Abstract
The transition of the 90S to the pre-40S pre-ribosome is a decisive step in eukaryotic small subunit biogenesis leading to a first pre-40S intermediate (state Dis-C or primordial pre-40S), where the U3 snoRNA keeps the nascent 18S rRNA locally immature. We in vitro reconstitute the ATP-dependent U3 release from this particle, catalyzed by the helicase Dhr1, and follow this process by cryo-EM revealing two successive pre-40S intermediates, Dis-D and Dis-E. The latter has lost not only U3 but all residual 90S factors including the GTPase Bms1. In vitro remodeling likewise induced the formation of the central pseudoknot, a universally conserved tertiary RNA structure that comprises the core of the small subunit decoding center. Thus, we could structurally reveal a key tertiary RNA folding step that is essential to form the active 40S subunit.
INTRODUCTION
Ribosomes are universally conserved macromolecular machines that consist of a small and large subunit, which together with translation factors, tRNAs and mRNA catalyze the synthesis of proteins in every cell. In eukaryotes, the ribosomal small (40S) and large (60S) subunits are assembled from precursor rRNA (pre-rRNA) and ribosomal proteins (RPs) during a multistep process that occurs in a hierarchical manner starting in the nucleolus and ending in the cytoplasm. This process involves the dynamic development of different pre-ribosomal particles, with each intermediate containing their specific set of ribosome assembly factors and in some cases also specific snoRNAs (1,2). How the complex interplay of >200 assembly factors with the dynamically evolving pre-ribosomes eventually leads to mature ribosomes has been extensively studied in the past by various approaches, especially by biochemical isolation of pre-ribosomal particles, but there are limitations because of the transient nature of many of these assembly intermediates. Nevertheless, cryo-EM analysis of a number of pre-ribosomal particles isolated from various organisms has provided insights into the conserved structural organization of the chronologically ordered assembly intermediates along the pathway, including the huge 90S pre-ribosome or SSU processome as the first stable precursor to the small subunit (3–6). This 90S particle is associated with ∼50 assembly factors, but it also contains the U3 small nucleolar ribonucleoprotein particle (U3 snoRNP) as a structural module (7–10), which via single-stranded regions in the U3 snoRNA hybridizes to two sites in the 5′ external transcribed spacer (5′ ETS) and to further two sites in the 18S rRNA part (11–13). In the past years, various 90S (also called SSU processome) particles from yeast and Chaetomium thermophilum, were analyzed by cryo-EM, which reveled at high structural resolution these four contact sites, called Box A’ and Box A hybridizing to 18S rRNA, and 5′ hinge and 3′ hinge binding to the 5′ ETS (8,14,15). Importantly, the two U3::18S rRNA hybrids prevent formation of the central pseudoknot (CPK), a hallmark tertiary rRNA structure in the mature 18S rRNA, which is part of the decoding center not only in the active 40S (12,16,17), but also prokaryotic 30S subunit (18,19).
Recently, several distinct pre-ribosomal intermediates formed during the 90S-to-pre-40S transition, called Dis-A, Dis-B and Dis-C, could be purified from cells and structurally determined, which shed first light on how the small subunit gradually evolves from the 90S pre-ribosome (4). Interestingly, the Dis-C (termed primordial pre-40S) intermediate still carried the U3 snoRNA together with a few residual 90S factors (i.e. Bms1-Rcl1, Mpp10-Imp4-Sas10, Utp24, Noc4-Nop14-Emg1) that are not present in the so far analyzed succeeding pre-40S particles. Importantly, in Dis-C particle the two interactions of U3 with the 5′ ETS were already lost, but U3 was still hybridizing to the 18S rRNA sites (Box A duplex). Moreover, the Dis-C particle also contained the DEAH-box RNA helicase Dhr1, which has been implicated in the removal of the U3 snoRNA during small subunit biogenesis (17,20) and supported by experimental findings showing that Dhr1 can trigger in vitro unwinding of an engineered U3–18S duplex with minimal nucleotides 4–50 of U3 and 6–22 of 18S rRNA (17).
Markedly, Dhr1 was found to adopt different conformations during the 90S-to-pre-40S transition. In state Dis-A, Dhr1 was seen in an ADP-bound open conformation, but when relocated in state Dis-C, it acquired a closed apo-state conformation in a 3′-to-5′ engagement with its U3 snoRNA substrate. In this position, the ß-barrel-like domain of Dhr1 interacted with an interface formed by 18S pre-rRNA, Utp14 and Pno1/Dim2, which could be the constellation to activate the Dhr1 ATPase (20–22). Thus, it appeared possible that isolated Dis-C particles could be triggered to further mature by Dhr1-catalyzed ATP hydrolysis.
MATERIALS AND METHODS
Yeast strains and plasmid constructs
The S. cerevisiae strains W303 (genotype: ade2-1, trp1-1, leu2-3,112, his3-11,15, ura3-1, can1-100), Noc4-TAP-Dhr1 K420A-Flag (genotype: NOC4-TAP::HIS3, pRS314 Dhr1 K420A-Flag, W303) (this study), Noc4-TAP–Dhr1-Flag (4) and Dhr1-TAP–Dim1-Flag (4) were used in this study. Genomic tagging was performed as described previously (23). For plasmid construction, E. coli DH5α strain (Thermo Fisher Scientific) was used.
Split-tag tandem affinity-purification
Yeast strains used for split-tag tandem affinity purification, were harvested during the logarithmic growth phase. Frozen yeast cells were mechanically disrupted by a cryogenic grinding mill (Retsch MM400) in lysis buffer [60 mM Tris–HCl pH 8.0, 40 mM KCl, 50 mM NaCl, 2 mM MgCl2, 5% glycerol, 1 mM DTT, 0.1% NP-40, EDTA-free protease inhibitors, 0.013 U/μl RiboLock RNase Inhibitors (Thermo Scientific)]. The lysate was cleared twice by centrifugation (10 min at 5000 rpm followed by 20 min at 17000 rpm, 4°C) and transferred to immunoglobulin G Sepharose 6 Fast Flow beads (GE Healthcare) for 3 h at 4°C, followed by washing with buffer (60 mM Tris–HCl pH 8.0, 40 mM KCl, 15 mM NaCl, 2mM MgCl2, 5% glycerol, 1 mM DTT, 0.01% NP-40). Bound proteins were eluted from beads by TEV cleavage at 16°C for 45 min (buffer supplemented with a final concentration of 1 U/μl RiboLock RNase inhibitors). For the second affinity purification step, the eluate was loaded onto Flag-agarose beads (Anti-Flag M2 Affinity Gel, Sigma–Aldrich) and incubated for 1 h at 4°C. Beads were washed and eluted with buffer containing Flag peptide. The elution buffer for the cryo-EM analysis contained 60 mM Tris–HCl (pH 8.0), 50 mM NaCl, 5 mM MgCl2, 2% glycerol, 0.01% NP-40 and 1 mM DTT.
In vitro ATP assay
The final eluate of split-tag tandem affinity purifications was divided into two equal aliquots. Samples were supplemented with MgCl2 to a final concentration of 3 mM and either AMP-PNP or ATP to a final concentration of 1 mM. Samples were incubated at 10°C for 25 min and immediately loaded onto a sucrose gradient for separation of pre-ribosomes from released factors and RNAs.
Sucrose gradient ultracentrifugation
Eluates from the in vitro ATP assay were loaded onto a linear 10–40% (w/v) sucrose gradient containing a buffer of 60 mM Tris–HCl (pH 8.0), 50 mM NaCl, 2 mM MgCl2, 0.003% NP-40 and 1 mM DTT and centrifuged for 16 h at 27 000 rpm at 4°C. The sucrose gradient was fractionated into 10 fractions and each fraction was split for RNA analysis (see below) or protein analysis. Protein samples were precipitated by 10% trichloroacetic acid (TCA), and TCA pellets resuspended in SDS sample buffer. Resuspended fractions were analyzed by 4–12% gradient polyacrylamide gel electrophoresis (NuPAGE, Invitrogen), followed by staining with colloidal Coomassie Blue (Roti-Blue, Roth).
RNA extraction and northern analysis
The RNA was isolated from the sucrose gradient fractions by phenol/chloroform and precipitated with ethanol as previously described (4,5). The extracted RNA was loaded onto an 8% polyacrylamide/8M urea gel to resolve smaller RNA species (U3 snoRNA) after denaturation with formaldehyde or loaded onto a 1.2% agarose gel to resolve larger pre-rRNA species (23S, 21S and 20S) after denaturation with glyoxal. Followed by blotting to positively charged nylon membranes and UV crosslinking, the following 5′-end 32P-labeled DNA oligonucleotide probes were used for northern analysis: 5′-GGTTATGGGACTCATCA-3′ (probe for U3) and 5′-CGGTTTTAATTGTCCTA-3′ (OMK002; probe for 23S, 21S, 20S).
Electron microscopy and image processing
Purified samples (3.5 μl) were applied to pre-coated (2 nm) R3/3 holey-carbon-supported copper grids (Quantifoil), blotted for 2–3 s at 4°C, and plunge-frozen in liquid ethane using an FEI Vitrobot Mark IV. Data were collected on a Titan Krios cryo-electron microscope operating at 300 keV. All data were collected with a pixel size of 1.047 Å/pixel and within a defocus range of −0.8 to −2.5 μm using a K2 Summit direct electron detector under low-dose conditions, with a total dose of 44 e−/Å2. Original image stacks were dose-weighted, aligned, summed, and drift-corrected using MotionCor2 (24). Contrast-transfer function (CTF) parameters and resolutions were estimated for each micrograph using CTFFIND4 and GCTF, respectively (25,26). Micrographs with an estimated resolution of less than 5 Å and an astigmatism of less than 5% were manually screened for contamination or carbon rupture.
In the end, a total of 6218 good micrographs are selected for the Dhr1-Dim1 sample treated with ATP, while a total of 6428 good micrographs for the control. Particle picking was carried out using Gautomatch without reference, resulting in 552 020 and 885 032 particles for the ATP treatment and control sample, respectively. Particle extraction was done in Relion 3.1 (27), after that all the particles were imported into cryoSPARC (28) for 2D classification and 3D classification until high resolution pre-40S particle with minimal orientation bias was reached, as shown in Supplementary Figure S2. These particles were imported back to Relion 3.1 to do the final 3D classification (or focused 3D classification), CTF refinement, 3D refinement, Postprocessing and Local resolution filter.
Model building and refinement
The structure of the yeast primordial pre-40S ribosome (PDB IDs: 6ZQG) (4) was used as initial references to generate the final models for the state Dis-D and Dis-E. In general, the model of state Dis-C was rigid body fitted using Coot (29), followed by adjustment, such as removing the entire 40S Head part. Assembly factors that are not present in state Dis-D (such as, Utp14, Utp24, Rcl1, Dhr1 and U3 snoRNA) and in state Dis-E (such as Bms1, Utp3/Sas10 and Mpp10) were manually removed afterward. The matured rRNA helix h27, and different conformation of the rRNA helices h1-h3 and h5 were de novo built in Coot (29). The new immature rRNA helix h18 was built based on the shape of the density map and the secondary structure prediction.
The final models for the states Dis-D and Dis-E were real-space refined with secondary structure restraints using the PHENIX suite (30). The final model evaluation was performed with MolProbity (31). Maps and models were visualized, and figures were created with ChimeraX (32).
RESULTS AND DISCUSSION
In vitro U3 snoRNA release from primordial pre-40S
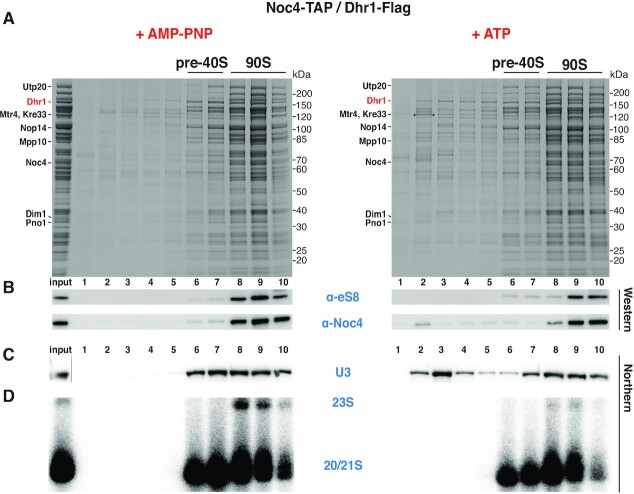
To investigate whether isolated Dis-C particles (also called primordial pre-40S) can be induced to mature into the next stage of pre-40S particles, we developed an in vitro assay for the U3 release from the Dis-C particles and followed this possibility both biochemically and by cryo-EM analysis. For this in vitro reaction, we isolated pre-ribosomes in the 90S-to-pre-40S transition stage via split-tag purification based on the Noc4-Dhr1 baits as previously described (4). The addition of either ATP or the non-hydrolysable ATP analog AMP-PNP (mock control) allowed us to monitor energy-dependent maturation steps of these pre-ribosomes carrying the functional wild-type Dhr1, which might induce such reactions due to its ATP-dependent helicase activity (Figure 1; for mutant Dhr1, see below). Following sucrose gradient ultracentrifugation combined with SDS-polyacrylamide gel electrophoresis, Dhr1-containing pre-ribosomes were resolved in two main populations, the pre-40S pool (fractions 6 and 7) and the 90S pool (fractions 8 to 10), but no drastic difference in the overall pattern of the associated assembly factors was observed, neither by ATP nor AMP-PNP treatment (Figure 1A). However, we noticed that upon ATP but not AMP-PNP incubation a fraction of Dhr1 was liberated from the pre-ribosomes and appeared in the upper part of the sucrose gradient (Figure 1A). Concomitantly, ATP but not AMP-PNP induced the release of a pool of U3 snoRNA from the pre-ribosomal fractions, again detected on the top of the sucrose gradient (Figure 1C). It appears that the U3 snoRNA was predominantly dissociated from the Dis-C particles, while the amount of U3 snoRNA associated with the 90S pool was less affected by the ATP treatment (Figure 1C). To rule out that ATP incubation has caused destabilization of pre-ribosomal particles triggering unspecific RNA dissociation (including U3 release), we probed for the stability and possible dissociation of the ribosomal protein (eS8/Rps8) and the 90S factor Noc4 by Western, as well as the integrity and dissociation of the 23S, 21S and 20S pre-rRNA species by Northern, which however in both cases were neither altered nor released (Figure 1B and D).
Figure 1.
ATP-induced U3 snoRNA release from isolated primordial pre-40S particles. Analysis of Noc4-TAP–Dhr1-Flag affinity-purified pre-ribosomes by sucrose gradient centrifugation, followed by Northern blotting or SDS-polyacrylamide gel electrophoresis, Coomassie staining and Western blotting. Samples were treated with AMP-PNP (left panel) or ATP (right panel). (A) SDS-PAGE of sucrose gradient fractions containing the pre-40S and 90S intermediates. Indicated assembly factors were identified by mass spectrometry (the band marked with an asterisk was identified as Mtr4). (B) Western blot using anti-eS8/Rps8 and anti-calmodulin (the calmodulin tag was still present on the Noc4 bait) antibodies. (C, D) Northern blot analysis of the gradient fractions using specific probes for the U3 snoRNA (C) and the 20S, 21S, 23S pre-rRNA species (D). In the case of the northern (C), a cut has been made between the input lane and lane 1, which is indicated by a grey line. We note a 23S pre-rRNA decrease in the ATP-dependent in vitro reaction. The reason for this is not clear, but the 23S pre-rRNA may have been processed to 21S pre-rRNA by A1 cleavage, stimulated by an ATP-dependent reaction involving an ATP-dependent enzyme such as Mtr4 or Dhr1, which are both present on the employed Noc4-Dhr1 particles (5).
Dhr1 helicase mutant defective in U3 snoRNA release from the pre-40S
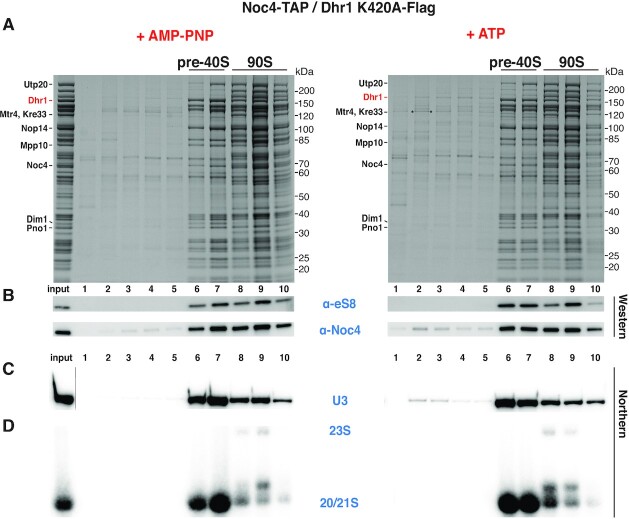
To directly show that the ATPase activity of Dhr1 has triggered in vitro U3 release from the Dis-C particles, we performed similar experiments as described above using pre-ribosomes carrying a mutant form of Dhr1 unable to bind and hydrolyze ATP. For this purpose, we performed split-tag affinity-purification from a yeast strain that carried chromosomally integrated NOC4-TAP for the first, and the Flag-tagged Dhr1 K420A mutant defective in ATP-binding and hydrolysis (17) for the second purification step. Evidently, treatment of these Noc4-Dhr1 K420A mutant particles with ATP no longer induced in vitro the release of the U3 snoRNA and only very little was detected on the top of the sucrose gradient (Figure 2). These observations suggested that intact Dhr1 helicase activity is required for the timely release of the U3 snoRNA during the 90S-to-pre-40S transition, which agrees well with previous findings that showed accumulation of a pool of pre-40S particles with trapped 90S factors in dhr1 mutant cells (17). Taken together, our data have shown that the Dhr1 helicase requires ATP to stimulate in vitro the release of the U3 snoRNA from primordial pre-40S particles, which in consequence could trigger further progression in the pre-40S maturation pathway.
Figure 2.
Inhibition of ATP-induced U3 snoRNA release from primordial pre-40S particles isolated via catalytically inactive Dhr1 K420A mutant. Analysis of mutant Noc4-TAP–Dhr1 K420A-Flag affinity-purified pre-ribosomes by sucrose gradient centrifugation, followed by Northern blotting or SDS–PAGE, Coomassie staining and Western blotting. Samples were treated with AMP-PNP (left panel) or ATP (right panel). (A) SDS-PAGE of sucrose gradient fractions containing the pre-40S and 90S intermediates. Indicated assembly factors were identified by mass spectrometry (the band marked with an asterisk was identified as Mtr4). (B) Western blot using anti-eS8/Rps8 and anti-calmodulin (the calmodulin tag was still present on the Noc4 bait) antibodies. (C,D) Northern blot analysis of the gradient fractions using specific probes for the U3 snoRNA (C) and the 20S, 21S, 23S pre-rRNA species (D). In the case of the Northern (C), a cut has been made between the input lane and lane 1, which is indicated by a grey line.
Cryo-EM of in vitro matured primordial pre-40S
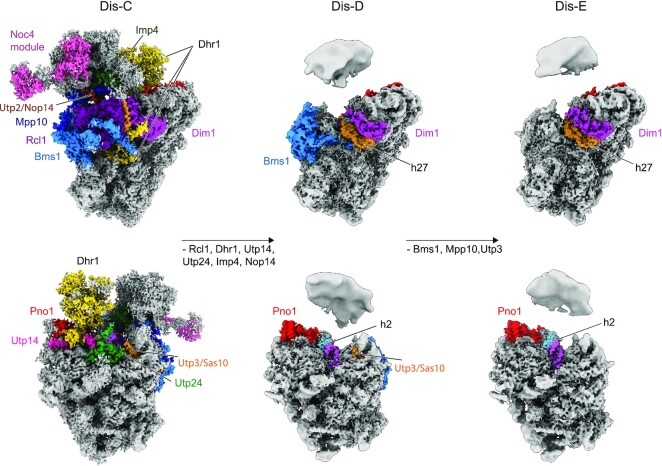
To analyze the in vitro induced maturation of Dis-C particles by structural means, we isolated particles in the 90S > pre-40S transition stage via split-tag purification using the Dhr1-Dim1 bait combination, because the yield of the Dis-C intermediate is higher in this preparation when compared to the Noc4-Dhr1 purification (4). However, both preparations responded similarly in the in vitro assay with a good release of the U3 snoRNA upon ATP treatment (Supplementary Figure S1). Hence, the Dhr1-Dim1 sample was split into two equal parts with one half treated with ATP at 10°C for 25 minutes, while to the other half, serving as mock control, no ATP was added. Two cryo-EM datasets were collected using exactly the same parameters, however only the ATP-treated sample revealed novel pre-40S states, whereas the control recapitulated the previously published findings (4). Overall, two new states (termed Dis-D and Dis-E) could be refined to an average resolution of 3.3 Å and 3.5 Å, respectively (Figure 3, Supplementary Figures S2 and S3, and Supplementary Tables S1 and S2). However, similar to the previously observed state Dis-C, both new states displayed a relatively stable and well-resolved 40S body with lower local resolution in the periphery, whereas very low local resolution indicated even more flexibility of the 40S head. Compared to state Dis-C, both new states showed further maturation of the 18S rRNA as well as a distinct assembly factor composition (Figure 3 and Supplementary Figure S4). Thus, we could arrange them into a plausible order, representing the Dhr1-driven maturation events following the primordial pre-40S state Dis-C.
Figure 3.
Cryo-EM analysis of the in vitro maturation of the primordial pre-40S driven by Dhr1. Two different views show the further maturation of the primordial pre-40S (PDB: 6ZQG, state Dis-C, left) to state Dis-D (middle) and Dis-E (right). Compositional changes are indicated in the middle row and all the AFs were color-coded. In order to show the body and head of the pre-40S in state Dis-D and Dis-E all together, different contour levels were used.
In vitro remodeling induces formation of the central pseudoknot
One of the key features in the first state, Dis-D, is the formation of helices h2 and h27 of the 18S rRNA facilitated by the release of U3 (Figure 3 and Supplementary Figure S4). During this snoRNA dissociation, the final contact between U3 and the 18S rRNA (Box A duplex) in the Dis-C state was dissolved in the in vitro reaction upon the addition of ATP and function of the Dhr1 helicase, consistent with the biochemical data (see Figure 1C). Maturation of the 18S rRNA was accompanied by dramatic changes in the composition of assembly factors. (i) The Dhr1 helicase and its nearby regulator Utp14 (21) were released along with the U3 snoRNA. It is possible that the proximity of these two proteins in Dis-C particles (4) might not be a coincidence, but in this way Utp14 could have been involved in activation of Dhr1 upon addition of ATP. (ii) Concomitantly, the N-terminus of Dhr1 dissociated, thereby freeing the h44 interface on the 5′ domain of the 18S rRNA. (iii) U3 snoRNA release promoted the formation of the CPK (i.e. formation of h1 and h2, see next section) resulting in the loss of Utp24, which was stably associated with h1 of the 18S rRNA in state Dis-C. (iv) Rcl1, which occupies the mature position of helix h27 of the 18S rRNA in state Dis-C, was no longer detectable. (v) Imp4, which coordinates the 40S head with the body in state Dis-C, was not visible in the states Dis-D and Dis-E, indicating that it was dislodged. (vi) When reaching state Dis-E, the entire Bms1 module together with Mpp10 and Sas10/Utp3 were released from the particle resulting in complete maturation of h1 to h3 within the 5′ domain of the 18S rRNA with the exception of rRNA helix h18 (Figure 3 and Supplementary Figure S4). At this point, state Dis-E is characterized by a complete shedding of all the remaining 90S assembly factors and thus represents the first canonical pre-40S particle. Taking together, cryo-EM has uncovered that in the presence of ATP the Dhr1 helicase can drive in vitro the maturation of the primordial pre-40S ribosome to novel transition intermediates that so far could not be isolated from cells.
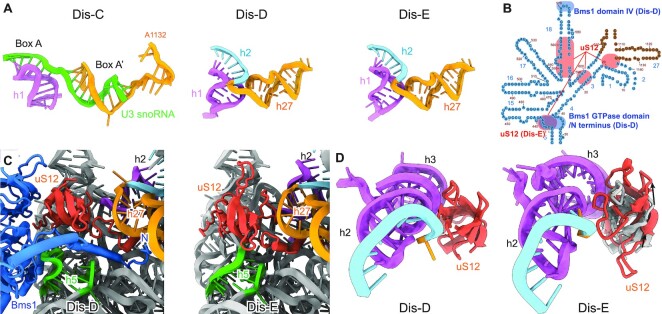
Importantly, our in vitro assay allowed observing the formation of the fully matured CPK in the pre-40S particles upon activation of the Dhr1 helicase. The Dhr1-driven extraction of the U3 snoRNA, which occupied the h2 hybridization region near h1 and h27 of the 18S rRNA, resulted in the formation of the rRNA helices h2 and h27 in state Dis-D (Figure 4A and Supplementary Figure S5A). However, h1 to h3 of the 18S rRNA in state Dis-D still required further conformational changes (including a single base shift) in state Dis-E to adopt their final mature conformation, a prerequisite for this is the correct positioning of ribosomal protein uS12. In state Dis-D, Bms1 was still associated with the immature h5 of the 18S rRNA together with its extended N-terminus next to uS12. Only after complete release of Bms1 in state Dis-E, h5 of the 18S rRNA could adopt its mature conformation, thereby allowing the globular domain of uS12 to slide into its mature binding site (Figure 4B, C). The movement and changed interaction of uS12 with the connecting region between h1 and h3 of the 18S rRNA apparently results in the stabilization of the base pairing of this region that finally establishes the central pseudoknot in its mature state (Figure 4B, D).
Figure 4.
Maturation of the central pseudoknot (CPK) and helix h18. (A) Conformations of the CPK region in state Dis-C (top), Dis-D (middle) and Dis-E. (B) Secondary structure of the immature 18S rRNA focusing on the central pseudoknot region. The Bms1 and uS12 interaction regions (including overlap region in states Dis-D and Dis-E) are shadowed by light blue and orange, respectively. (C) Compared to state Dis-D (left), the dissociation of Bms1 allows rRNA helix h5 to adopt its mature conformation and the relocation of uS12 to its mature position in state Dis-E (right). (D) Compared to state Dis-D (left), the relocation of uS12 in state Dis-E (right) finally promotes the maturation of the CPK. The position of uS12 in state Dis-D is shown in Dis-E colored in gray.
We found that the U3 snoRNA extraction by Dhr1 caused additional consequences for the pre-40S maturation mediated by a cascade of structural rearrangements of the rRNA region illustrated as 2D diagram (Figure 4B). The helix h18 of the 18S rRNA was observed in a new but still immature conformation stabilized in the state Dis-D, which is most probably the result of central pseudoknot formation. In state Dis-C, the immature h18 is distant from h1 of the 18S rRNA and stabilized by 90S factors Utp2/Nop14, Bms1 and Imp4. With the remodeling in the CPK region, helix h1 could now directly interact with h18 via a contact of an internal loop of h18, thereby rearranging the conformation of state Dis-C. The new conformation of h18 would no longer allow the interactions with the nearby 90S factors, thus contributing to the release of these factors during the Dis-C to Dis-E transitions (Supplementary Figure S5B). Interestingly, the conformational switch of helix h18 leads also to conformational switch of Bms1 by pushing domain IV of Bms1 outwards. Bms1 remains in essentially the same GTP-bound conformation all the way from 90S formation to the primordial pre-40S. A plausible scenario would be that this conformational switch during Dis-C to Dis-D transition allosterically activates the GTPase, which after GTP hydrolysis may cause the release of Bms1 in state Dis-D.
In conclusion, in this study we could reconstitute in vitro a long-sought maturation step during eukaryotic pre-ribosome synthesis. This revealed in detail the conversion of the primordial pre-40S into its next intermediates, which was accompanied by the shedding of the remaining 90S factors and formation of the central pseudoknot, a key tertiary RNA structure of the active mature 40S subunit. In vivo, it may not be possible to watch this transition in such a great detail, due to the dynamic nature of this process, which appears to be controlled by the energy state of the cell and possibly by linkage to other cellular pathways such as nutrient availability, gene expression or cell cycle progression. In addition, our cryo-EM structures provided information on the molecular basis of this transition with the local resolution also in the periphery mostly better than 10 Å allowing for a reliable model fitting. It thereby gave mechanistic insight into how release of U3 from the primordial pre-40S is coupled with a stepwise central pseudoknot formation, which is spontaneous and made irreversible by the integration of the ribosomal protein uS12 at the rearranged h2-h27 rRNA helices. Moreover, the Dhr1-driven release of the U3 snoRNA showed that the 40S head started to adopt a more compact conformation over the 40S body, however it still remained highly flexible. Thus, in the next maturation steps, pre-40S assembly factors need to relocate and therefore fix the largely immature pre-40S head before export into the cytoplasm. At the same time, although the 5′ domain was almost mature, h18 of the 18S rRNA adopted another new immature conformation. However, it is known that in later steps, a classical later pre-40S factor, Tsr1, will associate at the position previously occupied by Bms1. Thus, we predict that the maturation of h18 is coordinated with Tsr1 assembly to initiate the next phase in nuclear pre-40S maturation (see accompanying manuscript by Cheng et al., 2022).
DATA AVAILABILITY
EM densities have been deposited in the electron microscopy data bank (EMDB) and the coordinates of the EM-based models have been deposited in the Protein Data Bank (PDB) for the reported structures under accession codes: Dis-D (PDB: 7WTL, EMDB: EMD-32790); Dis-E (PDB: 7WTM, EMDB: EMD-32791). All other data needed to evaluate the conclusions in the paper are present in Supplementary Data.
Supplementary Material
ACKNOWLEDGEMENTS
Technical support for this work was provided by Charlotte Ungewickell and Susanne Rieder.
Contributor Information
Jingdong Cheng, Gene Center and Department of Biochemistry, University of Munich LMU, Feodor-Lynen-Str. 25, 81377 Munich, Germany; Institutes of Biomedical Science, Shanghai Key Laboratory of Medical Epigenetics, International Co-laboratory of Medical Epigenetics and Metabolism (Ministry of Science and Technology), Fudan University, Dong’an Road 131, 200032, Shanghai, China.
Giuseppe La Venuta, BZH, University of Heidelberg, Im Neuenheimer Feld 328, 69120 Heidelberg, Germany.
Benjamin Lau, BZH, University of Heidelberg, Im Neuenheimer Feld 328, 69120 Heidelberg, Germany.
Otto Berninghausen, Gene Center and Department of Biochemistry, University of Munich LMU, Feodor-Lynen-Str. 25, 81377 Munich, Germany.
Roland Beckmann, Gene Center and Department of Biochemistry, University of Munich LMU, Feodor-Lynen-Str. 25, 81377 Munich, Germany.
Ed Hurt, BZH, University of Heidelberg, Im Neuenheimer Feld 328, 69120 Heidelberg, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
ERC [ADG 741781 GLOWSOME to E.H.] from the DFG [BE 1814/15-1 to R.B.]; European Research Council grant ADG under the European Union's Horizon 2020 research and innovation program [885711-Human-Ribogenesis to R.B.]. Funding for open access charge: ERC [ADG 741781 GLOWSOME to E.H.].
Conflict of interest statement. None declared.
This paper is linked to: doi:10.1093/nar/gkac961.
REFERENCES
- 1. Bassler J., Hurt E.. Eukaryotic ribosome assembly. Annu. Rev. Biochem. 2019; 88:281–306. [DOI] [PubMed] [Google Scholar]
- 2. Klinge S., Woolford J.L. Jr. Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 2019; 20:116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Singh S., Vanden Broeck A., Miller L., Chaker-Margot M., Klinge S.. Nucleolar maturation of the human small subunit processome. Science. 2021; 373:eabj5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cheng J., Lau B., La Venuta G., Ameismeier M., Berninghausen O., Hurt E., Beckmann R.. 90S pre-ribosome transformation into the primordial 40S subunit. Science. 2020; 369:1470–1476. [DOI] [PubMed] [Google Scholar]
- 5. Lau B., Cheng J., Flemming D., La Venuta G., Berninghausen O., Beckmann R., Hurt E.. Structure of the maturing 90S Pre-ribosome in association with the RNA exosome. Mol. Cell. 2021; 81:293–303. [DOI] [PubMed] [Google Scholar]
- 6. Cheng J., Bassler J., Fischer P., Lau B., Kellner N., Kunze R., Griesel S., Kallas M., Berninghausen O., Strauss D.et al.. Thermophile 90S Pre-ribosome structures reveal the reverse order of co-transcriptional 18S rRNA subdomain integration. Mol. Cell. 2019; 75:1256–1269. [DOI] [PubMed] [Google Scholar]
- 7. Kornprobst M., Turk M., Kellner N., Cheng J., Flemming D., Kos-Braun I., Kos M., Thoms M., Berninghausen O., Beckmann R.et al.. Architecture of the 90S Pre-ribosome: a structural view on the birth of the eukaryotic ribosome. Cell. 2016; 166:380–393. [DOI] [PubMed] [Google Scholar]
- 8. Sun Q., Zhu X., Qi J., An W., Lan P., Tan D., Chen R., Wang B., Zheng S., Zhang C.et al.. Molecular architecture of the 90S small subunit pre-ribosome. Elife. 2017; 6:e22086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chaker-Margot M., Barandun J., Hunziker M., Klinge S.. Architecture of the yeast small subunit processome. Science. 2017; 355:eaal1880. [DOI] [PubMed] [Google Scholar]
- 10. Hunziker M., Barandun J., Petfalski E., Tan D., Delan-Forino C., Molloy K.R., Kim K.H., Dunn-Davies H., Shi Y., Chaker-Margot M.et al.. UtpA and UtpB chaperone nascent pre-ribosomal RNA and U3 snoRNA to initiate eukaryotic ribosome assembly. Nat. Commun. 2016; 7:12090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beltrame M., Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992; 11:1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hughes J.M.X. Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J. Mol. Biol. 1996; 259:645–654. [DOI] [PubMed] [Google Scholar]
- 13. Sharma K., Tollervey D. Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol. Cell. Biol. 1999; 19:6012–6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheng J., Kellner N., Berninghausen O., Hurt E., Beckmann R.. 3.2-A-resolution structure of the 90S preribosome before A1 pre-rRNA cleavage. Nat. Struct. Mol. Biol. 2017; 24:954–964. [DOI] [PubMed] [Google Scholar]
- 15. Barandun J., Chaker-Margot M., Hunziker M., Molloy K.R., Chait B.T., Klinge S.. The complete structure of the small-subunit processome. Nat. Struct. Mol. Biol. 2017; 24:944–953. [DOI] [PubMed] [Google Scholar]
- 16. Granneman S., Kudla G., Petfalski E., Tollervey D. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:9613–9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sardana R., Liu X., Granneman S., Zhu J., Gill M., Papoulas O., Marcotte E.M., Tollervey D., Correll C.C., Johnson A.W.. The DEAH-box helicase dhr1 dissociates U3 from the pre-rRNA to promote formation of the central pseudoknot. PLoS Biol. 2015; 13:e1002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brink M.F., Verbeet M.P., de Boer H.A.. Formation of the central pseudoknot in 16S rRNA is essential for initiation of translation. EMBO J. 1993; 12:3987–3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Poot R.A., van den Worm S.H., Pleij C.W., van Duin J.. Base complementarity in helix 2 of the central pseudoknot in 16S rRNA is essential for ribosome functioning. Nucleic Acids Res. 1998; 26:549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Choudhury P., Hackert P., Memet I., Sloan K.E., Bohnsack M.T.. The human RNA helicase DHX37 is required for release of the U3 snoRNP from pre-ribosomal particles. RNA Biol. 2019; 16:54–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhu J., Liu X., Anjos M., Correll C.C., Johnson A.W.. Utp14 recruits and activates the RNA helicase dhr1 to undock U3 snoRNA from the preribosome. Mol. Cell. Biol. 2016; 36:965–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sturm M., Cheng J., Bassler J., Beckmann R., Hurt E.. Interdependent action of KH domain proteins krr1 and dim2 drive the 40S platform assembly. Nat. Commun. 2017; 8:2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E.et al.. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004; 21:947–962. [DOI] [PubMed] [Google Scholar]
- 24. Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A.. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017; 14:331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016; 193:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rohou A., Grigorieff N.. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015; 192:216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H.. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018; 7:e42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A.. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017; 14:290–296. [DOI] [PubMed] [Google Scholar]
- 29. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D, Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 30. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W.et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D, Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen V.B., Arendall W.B. 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C.. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D, Biol. Crystallogr. 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E.. UCSF chimerax: meeting modern challenges in visualization and analysis. Protein Sci. 2018; 27:14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
EM densities have been deposited in the electron microscopy data bank (EMDB) and the coordinates of the EM-based models have been deposited in the Protein Data Bank (PDB) for the reported structures under accession codes: Dis-D (PDB: 7WTL, EMDB: EMD-32790); Dis-E (PDB: 7WTM, EMDB: EMD-32791). All other data needed to evaluate the conclusions in the paper are present in Supplementary Data.