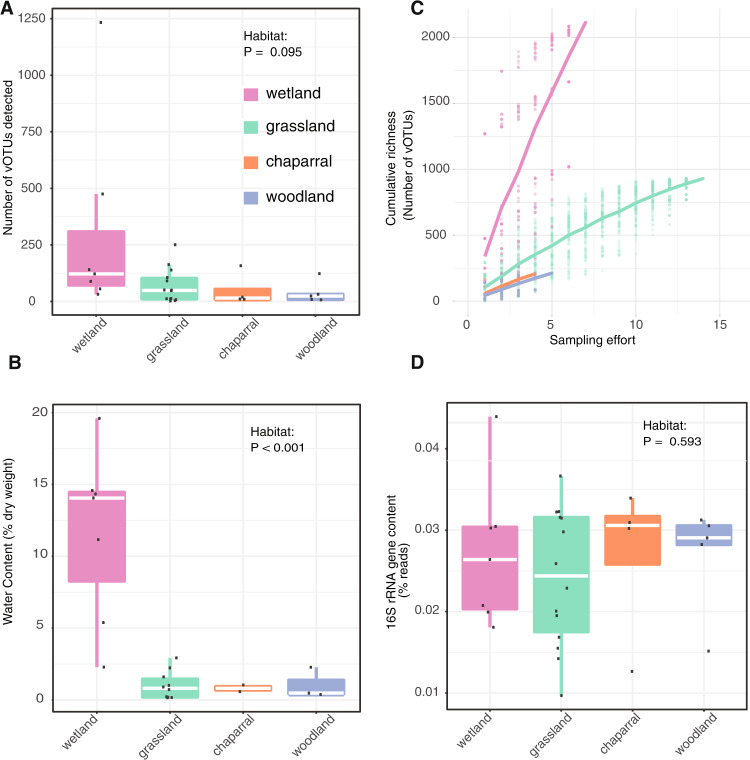
Multiple lines of evidence suggest that wetter soils harbored greater viral diversity than drier soils. We recovered the most vOTUs from wetlands, both in total (56% of all vOTUs were from wetlands) and per virome (on average, 307 vOTUs were recovered per wetland virome, compared to 116 from all habitats) (Fig. 1A). Unsurprisingly, wetlands had significantly greater moisture content than other habitats (Fig. 1B; ANOVA followed by Tukey multiple comparisons of means, p < 0.001), especially considering that soil samples were collected towards the end of the Mediterranean climate dry season, meaning that most habitats had not received precipitation for the preceding ~6 months. Although viral richness was highest in wetlands, this was not statistically significant (ANOVA model richness ~ habitat, p = 0.095). Still, nonparametric tests, which account for nonlinear correlations, revealed a significantly positive correlation between viral richness and soil moisture content (Spearman rho = 0.33, p = 0.036; Kendall tau = 0.22, p = 0.045). Viral community beta-diversity was also related to soil chemical properties overall (Mantel test, R2 = 0.43, p = 0.009; Table S1), while distance between sites only accounted for 5% of the variation (Partial Mantel test, R2 = 0.38, p = 0.009). Taken together, viral diversity was generally highest in wet soils.
Fig. 1. Evidence for higher viral richness in wetter soils.
Comparisons between habitats of (A) viral richness (number of identified vOTUs with coverage along at least 75% of the contig in a given virome), dots represent richness in individual viromes, (B) accumulation curves of cumulative vOTU richness as sampling effort increased, dots represent cumulative richness at each sampling effort across 100 permutations of virome order; the overlaid lines display the mean cumulative richness per habitat, (C) water content, calculated as (wet weight—dry weight) divided by dry weight, and (D) bacterial 16S rRNA gene content in the viromes, based on percent of viromic reads mapping to 16S rRNA reference genes. VIBRANT [21] was used to identify 3,490 viral contigs in our assemblies, and these viral contigs were clustered at 95% average nucleotide identity (ANI) into 3432 viral operational taxonomic units (vOTUs). For (A), (B), (D), raw data are plotted on top of the box plots, with white lines showing the median, boxes indicating 75% of the data, whiskers extending to 90%, and points beyond the whiskers indicating outliers.
We next wondered whether differences in sampling effort or bacterial content in the viromes could have produced the observed diversity patterns. For example, viral diversity could have appeared artificially higher in wetland habitats if wetland viral diversity was lower but well-sampled, compared to other habitats with higher diversity but poorer sampling. In a comparison of accumulation curves, such a pattern would include a more horizontal slope in the wetlands and a more steeply increasing vertical slope over sampling effort in the other habitats. We tested this by comparing accumulation curves across habitats, which revealed the opposite trend: wetlands exhibited the most steeply increasing slope and were thus likely to be the most under-sampled, in terms of true viral diversity (Fig. 1C). Given that relic DNA and small bacteria can pass through 0.22 µm filters, bacterial sequences are known to be present in viromes [18]. Although we had no specific reason to expect differences in the ratio of bacterial to viral DNA content among habitats, we wanted to exclude the possibility that viral diversity appeared artificially higher in wetlands due to a lower proportion of bacterial content in wetland viromes compared to other habitats. To compare bacterial content in viromes across habitats, 16S rRNA gene fragments were recovered from raw reads (Fig. 1D). The percentage of 16S rRNA gene sequences in each virome ranged from 0.01 to 0.044% (consistent with prior reports of 0.028% bacterial 16S rRNA gene content in similarly prepared viromes from agricultural soils [18]) and did not differ significantly by habitat (ANOVA, P = 0.595). Thus, viral diversity estimates did not seem to be disproportionately skewed by sampling effort or the presence of non-viral sequences in viromes. The higher diversity of viruses in wetlands compared to the other, drier habitats could be due to higher bacterial activity and therefore more susceptible hosts, leading to higher viral activity and diversity. Additionally, better soil connectivity in wet soil, leading to higher dispersion of free viruses and hosts, could increase the chances of virus-host encounters.
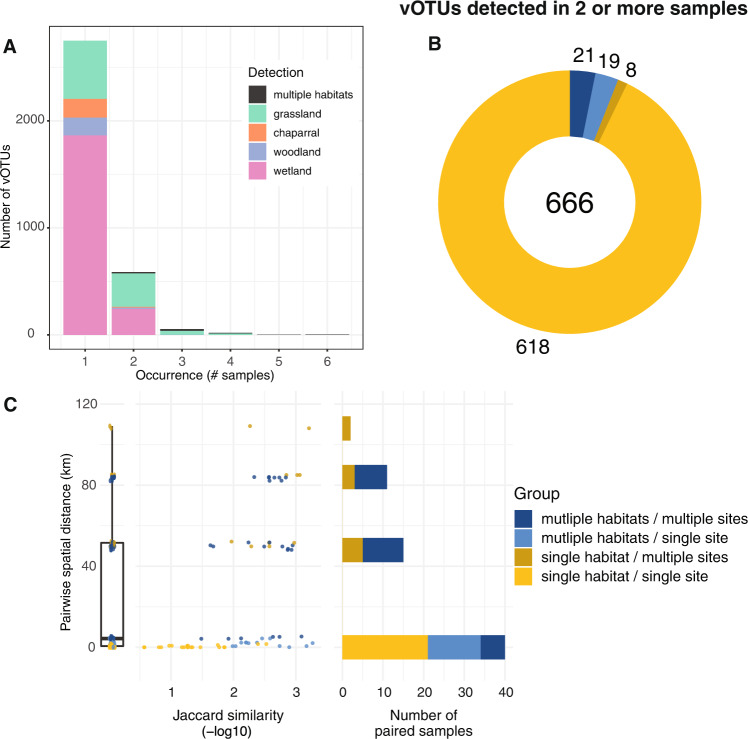
Perhaps the most striking result from this study was the uniqueness of each soil viral community. This feature has been previously highlighted in viruses identified in other environments, such as highly oligotrophic water [19] and hydraulic fracturing wells [8]. The majority of vOTUs (81%) in this regional study were only detected in a single virome (Fig. 2A, Fig. S3A). Of the 666 vOTUs detected in at least two viromes, 93% were detected in viromes from the same habitat and site (Fig. 2B, Fig. S3B). The most similar viral communities were from the same habitat and site (i.e., biological replicates) less than 1 Km apart (Fig. 2C). Within the same site, viral communities were less similar between habitats than within the same habitat. Additionally, viromes from the same habitat at different, nearby sites (within 6 Km) did not share any vOTUs, suggesting substantial differences in viral communities over local distances both within and between habitats. At greater distances, community similarity generally decreased, even between viromes from the same habitat (Fig. 2C). Still, 21 vOTUs were detected in multiple habitats across multiple sites (Fig. 2B), and some vOTUs were shared between the two farthest sites (109 Km apart, Fig. 2C), suggesting some degree of regional conservation of viral populations. Overall, results suggest substantial differences in soil viral community composition in the same habitat on the scale of meters, greater similarity of viral communities in close proximity and under similar environmental conditions, and a small number of vOTUs shared over regional distances.
Fig. 2. Substantial differences in vOTU detection and viral community composition between soil viromes, both within and across habitats.
A vOTU detection patterns, in terms of the number of viromes (samples) in which each vOTU was detected (x-axis) and the number of vOTUs exhibiting a given detection pattern (y-axis), with stacked bar colors indicating the habitat(s) in which each vOTU was detected, (B) vOTUs detected in more than one virome, colored by their detection patterns across sites and habitats (legend to the right of panel C), numbers indicate total vOTUs (center) and vOTUs per detection pattern, and (C) Pairwise community compositional similarity (x-axis) by geographic distance (y-axis) between viromes, colored according to detection patterns across habitats and sites (legend on the far right). Left graph: box plot of pairwise distances for all viromes (condensed x-axis). Middle graph: Each point represents the -log10 Jaccard similarity between two viromes along the x-axis, with lower values (left side of the x-axis) indicating greater viral community similarity. Right graph: Total number of virome pairs included at each geographic distance. Paired viromes were only included if they shared at least one vOTU.
Although soil viral communities are woefully under-sampled, making sweeping generalizations premature, results from this study and others converge on a picture of high local viral diversity, with communities differing substantially over space and by habitat, with little regional co-occurence of viral ‘species’ (vOTUs). Viral community distance-decay relationships have been observed across an 18-m long agricultural field [14] and in a 200 m2 grassland [20], consistent with the meters-scale differences in viral community composition between replicates from the same habitat observed in this study. However, the proportion of vOTUs shared over meters varied across these studies, with many vOTUs shared across the agricultural field but most vOTUs not shared between samples ~10 m apart in this study and in the grassland field, suggesting the potential for different spatial patterns in natural and managed soils, perhaps due to different amounts of soil mixing by management practices, such as tilling. Similarly substantial differences among viral communities on a regional scale were also identified in a study of grassland and peatland RNA viromes, which shared few viruses between samples [16]. However, ter Horst et al. showed that 4% of the vOTUs from a Minnesota, USA peatland were shared in other peatlands, often on different continents [15], consistent with the recovery of a small number of vOTUs shared over >100 Km distances here. Taken together, we propose that soil viral communities often display high heterogeneity within and among habitats, presumably due to a combination of host adaptations and microdiversity, dispersal limitation, and fluctuating environmental conditions over space and time.
Supplementary information
Acknowledgements
We thank the University of California, Davis Natural Reserves site directors and staff, particularly Shane Waddell, Catherine Koehler, Jeffrey Clary, Sarah Oktay, Suzanne Olyarnik, and Jacqueline Sones, for facilitating site access, showing us around the field sites, and providing logistical support for this work. We thank the three anonymous reviewers for their helpful and constructive suggestions.
Author contributions
AMH, CSM, SEG, and JBE collected the samples. DMD measured water content and sent samples for soil chemistry analysis. WCAB processed soils for viromics and extracted DNA. AMH performed the initial sequencing data processing and identification of viral contigs and 16S rRNA gene sequences. CSM and ETS wrote R scripts for data analysis. ETS, CSM, DMD, and AMH analyzed the data. ETS, JBE, and DMD wrote the manuscript. ETS and JBE edited and revised the manuscript. JBE supervised the project and provided resources. All authors edited and approved the final version of the manuscript.
Funding
Funding for this work was provided by new lab start-up to JBE from the University of California, Davis College of Agricultural and Environmental Sciences and Department of Plant Pathology, as well as by the U.S. Department of Energy (DOE), Office of Science, Office of Biological and Environmental Research (BER), Genomic Science Program, award number DE SC0021198 (grant to JBE).
Data availability
The datasets generated and/or analyzed in this study have been submitted to the NCBI sequence read archive (SRA) under BioProject number PRJNA831438 and will become available upon publication. The vOTU fasta sequences and R scripts are available on https://github.com/ellasiera/Nat_res_vOTUs.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Devyn M. Durham, Ella T. Sieradzki.
Supplementary information
The online version contains supplementary material available at 10.1038/s43705-022-00171-y.
References
- 1.Suttle CA. Viruses in the sea. Nature. 2005;437:356–61. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- 2.Emerson JB, Roux S, Brum JR, Bolduc B, Woodcroft BJ, Jang HB, et al. Host-linked soil viral ecology along a permafrost thaw gradient. Nat Microbiol. 2018;3:870–80. doi: 10.1038/s41564-018-0190-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williamson KE, Fuhrmann JJ, Wommack KE, Radosevich M. Viruses in soil ecosystems: an unknown quantity within an unexplored territory. Annu Rev Virol. 2017;4:201–19. doi: 10.1146/annurev-virology-101416-041639. [DOI] [PubMed] [Google Scholar]
- 4.Suttle CA. Marine viruses-major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–12. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 5.Trubl G, Kimbrel JA, Liquet-Gonzalez J, Nuccio EE, Weber PK, Pett-Ridge J, et al. Active virus-host interactions at sub-freezing temperatures in Arctic peat soil. Microbiome. 2021;9:208. doi: 10.1186/s40168-021-01154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trubl G, Jang HB, Roux S, Emerson JB, Solonenko N, Vik DR, et al. Soil viruses are underexplored players in ecosystem carbon processing. mSystems. 2018;3:e00076-18. doi: 10.1128/mSystems.00076-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roy K, Ghosh D, DeBruyn JM, Dasgupta T, Wommack KE, Liang X, et al. Temporal dynamics of soil virus and bacterial populations in agricultural and early plant successional soils. Front Microbiol. 2020;11:1494. doi: 10.3389/fmicb.2020.01494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daly RA, Roux S, Borton MA, Morgan DM, Johnston MD, Booker AE, et al. Viruses control dominant bacteria colonizing the terrestrial deep biosphere after hydraulic fracturing. Nat Microbiol. 2019;4:352–61. doi: 10.1038/s41564-018-0312-6. [DOI] [PubMed] [Google Scholar]
- 9.Srinivasiah S, Lovett J, Ghosh D, Roy K, Fuhrmann JJ, Radosevich M, et al. Dynamics of autochthonous soil viral communities parallels dynamics of host communities under nutrient stimulation. FEMS Microbiol Ecol. 2015;91:fiv063. doi: 10.1093/femsec/fiv063. [DOI] [PubMed] [Google Scholar]
- 10.Zablocki O, Adriaenssens EM, Cowan D. Diversity and ecology of viruses in hyperarid desert soils. Appl Environ Microbiol. 2016;82:770–7. doi: 10.1128/AEM.02651-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuzyakov Y, Mason-Jones K. Viruses in soil: Nano-scale undead drivers of microbial life, biogeochemical turnover and ecosystem functions. Soil Biol Biochem. 2018;127:305–17. doi: 10.1016/j.soilbio.2018.09.032. [DOI] [Google Scholar]
- 12.Wu R, Davison MR, Gao Y, Nicora CD, Mcdermott JE, Burnum-Johnson KE, et al. Moisture modulates soil reservoirs of active DNA and RNA viruses. Communications biology. 2021;4:1–11. doi: 10.1038/s42003-021-02514-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S, Sieradzki ET, Nicolas AM, Walker RL, Firestone MK, Hazard J, et al. Methane-derived carbon flows into host–virus networks at different trophic levels in soil. Proc Natl Acad Sci USA. 2021;118:e2105124118. doi: 10.1073/pnas.2105124118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santos-Medellin C, Zinke LA, Ter Horst AM, Gelardi DL, Parikh SJ, Emerson JB. Viromes outperform total metagenomes in revealing the spatiotemporal patterns of agricultural soil viral communities. ISME J. 2021;15:1956–70. doi: 10.1038/s41396-021-00897-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ter Horst AM, Santos-Medellín C, Sorensen JW, Zinke LA, Wilson RM, Johnston ER, et al. Minnesota peat viromes reveal terrestrial and aquatic niche partitioning for local and global viral populations. Microbiome. 2021;9:233. doi: 10.1186/s40168-021-01156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hillary LS, Adriaenssens EM, Jones DL, McDonald JE. RNA-viromics reveals diverse communities of soil RNA viruses with the potential to affect grassland ecosystems across multiple trophic levels. ISME Commun. 2022;2:1–10. doi: 10.1038/s43705-022-00110-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roux S, Adriaenssens EM, Dutilh BE, Koonin EV, Kropinski AM, Krupovic M, et al. Minimum information about an uncultivated virus genome (MIUViG) Nat Biotechnol. 2019;37:29–37. doi: 10.1038/nbt.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicolas AM, Jaffe AL, Nuccio EE, Taga ME, Firestone MK, Banfield JF. Soil candidate phyla radiation bacteria encode components of aerobic metabolism and co-occur with nanoarchaea in the rare biosphere of rhizosphere grassland communities. mSystems. 2021;6:e0120520. doi: 10.1128/mSystems.01205-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taboada B, Isa P, Gutiérrez-Escolano AL, Del Ángel RM, Ludert JE, Vázquez N, et al. The geographic structure of viruses in the Cuatro Ciénegas Basin, a unique oasis in Northern Mexico, reveals a highly diverse population on a small geographic scale. Appl Environ Microbiol. 2018;84:e00465-18. doi: 10.1128/AEM.00465-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santos-Medellín C, Estera-Molina K, Yuan M, Pett-Ridge J, Firestone MK, Emerson JB. Spatial turnover of soil viral populations and genotypes overlain by cohesive responses to moisture in grasslands. bioRxiv. [DOI] [PMC free article] [PubMed]
- 21.Kieft K, Zhou Z, Anantharaman K. VIBRANT: automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences. Microbiome. 2020;8:90. doi: 10.1186/s40168-020-00867-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed in this study have been submitted to the NCBI sequence read archive (SRA) under BioProject number PRJNA831438 and will become available upon publication. The vOTU fasta sequences and R scripts are available on https://github.com/ellasiera/Nat_res_vOTUs.