Abstract
Anaerobic ammonium oxidation (Anammox) bacteria are a group of extraordinary bacteria exerting a major impact on the global nitrogen cycle. Their phylogenetic breadth and diversity, however, are not well constrained. Here we describe a new, deep-branching family in the order of Candidatus Brocadiales, Candidatus Bathyanammoxibiaceae, members of which have genes encoding the key enzymes of the anammox metabolism. In marine sediment cores from the Arctic Mid-Ocean Ridge (AMOR), the presence of Ca. Bathyanammoxibiaceae was confined within the nitrate-ammonium transition zones with the counter gradients of nitrate and ammonium, coinciding with the predicted occurrence of the anammox process. Ca. Bathyanammoxibiaceae genomes encode the core genetic machinery for the anammox metabolism, including hydrazine synthase for converting nitric oxide and ammonium to hydrazine, and hydrazine dehydrogenase for hydrazine oxidation to dinitrogen gas, and hydroxylamine oxidoreductase for nitrite reduction to nitric oxide. Their occurrences assessed by genomes and 16S rRNA gene sequencings surveys indicate that they are present in both marine and terrestrial environments. By introducing the anammox potential of Ca. Bathyanammoxibiaceae and charactering their ideal niche in marine sediments, our findings suggest that the diversity and abundance of anammox bacteria may be higher than previously thought, and provide important insights on cultivating them in the future to not only assess their biogeochemical impacts but also constrain the emergence and evolutionary history of this functional guild on Earth.
Subject terms: Water microbiology, Biogeochemistry
Introduction
Anammox bacteria play critical roles in the global nitrogen cycle, especially in nitrogen loss in natural [1–6] and engineered environments [7, 8]. Currently known anammox bacteria are affiliated to five different genera in two families in the order Candidatus Brocadiales [per the nomenclature of Genome Taxonomy Database (GTDB; [9])]: Candidatus Brocadia, Candidatus Kuenenia, Candidatus Anammoxoglobus and Candidatus Jettenia [10] in the family of Candidatus Brocadiaceae, and Candidatus Scalindua in the family Candidatus Scalinduaceae. Considering that the phylogenetic breadth of functional guilds has been greatly expanded due to the explosion of available genomic data, such as bacteria involved in sulfate reduction [11] and prokaryotes responsible for ammonia oxidation [12–15], it remains unclear whether the currently characterized anammox lineages can fully represent all potential anammox bacterial diversity.
There is a general partitioning of the anammox bacteria between the marine and terrestrial environments. Anammox bacteria in the marine environment are dominated by Ca. Scalindua [16–20]. In contrast, anammox bacteria in the diverse terrestrial habitats hosted the other four lineages [4, 21], although Scalindua were also occasionally noted in in engineered [22] and freshwater systems [23]. The underlying mechanisms driving this ecological niche partitioning are not fully elucidated yet [24]. Recovering more deeply branching lineages of anammox bacteria would be valuable to illuminate this ecophysiological division and evolutionary history of this important functional guild in geological history.
Here we report a deep-branching family within the order of Ca. Brocadiales, which harbor members from both marine and terrestrial environments. Members of this family have a confined distribution within the nitrate-ammonium transition zones (NATZs) of marine sediments where the anammox reaction was predicted to occur, and their genomes encode the key enzymes of the anammox metabolism. Our study suggests that the diversity, abundance, and ecological impact of anammox bacteria may be higher than previously thought. The newly recognized family of anammox bacteria is also valuable to delineate the emergence and evolutionary history of anammox bacteria in the geological history.
Results and discussion
An uncharted deep-branching family within the order Brocadiales
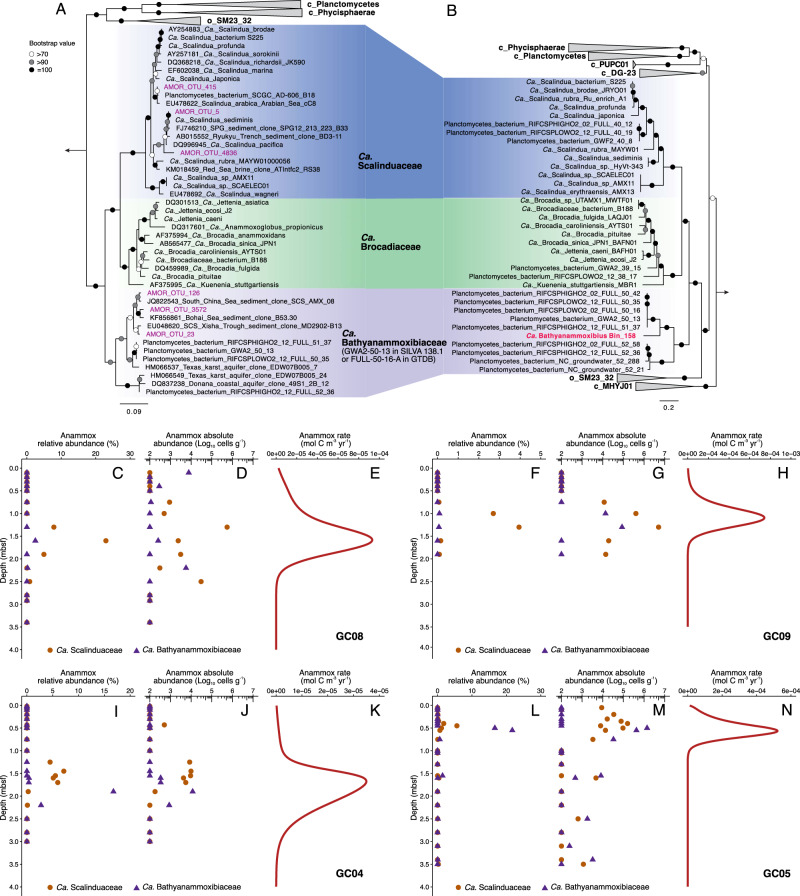
In the amplicon sequencing data of the four Arctic Mid-Ocean Ridge (AMOR) sediment cores (GC08, GC09, GC04, and GC05) reported in [20], there were three OTUs (OTU_23, OTU_126, and OTU_3572) that were classified as members of the order of Ca. Brocadiales, but were not affiliated with either of the two established anammox families (i.e., Ca. Brocadiaceae and Ca. Scalinduaceae). Phylogenetic analysis of 16S rRNA gene sequences of lineages from the entire Planctomycetes phylum indicated that these three OTUs fell into the basal branch in the order Ca. Brocadiales, representing a novel family (Fig. 1A). This family was named GWA2-50-13 in the SILVA 138.1 release [25]. Close relatives of these three OTUs were also detected in marine sediments of the South China Sea and Bohai Sea [26–28], while sequences from terrestrial habitats especially groundwater [29, 30] are also included within this family (Fig. 1A).
Fig. 1. Phylogeny and distribution of anammox bacteria in four AMOR sediment cores.
(A, N) Maximum-likelihood phylogenetic trees of bacteria in the order of Brocadiales based on the 16S rRNA gene (A) and 120 concatenated bacterial single-copy genes (B). The trees were inferred using IQ-TREE with GTR + F + R5 and LG + F + R7 as the best-fit evolutional model, and 1,000 times of ultrafast bootstrap iteration were applied to assess the robustness of both trees. The metagenome-assembled genome (MAG) recovered from the AMOR sediment is highlighted in red. The nomenclature used here following GTDB (https://gtdb.ecogenomic.org), while the new anammox family Ca. Bathyanammoxoceae (f__2-02-FULL-50-136 in GTDB 06-RS202 release) is named in this study. Bootstrap values of >70 are shown with symbols listed in the legend. The scale bars show estimated sequence substitutions per residue. C–N Vertical distribution of anammox bacteria and the predicted anammox reaction rates in AMOR sediment cores. The relative abundance of the two anammox families (B, E, H, K), Ca. Scalinduaceae and Ca. Bathyanammoxoceae, were determined by the sum of the OTUs assigned to these groups (showed in A), based on the 16S rRNA gene amplicon sequencing data reported in ref. [20]). The anammox reaction rates (D, G, J, M) were estimated by the reaction-transport model by ref. [20]. Panels (C–N) are modified from Fig. 2 of Ref. [20], an open access article distributed under Creative Commons Attribution License 4.0 (CC BY).
To further elucidate the identity and potential ecological functions of this bacterial family, we recovered a metagenome-assembled genome (MAG), Bin_158, from the existing metagenome sequencing data of core GC08 [20]. The genome size of this MAG is 2.2 Mbp, with 2368 genes distributing in 623 scaffolds. It is estimated to be of 74% completeness with 3.4% redundancy (Table S1). Phylogenomic analysis based on the concatenated 120 single-copy genes of bacteria indicates that this MAG is a member of the family FULL-50-16-A (corresponds to the GWA2-50-13 family of the 16S rRNA gene phylogeny, Fig. 1B), a deep-branching family within the order Ca. Brocadiales. This classification is also supported by the phylogenomic analysis based on the concatenated 14 ribosomal proteins (Fig. S1). In addition to Bin_158 recovered from marine subsurface sediments, this family also contains nine MAGs from the groundwater environment, including RIFCSPLOWO2_02_FULL_50_16 (called FULL_50_16 thereafter), RIFCSPLOWO2_12_FULL_50_35 (FULL_50_35), RIFCSPHIGHO2_02_FULL_50_42 (FULL_50_42), RIFCSPHIGHO2_12_FULL_52_36 (FULL_52_36), GWA2_50_13, RIFCSPHIGHO2_12_FULL_51_37 (FULL_51_37), and RIFCSPHIGHO2_02_FULL_52_58 (FULL_52_58) independently recovered from different groundwater wells of Rifle, Colorado [30], and NC_groundwater_1467_Ag_S-0.65um_52_288 (52_288) and NC_groundwater_1822_Pr3_B-0.1um_52_21 (52_21) from groundwater of Modesto, North California [31]. Despite that no 16S rRNA gene can be identified in Bin_158, the 16S rRNA gene sequences of FULL_51_37, FULL_50_35, and FULL_52_36 were placed into the family GWA2-50-13 (Fig. 1A), suggesting congruency between the two phylogenetic trees and that the GWA2-50-13 family of the 16S rRNA gene phylogeny should correspond to the FULL-50-16-A family genomes (Fig. 1). The 10 MAGs of the FULL-50-16-A family can be divided into two genera: 2-02-FULL-50-16-A and 2-12-FULL-52-36. Average amino acid identity (AAI) analysis indicated that Bin_158 has 76% AAI with members of 2-02-FULL-50-16-A (Fig. S2), which is above the AAI threshold (>65%) proposed for genomes within the same genus [32] and therefore should represent a new species in this genus. This classification is also supported by the automatic classification of GTDB-tk [33]. To highlight the genetic potential of the anammox metabolism of these new lineages (see below), we propose to name the genus 2-02-FULL-50-16-A as Candidatus Bathyanammoxibius gen. nov. (bathy-; referring to the deep origins of the genomes in this family; anammoxi-, derived from Neo-Latin noun anammox, referring to the anammox potential; bius; referring to life in the Latin language). Accordingly, we also propose a new family name, Candidatus Bathyanammoxibiaceae fam. nov., for the FULL-50-16-A family to harbor the genus Ca. Bathyanammoxibius.
Confined distribution and growth of Ca. Bathyanammoxibiaceae in the nitrate-ammonium transition zone
To explore the preferred niche(s) of Ca. Bathyanammoxibiaceae in marine sediments, we tracked its vertical distribution (represented by the sum of the three OTUs included in Fig. 1A) in the four AMOR sediment cores, based on the existing 16S rRNA gene amplicon sequencing data reported in [20] (the phylum level community structure of the total communities were shown in Fig. S4 therein). The relative abundances of Ca. Bathyanammoxibiaceae in the total communities of bacteria and archaea are higher than those of Ca. Scalinduaceae in cores GC04 and GC05 (Fig. 1I, L), but lower in cores GC08 and GC09 (Fig. 1C, F), with the relative abundance maxima (17% of the total communities in GC04 and 22% in GC05) detected within the NATZs (Fig. 1I, L). We also calculated the absolute abundances of Ca. Bathyanammoxibiaceae and Ca. Scalinduaceae in these cores by multiplying their relative abundances and the total cell numbers estimated from the qPCR quantification of 16S rRNA genes [20]. Similar to their relative abundances, the absolute abundances of these two families were highest within the NATZs (Fig. 1D, G, J, M). Based on reactive-transport modeling [20], the NATZ is where the anammox process is predicted to occur (Fig. 1E, H, K, N), and the confined distribution of Ca. Bathyanammoxibiaceae within the NATZs suggests that members of this family may be involved in the anammox process.
Considering that none of the Ca. Bathyanammoxibiaceae genomes contain genes responsible for bacterial flagellum synthesis (Fig. 2), Ca. Bathyanammoxibiaceae in the AMOR sediments are probably not able to migrate in the sediment pore space. The observed local subsurface peaks of Ca. Bathyanammoxibiaceae in the NATZs (Fig. 1D, G, J, M) represents another example of microbial in situ growth within energy-limited subseafloor sediments, adding to the growing body of such evidence previously reported for Ca. Scalindua [20], Thaumarchaeota [34, 35], and Atribacteria [36] in deep-sea sediments and various microbial taxa in estuarine sediments [37].
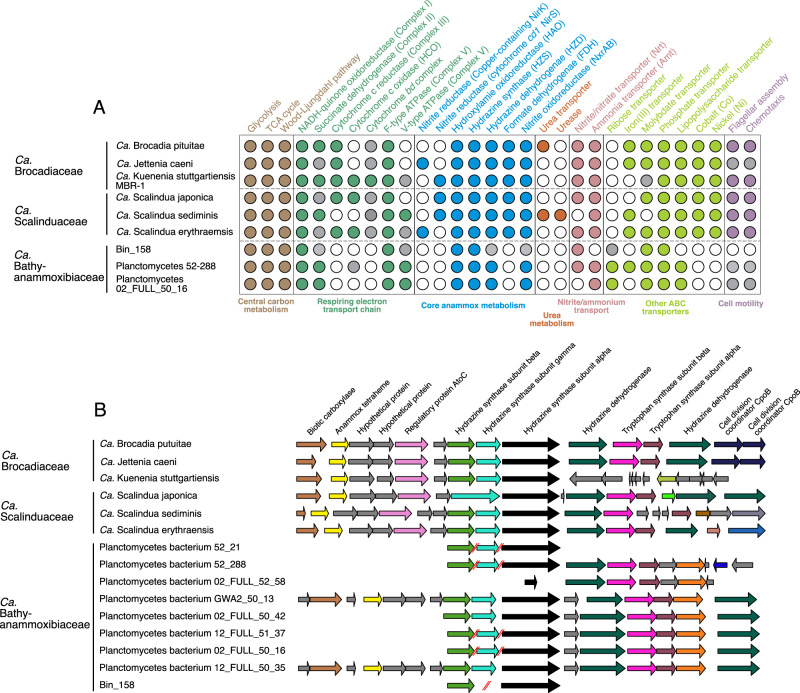
Fig. 2. Metabolic features of Ca. Bathyanammoxibiaceae bacteria.
A Comparison of metabolic potential of anammox bacteria from three different families. For each of the three anammox bacteria families, three genomes of high genome completeness were selected for the analysis. Filled circles represent the presence, grey circles denote the partial presence, and open circles indicate the absence of genes encoding the metabolic process. B Conservation of key genes encoding for the hydrazine metabolism in anammox bacteria genomes. The figure was produced in GeneSpy by comparing a region of 10 kb centered on the hydrazine synthase subunit alpha (HzsA). In addition to the three selected genomes from the families of Brocadiaceae and Scalinduaceae, all available genomes of the newly proposed family, Ca. Bathyanammoxibiaceae, were included in (B). The red double slash lines represent discontinuousness between contigs.
Members of Ca. Bathyanammoxibiaceae have the anammox capacity
In accordance with their confined distribution in the anammox zone, genome annotation indicates that members of Ca. Bathyanammoxibiaceae are capable of performing anammox, as they have the core genetic machinery for the anammox metabolism (Fig. 2A): hydrazine synthase for converting NO and NH4+ to hydrazine, hydrazine dehydrogenase for hydrazine oxidation to dinitrogen gas (N2), and nitrite oxidoreductase for nitrite oxidation [38]. The gene cluster encoding the most critical enzyme of anammox, hydrazine synthase, is conserved in eight of the ten Ca. Bathyanammoxibiaceae genomes (Fig. 2B), whereas its absence in the remaining two genomes (only a partial hydrazine synthase alpha subunit exists in FULL_52_58 and none of the subunits is present in FULL_52_36) can be attributed to the low genome completion levels (<73% complete; Table S1).
Ca. Bathyanammoxibiaceae genomes contain hydrazine synthases-encoding genes that are not only of similar gene arrangement but also similar structure to those of characterized anammox bacteria. Ca. Bathyanammoxibiaceae genomes encode the hydrazine synthase (HZS, constituted by the alpha, beta, and gamma subunits; Fig. 2B) as characterized anammox bacteria in the families Ca. Brocadiaceae and Ca. Scalinduaceae, except for S. brodae, S. profunda, and S. japonica in which the beta and gamma subunits are fused into a single polypeptide [39, 40]. Bin_158 recovered from AMOR sediments, has genes encoding only the hydrazine synthase subunits alpha (HzsA) and gamma (HzsC) on two separated contigs, probably due to the incomplete nature of this genome. Furthermore, the two heme-binding motifs (CXXCH) in the HZS alpha subunit for electron transfer and the 15-amino-acid loop in the HZS beta subunit that directs the trafficking of substrates and products of the alpha and gamma subunits, are also conserved in the Ca. Bathyanammoxibiaceae genomes (Fig. S3). These analyses strongly suggest that Ca. Bathyanammoxibiaceae have the hydrazine synthesis capacity, the most critical step of the anammox metabolism.
Phylogenetic analysis of the alpha (HzsA), beta (HzsB), and gamma (HzsC) subunits of the hydrazine synthase indicated that Ca. Bathyanammoxibiaceae formed a separate, basal branch from the families Ca. Brocadiceae and Ca. Scalinduaceae (Fig. S4). These three phylogenetic trees show similar topologies and are congruent with the above-described ones based on 16 rRNA gene, ribosomal proteins, and 120 single-copy genes (Fig. 1A, B), implying that anammox bacteria acquired this critical enzyme by vertical inheritance from the common ancestor of anammox bacteria rather than by horizontal gene transfer events.
Nitrite reduction responsible for the generation of nitric oxide from nitrite is also part of the core anammox metabolism [38], and can be carried out by the copper-containing nitrite reductase (NirK) or the cytochrome cd1-containing nitrite reductase (NirS). Members of Ca. Bathyanammoxibiaceae lack both of these two nitrite reductases (Fig. 2A), similar to some anammox bacteria in the genus of Ca. Brocadia [41]. However, recently Ferousi et al [42] revealed that anammox bacteria may also use a specific hydroxylamine oxidoreductase (HAO) (Kustc0457/458 in Kuenenia stuttgartiensis or HAO2 following the definition of [41]) to perform the nitrite reduction process. While Ca. Bathyanammoxibiaceae genomes do not encode HAO2, they encode 5–7 variants of HAO including 1–3 variants of HAO7 (Fig. S5). The branch of these HAO7 variants is the closest neighbor to HAO2 and share 47–53% amino acid identities with Kustc0457/458 (Fig. S5), but to what extent this allow Ca. Bathyanammoxibiaceae to have the nitrite reduction capacity is currently unknown.
Similar to the characterized anammox bacteria in the families Ca. Brocadiaceae and Ca. Scalinduaceae, members of Ca. Bathyanammoxibiaceae also encode the nitrite oxidoreductase (NXR), which in anammox bacteria is an essential enzyme with a tubular architecture functioning in the anammoxosome organelle [43]. Phylogenetic analysis of the nitrite oxidoreductase alpha subunit (NxrA) showed that members of Ca. Bathyanammoxibiaceae formed a separated cluster from characterized anammox bacteria in the families Ca.Brocadiaceae and Ca. Scalinduaceae, the latter formed a big cluster together with nitrite-oxidizing Nitrospira and Nitrospina and also some other diverse bacteria (Fig. S6). Again, this phylogenetic branching pattern among the three anammox bacteria families suggested that they gained this key enzyme through vertical inheritance.
Ca. Bathyanammoxibiaceae genomes encode the same central carbon metabolism pathways as other characterized anammox genomes. These include the Wood-Ljungdahl pathway (Fig. 2A), which could enable them to fix CO2 to generate acetyl-CoA and pyruvate, using electrons derived from the hydrazine oxidation [38, 44, 45]. Also included are the tricarboxylic acid (TCA) cycle and the gluconeogenesis pathway (Fig. 2A), which, respectively, can use acetyl-CoA and pyruvate produced from the Wood-Ljungdahl pathway to synthesize biomass precursors like amino acids and carbohydrates [44]. In addition, some Ca. Bathyanammoxibiaceae genomes contain genes for the ABC transporter of ribose (Fig. 2A), which could indicate that these bacteria can assimilate small carbohydrates using electrons derived from the hydrazine degradation. Similar to Ca. Scalindua sediminis, the three representative genomes of Ca. Bathyanammoxibiaceae encode both the F-type ATPase and V-type ATPase (Fig. 2A), which could be integrated into the two different ATP generation processes in these genomes. This may represent an adaptation strategy for members of Ca. Bathyanammoxibiaceae thrive in the energy-limiting subsurface.
Because they broadly occupy the same niche of Ca. Scalinduaceae, the NATZs, Ca. Bathyanammoxibiaceae may need surrounding denitrifying bacteria to provide them nitrite, one of the direct substrates of anammox bacteria [46], as previously proposed for Ca. Scalindua sediminis [20]. The ABC transporters for iron (Fe3+) and molybdate (MoO42−) seem to be conserved in all anammox genomes (Fig. 2A), which may be critical for these putative anammox bacteria in incorporating these metals as the co-factor of heme-containing cytochromes and molybdate-containing enzymes that are critical and widespread proteins in anammox bacteria [47]. Unlike Ca. Scalindua sediminis [20], members of Ca. Bathyanammoxibiaceae do not encode urease or cyanase, suggesting that they may not be capable of utilizing these alternative substrates.
A wide distribution of Ca. Bathyanammoxibiaceae in both marine and terrestrial environments
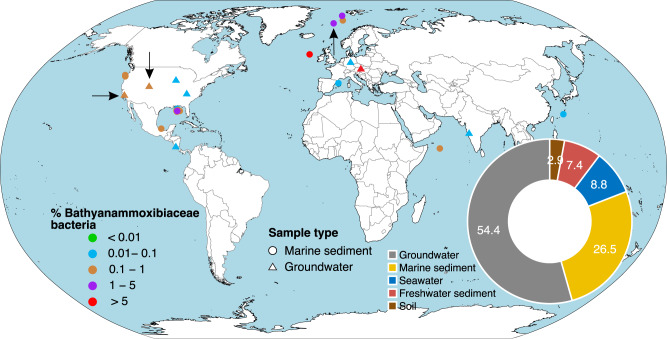
Ca. Bathyanammoxibiaceae is present in various ecosystems. Through the search against the available high-throughput sequencing data in IMNGS [48], we detected the presence of Ca. Bathyanammoxibiaceae in a total of 128 samples (Fig. 3). For marine sediments, in addition to AMOR sediments [e.g., [49] and Fig. 1], Ca. Bathyanammoxibiaceae account for >1% of the total communities in the surface sediments of the Gulf of Mexico [50] and Rockall Bank [51], and >0.1% of the total communities in subsurface sediments of the Hydrate Ridge, and the Indian and Pacific Ocean (Fig. 3). Consistent with the 9 MAGs recovered from the groundwater environment, Ca. Bathyanammoxibiaceae were also detected in groundwater in the US, off the Danube Riverbank in Austria [52], Central Germany [53], and India (Fig. 3).
Fig. 3. Global occurrence of Ca. Bathyanammoxibiaceae bacteria.
The occurrence and relative abundances of Ca. Bathyanammoxibiaceae were determined in the IMNGS database using the 16S rRNA gene sequences of GWA2_50_13 and FULL_52_36 as the queries. Sequences (>200 bp) with similarity higher than 95% to the queries were regarded as positive hits. At most of the sites, Ca. Bathyanammoxibiaceae was detected in multiple depths and the highest relative abundances are reported. The three sites where the available Ca. Bathyanammoxibiaceae MAGs originate from are indicated by black arrows. The insert pie chat shows the environmental distribution of the 16S rRNA gene sequences included in the GWA2-50-13 family in the SILVA 138.1 Release.
We also assessed the distribution of Ca. Bathyanammoxibiaceae by checking the origins of the 77 16S rRNA gene sequences included in the SILVA 138.1 Release, which were mainly derived from clone library-based studies. The majority of them were recovered from groundwater (54%) and marine sediment (26%) environments (Fig. 3), consistent with the dominance of groundwater and marine sediment as the habitat of Ca. Bathyanammoxibiaceae in this high-throughput sequencing data analysis. Sequences of Ca. Bathyanammoxibiaceae were also detected in coral reef carbonate sediments off Hawaii [54], South China Sea sediments [55], and Bohai Sea sediments [28]. In addition, we noticed that 9% of the sequences were from seawater (e.g., Deep brine pool in the Red Sea [56]), 7% from freshwater sediments, and 3% from soils (Fig. 3). These results suggested that Ca. Bathyanammoxibiaceae bacteria are potentially important players in the nitrogen cycle in a number of marine and terrestrial environments.
Conclusion and Outlook
Through genomic recovery and characterization, we report a new family of bacteria within the order of Ca. Brocadiales that have the genetic potential for the anammox metabolism. By geochemical characterization and ecological surveys, our study also provides valuable guidance (e.g., ideal inoculum and cultivation conditions) for future enrichment and cultivation efforts, which are critical to confirm the predicted function and activity of the bacteria in this family. When their anammox activity got confirmed, the genomes in the deeply branching Ca. Bathyanammoxibiaceae family may be valuable to delineate the evolutionary history of anammox in the nitrogen cycle [57]. Importantly, Ca. Bathyanammoxibiaceae contains members from both the marine and terrestrial environments, contrasting the partitioning between the previously recognized anammox families of Ca. Scalinduaceae and Ca. Brocadiaceae, and therefore represents an ancient lineage that may preserve some features of the anammox bacteria before their radiation to the marine and terrestrial environments.
In addition to the absence of the anammox function proof, there are also other prominent open questions pertaining to this family of bacteria. How are they competing (or cooperating) with the co-existing Ca. Scalinduaceae in the NATZ of marine sediments? Why are they apparently so widespread in the energy-limited subsurface? Addressing these questions and adding Ca. Bathyanammoxibiaceae to the important functional guild anammox may lead to a better understanding about the biogeochemical impacts and evolutionary history of anammox bacteria on Earth.
Materials and methods
Sampling collection and characterization
This study uses samples and data generated and reported in [20], in which the procedures of sample collection, processing, and data generation were thoroughly described. Briefly, the four sediment cores (2.0–3.6 m long) were retrieved by gravity coring from the seabed of ~2000 of water depth on the ridge flanks of the Arctic Mid-Ocean Ridge beneath the Norwegian-Greenland Sea. The retrieved cores were split to two halves (sampling and archiving halves), and the oxygen measurement (using a needle-type fiber-optic oxygen microsensor (optodes, PreSens)) and the subsampling of microbiology samples (using sterile 10-mL cutoff syringes) and porewater extraction were performed immediately on the sampling half using Rhizons samplers after the split. Nitrate, nitrite, and ammonium concentrations in the porewater were measured colorimetrically by a Quaatro 114 continuous flow analyzer (SEAL Analytical Ltd, Southampton, UK).
In each core, a downward flux of nitrate and an upward flux of ammonium were observed to be co-consumed in the middle part of the retrieved cores, which was named nitrate-ammonium transition zone (NATZ) [20]. The occurrence and rates of the anammox process in the NATZs of these cores were predicted using a reaction-transport model, which considers six reactions of carbon, nitrogen, oxygen, and manganese transformation during the early diagenesis of organic matter in marine sediments to simulate the measured porewater profiles of O2, NO3−, NH4+, dissolved inorganic carbon, and total organic carbon contents [20].
16S rRNA gene amplicon preparation, sequencing, and analysis
Total DNA in the sediment samples was extracted using the PowerLyze DNA extraction kits (MOBIO Laboratories, Inc.). Amplicon of the 16S rRNA gene were prepared using a two-round PCR amplification strategy with the “universal” primers of 515 F/806r [20]. The amplicon libraries were sequenced on an Ion Torrent Personal Genome Machine. As described in [20], sequencing reads were quality filtered and trimmed to 220 bp using the USEARCH v11.0.667 pipeline [58]. The taxonomic classification of OTUs was performed using the lowest common ancestor algorithm implemented in the python version of CREST4 (the latest version of CREST [59]) against the SILVA 138.1 Release [25]. This study focused on three OTUs that were classified as members of the order Ca. Brocadiales, but were not affiliated with either of known anammox families: Ca. Brocadiaceae and Ca. Scalinduaceae.
Genome binning and refinement
Metagenome sequencing data were generated and reported in [20]. The detailed procedures of the DNA extraction, library preparation, metagenome sequencing, raw data quality control, assembly, genome binning and refinement were also detailed therein [20]. Briefly, DNA were extracted from ~7 g sediment (0.7 g sediment in 10 individual lysis tubes) of four depths in core GC08. Metagenomic libraries were sequenced (2 × 150 bp paired-end) by an Illumina HiSeq 2500 sequencer at the Vienna Biocenter Core Facilities GmbH (Vienna, Austria). The quality of the raw sequencing data were first checked using FastQc v0.11.9 [60], and adapters were accordingly removed and reads were trimming based on the quality scores using Trimmomatic v0.39 [61]. The quality-controlled paired-end reads were de novo assembled into contigs using MEGAHIT v1.1.2 [62] with the k-mer length varying from 27 to 117. Contigs larger than 1000 bp were automatically grouped into genome bins using MaxBin2 v2.2.5 [63] with the default parameters. The quality of the obtained genome bins was assessed using the option “lineage_wf” of CheckM v.1.0.7 [64]. Genome bins of >50% completeness were manually refined using the tool gbtools v2.6.0 [65] based on the GC content, taxonomic assignments, and differential coverages of contigs across multiple samples. To generate the input data of the genome refinement, coverages of contigs in each sample were determined by mapping trimmed reads onto the contigs using BBMap v.37.61 [66]. Taxonomy classification of contigs were assigned by BLASTn [67] according to the taxonomy of the single-copy marker genes in contigs. SSU rRNA sequences in contigs were identified using Barrnap [68] and classified using VSEARCH (56) with the SILVA 132 release (57) as the reference. All refined MAGs were classified using GTDB-tk v1.5.0 [33] with the default setting. Only MAGs of ≥70% completeness with <5% redundancy were included for downstream analyses.
In this study, Bin_158 was found to be affiliated with a family without known anammox genera and was therefore subject to further analysis. To improve the quality of the genome of Bin_158, quality-trimmed reads of the sample GC08_160cm within the NATZ, were mapped onto the contigs using BBmap [66], and the successfully aligned reads were re-assembled using SPAdes v3.12.0 [69] with the k-mers of 21, 33, 55, and 77. After removal of contigs shorter than 1000 bp, the resulting scaffolds were visualized and manually re-binned using gbtools v2.6.0 [65] as described above. The quality of the resulting Bin_158 genome was checked using the CheckM v1.0.7 “lineage_wf” command again, based on the Planctomycetes marker gene set.
Genome annotation
Bin_158 were annotated together with the MAGs included in the family FULL-50-16-A in the GTDB Release 06-RS202 (https://gtdb.ecogenomic.org/). In addition, two closely relative MAGs not then included in the GTDB database, e.g., NC_groundwater_1467_Ag_S-0.65um_52_288 (52_288) and NC_groundwater_1822_Pr3_B-0.1um_52_21 (52_21) reported in [31], were also included in the genome annotation for comparison. Genes in these genomes were predicted using Prodigal [70]. Genome annotation was conducted using Prokka v1.13 [71], eggNOG [72], and also BlastKoala [73] using the KEGG database. The functional assignments of genes of interest were also confirmed using BLASTp [68] against the NCBI RefSeq database. The metabolic pathways were reconstructed using KEGG Mapper [74]. The gene arrangement around the hydrazine synthase genes were visualized using GeneSpy v1.2 [75], with the GFF files produced by Prokka as the input.
Phylogenetic analyses
To pinpoint the phylogenetic placement of Bin_158 and the relative genomes in the family FULL-50-16-A, we performed phylogenetic analyses for them together with high-quality genomes of the Planctomycetes phylum that was included in the GTDB Release 06-RS202. The 120 single-copy genes were identified, aligned, and concatenated using GTDB-tk v1.5.0 with the “classify_wf” command. The maximum-likelihood phylogenetic tree was inferred based on this alignment using IQ-TREE v1.5.5 [76] with LG + F + R7 the best-fit model selected by ModelFinder [77], and 1000 ultrafast bootstrap iterations using UFBoot2 [78]. To provide support to this phylogenomic analysis, we also performed the phylogenomic analysis based on the 14 syntenic ribosomal proteins (rpL2, 3, 4, 5, 6, 14, 16, 18, 22, and rpS3, 8, 10, 17, 19) that have been demonstrated to undergo limited lateral gene transfer [79]. These selected proteins were identified in Anvi’o v7.1 (67) using Hidden Markov Model (HMM) profiles and aligned individually using MUSCLE [80]. Alignment gaps were removed using trimAl [81] with the mode of “automated”. Individual alignments of ribosomal proteins were concatenated. The maximal likelihood phylogenetic tree was reconstructed using IQ-TREE v1.5.5 [76] with LG + R7 as the best-fit model.
A maximum-likelihood phylogenetic tree based on 16S rRNA genes was also constructed to highlight to the phylogenetic placement of the Ca. Bathyanammoxibiaceae family in the Planctomycetes phylum. To expand this family on the tree beyond the available genomes, the three Ca. Bathyanammoxibiaceae OTUs from the amplicon sequencing and their close relatives identified via BLASTn [82] in the NCBI database were also included. Sequences were aligned using MAFFT-LINSi [83] and the maximum-likelihood phylogenetic tree was inferred using IQ-TREE v1.5.5 [76] with GTR + F + R3 as the best-fit substitution model and 1000 ultrafast bootstraps, following the procedure described above.
For the phylogeny of HzsA (hydrazine synthase subunit alpha), the genomes of known anammox bacteria were downloaded from the NCBI database, annotated using Prokka v1.13 [71], and the HzsA amino acid sequences were extracted. Additional HzsA sequences of uncultured anammox deposited in the NCBI database were also identified using BLASTp [67] using the HzsA sequences of Ca. S. sediminis and Bin_158 as the query and the E-value of 10−6. Sequences were aligned using MAFFT-LINSi [83] and the maximum likelihood phylogenetic tree was inferred using IQ-TREE v1.5.5 following the procedure described above.
For the phylogeny of NxrA (nitrite oxidoreductase alpha subunit), the sequence of Bin_158 was used as the query in the BLASTp [82] search in the NCBI database (>50% similarity and E-value of 10−6) to identify its close relatives. These sequences were aligned using MAFF-LINSi [83] with reference sequences from ref. [84], and complemented with known nitrite-oxidizing bacteria. The alignment was then trimmed using trimAl [81] with the mode of “automated”. Maximum likelihood phylogenetic trees were reconstructed using IQ-TREE v1.5.5 [76] with the LG + C20 + F + G substitution model and 1,000 ultrafast bootstraps.
Global occurrence of Ca. Bathyanammoxibiaceae
We used the 16S rRNA gene sequence of Planctomycetes bacteria GWA2_50_13 and FULL_52_36 as the queries in the IMNGS (Integrated Microbial Next Generation Sequencing, https://www.imngs.org/) database [48] to explore the global occurrence of Ca. Bathyanammoxibiaceae. The similarity threshold was set at 95%, because (1) the inter-families identities in the order of Brocadiales were determined to be around 90%, and (2) generally species in the same families were suggested to share a 16S rRNA gene identity of 92% [32]. The sequence length threshold was set at 200 bp, because most of the sequences in the IMNGS database are short reads rather than full-length 16S rRNA gene sequences. Only hits with >5 sequences and >0.001 relative abundance were considered. To assess the distribution range of Ca. Bathyanammoxibiaceae based on sequence survey data prior to the emergence of Next Generation sequencing, we also parsed the information of the high-quality 16S rRNA sequences (n = 77) included in the GWA2-50-13 family of the SILVA 138.1 Release, and grouped the sample origins into the following categories: groundwater, marine sediment, seawater, freshwater sediment, and soil.
Supplementary information
Acknowledgements
We thank the chief scientist Rolf Birger Pedersen and the crew of R/V GO. Sars for the sediment coring opportunity from the AMOR area, Michael Melcher, Steffen Lydvo, and Gustavo Ramirez for sampling collection, Anita-Elin Fedøy, Thomas Pollak, Sophie Abby, and Christa Schleper for laboratory assistance and metagenome data generation. Computational resources were made possible through the BIOMIX computing cluster (Delaware INBRE grant NIGMS P20GM103446) at University of Delaware. We are grateful to the advices from Profs. Bernhard Schink and Aharon Oren on prokaryotic lineages naming. The comments from the anonymous reviewers have greatly improved the manuscript. This work was funded by the K.G. Jebsen Foundation and Trond Mohns Science Foundation (to SLJ). RZ and JFB were funded in part by the WM Keck Foundation. RZ was supported by MIT Molina Postdoctoral Fellowship.
Author contribution
RZ and SLJ conceived the study. RZ collected and analyzed the genome data. RZ, JFB and SLJ interpreted the results. RZ wrote the paper with inputs from all authors.
Data availability
All sequencing data used in this study are available in the NCBI Short Reads Archive under the project number PRJNA529480. In particular, raw metagenomic sequencing data are available in the NCBI database under the BioSample number SAMN11268106. The genome sequence of Bin_158 is available under the accession number JAFNKF000000000. All other genomes analyzed in this study are available in the NCBI database under the accession numbers listed in Table S1.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s43705-022-00125-4.
References
- 1.Dalsgaard T, Canfield DE, Petersen J, Thamdrup B, Acuna-Gonzalez J. N2 production by the anammox reaction in the anoxic water column of Golfo Dulce, Costa Rica. Nature. 2003;422:606–8. doi: 10.1038/nature01526. [DOI] [PubMed] [Google Scholar]
- 2.Kuypers MMM, Lavik G, Woebken D, Schmid M, Fuchs BM, Amann R, et al. Massive nitrogen loss from the Benguela upwelling system through anaerobic ammonium oxidation. Proc of the Natl Acad of Sci of the USA. 2005;102:6478–83. doi: 10.1073/pnas.0502088102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuypers MMM, Sliekers AO, Lavik G, Schmid M, Jorgensen BB, Kuenen JG, et al. Anaerobic ammonium oxidation by anammox bacteria in the Black Sea. Nature. 2003;422:608–11. doi: 10.1038/nature01472. [DOI] [PubMed] [Google Scholar]
- 4.Wang S, Zhu G, Zhuang L, Li Y, Liu L, Lavik G, et al. Anaerobic ammonium oxidation is a major N-sink in aquifer systems around the world. ISME J. 2020;14:151–63.. doi: 10.1038/s41396-019-0513-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu G, Wang S, Wang W, Wang Y, Zhou L, Jiang B, et al. Hotspots of anaerobic ammonium oxidation at land–freshwater interfaces. Nat Geosci. 2013;6:103–7. doi: 10.1038/ngeo1683. [DOI] [Google Scholar]
- 6.Devol AH. Denitrification, anammox, and N2 production in marine sediments. Ann Rev of Marine Sci. 2015;7:403–23.. doi: 10.1146/annurev-marine-010213-135040. [DOI] [PubMed] [Google Scholar]
- 7.Kuenen JG. Anammox bacteria: from discovery to application. Nat Rev Microbiol. 2008;6:320–6. doi: 10.1038/nrmicro1857. [DOI] [PubMed] [Google Scholar]
- 8.Kartal B, Kuenen JV, Van Loosdrecht M. Sewage treatment with anammox. Science. 2010;328:702–3. doi: 10.1126/science.1185941. [DOI] [PubMed] [Google Scholar]
- 9.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil PA, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 10.Jetten MS, Niftrik LV, Strous M, Kartal B, Keltjens JT, Op den Camp HJ. Biochemistry and molecular biology of anammox bacteria. Critical Rev in Biochem and Mol Biol. 2009;44:65–84. doi: 10.1080/10409230902722783. [DOI] [PubMed] [Google Scholar]
- 11.Anantharaman K, Hausmann B, Jungbluth SP, Kantor RS, Lavy A, Warren LA, et al. Expanded diversity of microbial groups that shape the dissimilatory sulfur cycle. ISME J. 2018;12:1715–28.. doi: 10.1038/s41396-018-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Kessel MAHJ, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJM, Kartal B, et al. Complete nitrification by a single microorganism. Nature. 2015;528:555–9. doi: 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, et al. Complete nitrification by Nitrospira bacteria. Nature. 2015;528:504–9. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mori JF, Chen L-X, Jessen GL, Rudderham SB, McBeth JM, Lindsay MBJ, et al. Putative mixotrophic nitrifying-denitrifying Gammaproteobacteria implicated in nitrogen cycling within the ammonia/oxygen transition zone of an oil sands pit lake. Front in Microbiol. 2019;10:2435. doi: 10.3389/fmicb.2019.02435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diamond S, Lavy A, Crits-Christoph A, Matheus Carnevali PB, Sharrar A, Williams KH, et al. Soils and sediments host Thermoplasmata archaea encoding novel copper membrane monooxygenases (CuMMOs) ISME J. 2022 doi: 10.1038/s41396-021-01177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nunoura T, Nishizawa M, Kikuchi T, Tsubouchi T, Hirai M, Koide O, et al. Molecular biological and isotopic biogeochemical prognoses of the nitrification-driven dynamic microbial nitrogen cycle in hadopelagic sediments. Environ Microbiol. 2013;15:3087–107. doi: 10.1111/1462-2920.12152. [DOI] [PubMed] [Google Scholar]
- 17.Woebken D, Lam P, Kuypers MM, Naqvi SWA, Kartal B, Strous M, et al. A microdiversity study of anammox bacteria reveals a novel Candidatus Scalindua phylotype in marine oxygen minimum zones. Environ Microbiol. 2008;10:3106–19. doi: 10.1111/j.1462-2920.2008.01640.x. [DOI] [PubMed] [Google Scholar]
- 18.Schmid MC, Risgaard-Petersen N, van de Vossenberg J, Kuypers MMM, Lavik G, Petersen J, et al. Anaerobic ammonium-oxidizing bacteria in marine environments: Widespread occurrence but low diversity. Environ Microbiol. 2007;9:1476–84. doi: 10.1111/j.1462-2920.2007.01266.x. [DOI] [PubMed] [Google Scholar]
- 19.Jaeschke A, Abbas B, Zabel M, Hopmans EC, Schouten S, Damste JSS. Molecular evidence for anaerobic ammonium-oxidizing (anammox) bacteria in continental shelf and slope sediments off northwest Africa. Limnol and Oceanogr. 2010;55:365–76.. doi: 10.4319/lo.2010.55.1.0365. [DOI] [Google Scholar]
- 20.Zhao R, Mogollón JM, Abby SS, Schleper C, Biddle JF, Roerdink DL, et al. Geochemical transition zone powering microbial growth in subsurface sediments. Proc of the Natl Acad of Sci. 2020;117:32617–26. doi: 10.1073/pnas.2005917117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Humbert S, Tarnawski S, Fromin N, Mallet M-P, Aragno M, Zopfi J. Molecular detection of anammox bacteria in terrestrial ecosystems: Distribution and diversity. ISME J. 2010;4:450–4. doi: 10.1038/ismej.2009.125. [DOI] [PubMed] [Google Scholar]
- 22.Schmid M, Walsh K, Webb R, Rijpstra WI, van de Pas-Schoonen K, Verbruggen MJ, et al. Candidatus “Scalindua brodae”, sp. nov., Candidatus “Scalindua wagneri”, sp. nov., two new species of anaerobic ammonium oxidizing bacteria. Syst and Appl Microbiol. 2003;26:529–38. doi: 10.1078/072320203770865837. [DOI] [PubMed] [Google Scholar]
- 23.Schubert CJ, Durisch‐Kaiser E, Wehrli B, Thamdrup B, Lam P, Kuypers MM. Anaerobic ammonium oxidation in a tropical freshwater system (Lake Tanganyika) Environ Microbiol. 2006;8:1857–63.. doi: 10.1111/j.1462-2920.2006.01074.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L, Okabe S. Ecological niche differentiation among anammox bacteria. Water Res. 2020;171:115468. doi: 10.1016/j.watres.2020.115468. [DOI] [PubMed] [Google Scholar]
- 25.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D6.. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han P, Klümper U, Wong A, Li M, Lin J-G, Quan Z, et al. Assessment of molecular detection of anaerobic ammonium-oxidizing (anammox) bacteria in different environmental samples using PCR primers based on 16s rRNA and functional genes. Appl Microbiol and Biotechnol. 2017;101:7689–702.. doi: 10.1007/s00253-017-8502-3. [DOI] [PubMed] [Google Scholar]
- 27.Li T, Wang P. Biogeographical distribution and diversity of bacterial communities in surface sediments of the South China Sea. J of Microbiol and Biotechnol. 2013;23:602–13. doi: 10.4014/jmb.1209.09040. [DOI] [PubMed] [Google Scholar]
- 28.Shehzad A, Liu J, Yu M, Qismat S, Liu J, Zhang X-H. Diversity, community composition and abundance of anammox bacteria in sediments of the north marginal seas of China. Microbes and Environ. 2016;31:111–20. [DOI] [PMC free article] [PubMed]
- 29.Gray CJ, Engel AS. Microbial diversity and impact on carbonate geochemistry across a changing geochemical gradient in a karst aquifer. ISME J. 2013;7:325–37. doi: 10.1038/ismej.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anantharaman K, Brown CT, Hug LA, Sharon I, Castelle CJ, Probst AJ, et al. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat Commun. 2016;7:13219. doi: 10.1038/ncomms13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He C, Keren R, Whittaker ML, Farag IF, Doudna JA, Cate JHD, et al. Genome-resolved metagenomics reveals site-specific diversity of episymbiotic CPR bacteria and DPANN archaea in groundwater ecosystems. Nat Microbiol. 2021;6:354–65. doi: 10.1038/s41564-020-00840-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konstantinidis KT, Rosselló-Móra R, Amann R. Uncultivated microbes in need of their own taxonomy. ISME J. 2017;11:2399–406.. doi: 10.1038/ismej.2017.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics. 2020;36:1925–7. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vuillemin A, Wankel SD, Coskun ÖK, Magritsch T, Vargas S, Estes ER, et al. Archaea dominate oxic subseafloor communities over multimillion-year time scales. Sci Adv. 2019;5:eaaw4108. doi: 10.1126/sciadv.aaw4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao R, Hannisdal B, Mogollon JM, Jørgensen SL. Nitrifier abundance and diversity peak at deep redox transition zones. Sci Rep. 2019;9:8633. doi: 10.1038/s41598-019-44585-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vuillemin A, Vargas S, Coskun ÖK, Pockalny R, Murray RW, Smith DC, et al. Atribacteria reproducing over millions of years in the Atlantic abyssal subseafloor. mBio. 2020;11:e01937–20.. doi: 10.1128/mBio.01937-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lloyd KG, Bird JT, Buongiorno J, Deas E, Kevorkian R, Noordhoek T, et al. Evidence for a growth zone for deep-subsurface microbial clades in near-surface anoxic sediments. Appl and Environm Microbiol. 2020;86:e00877–20.. doi: 10.1128/AEM.00877-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kartal B, de Almeida NM, Maalcke WJ, Op den Camp HJM, Jetten MSM, Keltjens JT. How to make a living from anaerobic ammonium oxidation. FEMS Microbiol Rev. 2013;37:428–61.. doi: 10.1111/1574-6976.12014. [DOI] [PubMed] [Google Scholar]
- 39.Dietl A, Ferousi C, Maalcke WJ, Menzel A, de Vries S, Keltjens JT, et al. The inner workings of the hydrazine synthase multiprotein complex. Nature. 2015;527:394–7. doi: 10.1038/nature15517. [DOI] [PubMed] [Google Scholar]
- 40.van de Vossenberg J, Woebken D, Maalcke WJ, Wessels H, Dutilh BE, Kartal B, et al. The metagenome of the marine anammox bacterium ‘Candidatus Scalindua profunda’ illustrates the versatility of this globally important nitrogen cycle bacterium. Environ Microbiol. 2013;15:1275–89.. doi: 10.1111/j.1462-2920.2012.02774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okubo T, Toyoda A, Fukuhara K, Uchiyama I, Harigaya Y, Kuroiwa M, et al. The physiological potential of anammox bacteria as revealed by their core genome structure. DNA Res. 2021;28:dsaa028. doi: 10.1093/dnares/dsaa028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferousi C, Schmitz RA, Maalcke WJ, Lindhoud S, Versantvoort W, Jetten MSM, et al. Characterization of a nitrite-reducing octaheme hydroxylamine oxidoreductase that lacks the tyrosine cross-link. J of Biol Chem. 2021;296:100476. doi: 10.1016/j.jbc.2021.100476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chicano TM, Dietrich L, de Almeida NM, Akram M, Hartmann E, Leidreiter F, et al. Structural and functional characterization of the intracellular filament-forming nitrite oxidoreductase multiprotein complex. Nat Microbiol. 2021;6:1129–39.. doi: 10.1038/s41564-021-00934-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lawson CE, Nuijten GHL, de Graaf RM, Jacobson TB, Pabst M, Stevenson DM, et al. Autotrophic and mixotrophic metabolism of an anammox bacterium revealed by in vivo 13C and 2H metabolic network mapping. ISME J. 2021;15:673–87.. doi: 10.1038/s41396-020-00805-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW, et al. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature. 2006;440:790–4. doi: 10.1038/nature04647. [DOI] [PubMed] [Google Scholar]
- 46.Mulder A, Vandegraaf AA, Robertson LA, Kuenen JG. Anaerobic ammonium oxidation discovered in a denitrifying fluidized-bed reactor. FEMS Microbiol Ecol. 1995;16:177–83.. doi: 10.1111/j.1574-6941.1995.tb00281.x. [DOI] [Google Scholar]
- 47.Kartal B, Keltjens JT. Anammox biochemistry: a tale of heme c proteins. Trends in Biochem Sci. 2016;41:998–1011. doi: 10.1016/j.tibs.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 48.Lagkouvardos I, Joseph D, Kapfhammer M, Giritli S, Horn M, Haller D, et al. IMNGS: a comprehensive open resource of processed 16s rRNA microbial profiles for ecology and diversity studies. Scientific Rep. 2016;6:33721. doi: 10.1038/srep33721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramírez GA, Jørgensen SL, Zhao R, D’Hondt S. Minimal influence of extracellular DNA on molecular surveys of marine sedimentary communities. Front in Microbiol. 2018;9:2969. doi: 10.3389/fmicb.2018.02969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Overholt WA, Schwing P, Raz KM, Hastings D, Hollander DJ, Kostka JE. The core seafloor microbiome in the Gulf of Mexico is remarkably consistent and shows evidence of recovery from disturbance caused by major oil spills. Environ Microbiol. 2019;21:4316–29.. doi: 10.1111/1462-2920.14794. [DOI] [PubMed] [Google Scholar]
- 51.Van Bleijswijk J, Whalen C, Duineveld G, Lavaleye M, Witte H, Mienis F. Microbial assemblages on a cold-water coral mound at the SE Rockall Bank (NE Atlantic): Interactions with hydrography and topography. Biogeosciences. 2015;12:4483–96.. doi: 10.5194/bg-12-4483-2015. [DOI] [Google Scholar]
- 52.Fiedler CJ, Schönher C, Proksch P, Kerschbaumer DJ, Mayr E, Zunabovic-Pichler M, et al. Assessment of microbial community dynamics in river bank filtrate using high-throughput sequencing and flow cytometry. Front in Microbiol. 2018;9:2887. doi: 10.3389/fmicb.2018.02887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar S, Herrmann M, Thamdrup B, Schwab VF, Geesink P, Trumbore SE, et al. Nitrogen loss from pristine carbonate-rock aquifers of the Hainich Critical Zone Exploratory (Germany) is primarily driven by chemolithoautotrophic anammox processes. Front in Microbiol. 2017;8:1951. doi: 10.3389/fmicb.2017.01951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rusch A, Gaidos E. Nitrogen‐cycling bacteria and archaea in the carbonate sediment of a coral reef. Geobiology. 2013;11:472–84. doi: 10.1111/gbi.12048. [DOI] [PubMed] [Google Scholar]
- 55.Li M, Hong Y, Cao H, Gu J-D. Community structures and distribution of anaerobic ammonium oxidizing and nirS-encoding nitrite-reducing bacteria in surface sediments of the South China Sea. Microb Ecol. 2013;66:281–96. doi: 10.1007/s00248-012-0175-y. [DOI] [PubMed] [Google Scholar]
- 56.Guan Y, Hikmawan T, Antunes A, Ngugi D, Stingl U. Diversity of methanogens and sulfate-reducing bacteria in the interfaces of five deep-sea anoxic brines of the Red Sea. Res in Microbiol. 2015;166:688–99. doi: 10.1016/j.resmic.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 57.Liao T, Wang S, Luo H Phylogenomics suggests the origin of obligately anaerobic anammox bacteria correlates with the great oxidation event. bioRxiv. 2021; 2021.07.07.451387.
- 58.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–8. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 59.Lanzen A, Jørgensen SL, Huson DH, Gorfer M, Grindhaug SH, Jonassen I, et al. CREST - Classification resources for environmental sequence tags. PLoS One. 2012;7:e49334. doi: 10.1371/journal.pone.0049334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andrews S FastQc: A quality control tool for high throughput sequence data. 2010; https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 61.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li DH, Liu CM, Luo RB, Sadakane K, Lam TW. Megahit: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–6. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 63.Wu Y-W, Simmons BA, Singer SW. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics. 2015;32:605–7. doi: 10.1093/bioinformatics/btv638. [DOI] [PubMed] [Google Scholar]
- 64.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seah BK, Gruber-Vodicka HR. Gbtools: Interactive visualization of metagenome bins in R. Front in Microbiol. 2015;6:1451. doi: 10.3389/fmicb.2015.01451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ernest Orlando Lawrence Berkeley National Laboratory, Berkeley, CA (US). BBMap: A fast, accurate, splice-aware aligner. Press Release 2014.
- 67.Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seemann T Barrnap. Online: https://github.com/tseemann/barrnap. In: Github, (2015).
- 69.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J of Computational Biol. 2012;19:455–77. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinform. 2014;30:2068–9. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 72.Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, et al. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44:D286–D93.. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J of Mol Biol. 2016;428:726–31.. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 74.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2011;40:D109–D14.. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Garcia PS, Jauffrit F, Grangeasse C, Brochier-Armanet C. GeneSpy, a user-friendly and flexible genomic context visualizer. Bioinformatics. 2018;35:329–31.. doi: 10.1093/bioinformatics/bty459. [DOI] [PubMed] [Google Scholar]
- 76.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol and Evol. 2015;32:268–74. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–9. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: Improving the ultrafast bootstrap approximation. Mol Biol and Evol. 2017;35:518–22.. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, et al. A new view of the tree of life. Nat Microbiol. 2016;1:16048. doi: 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- 80.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–3. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J of Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 83.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol and Evol. 2013;30:772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Koch H, Luecker S, Albertsen M, Kitzinger K, Herbold C, Spieck E, et al. Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc of the Natl Acad of Sci of the USA. 2015;112:11371–6. doi: 10.1073/pnas.1506533112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data used in this study are available in the NCBI Short Reads Archive under the project number PRJNA529480. In particular, raw metagenomic sequencing data are available in the NCBI database under the BioSample number SAMN11268106. The genome sequence of Bin_158 is available under the accession number JAFNKF000000000. All other genomes analyzed in this study are available in the NCBI database under the accession numbers listed in Table S1.