Abstract
The development of strategies for effectively manipulating and engineering beneficial plant-associated microbiomes is a major challenge in microbial ecology. In this sense, the efficacy and potential implications of rhizosphere microbiome transplant (RMT) in plant disease management have only scarcely been explored in the literature. Here, we initially investigated potential differences in rhizosphere microbiomes of 12 Solanaceae eggplant varieties and accessed their level of resistance promoted against bacterial wilt disease caused by the pathogen Ralstonia solanacearum, in a 3-year field trial. We elected 6 resistant microbiomes and further tested the broad feasibility of using RMT from these donor varieties to a susceptible model Solanaceae tomato variety MicroTom. Overall, we found the rhizosphere microbiome of resistant varieties to enrich for distinct and specific bacterial taxa, of which some displayed significant associations with the disease suppression. Quantification of the RMT efficacy using source tracking analysis revealed more than 60% of the donor microbial communities to successfully colonize and establish in the rhizosphere of recipient plants. RTM from distinct resistant donors resulted in different levels of wilt disease suppression, reaching up to 47% of reduction in disease incidence. Last, we provide a culture-dependent validation of potential bacterial taxa associated with antagonistic interactions with the pathogen, thus contributing to a better understanding of the potential mechanism associated with the disease suppression. Our study shows RMT from appropriate resistant donors to be a promising tool to effectively modulate protective microbiomes and promote plant health. Together we advocate for future studies aiming at understanding the ecological processes and mechanisms mediating rates of coalescence between donor and recipient microbiomes in the plant rhizosphere.
Subject terms: Ecology, Microbial ecology
Introduction
Public, veterinary, and plant health are increasingly challenged by pathogenic infections. The animal gut and the plant rhizosphere are systems colonized by numerous and diverse microbes [1, 2] that dynamically affect the host health [3, 4]. Similar to the gut system [5], the rhizosphere microbiome is tightly linked with the host developmental stage [6], and with levels of nutrient uptake [7]. These and other case-specific factors (e.g., host genetics, physiology) will determine the degree to which microbiomes mediate plant resistance to biotic [8] and abiotic stresses [9, 10]. While disease occurrence is often associated with microbiome dysbiosis in the gut [11] and at the rhizosphere [12], the assembly of specific beneficial taxa has been shown to successfully suppress pathogens in animals [13] and plants [8]. Some of these beneficial microbes, often termed probiotics for human health, act by inducing host immunity and/or by antagonizing specific pathogens [14]. Conversely, in plants, beneficial microbial taxa have mostly been associated with growth-promoting rhizobacteria (PGPR), albeit other beneficial functions (e.g., disease suppression) may not be included within this terminology [15].
Despite the multitude of commercial products of probiotics and PGPRs that are coming to the market, their successful effects on disease suppression are variable due to unstable colonization and forceful competition with other taxa within the resident microbiomes [16, 17]. To overcome this limitation and move beyond single strain/synthetic community inoculation, fecal microbiome transplantation—transferring the gut microbiome from healthy donors to diseased patient recipients—has been studied with cases of success for particular clinical diseases [18], such as Clostridium difficile infections [19] and cancer [20]. In the plant rhizosphere, this strategy has received relatively little attention, potentially due to the difficulty in applying it at large scales and in an effective manner [21]. However, transplanting ‘protective’ microbiomes from resistant to susceptible plants offer an opportunity to better understand the factors controlling the rhizosphere microbiome assembly. It takes a holistic approach of community coalescence and opens up new insights for prospective strategies aiming at controlling plant diseases [22].
Cultivation-dependent studies have shown that bacteria belonging to Pseudomonas [23, 24], Bacillus [25, 26], and other genera [27, 28] are able to endow disease suppression in soils [29–31]. Cultivation-independent studies, however, have advocated for a community-level approach aiming at promoting and establishing suppressive microbiomes in soils [21, 32]. These include examples in black root rot [33], common scab [34], Fusarium wilt [35], and bacterial wilt diseases [36]. A more classical approach has been based on the notion that the transplant of a disease-suppressive soil to a conducive soil based on a 1:9 ratio (w/w) can stimulate the plant resistance against Rhizoctonia solani infections. This has been validated and shown to enrich the abundance of 17 bacterial populations belonging to Proteobacteria, Firmicutes, and Actinobacteria [37]. Recently, the growth of a susceptible variety in soils previously cultivated with a resistant variety was reported to slow down the progression of bacterial wilt disease. This phenomenon was mostly associated with the enrichment of Flavobacterium in the rhizosphere microbiome [8]. Similarly, soils derived from healthy plants (in this case, enriched with Firmicutes and Proteobacteria) conferred a lower incidence of bacterial wilt in the subsequent plant cultivation [38]. Together, these studies provide mounting evidence to support rhizosphere microbiome transplants (RMT) as a plausible approach to protect plants against pathogen infections.
Advancing microbiome transplant strategies might take into account the ecological basis of community coalescence that is dynamically affected by the donor and recipient communities, as well as the environmental context [39]. First, the approach should seek to find appropriate donors for the RMT, since plants can actively select for a proportion of microbial taxa in rhizosphere [7, 40], leading to the difference in their resistance to soil-borne pathogens [8, 41]. Second, significant variability in inter-individual rhizosphere microbiomes mediated by factors such as growth stages [42], soil types and nutrients [43], and health status [44] should be taken into account. Third, the rate of colonization of transplanted microbiomes will be highly dependent on the local environment, similarities between plant species, and the outcome of biological interactions between taxa in the donor and recipient communities, thus resulting in coalescence between these systems [16].
Here, we used the soil-borne pathogen Ralstonia solanacearum as a model organism to study the microbiome of resistant and susceptible Solanaceae crop varieties. This pathogen is the causal agent of bacterial wilt disease affecting numerous economically important crops globally [45]. Eggplant varieties were used for donor selection since this species is one of the representative Solanaceae crops naturally infected by the model R. solanacearum pathogen [46] and recognized an important resistant resource for breeding for resistance against bacterial wilt disease in Solanaceae crops [47]. We started by profiling the landscape rhizosphere microbiomes both in resistant and susceptible plants in a 3-year field trial (Fig. 1). Then, we selected the resistant varieties as microbiome donors to test the efficiency of RMT in suppressing the occurrence of bacterial wilt disease on a susceptible (model) tomato variety MicroTom. We set a focus on answering three major questions: (1) What are the fine-scale differences in rhizosphere microbiomes between resistant and susceptible varieties? (2) To what extent the transplanted rhizosphere microbiomes from resistant donor plants are able to colonize susceptible recipient plants? (3) What are the underlying taxonomic and functional differences between ‘success’ and ‘failure’ of RMT mediating soil-borne disease suppression?
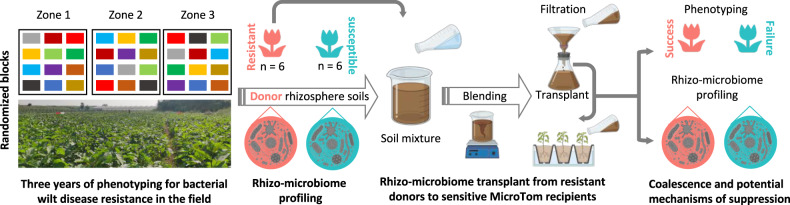
Fig. 1. Schematic representation of the RMT approach used in this study to investigate the mechanisms mediating wilt disease suppression in tomato plants.
The field site used in this study has been infected by R. solanacearum for more than 10 years.
Materials and methods
Field trials to assess disease resistance
A three-year phenotypic evaluation of bacterial wilt disease resistance of 12 eggplant varieties for donor selection (Table S1) was performed in the field. Eggplant is one of the repressive Solanaceae crop species naturally infected by the pathogen R. solanacearum [46] and recognized as an important resistant resource for Solanaceae crop resistance breeding against bacterial wilt disease [47]. The site has a history of soil infestation with R. solanacearum for more than 10 years and it is located at the Guangxi Agricultural Sciences Academy Lijian Scientific Research Bases, Guangxi, China (23.25° E, 108.06° N). Seeds of Solanaceae plant varieties were germinated in a greenhouse under controlled conditions (25 °C, 70% relative humidity) and under a natural light regime. About one and a half months after germination, the four-leaf stage plants were transplanted to the ‘diseased’ field in a randomized block design (ca. of 20 plants per block, 3 blocks). The system was maintained from April to July for three consecutive years 2017–2019. Temperatures in the field ranged between 26–35 °C (during the day) and 21–27 °C (during the night), and plants were cultivated for 3 months under open-field conditions. The disease progression was analyzed every two weeks until the fruit setting stage when bacterial wilt disease outbreaks. The disease incidence was defined as the proportion of severe bacterial disease symptoms (proportion of leaves wilted) in each block. In order to simplify the variation in resistance level of Solanaceae varieties, we used a binary classification of resistance level based on the cut-off of 50% (>50% of leaves wilted). This cut-off was established based on the average disease incidence of the three years. Thus, the average disease incidence above and below 50% was referred to as ‘susceptible’ and ‘resistant’ varieties, respectively.
Soil collection and rhizosphere sampling
The soil used in the greenhouse experiments was collected from a tomato field without detectable levels of the pathogen R. solanacearum, at the Nanjing Agricultural University Baima Teaching Scientific Research Base (119.18° E, 31.61° N), Nanjing, China. The top 10–20 cm of the soil was collected and sieved (3-mm sieve) to remove rocks and other debris. Seedlings of the 12 Solanaceae varieties were grown in sterilized jiffy substrates (Huaian Agricultural Technology Development Ltd) until they reached the four-leaf stage, and were further transplanted into pots containing 5 kg of soil. The rhizosphere samples were collected 4 weeks after transplanting. For that, excess soil on the roots was discarded by gently shaking the plants, and the remaining soil particles attached to the root surface were collected as rhizosphere soil [48]. Samples were homogenized and 0.5 g and 10 g of rhizosphere soils were subjected to rhizosphere microbiome profiling and to the transplant assay, respectively. The remaining sample amounts were cryopreserved in 5 mL of 30% glycerol at –80 °C. Each Solanaceae variety consisted of three plants.
Rhizosphere microbiome transplant and pathogen infection assays
We used a non-destructive rhizobox [38] to grow the recipient tomato plants (Solanum lycopersicum cv. MicroTom) and to perform the rhizosphere transplant experiment. This variety is susceptible to R. solanacearum and it is well-recognized as a model plant to study bacterial wilt disease [49]. The microbiome inoculum was based on a 10-times dilution of the rhizosphere soils (1:10 v/v) of each resistant variety (referred to as donor(s)). The rhizosphere microbiome inoculum was added to the root part of four-leaf stage MicroTom recipient plants, immediately after transplanting into the rhizobox. The rhizosphere soil samples within the small nylon bags (Taizhou Luqiao Yinfan Ltd.; see rhizobox design for details [38]) were harvested 4 weeks after the microbiome transplant and processed as described above. The self-transplant of the MicroTom rhizosphere microbiome (vMT) was used as control. After the collection of rhizosphere samples, the pathogen R. solanacearum (strain QL-Rs1115) was inoculated in all pots using a soil drenching method resulting in a final concentration of 5.0 × 106 CFU·g−1 of soil [48]. The disease development was daily monitored and quantified by the proportion of wilted leaves per plant [50]. ‘Failure’ and ‘success’ terminologies were used to refer to the proportion of wilted leaves similar or significantly lower than those observed in the self-transplant treatment (vMT referred to as the control treatment) 10 weeks after the pathogen inoculation. Statistics were performed using Student’s t tests.
Bacterial isolation and assessment of antimicrobial activity
To determine whether the success of RMT is due to the colonization by antagonistic bacteria from resistant donors on susceptible recipient plants, we isolated and tested the antimicrobial activity of rhizosphere bacterial isolates against the pathogen R. solanacearum. For that, ‘failure’ and ‘success’ rhizosphere soils were collected and homogenized independently and divided into three replicated samples. Approximately 200 isolates were randomly picked per replicate, resulting in a total of 997 bacterial isolates (486 and 513 isolates for ‘success’ and ‘failure’ groups, respectively). Isolation and purification of bacterial colonies were performed using tryptone soy agar (1.5 g L−1 tryptone, 0.5 g L−1 soytone, 0.5 g L−1 sodium chloride and 15 g L−1 agar, pH 7.0) at 30 °C as previously described [51]. All purified isolates were stored in 96-well microplates at −80 °C in 20% glycerol (v/v). The full-length sequences of the 16 S ribosomal RNA (rRNA) gene of all 997 bacterial isolates were sequenced with the primer pair 27 F (5′-GGTTACCTTGTTACGACTT-3′) and 1492 R (5′-AGAGTTTGATCCTGGCTCAG-3′) [52], and subjected to taxonomic identification using the online classifier tool of the Ribosomal Database Project (RDP) database (http://rdp.cme.msu.edu) [53]. These sequences were further aligned using MAFFT [54], and a ‘de novo’ phylogenetic tree was constructed using the FastTree2 based on the maximum-likelihood method [55]. The phylogenetic tree was further visualized using the iTOL web tool (https://itol.embl.de).
The antimicrobial activity of the bacteria isolates was estimated by testing the inhibition effect of their cell-free supernatants against the pathogen strain R. solanacearum QL-Rs1115 [42]. Briefly, overnight cultures of each isolate were filtered through a 0.22 μm filter to obtain sterile supernatants. A volume of 20 μL of supernatant was added to 180 μL of fresh casamino acid–peptone–glucose (CPG: casamino acid 1 g L−1, peptone 10 g L−1 and glucose 5 g L−1, pH 7.0) media containing the pathogen R. solanacearum at an initial OD600 of 0.05 as previously reported [42]. The pathogen R. solanacearum was set to grow for 24 h at 30 °C at 170 rpm, and the pathogen density was evaluated by OD600 values in a Max M5 Plate reader. Control treatments received 20 μl of CPG media instead of a bacterial supernatant. The pathogen inhibition effect of each bacterial cell-free supernatant was defined as the relative pathogen growth reduction compared to the control treatment.
Microbiome sequencing and analysis
The total DNA was extracted from 0.25 g of rhizosphere samples using the PowerSoil DNA Isolation Kit (Mobio Laboratories, Carlsbad, CA, USA), following the manufacturer’s protocol. The DNA quality and concentration were checked using a NanoDrop 1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). The DNA was subjected to bacterial 16S rRNA amplicon sequencing to determine the composition and diversity of bacterial communities. PCR amplifications were performed as follows: 95 °C for 2 min, followed by 25 cycles at 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, and a final extension at 72 °C for 5 min. PCRs were performed in triplicate in 20 μL mixture containing 4 μL of 5× FastPfu Buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu Polymerase, and 10 ng of template DNA. Amplicons were extracted from 2% agarose gels and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) according to the manufacturer’s instructions. Sequencing was carried out at the Shanghai Biozeron Biological Technology Co. Ltd, based on the V4 hypervariable region of the 16S rRNA gene using the primer pair 563F (5′-AYTGGGYDTAAAGVG-3′) and 802R (5′-TACNVGGGTATCTAATCC-3′). All sequences were processed using QIIME [56]. Bacterial OTUs were set at 97% of nucleotide identity using USEARCH [57]. The taxonomic affiliation of OTUs was carried out using the RDP database [53]. Community alpha-diversity was determined based on the Shannon diversity index (Shannon) and OTU richness using the vegan R package [58], after removing R. solanacearum sequences [44].
Tracking the microbial sources in recipient plants
To quantify the colonization rate of the transplanted microbiome of resistant donors into the recipient MicroTom plant rhizospheres, we tracked the most likely origin of microbial communities in the transplanted plants using the FEAST software [59]. FEAST uses a Bayesian approach to estimate the most probable proportion of the user-defined ‘source’ microbial communities in a given ‘sink’ community. For this analysis, samples from recipient MicroTom plants were set as ‘sinks’, and samples from the resistant donor plants as ‘source’. The ‘unknown’ proportion in recipient plant-rhizosphere microbiomes likely refers to the local colonization from the soil.
Statistics and data processing
Student’s t tests (two-sided) were used to compare the statistical significance between pairs of samples. Analysis of variance (ANOVA) and Tukey’s honest significant difference (HSD) tests were used to determine the statistical significance of multiple comparisons using the R agricolae package [60]. The microbial community composition was ordinated by principal coordinates analysis (PCoA) based on Bray-Curtis distances, and differences in rhizosphere soil samples between resistant and susceptible plant varieties were compared using the nonparametric permutational multivariate analysis of variance (PERMANOVA, P < 0.05, 999 permutations), using the Adonis function in the R package vegan [58]. Linear discriminant analysis and a significance test were used to explore the most discriminating OTUs between resistant levels using DESeq2 [61], edgeR [62], and LEfSe [63]. Three screening criteria were based on (1) fold change ≥ 4 (resistant relative to susceptible varieties), (2) linear discriminant analysis with a score ≥ 2 (resistant relative to susceptible varieties), and (3) significance test P < 0.05. Co-occurrence network analysis was performed following MENAP [64]. All differentially abundant OTUs based on DESeq2, edgeR, and LEfSe were retained for this analysis, and the count number of sequences was log-transformed and analyzed using a random matrix theory-based approach [65]. The edges (i.e., connections between OTUs) correspond to strong and significant correlations (positive or negative) between nodes (OTUs) [66]. We also used the NetShift method [67] to identify potential keystone taxa based on differences in network interactions between resistant and susceptible microbiomes. This method allows to quantify the directional changes in the individual node interactions by exploring: (1) whether there is a significant change in community patterns between resistant and susceptible plants, (2) whether there are major changes in associations of each constituent node in resistant and susceptible plants, (3) whether specific nodes have been important members in the community, and (4) whether there is an increase in specific node importance in resistant plants. Functional prediction of microbial communities was performed using PICRUSt2 (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States, version 2), allowing a putative inference of the functions encoded by these bacterial communities based on the 16S rRNA gene sequences [68]. The predicted functional annotations were based on the KEGG (Kyoto Encyclopedia of Genes and Genomes, e-value cut-off 10−5) database. The KEGG pathways at L3 level were used for comparing functional diversity (Shannon and richness), functional community composition (PCoA), and the discriminating functional genes (enriched vs depleted) as described above for OTU-based analysis.
Results
Differences in the rhizosphere microbiome and the association with wilt disease resistance
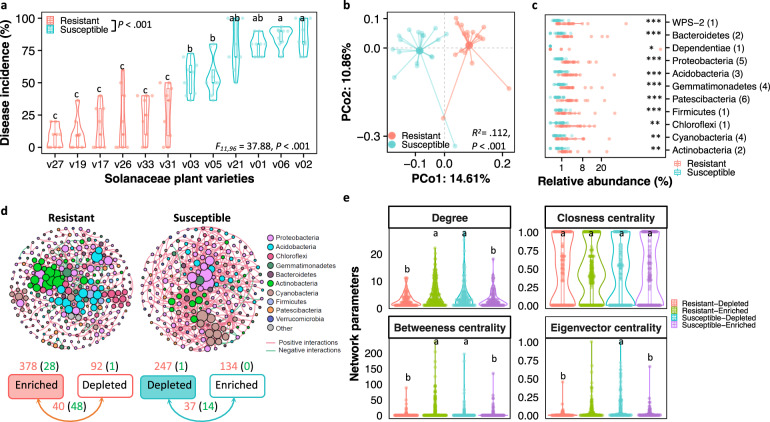
The three-year field trial revealed significant differences in bacterial wilt disease incidence across the 12 Solanaceae plant varieties (ANOVA: F11,96 = 37.9, P < 0.001, Fig. 2a). Based on the cut-off of 50% of average disease incidence, the resistance level was further classified into resistant (>50%) and susceptible (<50%) groups (see “Methods” for details). The disease incidence of susceptible varieties ranged from 54.7% to 86.1% with an average of 73.1%. Likewise, resistant varieties displayed a significantly lower disease incidence, ranging from 7.8% to 27.8%, with an average of 17.2% (Student’s t test: t = 16.1, df = 106.0, P < 0.001, Fig. 2a). Analyses of the rhizosphere microbiomes did not identify significant differences in alpha-diversity (Shannon index and OTU richness) across resistant and susceptible varieties (Supplementary Fig. 1a). However, the composition of the rhizosphere bacterial communities clustered into two distinct groups corresponding to resistant and susceptible groups (PERMANOVA: R2 = 0.112, P < 0.001, Fig. 2b). At the phylum level, Proteobacteria, Acidobacteria, Actinobacteria, Bacteroidetes, Chloroflexi, Cyanobacteria, and Gemmatimonadetes were the most dominant phyla accounting for 88.2% of the rhizosphere bacterial communities in total (86.9% and 89.5% in the resistant and susceptible varieties, respectively) (Supplementary Fig. 1b). The analysis of differentially abundant OTUs between resistant and susceptible groups revealed a total of 31 and 101 OTUs to be associated with resistant and susceptible plants, respectively (Supplementary Fig. 2a–e). These OTUs in the rhizosphere of resistant varieties were affiliated to the phyla Acidobacteria (3 OTUs, uncultured bacterial taxa within the family Acidobacteriaceae Subgroup 1), Actinobacteria (2 OTUs, Luedemannella sp. and an unclassified genus within the order Frankiales), Bacteroidetes (2 OTUs, uncultured genera within the orders Sphingobacteriales and Chitinophagales), Chloroflexi (an uncultured bacterium within the family Ktedonobacterales-JG30-KF-AS9), Cyanobacteria (2 OTUs, Tychonema sp.), Dependentiae (1 OTU, uncultured bacterium within the family Vermiphilaceae), Firmicutes (1 OTU, Pullulanibacillus sp.), Gemmatimonadetes (4 OTUs, Gemmatimonas sp. and Gemmatirosa sp.), Patescibacteria (4 OTUs, uncultured bacterial taxa within the order Saccharimonadales, and Candidatus Peribacteria), Proteobacteria (5 OTUs, Mizugakiibacter sp. and Bdellovibrio sp. and uncultured bacterial taxa within the orders Elsterales and Oligoflexales), and WPS-2 (Fig. 2c). Conversely, OTUs enriched in the rhizosphere of susceptible varieties belonged to the phyla of Bacteroidetes (18 OTUs), Cyanobacteria (25 OTUs), Patescibacteria (3 OTUs), Verrucomicrobia (9 OTUs), and other lower abundant phyla (proportion < 0.02, 46 OTUs) (Supplementary Fig. 2f). The co-occurrence analysis revealed the network of resistant plants to be more complex than that of susceptible plants. That is, displaying higher number of nodes and edges, greater average path lengths, and higher modularity (Fig. 2d, e). Besides, the network from resistant plants had more negative interactions (13.1%) than that of susceptible plants (3.2%) (Fig. 2d). The NetShift analysis indicated Pseudomonadaceae, Caulobacteraceae, and the A4b genera as potential keystone taxa in the rhizosphere microbiomes of resistant plants (Supplementary Fig. 3). Last, PICRUSt2 functional prediction suggested the resistant plant microbiomes have a higher genetic potential for the biosynthesis of indole alkaloid, flavone, and betalain (Supplementary Fig. 4h), all of which were positively associated with differentially abundant taxa in the rhizosphere microbiomes of resistant plants (Supplementary Fig. 4i).
Fig. 2. Plant resistance to bacterial wilt disease is mediated by the rhizosphere microbiome composition.
a Field trial assessment of wilt disease resistance across 12 Solanaceae varieties. Different lowercase letters indicate significant differences in disease incidence across varieties (HSD post hoc test: P < 0.05). b Principal coordinates analysis (PCoA) based on Bray-Curtis distances displaying a significant difference in the rhizosphere microbiomes between resistant and susceptible Solanaceae varieties (F1,35 = 4.3, R2 = 0.112, P < 0.001, PERMANOVA). c Differences in the relative abundances of bacterial phyla (average proportion > 0.02) based on discriminating OTUs (linear discriminant analysis score ≥ 2, fold change ≥ 4, and significance test P < 0.05). Numbers between parentheses denote the number of OTUs in the respective bacterial phylum. P values were calculated using pairwise Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001). d Co-occurrence networks are based on the rhizosphere microbiomes of resistant (left) and susceptible (right) plants. Pink and green numbers denote the number of positive and negative interactions between bacterial taxa, respectively. e Differences in network parameters (degree, closeness centrality, betweenness centrality, and eigenvector centrality) between networks based on the microbiomes of resistant and susceptible plants.
Quantifying the efficiency of RMT from donor to recipient plants
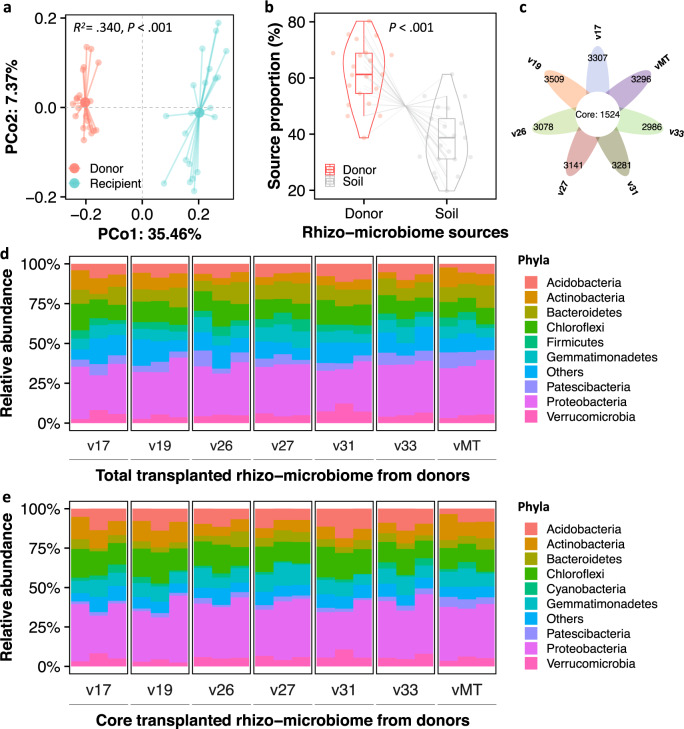
Four weeks after the RMT, no significant differences were found for bacterial community richness and diversity in the rhizosphere of donor and recipient plants (Supplementary Fig. 5a, b). However, the rhizosphere bacterial community compositions differed significantly between the two groups (PERMANOVA: R2 = 0.301, P < 0.001, Fig. 3a). Source tracking analysis revealed that between 45.3% and 68.3% (average of 60.8%) of bacterial OTUs in recipient plants were originated from the rhizosphere microbiome of the six different donor varieties (70.3% for vMT), i.e., referred to as the transplanted OTUs from donors compared to the unknown sources (Student’s t test: t = 16.1, df = 106.0, P < 0.001, Fig. 3b and Supplementary Fig. 6). The “unknown sources” of bacterial OTUs in the recipient rhizospheres refer to the potential colonization from the local microbiome in the soil, which accounted for an average proportion of 39.2% (ranging from 31.7% to 54.7%) (Fig. 3b and Supplementary Fig. 6). When compared to the self-transplanted OTUs in the MicroTom variety (vMT) (i.e., 3296 OTUs), we found the number of successfully transplanted OTUs to range from 2986 to 3509 according to different donor varieties. The sum results in a total of 5805 OTUs, of which 1524 represent the core transplanted OTUs (bacterial OTUs consistently detected in all donor varieties and recipient plants; Fig. 3c). Within the transplanted microbiomes, taxa belonging to the phyla Actinobacteria, Acidobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Gemmatimonadetes, Patescibacteria, Proteobacteria, and Verrucomicrobia accounted for 92.3% of the total (Fig. 3d). Similarly, taxa belonging to the phyla Acidobacteria, Actinobacteria, Bacteroidetes, Chloroflexi, Cyanobacteria, Gemmatimonadetes, Patescibacteria, Proteobacteria, and Verrucomicrobia accounted for 98.8% of the core (Fig. 3e). Worth mentioning, alpha-diversity parameters (Shannon index and OTU richness) of the total and core transplanted microbiomes in recipient plants were slightly lower than that at the donor plant varieties (Supplementary Fig. 5c–f).
Fig. 3. Differences in the RMT from resistant Solanaceae donors to the recipient MicroTom variety.
a Principal coordinates analysis (PCoA) based on Bray-Curtis distances displaying a significant difference in the rhizosphere microbiomes between donor and recipient plants (F1,35 = 4.3, R2 = 0.301, P < 0.001, PERMANOVA). b Source proportion of the donor and the soil in the assembly of the rhizosphere microbiome of recipient tomato MicroTom plants (vMT). Gray lines denote the track of RMT from donor to recipient plants. P values were calculated using Student’s t test. c Venn diagram displaying the total and the core bacterial OTUs transplanted from each donor variety to the recipient vMT. Relative abundances of bacterial phyla associated with the total (d) and core (e) transplanted rhizosphere microbiomes from each donor variety to the recipient vMT.
RMT promotes wilt disease resistance in recipient plants
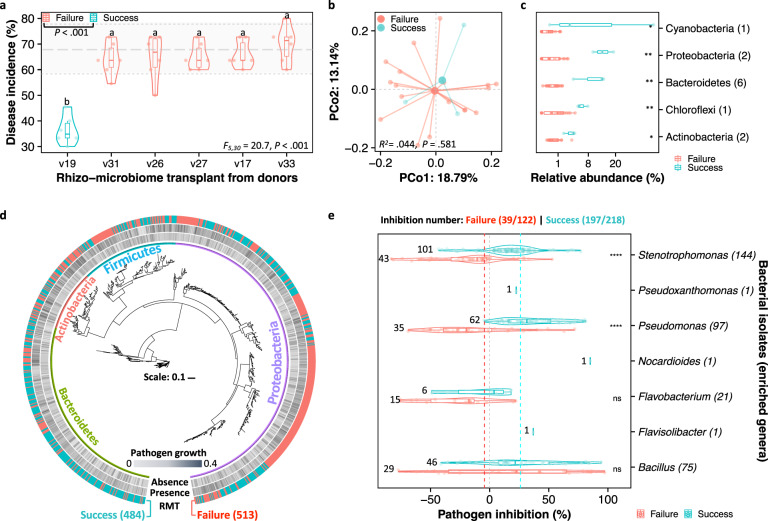
The RMT-greenhouse experiment revealed a significant difference in wilt disease incidence in MicroTom tomato plants that received RMT from 6 resistant donor varieties (ANOVA: F5,30 = 20.6, P < 0.001, Fig. 4a and Supplementary Fig. 7). Interestingly, one out of six RMT (v19: S. melongena var. Shf) completely suppressed the disease incidence at 10 weeks post-inoculation with the pathogen R. solanacearum (Fig. 4a and Supplementary Fig. 8). The RMT of four resistant donors (v17: S. anguivi var. wcb, v26: S. melongena var. GX-11, v31: S. melongena var. D11, v33: S. melongena var. D15) was found to slow down the wilting symptom progression without significantly differing from the control (diseased: self-transplant of vMT) treatment in the lag-phase of the disease curve (HSD test: P > 0.05, Supplementary Fig. 7b). Based on the cut-off of disease incidence in the vMT control (average of 68.2%), the recipient plants were further classified into ‘success’ and ‘failure’ with respect to RMT mediating wilt disease suppression (Supp. Fig. 7). The disease incidence of ‘failure’ recipients ranged from 64.1% to 69.8%, with an average of 66.3% (similar to the vMT, HSD test: P < 0.05). These values were significantly higher than that of ‘success’ transplanted recipients with an average disease incidence of 36.4% (Student’s t test: t = 11.5, df = 8.1, P < 0.001, Fig. 4a). The analysis of the rhizosphere microbiomes of ‘failure’ and ‘success’ recipients did not significantly differ in terms of alpha (Supp. Fig. 8a) and beta diversities (PERMANOVA: R2 = 0.044, P = 0.581, Fig. 4b). At the phyla level, these communities were mostly dominated by taxa belonging to Acidobacteria, Actinobacteria, Bacteroidetes, Chloroflexi, Cyanobacteria, Gemmatimonadetes, Patescibacteria, Proteobacteria, and Verrucomicrobia, collectively accounting for 94.2% of these communities (in detail, 94.6% and 94.1% in ‘success’ and ‘failure’ transplanted recipients, respectively; Supplementary Fig. 8b). A more detailed analysis revealed that a total of 159 and 170 potentially differential OTUs (Supplementary Fig. 9f, g), of which 12 and 11 core OTUs (Supplementary Fig. 9i, j) were found to be associated with ‘success’ and ‘failure’ transplanted recipients, respectively (Supp. Fig. 9). Taxa differentially occurring in the ‘success’ transplants were affiliated to Actinobacteria (2 OTUs, Nonomuraea sp. and Actinoplanes sp.), Bacteroidetes (6 OTUs, Lacibacter sp., Fluviicola sp. and uncultured bacterial taxa within the families Sphingobacteriales, Chitinophagales, and Ignavibacteria-OPB56), Chloroflexi (an uncultured bacterium within the family Roseiflexaceae), Cyanobacteria (an uncultured Microcoleus sp.), and Proteobacteria (2 OTUs, uncultured Aquicella sp. and a bacterium within the family Solimonadaceae) (Fig. 4c). Conversely, those associated with the ‘failure’ were affiliated to Bacteroidetes (an uncultured bacterium within the family Microscillaceae), Chloroflexi (2 uncultured bacterial taxa within the order Anaerolineae-SBR1031), Cyanobacteria (an uncultured bacterium within the family Chroococcidiopsaceae), Proteobacteria (5 OTUs, Caulobacter sp., Paraburkholderia sp., Sneathiellaceae-AT-s3-44 sp., Bdellovibrio sp. and an uncultured bacterium within the order Xanthomonadales), and Verrucomicrobia (2 OTUs, Lacunisphaera sp. and an unclassified bacterium within the family Pedosphaeraceae) (Supplementary Fig. 9e). Worth mentioning, the PICRUSt2 functional prediction did not identify any significant differences in predicted functional profiles of the rhizosphere microbiomes between the ‘success’ and ‘failure’ groups (Supplementary Fig. 10).
Fig. 4. Resistance to wilt disease promoted by the RMT is likely associated with the colonization of antagonistic bacterial taxa.
a Violin plot displaying the disease incidence across RMT from different resistant donors to the rhizosphere of the susceptible tomato variety MicroTom. Horizontal gray lines indicate the range of disease incidence in the control (MicroTom self-transplant, vMT). The terminologies ‘failure’ and ‘success’ denote the disease incidence similar to or significantly lower than that of the vMT, respectively. Different lowercase letters indicate significant differences in disease incidence among varieties (HSD post hoc test: P < 0.05). b Principal coordinates analysis (PCoA) based on Bray-Curtis distances displaying non-significant differences in the rhizosphere microbiomes between ‘success’ and ‘failure’ groups (F1,20 = 0.9, R2 = 0.044, P = 0.581, PERMANOVA). c Differences in relative abundances (mean proportion in each corresponding phylum > 0.01) of discriminating OTUs (linear discriminant analysis score ≥ 2, fold change ≥ 4, and significance test P < 0.05) associated with the ‘success’ group. P values were calculated using pairwise Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001). d Phylogenetic tree based on the obtained bacterial isolates and their respective influences on the growth of the pathogen R. solanacearum. The phylogenetic reconstruction was based on the Maximal likelihood method and included taxa belonging to the phyla Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria. e Test of the inhibitory effects of the bacterial isolates on the growth of the pathogen R. solanacearum. The violin plot displays the seven genera that also had differential abundance in the ‘success’ group of plants (Supplementary Fig. 9g). The numbers in the y axis denote the number of bacterial isolates per genera.
To explore potential antibacterial mechanisms associated with wilt disease suppression, we isolated a total of 997 bacteria from the rhizosphere of ‘success’ (484) and ‘failure’ (513) groups (Fig. 4d). The phylogenetic reconstruction based on the bacterial 16S rRNA gene sequences (Fig. 4d) revealed the isolates to encompass a total of 70 bacterial genera belonging to four main bacterial phyla, including Actinobacteria (90), Bacteroidetes (284), Firmicutes (135), and Proteobacteria (488) (Supplementary Fig. 11). Microplate-based antagonistic assays revealed 65.4% of these bacterial isolates to be able to inhibit the pathogen growth. A higher number of these potentially suppressive strains were obtained from the ‘success’ group (441 out of 484; 91.1%) compared to the ‘failure’ group (198 out of 513; 38.6%) (Supplementary Fig. 12). Besides, the pathogen inhibition of bacterial isolates originated from ‘success’ group (average at 26.1%) were significantly higher than that of bacteria isolated from the ‘failure’ group (average at −4.4%) (Student’s t test: t = 14.7, df = 897.0, P < 0.001; Supplementary Fig. 12a, b). Although a few bacterial isolates were matched to the taxonomic feature of core differential OTUs (Supp. Fig. 9j), 34.1% of them (340) were affiliated to the genera Nocardioides, Stenotrophomonas, Flavobacterium, Pseudomonas, Brevundimonas, Pseudoxanthomonas, Bacillus, and Flavisolibacter (Supp. Fig. 9g and Supplementary Fig. 12c). All of which were found to be potentially differentially abundant in the ‘success’ transplanted recipients (Fig. 4e, Supplementary Figs. 9g and 12c). Notably, these potentially suppressive isolates were either specific in the ‘success’ transplanted recipients (Nocardioides and Pseudoxanthomonas) and/or highly antagonistic (Stenotrophomonas and Pseudomonas; P < 0.001 by Student’s t test), when compared to isolates obtained from the ‘failure’ transplanted recipients (Fig. 4e). In addition, the ‘success’ recipients harbored other genera (e.g., Sphingobacterium and Arthrobacter) that displayed inhibitory effects on the growth of the pathogen (Student’s t test: P < 0.001; Supp. Fig. 12c).
Discussion
Here we tested the feasibility and efficiency of using RMT to actively manipulate the plant-rhizosphere microbiome towards promoting the suppression of an important soil-borne pathogen. We also unraveled the potential ecological mechanism mediating the disease-suppressive phenotype. The experimental design intentionally combined different plant species (resistance screening and susceptible host transplant validation) in order (i) to minimize the potential effects of negative plant-soil feedbacks (i.e., growing the same plant genotype successively in the soil can lead to higher disease incidence), and (ii) to prove the generality of our principle by showing the enrichment of suppressive taxa to be consistent across different plant species. Our data showed that the RMT from one (out of six) Solanaceae resistant variety was successful and significantly reduced the incidence of the bacterial wilt disease caused by the pathogen R. solanacearum by up to 47%. Besides, the RMTs from the other four resistant varieties were found to slightly delay the disease incidence and progression. We advanced our analysis by investigating potential antagonistic effects of rhizosphere bacterial isolates (ca. 997 isolates, 486 and 513 isolates for the ‘success’ and ‘failure’ groups, respectively, see “Methods” for details) suppressing the growth of the pathogen R. solanacearum using in vitro assays based on the inhibitory effect of cell-free supernatants. We acknowledge that caution is warranted in terms of extrapolating the outcome of this in vitro essay with in vivo performances of these isolated taxa. With that said, although non-significant difference was found between ‘success’ and ‘failure’ transplanted microbiomes, we showed that a total of 218 bacterial isolates matched the differentially abundant bacterial genera detected in the ‘success’ RMT plants. From this total, a large proportion of the isolates (197 out of 218) displayed in vitro antagonistic effects on the growth of the pathogen. This indicates that most likely the microbiome-mediated wilt disease suppression is mediated by a small fraction of the successfully transplanted microbial taxa rather than an overall suppressive community effect. These results are in line with previous studies showing that patterns of disease resistance against soil-borne pathogens are tightly linked with the occurrence/colonization of protective bacterial taxa in the soil [37, 69] and/or in the plant rhizosphere [8, 38].
It is worth discussing that despite we achieved a significant success only in one out of six cases for the RMT, other studies reported a similar percentage of success (ca. 10–20%). For example, it was shown that only 2 out of 18 (ca. 11%) transplanted soils effectively reduced the incidence of bacterial wilt disease at levels ranging from 20–60% in tomato plants [69]. Notably, susceptible plants were less wilted after being replanted in the soils previously cultivated with a resistant variety, displaying a 27.7% reduction in disease incidence [8]. This value was significantly higher in our study, reaching up to 47%, likely due to differences in experimental designs and material types. The functional failure of RMT can simply be related to our yet inability to properly manipulate the rhizosphere microbiome in different scenarios (e.g. soil types, plant developmental stage, genotype, etc.) [69]. Here we show that to quantitatively study our efficiency in RTM, the approach of community coalescence combined with source tracking analysis provides an opportunity to advance this ecological principle with direct practical implications (possibly also in the clinics). For comparison, lower success cases have been also observed in fecal transplant therapy in medical studies. For example, ca. of 10% to 30% of patients with melanoma were reported to gain clinical benefits by fecal microbiome transplant [20, 70], and a similar range of effectiveness ranging from 10% to 40% was reported for gastrointestinal disorders [71]. However, in more specific cases, such as the control of recurring infections caused by C. difficile, the success of fecal transplant therapy can reach up to >90% of the cases [19, 72]. We speculate that this variability in efficacy of microbiome transplant is likely related to the association between the disease and the dysbiosis caused in the microbiome [73], and edaphic changes imposed in the local environment (e.g., shifts in pH, nutrients, etc.). Collectively, these will be ultimately linked with the overall composition and status of the donors and recipient microbiomes [74].
In ecology, the transplant of entire communities into existing ones can better be envisioned in light of ‘community coalescence’ (sensu [39]). In brief, it has been conceptualized that several factors are likely to influence the outcome of community coalescence, including environmental conditions, the mixing ratios of donor and recipient communities, the biological interaction interface between taxa, and the subsequent temporal dynamics of community coalescence. In our research model, the pathogen R. solanacearum is known to cause a significant disruption of the resident microbiome [44], thus leading to dysbiosis in the rhizosphere microbiome of diseased plants [75]. In this case, the initial outbreak of the disease is also known to be determined by the pathogen’s ability to initially invade the soil and rhizosphere microbiomes [12, 38]. Besides, since the coalescence outcome may vary due to several factors and may also change in time, our results show that in four cases, the disease incidence was not significantly controlled but was rather delayed in terms of the progression and aggressiveness. Taken together, disentangling these underlying mechanisms associated with factors controlling the coalescence and thus the success of RMT might be a plausible way to advance studies focusing on soil-borne disease control based on rhizosphere microbiome manipulation, with potential theoretical implications for clinical settings.
It is well-known that the rhizosphere microbiome can form a biological layer of protection that promotes plant health [76]. This protection can be mediated by developing niche space for more fit competitions to thrive [42, 77] and directly antagonize pathogens [8, 38], and/or via indirect effects based on the activation of plant immune defenses [78, 79]. Despite we did not observe a more diverse bacterial community in the rhizosphere of resistant, compared to susceptible, Solanaceae varieties (as similarly observed elsewhere, Wei et al. [79]; Hu et al. [77]), we found resistant varieties to host a higher number of taxa displaying negative interactions within the network. These are potentially associated with strong competitive interactions in the microbiome that—to some extent—can prevent the pathogen invasion [80]. Besides, we also detected significant differences in the composition of the rhizosphere microbiomes of resistant and susceptible plants, which are consistent with previous studies [4], including those studying bacterial wilt [8], and Fusarium wilt [81]. These distinct microbiomes from resistant plants were suggested to host a potentially protective functional gene pool able to produce antagonistic secondary metabolites that control the pathogen growth. These include the indole alkaloid [82], flavone [83], and betalain [84]. These findings align with our results, in which some of these genes were also predicted to be enriched in the microbiomes of our resistant lines and associated with the differentially abundant taxa belonging to the phyla Acidobacteria, Actinobacteria, Bacteroidetes, Firmicutes, Gemmatimonadetes, Patescibacteria, Gammaproteobacteria. Within these phyla, several taxa, i.e., Stenotrophomonas [85], Flavobacterium [86], Pseudomonas [87], and Bacillus [88] within Actinobacteria, Firmicutes, Bacteroidetes, and Gammaproteobacteria have also been reported as important in plant protection and termed PGPRs in bacterial wilt disease management [8, 12, 77].
In summary, RMT is a promising approach for the effective manipulation of protective plant microbiomes, in this case, applied to control an important soil-borne disease. Although the concept of fecal transplant therapy is advancing in clinical medicine, the use of a similar approach for RMT is still limited for plant disease management [22]. We acknowledge that most of the challenges relate to (1) the problem of scale-up this potential application with a significant impact for large-scale agroecosystems, and (2) the difference in complexity of soil/rhizosphere microbiomes compared to the gut systems, and how these relate to cases of success. However, we posit that advancing research in RMT in light of the ecology of community coalesce might provide benefits beyond the application of this method for disease control. For instance, it might open up new avenues for root-microbiome manipulation and provide new insights into the mechanisms by which beneficial microbial taxa act within a community to promote plant health and protection.
Supplementary information
Acknowledgements
We thank Dr. Alexandre Jousset and Dr. Ye Tao for critical discussions. This research was financially supported by the National Natural Science Foundation of China (42090062, 42090064, 42007038, 41922053, 31972504 and 32170180), the National Key Research and Development Program of China (2021YFD1900100), and the Fundamental Research Funds for the Central Universities (KJQN202116-KJQN202117 and KYXK202009-KYXK202012), the Science and Technology Major Project of Guangxi (GuiKeAA20108005-3), the Natural Science Foundation of Jiangsu Province (BK20190518 and BK20200533), the China Postdoctoral Science Foundation (2019M651848) and technically supported by the Bioinformatics Center of Nanjing Agricultural University.
Author contributions
Author contributions following the CRediT taxonomy (https://casrai.org/credit/) are as follows: Conceptualization: GJ, ZW, YW, FDA; Resources: GJ, ZW, YW; Methodology: GJ, ZW, YW, FDA; Data curation: GJ, YLZ, GG, WL, WW; Formal analysis: GJ, YLZ, GG, WL, YJ, WW, YG; Funding acquisition: GJ, YZ, TY, ZW, YW, YX, QS; Investigation: GJ, YLZ, GG, WL, WW, TY, YZ, YX; Project administration: GJ, ZW, YW, FDA; Supervision: GJ, ZW, YW, FDA; Software: GJ, YZ, ZW, YW, FDA; Visualization: GJ, YLZ, ZW, YW, FDA, Writing—original draft: GJ; Writing—review & editing: GJ, YZ, ZW, YW, FDA.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yikui Wang, Email: ykwang@gxaas.net.
Zhong Wei, Email: weizhong@njau.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s43705-022-00094-8.
References
- 1.Mendes R, Raaijmakers JM. Cross-kingdom similarities in microbiome functions. ISME J. 2015;9:1905–7. doi: 10.1038/ismej.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg G, Rybakova D, Fischer D, Cernava T, Vergès M-CC, Charles T, et al. Microbiome definition re-visited: old concepts and new challenges. Microbiome. 2020;8:103. doi: 10.1186/s40168-020-00875-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall AB, Tolonen AC, Xavier RJ. Human genetic variation and the gut microbiome in disease. Nat Rev Genet. 2017;18:690–9. doi: 10.1038/nrg.2017.63. [DOI] [PubMed] [Google Scholar]
- 4.Trivedi P, Leach JE, Tringe SG, Sa T, Singh BK. Plant-microbiome interactions: from community assembly to plant health. Nat Rev Microbiol. 2020;18:607–21. doi: 10.1038/s41579-020-0412-1. [DOI] [PubMed] [Google Scholar]
- 5.Ramírez-Puebla ST, Servín-Garcidueñas LE, Jiménez-Marín B, Bolaños LM, Rosenblueth M, Martínez J, et al. Gut and root microbiota commonalities. Appl Environ Microbiol. 2013;79:2–9. doi: 10.1128/AEM.02553-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu T, Ke M, Lavoie M, Jin Y, Fan X, Zhang Z, et al. Rhizosphere microorganisms can influence the timing of plant flowering. Microbiome. 2018;6:231. doi: 10.1186/s40168-018-0615-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Liu Y-X, Zhang N, Hu B, Jin T, Xu H, et al. NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat Biotechnol. 2019;37:676–84. doi: 10.1038/s41587-019-0104-4. [DOI] [PubMed] [Google Scholar]
- 8.Kwak MJ, Kong HG, Choi K, Kwon SK, Song JY, Lee J, et al. Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat Biotechnol. 2018;36:1100–9. doi: 10.1038/nbt.4232. [DOI] [PubMed] [Google Scholar]
- 9.Li H, La S, Zhang X, Gao L, Tian Y. Salt-induced recruitment of specific root-associated bacterial consortium capable of enhancing plant adaptability to salt stress. ISME J. 2021;15:2865–82. doi: 10.1038/s41396-021-00974-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu L, Dong Z, Chiniquy D, Pierroz G, Deng S, Gao C, et al. Genome-resolved metagenomics reveals role of iron metabolism in drought-induced rhizosphere microbiome dynamics. Nat Commun. 2021;12:3209. doi: 10.1038/s41467-021-23553-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol. 2017;17:219–32. doi: 10.1038/nri.2017.7. [DOI] [PubMed] [Google Scholar]
- 12.Lee SM, Kong HG, Song GC, Ryu CM. Disruption of Firmicutes and Actinobacteria abundance in tomato rhizosphere causes the incidence of bacterial wilt disease. ISME J. 2021;15:330–47. doi: 10.1038/s41396-020-00785-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stacy A, Andrade-Oliveira V, McCulloch JA, Hild B, Oh JH, Perez-Chaparro PJ, et al. Infection trains the host for microbiota-enhanced resistance to pathogens. Cell. 2021;184:615–.e17. doi: 10.1016/j.cell.2020.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanders ME, Merenstein DJ, Reid G, Gibson GR, Rastall RA. Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat Rev Gastroenterol Hepatol. 2019;16:605–16. doi: 10.1038/s41575-019-0173-3. [DOI] [PubMed] [Google Scholar]
- 15.Bhattacharyya PN, Jha DK. Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J Microbiol Biotechnol. 2012;28:1327–50. doi: 10.1007/s11274-011-0979-9. [DOI] [PubMed] [Google Scholar]
- 16.Bashan Y, de-Bashan LE, Prabhu SR, Hernandez J-P. Advances in plant growth-promoting bacterial inoculant technology: formulations and practical perspectives (1998–2013) Plant Soil. 2014;378:1–33. doi: 10.1007/s11104-013-1956-x. [DOI] [Google Scholar]
- 17.Zmora N, Zilberman-Schapira G, Suez J, Mor U, Dori-Bachash M, Bashiardes S, et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell. 2018;174:1388–.e21. doi: 10.1016/j.cell.2018.08.041. [DOI] [PubMed] [Google Scholar]
- 18.Kaakoush NO. Fecal transplants as a microbiome-based therapeutic. Curr Opin Microbiol. 2020;56:16–23. doi: 10.1016/j.mib.2020.05.008. [DOI] [PubMed] [Google Scholar]
- 19.Kassam Z, Lee CH, Yuan Y, Hunt RH. Fecal microbiota transplantation for Clostridium difficile infection: systematic review and meta-analysis. Am J Gastroenterol. 2013;108:500–8. doi: 10.1038/ajg.2013.59. [DOI] [PubMed] [Google Scholar]
- 20.Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science. 2021;371:602–9. doi: 10.1126/science.abb5920. [DOI] [PubMed] [Google Scholar]
- 21.Weller DM, Raaijmakers JM, Gardener BBM, Thomashow LS. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu Rev Phytopathol. 2002;40:309–48. doi: 10.1146/annurev.phyto.40.030402.110010. [DOI] [PubMed] [Google Scholar]
- 22.Gopal M, Gupta A, Thomas GV. Bespoke microbiome therapy to manage plant diseases. Front Microbiol. 2013;4:355. doi: 10.3389/fmicb.2013.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raaijmakers JM, Bonsall RF, Weller DM. Effect of population density of Pseudomonas fluorescens on production of 2,4-diacetylphloroglucinol in the rhizosphere of wheat. Phytopathology. 1999;89:470–5. doi: 10.1094/PHYTO.1999.89.6.470. [DOI] [PubMed] [Google Scholar]
- 24.Mazurier S, Corberand T, Lemanceau P, Raaijmakers JM. Phenazine antibiotics produced by fluorescent pseudomonads contribute to natural soil suppressiveness to Fusarium wilt. ISME J. 2009;3:977–91. doi: 10.1038/ismej.2009.33. [DOI] [PubMed] [Google Scholar]
- 25.Siddiqui ZA, Shakeel U. Screening of Bacillus isolates for potential biocontrol of the wilt disease complex of pigeon pea (Cajanus cajan) under greenhouse and small-scale field conditions. J Plant Pathol. 2007;89:179–83. [Google Scholar]
- 26.Yadav K, Damodaran T, Dutt K, Singh A, Muthukumar M, Rajan S, et al. Effective biocontrol of banana fusarium wilt tropical race 4 by a bacillus rhizobacteria strain with antagonistic secondary metabolites. Rhizosphere. 2021;18:100341. doi: 10.1016/j.rhisph.2021.100341. [DOI] [Google Scholar]
- 27.Meng Q, Yin J, Rosenzweig N, Douches D, Hao JJ. Culture-based assessment of microbial communities in soil suppressive to potato common scab. Plant Dis. 2012;96:712–7. doi: 10.1094/PDIS-05-11-0441. [DOI] [PubMed] [Google Scholar]
- 28.Carrión VJ, Cordovez V, Tyc O, Etalo DW, de Bruijn I, de Jager VCL, et al. Involvement of Burkholderiaceae and sulfurous volatiles in disease-suppressive soils. ISME J. 2018;12:2307–21. doi: 10.1038/s41396-018-0186-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gómez Expósito R, de Bruijn I, Postma J, Raaijmakers JM. Current insights into the role of rhizosphere bacteria in disease suppressive soils. Front Microbiol. 2017;8:2529. doi: 10.3389/fmicb.2017.02529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raaijmakers JM, Mazzola M. Soil immune responses. Science. 2016;352:1392–3. doi: 10.1126/science.aaf3252. [DOI] [PubMed] [Google Scholar]
- 31.Bakker PAHM, Pieterse CMJ, de Jonge R, Berendsen RL. The soil-borne legacy. Cell. 2018;172:1178–80. doi: 10.1016/j.cell.2018.02.024. [DOI] [PubMed] [Google Scholar]
- 32.Schlatter D, Kinkel L, Thomashow L, Weller D, Paulitz T. Disease suppressive soils: new insights from the soil microbiome. Phytopathology. 2017;107:1284–97. doi: 10.1094/PHYTO-03-17-0111-RVW. [DOI] [PubMed] [Google Scholar]
- 33.Kyselková M, Kopecký J, Frapolli M, Défago G, Ságová-Marecková M, Grundmann GL, et al. Comparison of rhizobacterial community composition in soil suppressive or conducive to tobacco black root rot disease. ISME J. 2009;3:1127–38. doi: 10.1038/ismej.2009.61. [DOI] [PubMed] [Google Scholar]
- 34.Rosenzweig N, Tiedje JM, Quensen JF, Meng Q, Hao JJ. Microbial communities associated with potato common scab-suppressive soil determined by pyrosequencing analyses. Plant Dis. 2012;96:718–25. doi: 10.1094/PDIS-07-11-0571. [DOI] [PubMed] [Google Scholar]
- 35.Cha JY, Han S, Hong H-J, Cho H, Kim D, Kwon Y, et al. Microbial and biochemical basis of a Fusarium wilt-suppressive soil. ISME J. 2016;10:119–29. doi: 10.1038/ismej.2015.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, Zhang S, Jiang Q, Bai Y, Shen G, Li S, et al. Using community analysis to explore bacterial indicators for disease suppression of tobacco bacterial wilt. Sci Rep. 2016;6:36773. doi: 10.1038/srep36773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 38.Wei Z, Gu Y, Friman VP, Kowalchuk GA, Xu Y, Shen Q, et al. Initial soil microbiome composition and functioning predetermine future plant health. Sci Adv. 2019;5:eaaw0759. doi: 10.1126/sciadv.aaw0759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rillig MC, Antonovics J, Caruso T, Lehmann A, Powell JR, Veresoglou SD, et al. Interchange of entire communities: microbial community coalescence. Trends Ecol Evol. 2015;30:470–6. doi: 10.1016/j.tree.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 40.Deng S, Caddell DF, Xu G, Dahlen L, Washington L, Yang J, et al. Genome wide association study reveals plant loci controlling heritability of the rhizosphere microbiome. ISME J. 2021;15:3181–94. doi: 10.1038/s41396-021-00993-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mendes LW, Mendes R, Raaijmakers JM, Tsai SM. Breeding for soil-borne pathogen resistance impacts active rhizosphere microbiome of common bean. ISME J. 2018;12:3038–42. doi: 10.1038/s41396-018-0234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu J, Wei Z, Kowalchuk GA, Xu Y, Shen Q, Jousset A. Rhizosphere microbiome functional diversity and pathogen invasion resistance build up during plant development. Environ Microbiol. 2020;22:5005–18. doi: 10.1111/1462-2920.15097. [DOI] [PubMed] [Google Scholar]
- 43.Schreiter S, Ding G-C, Heuer H, Neumann G, Sandmann M, Grosch R, et al. Effect of the soil type on the microbiome in the rhizosphere of field-grown lettuce. Front Microbiol. 2014;5:144. doi: 10.3389/fmicb.2014.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei Z, Hu J, Gu Y, Yin S, Xu Y, Jousset A, et al. Ralstonia solanacearum pathogen disrupts bacterial rhizosphere microbiome during an invasion. Soil Biol Biochem. 2018;118:8–17. doi: 10.1016/j.soilbio.2017.11.012. [DOI] [Google Scholar]
- 45.Jiang G, Wei Z, Xu J, Chen H, Zhang Y, She X, et al. Bacterial wilt in China: History, current status, and future perspectives. Front Plant Sci. 2017;8:1549. doi: 10.3389/fpls.2017.01549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manda RR, Addanki VA, Srivastava S. Bacterial wilt of solanaceous crops. Int J Chem Stud. 2020;8:1048–57. doi: 10.22271/chemi.2020.v8.i6o.10903. [DOI] [Google Scholar]
- 47.Barik S, Reddy AC, Ponnam N, Kumari M, C AG, Reddy DCL, et al. Breeding for bacterial wilt resistance in eggplant (Solanum melongena L.): progress and prospects. Crop Prot. 2020;137:105270. doi: 10.1016/j.cropro.2020.105270. [DOI] [Google Scholar]
- 48.Wei Z, Yang X, Yin S, Shen Q, Ran W, Xu Y. Efficacy of Bacillus-fortified organic fertiliser in controlling bacterial wilt of tomato in the field. Appl Soil Ecol. 2011;48:152–9. doi: 10.1016/j.apsoil.2011.03.013. [DOI] [Google Scholar]
- 49.Park E-J, Lee S-D, Chung E-J, Lee M-H, Um H-Y, Murugaiyan S, et al. MicroTom - A model plant system to study bacterial wilt by Ralstonia solanacearum. Plant Pathol J. 2007;23:239–44. doi: 10.5423/PPJ.2007.23.4.239. [DOI] [Google Scholar]
- 50.Schandry N. A practical guide to visualization and statistical analysis of R. solanacearum infection data using R. Front Plant Sci. 2017;8:623. doi: 10.3389/fpls.2017.00623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gu S, Wei Z, Shao Z, Friman VP, Cao K, Yang T, et al. Competition for iron drives phytopathogen control by natural rhizosphere microbiomes. Nat Microbiol. 2020;5:1002–10. doi: 10.1038/s41564-020-0719-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Price MN, Dehal PS, Arkin AP. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 58.Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci. 2003;14:927–30. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 59.Shenhav L, Thompson M, Joseph TA, Briscoe L, Furman O, Bogumil D, et al. FEAST: fast expectation-maximization for microbial source tracking. Nat Methods. 2019;16:627–32. doi: 10.1038/s41592-019-0431-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mendiburu F de. agricolae: Statistical procedures for agricultural research. R package version 1.3–5. 2021.
- 61.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deng Y, Jiang Y-H, Yang Y, He Z, Luo F, Zhou J. Molecular ecological network analyses. BMC Bioinformatics. 2012;13:113. doi: 10.1186/1471-2105-13-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou J, Deng Y, Luo F, He Z, Tu Q, Zhi X. Functional molecular ecological networks. mBio. 2010;1:e00169–10.. doi: 10.1128/mBio.00169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ma B, Wang Y, Ye S, Liu S, Stirling E, Gilbert JA, et al. Earth microbial co-occurrence network reveals interconnection pattern across microbiomes. Microbiome. 2020;8:82. doi: 10.1186/s40168-020-00857-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuntal BK, Chandrakar P, Sadhu S, Mande SS. ‘NetShift’: a methodology for understanding ‘driver microbes’ from healthy and disease microbiome datasets. ISME J. 2019;13:442–54. doi: 10.1038/s41396-018-0291-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, et al. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. 2020;38:685–8. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choi K, Choi J, Lee PA, Roy N, Khan R, Lee HJ, et al. Alteration of bacterial wilt resistance in tomato plant by microbiota transplant. Front Plant Sci. 2020;11:1186. doi: 10.3389/fpls.2020.01186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davar D, Dzutsev AK, McCulloch JA, Rodrigues RR, Chauvin J-M, Morrison RM, et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science. 2021;371:595–602. doi: 10.1126/science.abf3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.D’Haens GR, Jobin C. Fecal microbial transplantation for diseases beyond recurrent Clostridium difficile infection. Gastroenterology. 2019;157:624–36. doi: 10.1053/j.gastro.2019.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gough E, Shaikh H, Manges AR. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis. 2011;53:994–1002. doi: 10.1093/cid/cir632. [DOI] [PubMed] [Google Scholar]
- 73.Durack J, Lynch SV. The gut microbiome: relationships with disease and opportunities for therapy. J Exp Med. 2019;216:20–40. doi: 10.1084/jem.20180448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Danne C, Rolhion N, Sokol H. Recipient factors in faecal microbiota transplantation: one stool does not fit all. Nat Rev Gastroenterol Hepatol. 2021;18:503–13. doi: 10.1038/s41575-021-00441-5. [DOI] [PubMed] [Google Scholar]
- 75.Jiang G, Wang N, Zhang Y, Zhang Y, Yu J, Zhang Y, et al. The relative importance of soil moisture in predicting bacterial wilt disease occurrence. Soil Ecol Lett. 2021;3:356–66.
- 76.Wei Z, Friman VP, Pommier T, Geisen S, Jousset A, Shen Q. Rhizosphere immunity: targeting the underground for sustainable plant health management. Front Agric Sci Eng. 2020;7:317–28. doi: 10.15302/J-FASE-2020346. [DOI] [Google Scholar]
- 77.Hu J, Wei Z, Friman VP, Gu SH, Wang X-F, Eisenhauer N, et al. Probiotic diversity enhances rhizosphere microbiome function and plant disease suppression. mBio. 2016;7:e01790–16.. doi: 10.1128/mBio.01790-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bakker PAHM, Doornbos RF, Zamioudis C, Berendsen RL, Pieterse CMJ. Induced systemic resistance and the rhizosphere microbiome. Plant Pathol J. 2013;29:136–43. doi: 10.5423/PPJ.SI.07.2012.0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wei Z, Yang T, Friman VP, Xu Y, Shen Q, Jousset A. Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health. Nat Commun. 2015;6:8413. doi: 10.1038/ncomms9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li M, Wei Z, Wang J, Jousset A, Friman V-P, Xu Y, et al. Facilitation promotes invasions in plant-associated microbial communities. Ecol Lett. 2019;22:149–58. doi: 10.1111/ele.13177. [DOI] [PubMed] [Google Scholar]
- 81.Mendes LW, Raaijmakers JM, de Hollander M, Mendes R, Tsai SM. Influence of resistance breeding in common bean on rhizosphere microbiome composition and function. ISME J. 2018;12:212–24. doi: 10.1038/ismej.2017.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rosales PF, Bordin GS, Gower AE, Moura S. Indole alkaloids: 2012 until now, highlighting the new chemical structures and biological activities. Fitoterapia. 2020;143:104558. doi: 10.1016/j.fitote.2020.104558. [DOI] [PubMed] [Google Scholar]
- 83.Sarbu LG, Bahrin LG, Babii C, Stefan M, Birsa ML. Synthetic flavonoids with antimicrobial activity: a review. J Appl Microbiol. 2019;127:1282–90. doi: 10.1111/jam.14271. [DOI] [PubMed] [Google Scholar]
- 84.Madadi E, Mazloum-Ravasan S, Yu JS, Ha JW, Hamishehkar H, Kim KH. Therapeutic application of betalains: a review. Plants. 2020;9:E1219. doi: 10.3390/plants9091219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ryan RP, Monchy S, Cardinale M, Taghavi S, Crossman L, Avison MB, et al. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat Rev Microbiol. 2009;7:514–25. doi: 10.1038/nrmicro2163. [DOI] [PubMed] [Google Scholar]
- 86.Kolton M, Erlacher A, Berg G, Cytryn E. The Flavobacterium genus in the plant holobiont: ecological, physiological, and applicative insights. In: Castro-Sowinski S, editor. Microbial models: from environmental to industrial sustainability. Singapore: Springer; 2016. p. 189–207.
- 87.Haas D, Défago G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol. 2005;3:307–19. doi: 10.1038/nrmicro1129. [DOI] [PubMed] [Google Scholar]
- 88.Fira D, Dimkić I, Berić T, Lozo J, Stanković S. Biological control of plant pathogens by Bacillus species. J Biotechnol. 2018;285:44–55. doi: 10.1016/j.jbiotec.2018.07.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.