Abstract
The mixoplankton green Noctiluca scintillans (gNoctiluca) is known to form extensive green tides in tropical coastal ecosystems prone to eutrophication. In the Arabian Sea, their recent appearance and annual recurrence have upended an ecosystem that was once exclusively dominated by diatoms. Despite evidence of strong links to eutrophication, hypoxia and warming, the mechanisms underlying outbreaks of this mixoplanktonic dinoflagellate remain uncertain. Here we have used eco-physiological measurements and transcriptomic profiling to ascribe gNoctiluca’s explosive growth during bloom formation to the form of sexual reproduction that produces numerous gametes. Rapid growth of gNoctiluca coincided with active ammonium and phosphate release from gNoctiluca cells, which exhibited high transcriptional activity of phagocytosis and metabolism generating ammonium. This grazing-driven nutrient flow ostensibly promotes the growth of phytoplankton as prey and offers positive support successively for bloom formation and maintenance. We also provide the first evidence that the host gNoctiluca cell could be manipulating growth of its endosymbiont population in order to exploit their photosynthetic products and meet critical energy needs. These findings illuminate gNoctiluca’s little known nutritional and reproductive strategies that facilitate its ability to form intense and expansive gNoctiluca blooms to the detriment of regional water, food and the socio-economic security in several tropical countries.
Subject terms: Environmental sciences, Water microbiology
Introduction
Green Noctiluca scintillans (hereinafter gNoctiluca) is one of the fastest biogeographically spreading marine planktonic organisms in the world’s oceans [1–5]. The dinoflagellate N. scintillans is an enigmatic species because it has the capability to occupy exclusively heterotrophic (red Noctiluca, rNoctiluca) as well as mixoplanktonic niches (gNoctiluca) [4]. The photoautotrophy exhibited by gNoctiluca is due to a seemingly loose but exclusive mutualistic partnership with the prasinophyte green alga Protoeuglena noctilucae [6–8]. This exclusive relationship between the host and free-swimming endosymbionts makes gNoctiluca different from other mixoplankton that have either permanent chloroplasts, or those that sequester (steal) chloroplasts (kleptoplasty). gNoctiluca also feeds on other protists especially diatoms as evidenced in the Arabian Sea [9] and in the Gulf of Thailand [10] as well as in laboratory experiments [11–13]. Conversely, the more cosmopolitan rNoctiluca, which is thought to acquire an orange-red coloration because of carotenoids from the plankton it feeds on [14], is exclusively heterotrophic, has no symbionts and is a voracious grazer [15–17]. Unlike rNoctiluca, which is globally ubiquitous [4, 18], gNoctiluca is restricted to the tropics and sub-tropics and particularly in coastal ecosystems that are subjected to protracted nutrient overloading and hypoxia, either due to domestic sewage outflow, or heavy use of synthetic fertilizers for agriculture [2, 9, 17, 19].
gNoctiluca’s emergence as a dominant bloom forming organism in the Arabian Sea during the winter monsoon is fairly recent [3, 5, 20–23] and no reports exist of these blooms in the northern Arabian Sea prior to 2000. Their sudden appearance has been linked to the intrusion of sub-oxic waters from an expanding Oxygen Minimum Zone [9, 24], which stimulates endosymbiont photosynthesis and changes in inorganic nutrient stoichiometry [25]. In addition, nutrient replenishment of oligotrophic waters from winter convective mixing [26] produces large diatom blooms [27–30] that serve as a food source for this mixoplankton. In the Sea of Oman, gNoctiluca blooms were also first seen around 1999 [31] and since then, have been appearing almost year round, and often, in association with the shoaling of hypoxic water [3, 32–34].
In the Arabian Sea, it is still inexplicable how gNoctiluca blooms, which first appear in late December and early January, are able to persist even until late March when waters turn oligotrophic, before their termination either by grazing by its major predators, salps and jellyfish, and/or lethal rising seawater temperatures [9, 35]. Along the coast of Oman, gNoctiluca alone or in combination with salps and jellyfish swarms are especially devastating to artisanal fisheries and freshwater supply from desalination plants [36]. While clearly there have been considerable advances in our understanding of the ecology of this organism, very little is known about the molecular and metabolic mechanisms that underpin its capacity to outgrow other phytoplankton and mixoplankton to form massive green tides. Several studies have examined the formation of Noctiluca blooms in various ecosystems, but most have focused on the red form (rNoctiluca) [1, 17, 20]. gNoctiluca on the other hand, is less studied, with its fundamental nutrition and reproduction mechanisms remaining elusive. Here we report findings from a field study that integrated ecological, physiological, and molecular analyses during gNoctiluca blooms in the Sea of Oman, which provide novel insights into its fundamental reproductive and nutritional strategies that contribute not only to the formation of gNoctiluca blooms but to their persistence even under conditions unfavorable to most other protists.
Materials and methods
Study areas, sample collection, species isolation and measurements
Our primary sampling site was Seeb Jetty, located in a shallow semi-enclosed bay in Muscat, Oman (Fig. S1), chosen because of its proximity to the shore laboratory, and because gNoctiluca blooms occur here with predictable regularity every year. We also sampled at 6 stations (ST0 to ST5) along a transect that extended 20 km offshore into the Sea of Oman (Fig. S1). Sampling offshore was guided by Level 2 Aqua-MODIS satellite images of the previous day, which showed high Chl a concentrations (>10 mg m−3) and blooms in offshore flowing filaments associated with a cyclonic eddy (Fig. S1). Sampling at Seeb Jetty was undertaken daily from the 2nd to 11th Feb. 2018, usually at high tide each day, to monitor the development of the gNoctiluca bloom.
After recording surface seawater temperatures, 10 L of surface seawater were collected in a polycarbonate carboy that had been previously washed with 5% technical grade HCl and rinsed with deionized water. Water samples were immediately covered with a black sheet to shield the biological constituents from light, and immediately transported to the laboratory for further analysis as described below.
In the laboratory, the carboy was gently mixed, and duplicate samples of 50 mL were immediately fixed with two drops of glutaraldehyde for microscopy and FlowCAM [12] determination of cell counts and phytoplankton taxonomy. Glutaraldehyde was chosen over buffered Formalin + Lugol’s iodine as it preserves the gNoctiluca cells intact. Aliquots of 50–300 mL of seawater sample were collected daily, and filtered onto 0.7 µm Whatman GF/F. Filters were stored at −20 °C for measurements of phytoplankton Chl a extracted from the filters in cold 90% acetone and fluorometrically determined in a Turner Designs Trilogy fluorometer pre-calibrated with standard Chl a (Sigma-Aldrich). Phaeopigments were estimated by acidifying the sample with 1 M HCl.
For analysis of dissolved nutrients (nitrite, nitrate, ammonium and dissolved inorganic phosphate), 50 mL of seawater was gently filtered through a 0.22 µm Nucleopore syringe filter and stored at −20 °C until analysis in a SmartChem (Westborough, MA, USA) discreet nutrient auto-analyzer. Dissolved inorganic phosphorus measurements were determined by colorimetric method for orthophosphate using the USEPA method 365.1. Nitrogen (nitrate, nitrite, ammonium and urea) measurements were based on American Public Health Association’s protocols.
Spearman’s rank correlation coefficient (corrplot library in R) [37] was undertaken to infer the relationships between nutrients (nitrate, nitrite, ammonium and phosphate), cell counts and Chl a in two phytoplankton fractions (whole and <200 µm).
Growth rates of P. noctilucae in symbiosis with gNoctiluca and as a free-living culture
These experiments were undertaken in the laboratory using isolates of gNoctiluca with its symbiont P. noctilucae and as well as P. noctilucae isolated from gNoctiluca cells and cultured independent of the host. About 2000 gNoctiluca cells growing exponentially in f/20 media, were transferred into a set of triplicate 2 L polycarbonate bottles. Ten mL of exponentially growing cells of P. noctilucae were transferred into another set of triplicate 2 L polycarbonate bottles containing f/20 media. Both sets of bottles were incubated for 10 days under identical conditions at 26.5 °C under a light:dark cycle of 14 L:10 D with a photon flux of about 200 µmol photons m−2 s−1. Growth rates were based on changes in Chl a concentration recorded during the mid-exponential phase of cells for each flask.
Sample collection, RNA isolation, and high-throughput metatranscriptome sequencing
For transcriptomic analyses, duplicate field samples (collected on 2nd Feb.) were filtered onto Nucleopore 3 µm pore size polycarbonate filters, immediately fixed in 1 mL Trizol and transferred into liquid Nitrogen. In parallel, to profile gene expression in the aposymbiotic alga P. noctilucae growing in the free-living mode, 50 mL (~1 × 106 cells) aliquots of P. noctilucae were filtered onto 3 µm Nucleopore membrane filters, and transferred into a 2 mL microcentrifuge tube containing 1 mL Trizol and immediately placed in liquid nitrogen.
To isolate RNA, both sets of samples were disrupted using a FastPrep-24 bead mill (MP Biomedicals, Solon, OH, United States) prior to RNA extraction as previously reported for field samples [38]. Total RNA was extracted following protocols detailed in Lin et al. [39] and Luo et al. [40] and quantified using a NanoDrop-2000 Spectrophotometer (Thermo Fisher Scientific, United States). For RNA high-throughput sequencing, 1 µg of total RNA was used for mRNA enrichment using NEBNext Poly (A) mRNA Magnetic Isolation Module (New England Biolabs, United States). mRNA was then fragmented and used as template to synthesize double-strand cDNA. Fragments with specific size (~400 bp) were chosen for sequencing on an Illumina HiSeq 4000 instrument according to the protocol of Zhang et al. (2019) [38].
Bioinformatic analysis
Raw reads were trimmed to eliminate low quality reads and reads with adaptor using Trimmomatic V0.30 [41]. The resultant clean reads were assembled de novo in Trinity 2.0, and clustered to unigenes using TGICL (TIGR Gene Indices clustering tools, V2.1) [42]. These unigenes were annotated against NCBI non-redundant (nr) database, Swiss-Prot and Pfam database with a cutoff of 1e−5 and mapped to KEGG pathways for further analysis. Meanwhile, to quantify the expression level of genes in gNoctiluca, clean reads were mapped to the Noctiluca database in MMETSP0253 using Bowtie2 (parameters: -sensitive). Gene expression was quantified by normalizing the mapped reads to total transcriptomic reads of gNoctiluca and presented as FPKM value (Fragments Per Kilobases per Million mapped reads). Most highly expressed genes (HEGs) (top 25%) were further examined for their functions potentially related to bloom development.
Genes differentially expressed between gNoctiluca bloom and cultured rNoctiluca
Besides the analysis of most HEGs, we also investigated the genes that were upregulated during bloom development. This would be ideally undertaken by comparing the bloom samples with pre-bloom samples. Unfortunately, our pre-bloom samples contained too few gNoctiluca cells to obtain meaningful data (most gNoctiluca genes showed zero expression level). As a compromise, we compared the bloom samples with cultured Noctiluca samples (MMETSP), assuming that the culture condition was not quite a bloom condition. Differentially expressed genes (DEGs) between our metatranscriptome (OMF02) and transcriptome data (SRR1296929) were analyzed using edgeR V3.28.1 [43]. The moderate biological coefficient of variation of 0.2 was used to estimate dispersion. DEGs of edgeR were screened with a threshold of FDR ≤ 0.05 and the log2(Fold change) ≥1.
Nutrient release experiments
gNoctiluca cells were collected from blooms at two locations, one at Seeb Jetty on 3rd Feb. 2018 and the second from ST0 on 13th Feb. 2018. For these and the following experiments, all 500 mL clear polycarbonate bottles were acid-washed and soaked overnight with 5% laboratory grade HCl, rinsed with Milli-Q distilled water, and autoclaved prior to use.
For each experiment, 500 mL aliquots of whole water samples were gently pre-filtered through a sterilized 200 µm mesh net. The filtrate was discarded and the gNoctiluca cells retained on the net were gently washed with 0.22 µm pre-filtered seawater (FSW) from the study site to eliminate other species, and then carefully transferred into 4 × 500 mL polycarbonate bottles containing FSW. After sampling for initial nutrient (urea, ammonium, nitrate, nitrite and phosphate) concentrations, 2 bottles were covered with four layers of aluminum foil (dark treatment), while 2 were left uncovered (light treatment). All 4 bottles were incubated under ~70 µmol photons m−2 s−1, and a photoperiod of 14 L:10 D, for 8 days at 26.5 °C without adding nutrients or prey. 25 mL aliquots were drawn from each bottle for the first 2 days and every second day over the next 5 days. The aliquots were gently filtered through a 0.22 µm Nucleopore syringe filter and the filtrate was stored at −20 °C for nutrient analysis in the SmartChem auto-analyzer as described above.
Nitrogenous nutrient (ammonium and urea) and carbon uptake experiments
Nitrogenous and carbon uptake experiments were also undertaken on seawater samples from the Seeb Jetty on 3rd and 7th Feb as well as from samples collected from the 6 offshore stations ST0- ST5. The experiments were conducted using whole seawater samples, which included gNoctiluca and phytoplankton, and samples pre-filtered through a 200 µm Nybolt mesh to remove gNoctiluca. Microscopic examination showed this fraction (<200 µm fraction) consisted mostly of diatoms but also some small-sized gNoctiluca that may have passed through the mesh. Any large grazers visible in the whole water sample were removed with a plastic Pasteur pipette before the sample was transferred into bottles. Prior to start of the experiment, separate 50 mL aliquots of whole water samples were syringe filtered (0.22 µm Nucleopore) and the filtrate was stored at −20 °C for nutrient analyses.
The two fractions (whole water and <200 µm) were then dispersed into triplicate 250 mL acid-washed, Milli-Q water rinsed and autoclaved polycarbonate bottles. The bottles were amended with 15N labeled NH4Cl (98% atom 15N) such that final 15N-NH4 concentrations were 1 µM. A similar set was spiked separately with 1 µM 15N labelled urea (98% atom 15N). A third set was spiked with 1 µM 13C labelled bicarbonate (98% atom 13C). All bottles were sealed and placed in the incubator under same conditions as the nutrient release experiments described above. We were not able to conduct experiments with 15N labeled nitrate because it was unavailable at the time of the experiment.
After 24 h of the incubation, samples were gently filtered onto pre-combusted (400 °C for 1 h) Whatman GF/F filters, washed repeatedly with FSW to rid the filters of residual non-assimilated 15N nitrogenous nutrients and 13 C bicarbonate, dried in an oven at 60 °C, and stored in a desiccator until the time of analysis. Nitrogen and carbon content as well as 15N and 13 C isotopic enrichment on the filters were measured on a Thermo Delta V Plus continuous flow stable isotope ratio mass spectrometer equipped with a Costech 4050 Elemental Analyzer [44]. Nitrogen and carbon uptake rates were calculated following standard protocols [45] after correcting for residual isotope on the filters from the control samples.
Nitrogenous and carbon uptake rates of gNoctiluca were estimated by subtracting the uptake rates of the <200 µm fraction from that of the whole seawater samples. The difference represented the lower bound of gNoctiluca uptake rates, because some small-sized gNoctiluca passed into the <200 µm fraction, with the rates in the whole water being the upper bound.
Results
gNoctiluca bloom dynamics
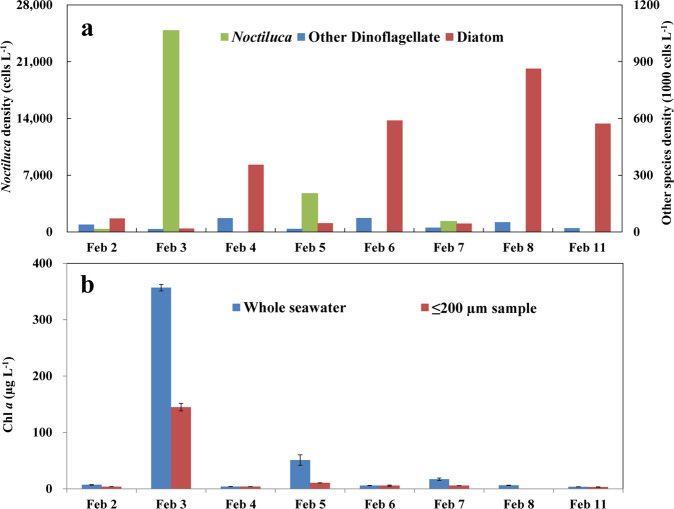
The study was undertaken from 2nd Feb. to 11th Feb., 2018 at Seeb Jetty, Muscat, a sheltered bay along the coast of the Sultanate of Oman where gNoctiluca was in an early stage of bloom formation (Fig. S1). After a slow start, gNoctiluca cell densities rose from 3.9 × 102 cells L−1 on 2nd Feb. to 2.5 × 104 cells L−1 on the following day (Fig. 1a), resulting in a bloom with a high cell-specific growth rate of 1.86 d−1 that dramatically changed the color of the previously clear bay waters to green. On the next day (4th Feb.), the color of the waters abruptly changed to golden yellow, and gNoctiluca cell numbers dropped to almost negligible. However, on 5th Feb., the waters turned light green and gNoctiluca cell concentrations rose once again with cell counts of 4.8 × 103 cells L−1. This boom-and-bust cycle repeated itself on the 7th Feb. although the cell numbers were lower (1.3 × 103 cells L−1), following which gNoctiluca counts remained low until their reappearance in small numbers on the 11th of Feb. (28 cells L−1). Microscopy and FlowCAM analyses also showed the presence of diatoms and dinoflagellates, but cell counts of both groups were always very low when gNoctiluca counts were high (Fig. 1a). Diatom counts were highest on the 8th of Feb. (8.6 × 105 cells L−1), likely due to reduced grazing pressure from gNoctiluca and a sudden increase in nutrients in the water column from death and decay of gNoctiluca cells (Fig. S2a). Chl a concentrations in whole water samples exhibited a strong positive correlation with gNoctiluca abundance (Fig. 1b; Fig. S3) indicating a healthy population of photosynthesizing symbionts, P. noctilucae, in the host cell. Throughout the study period, seawater temperature was fairly stable (22.5 ± 0.3 °C), excluding the contribution of temperature to the bloom dynamics.
Fig. 1. gNoctiluca and phytoplankton dynamics in the study period during a bloom at Seeb Jetty, Muscat, Sultanate of Oman.
a Contrasting cell abundances (cells L−1) between gNoctiluca and phytoplankton functional groups. b Chlorophyll a (Chl a) concentration (µg L−1) for whole seawater and ≤200 µm size fraction (excluding most gNoctiluca cells). Error bar stands for standard deviation.
Active sexual reproduction during bloom development
Noctiluca is known to reproduce asexually and sexually, and a high number of gametes observed in the host cells (progametes) suggest a potentially vital role of sexual reproduction as an accelerator for bloom formation [10, 46–49]. We also observed gametogenesis inside gNoctiluca, and zoospores (by morphology mixture of zygotes and released mature gametes) were especially abundant when the bloom transitioned from green to golden yellow (Figs. S4 and S5). Because it is very challenging to distinguish zygotes from free-swimming gametes in preserved samples, counting the abundance of zygotes was not possible. We used an indirect method, i.e., estimating the percentage of gNoctiluca cells that were in gametogenesis and multiplying it by the number of gametes each gametocyte produces previously reported [10] for Noctiluca, then dividing the value by two (two gametes produces one zygote). We observed that the percentages of gNoctiluca experiencing sexual reproduction on Feb 2, Feb 4, and Feb 6 were clearly more than that on Feb 3, Feb 5, and Feb 7 (Fig. S6), when the peak of gNoctiluca density appeared. We estimated that ~50% of gNoctiluca carried progametes on Feb 2, and according to previous reports of ~1024 gametes produced by one Noctiluca gametocyte, this potentially gave a maximum of 256 doubling of cells per day, which roughly agrees with our observed growth rate described above.
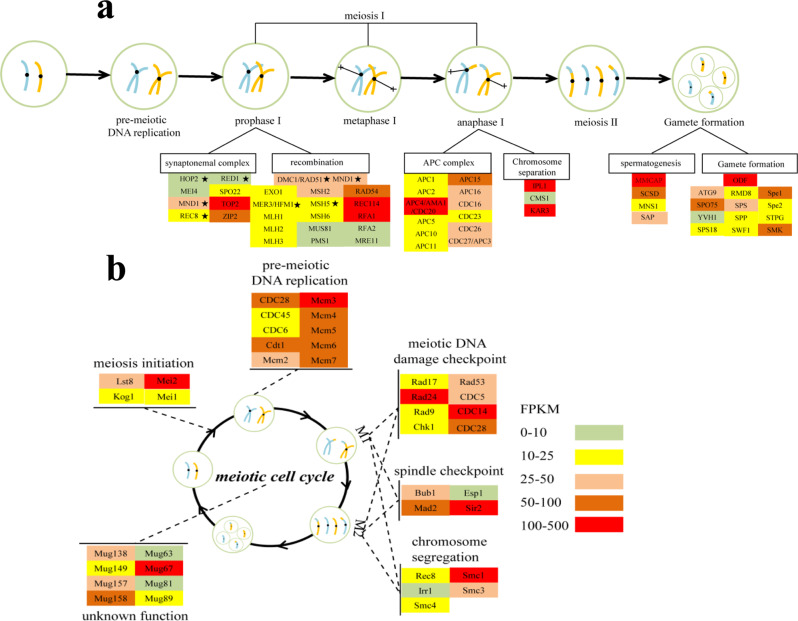
Consistent with our field observations of zygotes, transcriptomic data of gNoctiluca from Seeb Jetty on 2nd Feb., immediately before the bloom, showed 97 meiosis-related genes (Fig. 2), most of which were HEGs. As our metatranscriptomics sequences were annotated against previously reported Noctiluca transcriptome data (MMETSP0253; see Materials and Methods) with strict parameter setting, these genes (and all others discussed in this paper) were clearly from gNoctiluca. Further analyses indicated that seven of them were eukaryotic meiosis-specific genes: HOP2, MND1, REC8, RED1, DMC1, MER3/HFM1 and MSH [50–52]. Among these, HOP2 and MND1 interact with DMC1 by constituting a heterodimeric complex [53], promoting the interhomolog meiotic recombination [54]. REC8 is a meiosis-specific cohesin subunit [55] and RED1 is a key meiotic prophase factor [56], both known to play an important role in meiosis I in typical sexual organisms. MER3/HFM1 and MSH4/MSH5 are components of ZMM proteins (also known as synapsis initiation complex) that directly promote meiotic recombination (Fig. 2a) [57]. Some other meiosis-specific genes documented in model sexual eukaryotes, such as HOP1, MSH4, SPO11 and ZIP1, were not detected, probably because gNoctiluca does not use these genes or has evolutionarily replaced them with alternatives. For instance, genes MRE11 and EXO1 that we identified in gNoctiluca are known to perform the same function as SPO11, which is to repair meiotic DNA double-strand breaks (DSBs) [58].
Fig. 2. Active sexual reproduction in gNoctiluca during the bloom.
a Expression profile of genes associated with major events in the gametogenesis process in gNoctiluca. b Expression profile of meiosis regulatory genes. Asterisks depict eukaryotic meiosis-specific core genes. Color bar and scale in a denotes gene expression level (normalized by FPKM), which is also applicable to b.
Also represented in the transcriptomes was the anaphase-promoting complex (APC) (Fig. 2a), which is important in both meiotic and mitotic cell cycles as it induces the degradation of several factors that act on spindle-pole separation and spindle disassembly [59]. Among the several subunits of APC detected, APC15/MND2 is essential to meiosis but not required for mitosis [60]. We also identified KAR3 and IPL1, which are vital for homologous chromosome synapsis during meiosis [61, 62], both of which were highly expressed (i.e., in the HEGs group).
Furthermore, transcriptomes contained many genes associated with gamete formation such as MMCAP that functions in spermatogenesis [63], as well as MNS1, SAP and SCSD. Interestingly, several of these genes (SPE, STPG, ODF, SPO75, SPS18, RMD8, SPS and SWF1) are known to be involved in the structure of sperm flagella and sporulation in mice [64, 65] and yeast [66, 67]. We also identified a series of meiosis regulatory genes (Fig. 2b), of which, Mei2, was highly expressed (FPKM = 190.03). Mei2 is a regulatory RNA-binding protein which plays a vital role in the meiosis initiation [68, 69] and was originally discovered in yeast, then in plants, and more recently in several algal species including Fugacium kawagutii [70], Scrippsiella trochoidea [71], Prorocentrum donghaiense [38], Heterosigma akashiwo [72], and Prorocentrum cordatum [73]. Other highly expressed meiosis genes found in the gNoctiluca bloom included mini chromosome maintenance proteins (MCM2-7), which plays an important role in DNA replication during pre-meiotic S phase [74]. CDC14 and CDC28, Mad2, Rad24 and Smc1, which regulate M phase, spindle checkpoint and DNA damage checkpoint, respectively [67]. Finally, several meiotically up-regulated genes, originally described in yeast (Mug, Fig. 2b), were also actively expressed in the gNoctiluca bloom.
Moreover, we also compared the expression level of some meiosis genes in our bloom samples with that in the previously reported rNoctiluca laboratory culture samples (MMETSP0253). Among the genes common in the two sets of samples, four meiosis-specific genes (MND1, REC8, DMC1 and MSH5) were more highly expressed in our sample than in the cultured sample, and some meiosis-related genes such as Mei2 also exhibited significantly higher expression (P < 0.05) (Fig. S7). These above findings clearly demonstrate the active sexual reproduction (meiosis to produce gametes) during the gNoctiluca bloom, suggesting its vital role in driving the bloom.
Positive nutrient feedback loop associated with the bloom
It is generally believed that mixoplankton turn to phagotrophy when nutrients are depleted in the environment [7]. During our study period, P and N nutrient concentrations at Seeb Jetty were non-limiting and showed a striking increase. Urea, ammonium, nitrate and phosphate concentrations were moderate (3.5, 2.5, 2.8 and 0.74 µM respectively) on 2nd Feb., but rose by approximately tenfold (34.1, 32.8, 30.9 and 18.2 µM, respectively) on 3rd Feb. (Fig. S2a), when the gNoctiluca bloom appeared (Fig. 1a). Similarly, at the sampling station ST0 in the coastal waters of the Sea of Oman, where the bloom was also observed, urea, ammonium, nitrate and phosphate concentrations were also high (13.23, 17.55, 3.01 and 8.68 µM) (Fig. S2b). Nutrients at non-bloom sites remained at moderate levels. Thus, the elevated nutrient concentrations in bloom waters were most likely from gNoctiluca.
With previous information that rNoctiluca and gNoctiluca accumulate high concentrations of ammonium and urea within their central cytoplasm [12, 75], we conducted experiments to examine if gNoctiluca was the source of the nutrients measured in the water. When gNoctiluca cells from two separate blooms were incubated in 0.22 µm-filtered seawater in light and dark bottles for 5–8 days without addition of nutrient or prey, we measured steady increases of nutrients in the media during the incubation. Within a day, nutrients increased in both treatments by 132–281 µM for ammonium, 23.6–50.3 µM for urea, 2.5–18.1 µM for nitrate and 39.4–68.1 µM for phosphate (Fig. S8a). Of the total released nitrogenous nutrients (sum of nitrite, nitrate, ammonium, and urea), ammonium accounted for >78% (Fig. S8b). For such an enormous amount of ammonium to be released, it firstly had to accumulate within gNoctiluca, either via active cell membrane transport and/or ammonium regeneration from digesting and metabolizing of ingested prey.
Nitrogen uptake measurements of gNoctiluca with 15N labeled ammonium or 15N labeled urea (Fig. S9) clearly showed gNoctiluca’s N-nutrient uptake and its preference for ammonium over urea. We also saw higher uptake rates of ammonium and urea at bloom sites relative to non-bloom sites (Fig. S9). In accordance with the uptake data, both ammonium and urea transport genes (AMT and URT respectively) were actively expressed in gNoctiluca, but the expression of AMT (FPKM = 443.3) was four times higher than that of URT (FPKM = 101.1) (Fig. 3). Although we were unable to conduct parallel 15N-nitrate uptake incubations, nitrate transporter genes in gNoctiluca were absent in the transcriptome, implying lower preference for or inability to use nitrate as a nutrient source.
Fig. 3. Highly expressed genes (top 25%) and their related metabolic processes in gNoctiluca.
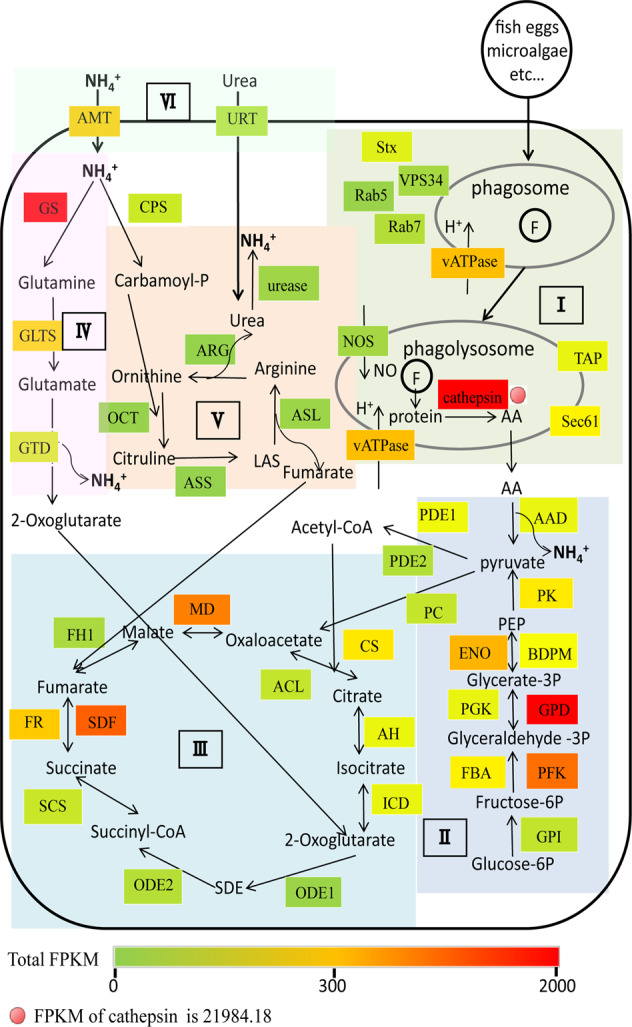
I Phagocytosis metabolism. II Glycolysis. III TCA cycle. IV Ammonium metabolism. V Urea cycle. VI Nitrogen transport. Color bar denotes gene expression level (normalized by FPKM). Note that the expression of cathepsin was so high (FPKM = 21984.18) that it far exceeds the color scale used to properly indicate the differential expression of other genes. Stx Syntaxin, Rab5 Ras-related protein Rab5, VPS34 Phosphatidylinositol, 3-kinase vps34, vATPase V-type proton ATPase, Rab7 Ras-related protein Rab7, NOS Nitric oxide synthase, Sec61 Protein transport protein Sec61, TAP ABC transporter B family, AAD amino acid deaminase, GPI Glucose-6-phosphate isomerase, PFK Phosphofructokinase, FBA Fructose-bisphosphate aldolase, GPD Glyceraldehyde-3-phosphate dehydrogenase, PGK Phosphoglycerate kinase, BDPM 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase, ENO Enolase, PK Pyruvate kinase, PDE1 Pyruvate dehydrogenase E1 component, PDE2 Pyruvate dehydrogenase E2 component, PC Pyruvate carboxylase, CS Citrate synthase, ACL ATP-citrate (pro-S-)-lyase, AH Aconitate hydratase, ICD Isocitrate dehydrogenase [NADP], ODE1 2-oxoglutarate dehydrogenase complex component E1, ODE2 2-oxoglutarate dehydrogenase complex component E2, SCS Succinyl-CoA synthetase, FR Fumarate reductase, SDF Succinate dehydrogenase [ubiquinone] flavoprotein subunit, FH1 Fumarate hydratase class I, MD Malate dehydrogenase, AMT Ammonium transporter, GS Glutamine synthetase, GLTS Glutamate synthase, GTD NAD-specific glutamate dehydrogenase, CPS Carbamoyl-phosphate synthase [ammonia], OCT Ornithine carbamoyltransferase, ASS Argininosuccinate synthase, ASL Argininosuccinate lyase, ARG Arginase.
Our metatranscriptomes also indicated active grazing as a major source of accumulated N-nutrients in gNoctiluca. A large fraction of genes involved in phagocytosis and nitrogenous molecule metabolism was highly expressed (Table S1, Fig. 3). One of these genes encodes the digestive enzyme cathepsin, which cleaves proteins and peptides [76], potentially producing amino acids, and it was the most highly expressed gene family found in this study with a total FPKM of 21,984.18 (Fig. 3). Furthermore, the HEG pool also contained genes encoding amino acid deaminases (FPKM = 243.49) that act on amino acids to produce ammonium. In addition, NAD-specific glutamate dehydrogenase (FPKM = 196.30) and urease (FPKM = 36.6), catalyzing reactions that generate ammonium from glutamate and urea, respectively, were also highly expressed (Fig. 3).
Phagocytosis mainly consists of three processes, which together include motility, prey capture, phagosome maturation and prey digestion [77, 78]. Genes that encode actin and tubulin, the structural components of cytoskeleton, were most abundant and most highly expressed (SI Appendix, Table S1). This set of HEGs also included some motility-related genes that encode kinesin, myosin, dynein and flagella-related proteins. Also highly expressed in our transcriptome were MRCK, ARP2/3 complex, calreticulin and calnexin, known to be associated with particle uptake [77, 79]. Of these, the ARP2/3 complex and calreticulin were HEGs. Nine other HEGs involved in the maturation of phagosome, including small GTPase Rab5 and PIK3C3, as well as vATPase that act to regulate phagosome acidification were also recorded. Syntaxin and Sec22 as parts of SNARE complex (soluble N-ethylmaleimide-sensitive factor activating protein receptor), which are responsible for endosome membrane fusion and vesicle trafficking, were also highly expressed. The two other HEGs, TAP and Sec61, are involved in regulation of transmembrane transport in phagolysosome. A range of HEGs involved in the production of ammonium by phagocytosis (Fig. 3) complemented the increased concentration of phaeopigments (chlorophyll digestion products) in the bloom samples (Fig. S10), indicating that grazing on phytoplankton was likely the dominant energy and nutrition source for gNoctiluca bloom formation and the dominant mode of ammonium accumulation within gNoctiluca cells.
It is believed that accumulation of ammonium within the symbiosome of gNoctiluca helps support photosynthesis of the endosymbiont P. noctilucae [2, 9], which is evident from 13C-HCO3 tracer uptake experiments (Fig. S9). However, we speculate that ammonium accumulation within gNoctiluca may stimulate sexual reproduction in gNoctiluca. Although for most dinoflagellates, environmental stresses such as nutrient deficiency and temperature are the most common triggers [49, 80], Noctiluca is clearly an exception because its active gametogenesis occurred during actively growing blooms. Furthermore, ammonium is indeed able to induce sexual reproduction and gamete formation in the diatom Thalassiosira pseudonana, as evidenced by elevated expression of meiosis genes, triggering sexual morphologies, including oogonia, auxospores and spermatogonia [81]. This is consistent with our observations in gNoctiluca, where we also observed high expression of gamete-forming genes and active gamete formation during the bloom in conjunction with an incredibly large amount of ammonium released by gNoctiluca. If verified experimentally, this finding will have profound implications in dinoflagellate bloom ecology.
High photosynthetic activity but inactive cell division in endosymbionts in-hospite: potential host manipulation for photosynthates
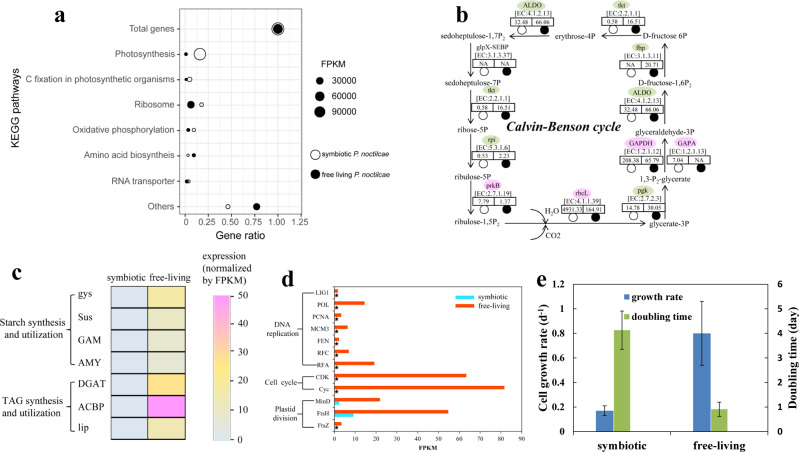
In mutualistic relationships between algae and animals (e.g., Symbiodiniaceae and Cnidaria), the host manipulates the nutrient supply of endosymbionts to maximize the production and translocation of photosynthates (organic carbon) and minimize symbionts’ reproduction [82–84]. In protist symbiosis, little has been explored about host manipulating symbionts except for enslaving kleptoplasts. However, in the Acantharia-Phaeocystis symbiosis, the symbiotic Phaeocystis were blocked from cell division and carbon storage, while their carbon uptake increased 150-fold and genes involved in photosynthesis and carbon fixation were upregulated, a clear case of host manipulating the symbiont [84]. Although, in the case of gNoctiluca and P. noctilucae, the exact mode of the mutualistic interactions is still unclear, our transcriptomic data showed that light reactions of endosymbiont photosynthesis were extremely active during the bloom, accounting for 89% of FPKM of all mapped genes (Fig. 4a). RbcL and GAPA, both critical genes for photosynthetic carbon fixation (dark reactions), were also highly expressed (Fig. 4b). This enhanced carbon fixation and carbon storage typically benefits the algae, but could also benefit the host. Intriguingly, biosynthesis and degradation of carbon storage substances such as triacylglycerol (TAG) and starch were transcriptionally inactive in the symbiotic alga (Fig. 4c), suggesting that most of the fixed carbon was not converted into lipid storage in the symbionts. Furthermore, genes related to DNA replication and cell division were markedly less expressed in the P. noctilucae living inside gNoctiluca as compared to free-living counterparts (Fig. 4d), consistent with the possibility that the host depresses symbiont population growth. Strikingly, results from our physiological experiments also revealed significantly lower growth rates for P. noctilucae in-hospite as compared to its free-living counterparts (Fig. 4e). In contrast, genes involved in plastid division, which is generally synchronized with cell division in unicellular algae, exhibited higher expression relative to the cell division genes (Fig. 4d). Given that plastid proliferation can enhance photosynthesis efficiency [85], this result agrees with the idea that gNoctiluca promotes photosynthesis of P. noctilucae in-hospite, but slows their reproduction. This would benefit the host by gaining an additional energy source to supplement that from grazing especially during the bloom when a large amount of energy is required for the explosive proliferation of gNoctiluca.
Fig. 4. Transcriptomic proofs of host manipulation for photosynthates.
a Enrichment of KEGG pathways in both in-hospite and free-living P. noctilucae, indicating photosynthesis was the most dominant metabolic activity in in-hospite P. noctilucae. b Expression of genes involved in Calvin-Benson cycle. Pink (green) denotes higher (lower) expression in in-hospite P. noctilucae. Expression level (FPKM) of each gene in both in-hospite and free-living P. noctilucae are indicated in the relevant boxes. c Low starch and TAG metabolisms in the in-hospite P. noctilucae. d Low cell division and relatively high plastid division in the in-hospite P. noctilucae. Asterisks depict the absence of genes in the metatranscriptomics data. e Low growth rate (d−1) and longer cell doubling time (days) of the in-hospite P. noctilucae compared to free-living counterparts under laboratory incubation. Error bar stands for standard deviation.
Discussion
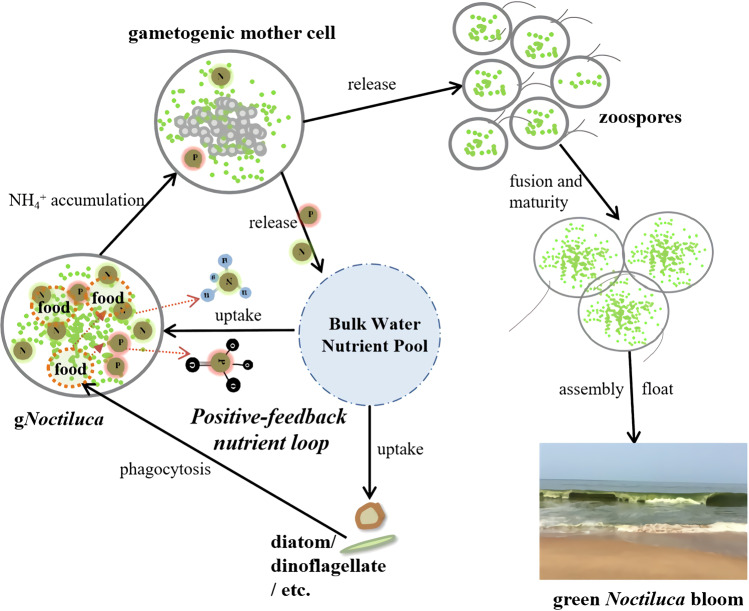
Emerging from the present study is a sketch of a hitherto poorly understood process and mechanisms underpinning the rise of a gNoctiluca bloom (Fig. 5). It begins with nutrients (N and P), which are essential for algal growth and the formation of algal blooms [86–88]. Our Spearman correlation analysis showed significant positive correlations (P < 0.001) between nutrients, especially ammonium, urea and phosphate, and gNoctiluca abundance at both the Seeb Jetty and along the transect into the offshore waters of the Sea of Oman (Fig. S3). While algal blooms are usually stimulated by dissolved inorganic and dissolved organic nutrients from the environment [89], we have found that the major source of N and P nutrients for the development of the gNoctiluca bloom was in part derived from external uptake, but likely more from grazing on phytoplankton prey [9, 12]. Grazing is most likely an ancestral trait of Noctiluca as the exclusively heterotroph rNoctiluca is more prevalent in the world’s oceans and its elemental growth yield and excretion of N and P are highly affected by prey nutritional quality [90, 91]. In gNoctiluca, prey metabolism and nutrient uptake occur in tandem leading to the build-up of ammonium in the central cytoplasm of gNoctiluca [12], a process better studied in the heterotrophic rNoctiluca, which also has been shown to accumulate high concentrations of ammonium and excrete it into the surrounding waters [92–95]. Our transcriptomic data also indicated the active metabolism that generated ammonium. When released into the external environment, these nutrients could promote the population growth of its prey (e.g., diatoms). This recurrent process by which nutrients first accumulate in gNoctiluca cells primarily via grazing, and are then released into the ambient environment during the sexual reproduction phase, to be then used by diatoms or dinoflagellates, constitutes a positive feedback loop that can theoretically support gNoctiluca proliferation and bloom persistence over extended periods even under nutrient-poor conditions as in the cases of the Sea of Oman and the Arabian Sea.
Fig. 5. Schematic of a “positive feedback loop” nutrition strategy and sexual reproduction that promote the formation and maintenance of gNoctiluca blooms.
The process by which nutrients first accumulate in gNoctiluca cells primarily via grazing, and are then released into the ambient environment during the sexual reproduction phase, to be then used by diatoms or dinoflagellates and ultimately back to support the growth of gNoctiluca, constitutes a positive feedback nutrient loop. Accumulation of ammonium may potentially stimulate the initiation of sexual reproduction of gNoctiluca, contributing to the outbreak of bloom.
The next step toward a bloom outbreak is a high level of cell proliferation and population growth. This appears to be achieved mainly through sexual reproduction. Specific growth rates attributable to asexual reproduction (binary division) of gNoctiluca are typically 0–0.28 d−1 [10], which would result in population growth of maximally 4 (=e0.28*5) folds in 5 days. This growth rate is far lower than 1.86 d−1, which occurred in the bloom event that we investigated. For sexual reproduction, gNoctiluca cell can generate 1024 gametes within a day [10], and typically every two gametes fuse to produce a trophont cell. This meiosis-driven reproduction can lead to an explosive growth and an abrupt bloom outbreak. Our transcriptomic data as well as microscopic images clearly show that this mode of reproduction is an essential part of the bloom formation. The potential of sexual reproduction to promote population (numeric) growth and bloom formation or sustenance has recently been proposed for other dinoflagellates as well [96].
The nutrient positive feedback loop can ensure an adequate supply of nitrogenous nutrients and energy that is essential for gamete production and outbreaks of gNoctiluca blooms. Although it has been reported that sexual reproduction in Noctiluca may be triggered by phagotrophic mode of nutrition [10, 49] and high density of ammonium [97], to this date, evidence in support of this notion has been circumstantial. Our transcriptome data, combined with the physiological results, provide insights that accumulation of ammonium derived primarily from metabolism of ingested prey might be responsible for inducing sexual reproduction in gNoctiluca, and the release of the gametes might accompany the release of the accumulated nutrients. Controlled experiments involving prey density and/or high ammonium concentrations in the growth medium will need to be undertaken to verify this possibility in the future. Furthermore, the transcriptomic evidence of enhanced photosynthesis yet depressed cell reproduction of P. noctilucae in-hospite leads us to hypothesize that gNoctiluca is capable of manipulating its endosymbiotic population (Fig. 4), enslaving them to be energy (ATP) and organic carbon providers. This additional source of energy that the host is able to obtain can potentially boost sexual reproduction accelerating bloom development. Similar host manipulation of symbionts has been reported in corals [82–84] and in the Acantharia-Phaeocystis symbiosis [98].
Although our field observations as well as transcriptomic analysis provide some valuable and insightful information about bloom formation in gNoctiluca, further studies involving metabolomics and advanced instrumentation such as nano-SIMS [99] or Single-Cell-Raman Scattering Microspectrometric Imaging [100] that allow probing of in-hospite host-symbiont C, N, P metabolite exchanges, will shed additional light on biochemical processes and mechanisms governing gNoctiluca bloom outbreaks. This information is important not only because there is growing evidence that gNoctiluca blooms may be expanding their range as a result of climate change and the spread of eutrophication and hypoxia in the global oceans, but also because of evidence showing that gNoctiluca blooms can: (1) short-circuit the food chain leading to the production of swarms of jellyfish and salps in place of fish, and (2) create water quality issues that impact freshwater, food, economic security and well-being of hundreds of millions of people living around impacted areas such as the Sea of Oman and the Arabian Sea.
Supplementary information
Acknowledgements
We appreciate Dr Xiaojian Jiang for assistance with samples collection. We are deeply thankful to the Department of Marine Sciences and Fisheries, Sultan Qaboos University, Muscat, Oman for providing access to facilities and all the help rendered during the fieldwork. HL, JT, and SL were supported by the National Natural Science Foundation of China (grants #42276096), the Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) (grant #2018SDKJ0406-3), and the Gordon and Betty Moore Foundation (GBMF grant #4980.01). JIG and HRG were supported by the Gordon and Betty Moore Foundation, US NSF (grant #2019983), NASA (grants #NNX17AG66G-ECO4CAST and NASA-CMS-80NSSC20K0014) and the Sultan Qaboos Cultural Centre, Washington, DC. HL was supported by the China Scholarship Council to conduct research at Columbia University and University of Connecticut where parts of this work were completed. We acknowledge the Ocean Biology Processing Group of NASA Goddard Space Flight Center for providing Moderate-resolution Imaging Spectroradiometer (MODIS) Aqua L2 data.
Author contributions
JIG and SL conceived the study. HL, JIG and KAH collected and processed samples. HL performed experiments with help from JIG, CT and CK. Data analysis was performed by HL and JW. HL, SL, JIG, and HRG wrote the paper with input from all authors.
Data availability
The datasets generated during and/or analyzed during the current study are available in the National Center for Biotechnology Information (NCBI) under the accession number PRJNA774540 (gNoctiluca and symbiotic Protoeuglena noctilucae) and PRJNA774510 (free-living P. noctilucae).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Joaquim I. Goes, Email: jig@ldeo.columbia.edu
Senjie Lin, Email: senjie.lin@uconn.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s43705-022-00187-4.
References
- 1.Al-Azri A, Al-Hashmi K, Goes J, Gomes H, I.Rushdi A, Al-Habsi H, et al. Seasonality of the bloom-forming heterotrophic dinoflagellate Noctiluca scintillans in the Gulf of Oman in relation to environmental conditions. Int J Oceans Oceanogr. 2007;2:51–60. [Google Scholar]
- 2.Goes JI, Gomes H, Al-Hashimi K, Buranapratheprat A. Ecological Drivers of Green Noctiluca blooms in two monsoonal-driven ecosystems. In: Glibert PM, Berdalet E, Burford MA, Pitcher GC, Zhou M, editors. Global Ecology and Oceanography of Harmful Algal Blooms. Cham:Springer International Publishing; 2018. p. 327–36.
- 3.Gomes HR, Goes JI, Matondkar SGP, Parab SG, Al-Azri ARN, Thoppil PG. Blooms of Noctiluca miliaris in the Arabian Sea-An in situ and satellite study. Deep Sea Res Part I. 2008;55:751–65. doi: 10.1016/j.dsr.2008.03.003. [DOI] [Google Scholar]
- 4.Harrison PJ, Furuya K, Glibert PM, Xu J, Liu HB, Yin K, et al. Geographical distribution of red and green Noctiluca scintillans. Chinese J Oceanol Limnol. 2011;29:807–31. doi: 10.1007/s00343-011-0510-z. [DOI] [Google Scholar]
- 5.Madhu NV, Jyothibabu R, Maheswaran PA, Jayaraj KA, Achuthankutty CT. Enhanced chlorophyll a and primary production in the northern Arabian Sea during the spring intermonsoon due to green Noctiluca scintillans bloom. Marine Biol Res. 2012;8:182–8. doi: 10.1080/17451000.2011.605143. [DOI] [Google Scholar]
- 6.Mitra A, Flynn KJ, Tillmann U, Raven JA, Caron D, Stoecker DK, et al. Defining planktonic protist functional groups on mechanisms for energy and nutrient acquisition: incorporation of diverse mixotrophic strategies. Protist. 2016;167:106–20. doi: 10.1016/j.protis.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Stoecker DK, Hansen PJ, Caron DA, Mitra A. Mixotrophy in the marine plankton. Ann Rev Marine Sci. 2017;9:311–35. doi: 10.1146/annurev-marine-010816-060617. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, Lin X, Goes JI, Lin S. Phylogenetic analyses of three genes of Pedinomonas noctilucae, the green endosymbiont of the marine dinoflagellate Noctiluca scintillans, reveal its affiliation to the order marsupiomonadales (chlorophyta, pedinophyceae) under the reinstated name Protoeuglena noctilucae. Protist. 2016;167:205–16. doi: 10.1016/j.protis.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Gomes HR, Goes JI, Matondkar SGP, Buskey EJ, Basu S, Parab S, et al. Massive outbreaks of Noctiluca scintillans blooms in the Arabian Sea due to spread of hypoxia. Nat Commun. 2014;5:1–8. [DOI] [PubMed]
- 10.Sriwoon R, Pholpunthin P, Lirdwitayaprasit T, Kishino M, Furuya K. Population dynamics of green Noctiluca scintillans (Dinophyceae) associated with the monsoon cycle in the upper Gulf of Thailand. J Phycol. 2008;44:605–15. doi: 10.1111/j.1529-8817.2008.00516.x. [DOI] [PubMed] [Google Scholar]
- 11.Hansen PJ, Miranda L, Azanza R. Green Noctiluca scintillans: a dinoflagellate with its own greenhouse. Marine Ecol Prog Series. 2004;275:79–87. doi: 10.3354/meps275079. [DOI] [Google Scholar]
- 12.Gomes HR, McKee K, Mile A, Thandapu S, Al-Hashmi K, Jiang X, et al. Influence of light availability and prey type on the growth and photo-physiological rates of the mixotroph Noctiluca scintillans. Front Marine Sci. 2018;5:374.
- 13.Furuya K, Saito H, Sriwoon R, Omura T, Furio E, Borja V, et al. Vegetative growth of Noctiluca scintillans containing the endosymbiont Pedinomonas noctilucae. Afr J Marine Sci. 2006;28:305–8. doi: 10.2989/18142320609504167. [DOI] [Google Scholar]
- 14.Balch WM, Haxo FT. Spectral properties of Noctiluca miliaris Suriray, a heterotrophic dinoflagellate. J Plankton Res. 1984;6:515–25. doi: 10.1093/plankt/6.3.515. [DOI] [Google Scholar]
- 15.Elbrachter M, Qi Y-Z. Aspects of Noctiluca (Dinophyceae) population dynamics. In: Andersen DM, Cembella A, Hallegraeff GM, editors. Physiological Ecology of harmful algal blooms. Berlin: NATO ASI Series;1998. p. 315–35.
- 16.Fonda-Umani S, Beran A, Parlato S, Virgilio D, Zollet T, Olazabal AD, et al. Noctiluca scintillans MACARTNEY in the Northern Adriatic Sea: long-term dynamics, relationships with temperature and eutrophication, and role in the food web. J Plankton Res. 2004;26:545–61. doi: 10.1093/plankt/fbh045. [DOI] [Google Scholar]
- 17.Nakamura Y. Growth and grazing of a large heterotrophic dinoflagellate, Noctiluca scintillans, in laboratory cultures. J Atmos Sci. 1998;20:1711–20. [Google Scholar]
- 18.Gomes HR, Goes JI, Al-Hashimi K, Al-Kharusi L. Global distribution and range expansion of green vs red Noctiluca Scintillans. In: Rao DS, editor. Dinoflagellates: morphology, life history, and ecological significance. New York: Nova Publishers;; 2020. pp. 499–525. [Google Scholar]
- 19.Morimoto A, Mino Y, Buranapratheprat A, Kaneda A, Tong-U-Dom S, Sunthawanic K, et al. Hypoxia in the Upper Gulf of Thailand: hydrographic observations and modeling. J Oceanogr. 2021;77:859–77.
- 20.Xiang C, Tan Y, Zhang H, Liu J, Ke Z, Li G. The key to dinoflagellate (Noctiluca scintillans) blooming and outcompeting diatoms in winter off Pakistan, northern Arabian Sea. Sci Total Environ. 2019;694:133396. doi: 10.1016/j.scitotenv.2019.07.202. [DOI] [PubMed] [Google Scholar]
- 21.Parab SG, Matondkar SGP, Gomes HR, Goes JI. Monsoon driven changes in phytoplankton populations in the eastern Arabian Sea as revealed by microscopy and HPLC pigment analysis. Continental Shelf Res. 2006;26:2538–58. doi: 10.1016/j.csr.2006.08.004. [DOI] [Google Scholar]
- 22.Chaghtai F, Saifullah SM. On the occurrence of green Noctiluca Scintillans blooms in coastal waters of Pakistan, North Arabian Sea. Pakistan J Botany. 2006;38:893–8. [Google Scholar]
- 23.Prakash S, Ramesh R, Sheshshayee MS, Dwivedi RM, Raman M. Quantification of new production during a winter Noctiluca scintillans bloom in the Arabian Sea. Geophys Res Lett. 2008;35:L08604. doi: 10.1029/2008GL033819. [DOI] [Google Scholar]
- 24.Lachkar Z, Lévy M, Smith S. Intensification and deepening of the Arabian Sea oxygen minimum zone in response to increase in Indian monsoon wind intensity. Biogeosciences. 2018;15:159–86. doi: 10.5194/bg-15-159-2018. [DOI] [Google Scholar]
- 25.Goes JI, Tian H, Gomes HR, Anderson OR, Al-Hashmi K, Derada S, et al. Ecosystem state change in the Arabian Sea fuelled by the recent loss of snow over the Himalayan-Tibetan Plateau region. Sci Rep. 2020;10:7422. doi: 10.1038/s41598-020-64360-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banse K, English DC. Geographical differences in seasonality of CZCS derived phytoplankton pigment in the Arabian Sea for 1978-1986. Deep Sea Res II. 2000;47:1623–77. doi: 10.1016/S0967-0645(99)00157-5. [DOI] [Google Scholar]
- 27.Tarran GA, Burkill PH, Edwards ES, Woodward EMS. Phytoplankton community structure in the Arabian Sea during and after the SW monsoon, 1994. Deep Sea Res Part II. 1999;46:655–76. doi: 10.1016/S0967-0645(98)00122-2. [DOI] [Google Scholar]
- 28.Sawant SS, Madhupratap M. Seasonality and composition of phytoplankton in the Arabian Sea. Curr Sci. 1996;71:869–73. [Google Scholar]
- 29.Latasa M, Bidigare RR. A comparison of phytoplankton populations of the Arabian Sea during the Spring Intermonsoon and Southwest Monsoon of 1995 as described by HPLC-analyzed pigments. Deep Sea Res Part II. 1998;45:2133–70. doi: 10.1016/S0967-0645(98)00066-6. [DOI] [Google Scholar]
- 30.Garrison DL, Gowing MM, Hughes MP. Nano- and microplankton in the northern Arabian Sea during the Southwest Monsoon, August-September 1995 A US-JGOFS study. Deep Sea Res II. 1998;45:2269–99. doi: 10.1016/S0967-0645(98)00071-X. [DOI] [Google Scholar]
- 31.Thangaraja M, Al-Aisry A, Al-Kharusi L. Harmful algal blooms and their impacts in the middle and outer ROPME sea area. Int J Oceans Oceanogr. 2007;2:85–98. [Google Scholar]
- 32.Al-Hashmi KA, Smith SL, Claereboudt M, Piontkovski SA, Al-Azri A. Dynamics of potentially harmful phytoplankton in a semi-enclosed bay in the Sea of Oman. Bullet Mar Sci. 2015;91:141–66. doi: 10.5343/bms.2014.1041. [DOI] [Google Scholar]
- 33.Al-Azri AR, Al-Hashmi KA, Al-Habsi H, Al-Azri N, Al-Khusaibi S. Abundance of harmful algal blooms in the coastal waters of Oman: 2006–2011. Aquatic Ecosyst Health Manag. 2015;18:269–81. doi: 10.1080/14634988.2015.1027131. [DOI] [Google Scholar]
- 34.Gomes H, Goes JI, Matondkar SGP, Parab SG, Al-Azri A, Thoppil PG. Unusual blooms of the green Noctiluca Miliaris (Dinophyceae) in the Arabian Sea during the Winter Monsoon. In: JD Wiggert, Hood RR, Naqvi SWA, Smith SL, Brink KH, editors. Indian Ocean:biogeochemical processes and ecological variability. AGU Book Series. Geophysical Mongraph 185: American Geophysical Union; 2009. p. 347–63.
- 35.Baliarsingh SK, Lotliker AA, Sudheesh V, Samanta A, Das S, Vijayan AK. Response of phytoplankton community and size classes to green Noctiluca bloom in the northern Arabian Sea. Marine Pollut Bullet. 2018;129:222–30. doi: 10.1016/j.marpolbul.2018.02.031. [DOI] [PubMed] [Google Scholar]
- 36.Al-Azri A, Piontkovski S, Al-Hashmi K, Al-Gheilani H, Al-Habsi H, Al-Khusaibi S, et al. The occurrence of algal blooms in Omani coastal waters. Aquatic Ecosyst Health Manag. 2012;15:56–63. doi: 10.1080/14634988.2012.672151. [DOI] [Google Scholar]
- 37.Zar JH. Spearman rank correlation: overview. Wiley StatsRef: Statistics Reference Online. 2014; 10.1002/9781118445112.stat05964.
- 38.Zhang Y, Lin X, Shi X, Lin L, Luo H, Li L, et al. Metatranscriptomic signatures associated with phytoplankton regime shift from diatom dominance to a dinoflagellate bloom. Front Microbiol. 2019;10:590. [DOI] [PMC free article] [PubMed]
- 39.Lin S, Zhang H, Zhuang Y, Tran B, Gill J. Spliced leader–based metatranscriptomic analyses lead to recognition of hidden genomic features in dinoflagellates. Proc Natl Acad Sci. 2010;107:20033–8. doi: 10.1073/pnas.1007246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo H, Lin X, Li L, Lin L, Zhang C, Lin S. Transcriptomic and physiological analyses of the dinoflagellate Karenia mikimotoi reveal non-alkaline phosphatase-based molecular machinery of ATP utilisation. Environ Microbiol. 2017;19:4506–18. doi: 10.1111/1462-2920.13899. [DOI] [PubMed] [Google Scholar]
- 41.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pertea G, Huang X, Liang F, Antonescu V, Sultana R, Karamycheva S, et al. TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics. 2003;19:651–2. doi: 10.1093/bioinformatics/btg034. [DOI] [PubMed] [Google Scholar]
- 43.Robinson MD, McCarthy DJ, Smyth GK. EdgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duernberger KA, Tobias CR, Mallin MA. Ecosystem scale nitrification and watershed support of tidal creek productivity revealed using whole system isotope tracer labeling. Limnol Oceanogr. 2018;63:2110–25. doi: 10.1002/lno.10927. [DOI] [Google Scholar]
- 45.Knap A, Michaels A, Close A, Ducklow H, Dickson A. Protocols for the Joint Global Ocean Flux Study (JGOFS) core measurements. Bergen, Norway: Scientific Committee on Oceanic Research, International Council of Scientific Unions. Intergovernmental Oceanographic Commission; 1996. Report No.: 19.
- 46.Fukuda Y, Endoh H. New details from the complete life cycle of the red-tide dinoflagellate Noctiluca scintillans (Ehrenberg) McCartney. Eur J Protistol. 2006;42:209–19. doi: 10.1016/j.ejop.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 47.Furuya K, Saito H, Rujinard S, Vijayan AK, Omura T, Furio EE, et al. Persistent whole-bay red tide of Noctiluca scintillans in Manila Bay, Philippines. Coastal Marine Sci. 2006;30:74–9. [Google Scholar]
- 48.Sathish T, Thomas LC, Padmakumar KB. Vegetative and sexual reproduction of bloom-forming dinoflagellate Noctiluca Scintillans (Ehrenberg) McCartney from tropical cochin estuary (southwest coast of India): in-situ and laboratory studies. Thalassas: Int J Marine Sci. 2021;37:31–7. doi: 10.1007/s41208-020-00247-3. [DOI] [Google Scholar]
- 49.Zingmark RG. Sexual reproduction in the dinoflagellate Noctiluca miliaris Suriray. J Phycol. 1970;6:122–6. [Google Scholar]
- 50.Chi J, Parrow MW, Dunthorn M. Cryptic Sex in Symbiodinium (Alveolata, Dinoflagellata) is supported by an inventory of meiotic genes. J Eukaryotic Microbiol. 2014;61:322–7. doi: 10.1111/jeu.12110. [DOI] [PubMed] [Google Scholar]
- 51.Malik S-B, Pightling AW, Stefaniak LM, Schurko AM, Logsdon JM., Jr An expanded inventory of conserved meiotic genes provides evidence for sex in Trichomonas vaginalis. PLOS ONE. 2008;3:e2879. doi: 10.1371/journal.pone.0002879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schurko AM, Logsdon JM., Jr Using a meiosis detection toolkit to investigate ancient asexual “scandals” and the evolution of sex. BioEssays. 2008;30:579–89. doi: 10.1002/bies.20764. [DOI] [PubMed] [Google Scholar]
- 53.Chan Y-L, Brown MS, Qin D, Handa N, Bishop DK. The third exon of the budding yeast meiotic recombination gene HOP2 is required for calcium-dependent and recombinase Dmc1-specific stimulation of homologous strand assimilation. J Biol Chem. 2014;289:18076–86. doi: 10.1074/jbc.M114.558601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pezza RJ, Voloshin ON, Vanevski F, Camerini-Otero RD. Hop2/Mnd1 acts on two critical steps in Dmc1-promoted homologous pairing. Genes Dev. 2007;21:1758–66. doi: 10.1101/gad.1562907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sakuno T, Watanabe Y. Studies of meiosis disclose distinct roles of cohesion in the core centromere and pericentromeric regions. Chromo Res. 2009;17:239–49. doi: 10.1007/s10577-008-9013-y. [DOI] [PubMed] [Google Scholar]
- 56.Rockmill B, Roeder GS. RED1: a yeast gene required for the segregation of chromosomes during the reductional division of meiosis. Proc Natl Acad Sci. 1988;85:6057–61. doi: 10.1073/pnas.85.16.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lynn A, Soucek R, Börner GV. ZMM proteins during meiosis: Crossover artists at work. Chromo Res. 2007;15:591–605. doi: 10.1007/s10577-007-1150-1. [DOI] [PubMed] [Google Scholar]
- 58.Hodgson A, Terentyev Y, Johnson RA, Bishop-Bailey A, Angevin T, Croucher A, et al. Mre11 and Exo1 contribute to the initiation and processivity of resection at meiotic double-strand breaks made independently of Spo11. DNA Repair. 2011;10:138–48. doi: 10.1016/j.dnarep.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 59.Castro A, Bernis C, Vigneron S, Labbé J-C, Lorca T. The anaphase-promoting complex: a key factor in the regulation of cell cycle. Oncogene. 2005;24:314–25. doi: 10.1038/sj.onc.1207973. [DOI] [PubMed] [Google Scholar]
- 60.Hall IM, Noma K, Grewal S. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc Natl Acad Sci. 2003;100:193–8. doi: 10.1073/pnas.232688099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bascom-Slack CA, Dawson DS. The yeast motor protein, Kar3p, is essential for meiosis I. J Cell Biol. 1997;139:459–67. doi: 10.1083/jcb.139.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meyer RE, Kim S, Obeso D, Straight PD, Winey M, Dawson DS. Mps1 and Ipl1/Aurora B act sequentially to correctly orient chromosomes on the meiotic spindle of budding yeast. Science. 2013;339:1071–4. doi: 10.1126/science.1232518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsuchida J, Nishina Y, Wakabayashi N, Nozaki M, Sakai Y, Nishimune Y. Molecular cloning and characterization of meichroacidin (male meiotic metaphasechromosome-associatedacidic protein) Dev Biol. 1998;197:67–76. doi: 10.1006/dbio.1998.8885. [DOI] [PubMed] [Google Scholar]
- 64.Chan SW, Fowler KJ, Choo KA, Kalitsis P. Spef1, a conserved novel testis protein found in mouse sperm flagella. Gene. 2005;353:189–99. doi: 10.1016/j.gene.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 65.Herrmann BG, Koschorz B, Wertz K, McLaughlin KJ, Kispert A. A protein kinase encoded by the t complex responder gene causes non-mendelian inheritance. Nature. 1999;402:141–6. doi: 10.1038/45970. [DOI] [PubMed] [Google Scholar]
- 66.Thevissen K, Ayscough KR, Aerts AM, Du W, De Brucker K, Meert EM, et al. Miconazole induces changes in actin cytoskeleton prior to reactive oxygen species induction in yeast. J Biol Chem. 2007;282:21592–7. doi: 10.1074/jbc.M608505200. [DOI] [PubMed] [Google Scholar]
- 67.Morse D. A transcriptome-based perspective of meiosis in dinoflagellates. Protist. 2019;170:397–403. doi: 10.1016/j.protis.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 68.Kaur J, Sebastian J, Siddiqi I. The Arabidopsis-mei2-like genes play a role in meiosis and vegetative growth in Arabidopsis. Plant Cell. 2006;18:545–59. doi: 10.1105/tpc.105.039156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mukherjee K, Futcher B, Leatherwood J. mmi1 and rep2 mRNAs are novel RNA targets of the Mei2 RNA-binding protein during early meiosis in Schizosaccharomyces pombe. Open Biol. 2018;8:180110. doi: 10.1098/rsob.180110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin S, Cheng S, Song B, Zhong X, Lin X, Li W, et al. The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science. 2015;350:691–4. doi: 10.1126/science.aad0408. [DOI] [PubMed] [Google Scholar]
- 71.Deng Y, Hu Z, Shang L, Peng Q, Tang YZ. Transcriptomic analyses of Scrippsiella trochoidea reveals processes regulating encystment and dormancy in the life cycle of a dinoflagellate, with a particular attention to the role of abscisic acid. Front Microbiol. 2017;8:2450. [DOI] [PMC free article] [PubMed]
- 72.Ji N, Huang J, Zhang Z, Zhou L, Shen X, Lin S. Identification and expression analysis of meiosis-related genes in the harmful alga Heterosigma akashiwo (Raphidophyceae) Harmful Algae. 2020;92:101736. doi: 10.1016/j.hal.2019.101736. [DOI] [PubMed] [Google Scholar]
- 73.Berdieva MA, Pozdnyakov IA, Kalinina VO, Skarlato SO. Putative meiotic toolkit in the dinoflagellate Prorocentrum cordatum: additional evidence for sexual process from transcriptome. J Eukaryotic Microbiol. 2021;68:e12845. doi: 10.1111/jeu.12845. [DOI] [PubMed] [Google Scholar]
- 74.Lindner K, Gregán J, Montgomery S, Kearsey SE. Essential role of MCM proteins in premeiotic dna replication. Mol Biol Cell. 2002;13:435–44. doi: 10.1091/mbc.01-11-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Okaichi T, Ochi T, Tada K, Urai T, Wissesang S, Ishimaru T, et al. (eds). Isolation and culture of Pedinomonas noctilucae, a symbiotic of Noctiluca scintillans of Gulf of Thailand. Second WESTPAC Symposium. Penang, Malaysia;1991. p. 166–176.
- 76.Buttle DJ, Mort JS (eds). Cysteine proteases. Encyclopedia of Biological Chemistry. 2nd ed. Cambridge, MA, USA: Academic Press; 2013. p. 589–92.
- 77.Levin R, Grinstein S, Canton J. The life cycle of phagosomes: formation, maturation, and resolution. Immunol Rev. 2016;273:156–79. doi: 10.1111/imr.12439. [DOI] [PubMed] [Google Scholar]
- 78.Uribe-Querol E, Rosales C. Control of phagocytosis by microbial pathogens. Front Immunol. 2017;8:1368. [DOI] [PMC free article] [PubMed]
- 79.Müller-Taubenberger A, Lupas AN, Li H, Ecke M, Simmeth E, Gerisch G. Calreticulin and calnexin in the endoplasmic reticulum are important for phagocytosis. EMBO J. 2001;20:6772–82. doi: 10.1093/emboj/20.23.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Spector DL (Eds). Dinoflagellates. Academic Press, London, 2012.
- 81.Moore ER, Bullington BS, Weisberg AJ, Jiang Y, Chang J, Halsey KH. Morphological and transcriptomic evidence for ammonium induction of sexual reproduction in Thalassiosira pseudonana and other centric diatoms. PLOS ONE. 2017;12:e0181098. doi: 10.1371/journal.pone.0181098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Davy SK, Allemand D, Weis VM. Cell biology of Cnidarian-dinoflagellate symbiosis. Microbiol Mol Biol Rev. 2012;76:229–61. doi: 10.1128/MMBR.05014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krueger T, Horwitz N, Bodin J, Giovani ME, Escrig S, Fine M, et al. Intracellular competition for nitrogen controls dinoflagellate population density in corals. Proc Royal Soc B: Biol Sci. 2020;287:20200049. doi: 10.1098/rspb.2020.0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xiang T, Lehnert E, Jinkerson RE, Clowez S, Kim RG, Denofrio JC, et al. Symbiont population control by host-symbiont metabolic interaction in Symbiodiniaceae-cnidarian associations. Nat Commun. 2020;11:108. doi: 10.1038/s41467-019-13963-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xiong D, Huang J, Peng S, Li Y. A few enlarged chloroplasts are less efficient in photosynthesis than a large population of small chloroplasts in Arabidopsis thaliana. Sci Rep. 2017;7:5782. doi: 10.1038/s41598-017-06460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Glibert PM, Anderson DM, Gentien P, Granéli E, Sellner KG. The global, complex phenomena of harmful algal blooms. Oceanography. 2005;18:130–41. [Google Scholar]
- 87.Glibert PM, Seitzinger S, Heil CA, Burkholder JM, Parrow MW, Codispoti LA, et al. The role of eutrophication in the global proliferation of harmful algal blooms new perspectives and new approaches. Oceanography. 2005;18:198–209. doi: 10.5670/oceanog.2005.54. [DOI] [Google Scholar]
- 88.Lin S, Litaker RW, Sunda WG. Phosphorus physiological ecology and molecular mechanisms in marine phytoplankton. J Phycol. 2016;52:10–36. doi: 10.1111/jpy.12365. [DOI] [PubMed] [Google Scholar]
- 89.Davidson K, Gowen RJ, Harrison PJ, Fleming LE, Hoagland P, Moschonas G. Anthropogenic nutrients and harmful algae in coastal waters. J Environ Manag. 2014;146:206–16. doi: 10.1016/j.jenvman.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 90.Tiselius P, Kiørboe T. Colonization of diatom aggregates by the dinoflagellate Noctiluca scintillans. Limnol Oceanogr. 1998;43:154–9.
- 91.Zhang S, Liu H, Glibert PM, Guo C, Ke Y. Effects of prey of different nutrient quality on elemental nutrient budgets in Noctiluca scintillans. Sci Rep. 2017;7:7622. doi: 10.1038/s41598-017-05991-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Okaichi T, Nishio S. Identification of ammonia as the toxic principle of red tide Noctiluca miliaris. Bullet Plankton Soc Japan. 1976;23:75–80. [Google Scholar]
- 93.Pithakpol S (ed). Nutrient regeneration during Noctiluca scintillans red tide in Harima Nada, the Seto Inland Sea, Japan. Proceedings of the 15th Ocean Engineering Symposium;2000. p. 127–134.
- 94.Ara K, Nakamura S, Takahashi R, Shiomoto A, Hiromi J. Seasonal variability of the red tide-forming heterotrophic dinoflagellate Noctiluca scintillans in the neritic area of Sagami Bay, Japan: its role in the nutrient-environment and aquatic ecosystem. Plankton Benthos Res. 2013;8:9–30. doi: 10.3800/pbr.8.9. [DOI] [Google Scholar]
- 95.Montani S, Pithakpol S, Tada K. Nutrient regeneration in coastal seas by Noctiluca scintillans, a red tide-causing dinoflagellate. J Marine Biotechnol. 1998;6:224–8. [PubMed] [Google Scholar]
- 96.Lin S, Yu L, Wu X, Li M, Zhang Y, Luo H, et al. Active meiosis in dinoflagellate blooms: a ‘sex for proliferation’ hypothesis. Harmful Algae. 2022;118:102307. doi: 10.1016/j.hal.2022.102307. [DOI] [PubMed] [Google Scholar]
- 97.Cheng-xu Z, Xiao-jun Y. New observations on the meiotic process in the marine dinoflagellate Noctiluca scintillans (Noctilucales, dinophyceae) Chinese J Oceanol Limnol. 2002;20:67–73. doi: 10.1007/BF02846613. [DOI] [Google Scholar]
- 98.Uwizeye C, Mars Brisbin M, Gallet B, Chevalier F, LeKieffre C, Schieber NL, et al. Cytoklepty in the plankton: a host strategy to optimize the bioenergetic machinery of endosymbiotic algae. Proc Natl Acad Sc. 2021;118:e2025252118. doi: 10.1073/pnas.2025252118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Foster RA, Sztejrenszus S, Kuypers MMM. Measuring carbon and N2 fixation in field populations of colonial and free-living unicellular cyanobacteria using nanometer-scale secondary ion mass spectrometry. J Phycol. 2013;49:502–16. doi: 10.1111/jpy.12057. [DOI] [PubMed] [Google Scholar]
- 100.Wang Y, Huang WE, Cui L, Wagner M. Single cell stable isotope probing in microbiology using Raman microspectroscopy. Cur Opinion Biotechnol. 2016;41:34–42. doi: 10.1016/j.copbio.2016.04.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available in the National Center for Biotechnology Information (NCBI) under the accession number PRJNA774540 (gNoctiluca and symbiotic Protoeuglena noctilucae) and PRJNA774510 (free-living P. noctilucae).