Abstract
Novel methods for adapting DNA vaccine technology to the prevention of mucosal diseases are greatly needed. Here we show that regulated expression of phage lambda lysis genes S and R causes dramatic lysis of both Vibrio cholerae and Salmonella enterica serovar Typhimurium cells with concomitant release of plasmid DNA into the surrounding media. We also used single and double DNase mutant strains to show that secreted V. cholerae DNases can adversely affect the integrity of DNA molecules released upon lysis. These results suggest that incorporation of lambda SR-mediated lysis constructs and DNA stabilizing mutations into candidate live attenuated bacterial vaccines offers a promising approach for the development of effective mucosal DNA delivery vectors for humans.
Vaccines rank amongst the most effective public health tools for lowering the incidence of infectious diseases. It has recently been shown that administration of nucleic acids, typically DNA, to animals can elicit both humoral and cytotoxic T-lymphocyte responses to a variety of antigens (4). A chief aim of vaccine researchers is to expand this technique of DNA immunization to the prevention of mucosal infections. Diseases of the mucosa—defined as the epithelial surfaces of the gastrointestinal, respiratory, and urogenital tracts—are responsible for a substantial portion of the world's infectious disease morbidity and mortality. For example, respiratory infections and diarrheal diseases alone currently claim more than 10 million lives each year (17). Protection against diseases of the mucosal surface is thought to occur by a process that involves local antigen exposure followed by induction of humoral antibodies, particularly secretory immunoglobulin A (15). However, little work on how to induce mucosal immune responses by DNA-based vaccination has been done.
An attractive concept for delivering DNA vaccines to the mucosal epithelium is the use of live attenuated bacterial vectors. Sizemore et al. (18) first reported the successful use of an attenuated Shigella flexneri mutant as a delivery system for DNA. More recently, Dietrich et al. (3) have explored the use of Listeria monocytogenes for the same application. Because both of these organisms have an inherent ability to invade the cells of the mucosal epithelium, they are attractive delivery vectors; however, concerns about reactogenicity (i.e., adverse events in vaccinees) also make both organisms problematic for human use (13). In contrast, live, attenuated vaccines derived from Vibrio cholerae and Salmonella species represent promising mucosal vaccines that have presented far fewer reactogenicity problems in humans (1, 11, 12). These organisms have the ability to enter the M cells of the gut epithelium and induce mucosal immune responses (15) but have not been explored as DNA delivery vectors.
A key step required for this application is to devise methods that allow the programmed lysis of these organisms in a way that would allow bacterial cells to deliver DNA constructs to the eukaryotic cell cytosol. Here we have investigated whether phage lysis proteins, the S and R gene products of phage lambda (9), can be used to lyse V. cholerae or Salmonella enterica serovar Typhimurium cells.
Construction and testing of inducible lysis plasmid in different strain backgrounds.
Phage lambda encodes two proteins (S and R) which are sufficient to cause bacterial cell lysis (8, 9, 20). In Escherichia coli, the protein product of the S gene is termed “holin” and oligomerizes to form a channel in the bacterial inner membrane. The R gene encodes a transglycosylase which gains access to the periplasm via this channel and then proceeds to degrade the cell wall, causing lysis. The RZ gene product has no known function but is part of most E. coli “SR lysis cassettes” that have been studied previously (20). To test the effect of these lysis proteins on various bacterial species, we constructed a plasmid (pVJ4) that expressed the S, R, and RZ genes under the control of the arabinose-inducible promoter PBAD (10). This plasmid was constructed by cloning the 1.5-kbp EcoRI-HindII fragment of plasmid pRY100 (generously provided by Ry Young, Texas A&M University, College Station, Tex.), containing S, R, and RZ genes, into plasmid pBAD18 (10). Plasmid pVJ4 and control plasmid pBAD18 or pBAD24 were introduced into E. coli DH5α, V. cholerae O395, and Salmonella serovar Typhimurium CDC 6516-60 (Table 1), and the resultant six strains were tested for arabinose-induced cell lysis.
TABLE 1.
Strains used in this study
| Strain | Relevant genotype or phenotype |
|---|---|
| E. coli | |
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 |
| β2155 | thrB1004 pro thi strA hsdS lacZΔM15 (F′ lacZΔM15 lacIqtraD3 proA+proB+) ΔdapA::erm (Ermr) pir::RP4 [::kan (Kmr) from SM10] |
| V. cholerae | |
| O395 | Classifcal biotype; serotype Ogawa; Smr |
| N16961 | El Tor biotype; serotype Inaba |
| V33 | El Tor biotype; serotype Ogawa; O17; Smr |
| V754 | dns derivative of V33 |
| V62 | El Tor biotype; serotype Ogawa; Rifr derivative of 1621 |
| V751 | dns/xds derivative of V62 |
| Salmonella serovar Typhimurium CDC 6516-60 | Previously classified as Salmonella choleraesuis subsp. choleraesuis; serotype Typhimurium |
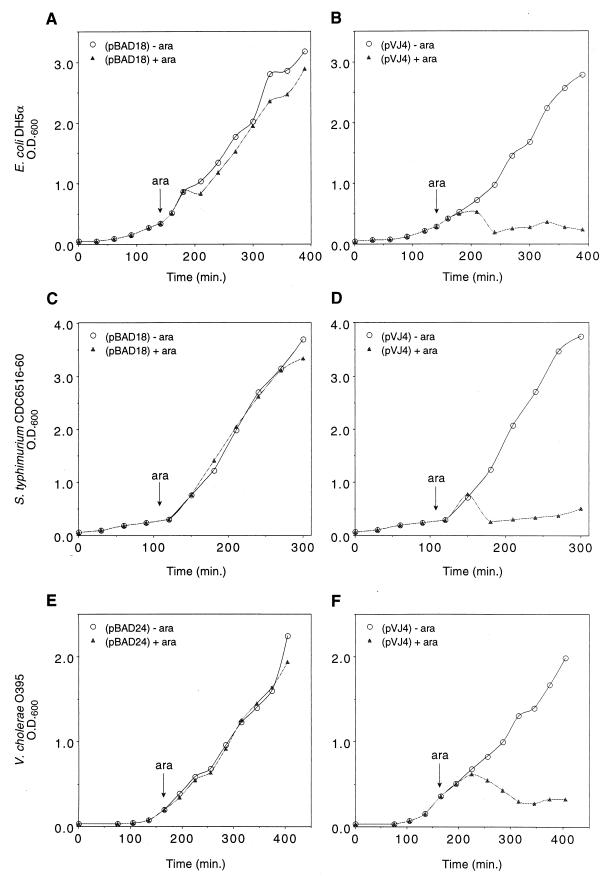
Each strain was grown in Luria broth (LB) at 37°C until mid-logarithmic phase (optical density at 600 nm [OD600] of approximately 0.35), at which time arabinose was added to a final concentration of 0.2%. Optical densities of the cultures were monitored to assess cell lysis (Fig. 1). Approximately 1 h after induction with arabinose, the OD600 of a DH5α(pVJ4) culture fell from a level of 0.5 to 0.2. In the absence of arabinose, the growth of DH5α(pVJ4) was unimpeded, confirming that arabinose induction had elicited cell lysis. Similar results were obtained with Salmonella serovar Typhimurium cells carrying these two plasmids. CDC 6516-60 carrying control plasmid pBAD18 neither lysed nor showed any growth inhibition in the presence of arabinose (Fig. 1C). In contrast, the OD600 of the CDC 6516-60(pVJ4) culture decreased dramatically after the addition of arabinose and thereafter did not increase further (Fig. 1D). Like the other bacterial hosts, V. cholerae O395 displayed no lysis upon induction with arabinose when carrying control plasmid pBAD24 (Fig. 1E). However, O395(pVJ4) responded to addition of arabinose with steady lysis and eventual growth inhibition (Fig. 1F). The kinetics of V. cholerae lysis were again similar to those of the other two hosts, with significant lysis apparent approximately 60 min after induction. Similar results were also observed when pVJ4 was introduced into other strains of V. cholerae (data not shown). Taken together, these results suggest that the phage lambda phage lysis genes S and R function well in all three gram-negative hosts when expressed from the PBAD promoter.
FIG. 1.
Effects of induced phage lysin expression on growth of bacterial cells. E. coli DH5α cells carrying either pBAD18 (A) or pVJ4 (B) were grown in the absence (circles) or presence (triangles) of 0.02% arabinose. Growth was monitored by optical density measurements. Growth of Salmonella serovar Typhimurium CDC 6516-60 (C and D) and V. cholerae O395 (E and F) were similarly quantitated.
Measurement of DNA released by cell lysis.
Given that arabinose induction of the three different strains carrying pVJ4 caused the apparent cell lysis, we next addressed whether the lysis process mediated by S and R was compatible with the release of intact plasmid DNA molecules into the surrounding media. Towards this end, nucleic acids were purified from the extracellular media of these lysed cell cultures. Southern blot analysis confirmed that intact plasmid was being released by the SR-mediated lysis process (data not shown). To better assess how much plasmid DNA was being released by these three different bacterial species, DNA from cell free culture supernatant fluids was purified and then transformed into E. coli under standard conditions. The number of transformants per CFU per milliliter provided a quantitative assessment of the level of intact plasmid DNA that was released by arabinose induction of control (pBAD18) and pVJ4-carrying cells.
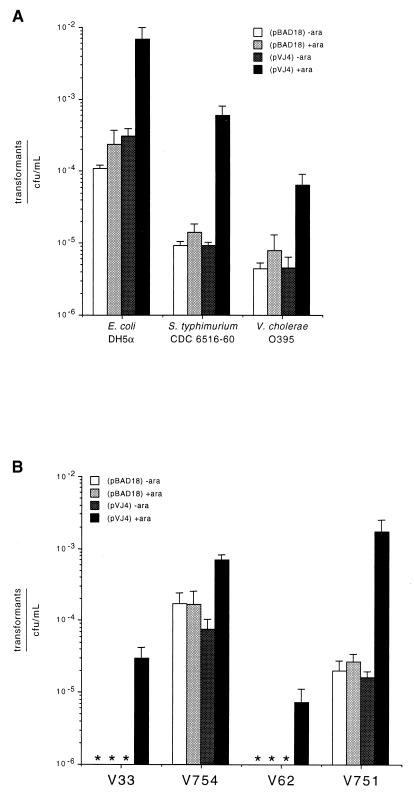
As shown in Fig. 2A, the amount of intact plasmid DNA released from E. coli control strain DH5α(pBAD18) was not substantially different in the presence or absence of arabinose. In contrast, cultures of DH5α(pVJ4) that were induced with arabinose showed an approximately 22-fold increase in the level of plasmid DNA released compared to the uninduced culture (Fig. 2A).
FIG. 2.
(A) Quantitation of transformation efficiency of released plasmids. Plasmid purified from culture supernatants was transformed into electrocompetent E. coli, and the resulting numbers of transformants were counted. Ratios of transformants to CFU of the culture per milliliter at the time of lysis were calculated for the three strains under study. (B) Analysis of the effects of DNase mutations on efficiency of plasmid release. Ratios of transformants to CFU of cultures of V754 and V751 as well as their isogenic wild-type strains per milliliter were determined as before. Values represent means of triplicate experiments; error bars represent standard errors of the means.
Similar results were observed with S. typhimurium CDC 6516-60(pVJ4) and control strains. Induction of S. typhimurium CDC 6516-60(pVJ4) caused approximately a 66-fold increase in the release of plasmid DNA. Uninduced cultures of S. typhimurium carrying these plasmids showed lower levels of background transformants than did DH5α cultures, and the peak level of transformants reached after induced cell lysis was also lower than that for DH5α (Fig. 2A). This lower efficiency of release may be due to the fact that E. coli is the natural host of phage lambda and the lysis gene products may simply cause more complete lysis of E. coli cells than of Salmonella serovar Typhimurium cells, potentially leading to less “trapping” of plasmid DNA in E. coli cell debris.
Arabinose induction of V. cholerae O395(pVJ4) caused an approximately 15-fold increase in plasmid DNA release (Fig. 2A). However, the peak ratio of transformants to CFU per milliliter observed in O395 was approximately 1 log lower than that seen in Salmonella serovar Typhimurium and 2 logs lower than that observed in E. coli. Although a lower lysis efficiency may have been responsible for the reduced plasmid release observed from V. cholerae, we hypothesized that extracellular DNase produced by V. cholerae may have been responsible for reducing the plasmid yield in these experiments (6, 7). Growth of the three different strains on DNase test agar confirmed that V. cholerae O395 was producing much more DNase than either E. coli or Salmonella serovar Typhimurium (data not shown).
Effects of V. cholerae DNases on the levels of DNA release.
To prove that the degradative action of DNase produced by V. cholerae O395 was reducing the level of intact plasmid in our lysis experiments, we repeated these with two V. cholerae strains that carried mutations that affect the production of DNase (kindly provided by Paul Manning). Strain V754 carries a mutation in the dns gene, one of the two genes implicated in V. cholerae DNase secretion (7), while strain V751 is a double-knockout strain lacking both dns and xds, the structural gene for a major V. cholerae DNase (16). The parental strains of V754 and V751 are strains V33 and V62, respectively. Plasmid pVJ4 or control plasmid pBAD18 was introduced into these four strains, and release of plasmid DNA was monitored with and without arabinose inductions by quantitative transformation assays. As shown in Fig. 2B, approximately 24-fold more plasmid was released by strain V754 than by its parental strain, V33, suggesting that the dns mutation does increase the yield of intact plasmid released by arabinose-induced lysis. Furthermore, strain V751 released approximately 240 times more plasmid after arabinose induction than its parental strain V62 (Fig. 2B). Because strain V751 has a more severe defect in the secretion of extracellular DNase (due to its double mutations in both dns and xds (6, 16), these results indicate that the presence of extracellular DNases is detrimental to the recovery of DNA released by arabinose induction of lysis plasmid pJV4. Interestingly, the peak ratio of transformants to number of CFU per milliliter achieved with the double DNase-knockout strain V751 was the highest for all V. cholerae strains tested and higher than that seen with Salmonella serovar Typhimurium. It is not known whether the Salmonella strains examined here make DNases, but our results strongly suggest that deletion of DNase genes might improve the utility of live, bacterial DNA delivery vectors derived from this or other bacterial species.
The results presented here indicate that the lambda phage lysis genes S and R can be used as a means of achieving controlled cell lysis of diverse bacterial species, including V. cholerae. By analogy, other phage lysis genes might also work well in this application (5). It is generally thought that bacterial DNA delivery systems will need some sort of programmed cell lysis system in order to efficiently deliver DNA to eukaryotic cells. For example, Sizemore et al. (18) utilized an attenuating mutation in the S. flexneri asd gene, whose protein product is necessary for the synthesis of diaminopimelic acid, an essential component of the cell wall. Starvation for diaminopimelic acid in vivo eventually causes cell lysis. While such a strategy is clever, one can imagine that this strategy for in vivo lysis might cause premature or anatomically incorrect release of DNA constructs. In contrast, the use of lysis genes to program cell lysis provides in theory the ability to finely regulate cell lysis by using in vivo induced promoters that are expressed only in the mucosa-associated lymphoid tissue or in antigen-presenting cells. For example, a recent report by Dietrich et al. (3) demonstrated the successful use of the PactA promoter of L. monocytogenes to drive expression of a lysis gene, thereby achieving lysis of the vector only after invasion of target host macrophages and entry into the cytoplasmic compartment. A variety of techniques for identifying in vivo induced promoters for both Vibrio and Salmonella exist (2, 14, 19). It is also likely that different bacterial DNA delivery systems will each require their own additional genetic engineering to optimize efficiency and safety. For example, we found that mutations that decreased the level of extracellular DNase expressed by V. cholerae greatly improved the recovery of intact plasmid DNA. Whether such secondary mutations will improve the efficiency of delivery of functional DNA constructs to antigen-presenting cells remains to be determined.
Acknowledgments
We thank members of the Mekalanos laboratory for helpful discussion and critical review. V. cholerae strains V33, V754, V62, and V751 were generously provided by P. Manning. V.J. also thanks Timothy Clackson, Ariad Pharmaceuticals, for insightful suggestions and advice.
This work was supported by DARPA grant N65236-97-1-5813.
REFERENCES
- 1.Camilli A, Mekalanos J J. Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol Microbiol. 1995;18:671–683. doi: 10.1111/j.1365-2958.1995.mmi_18040671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coster T S, Killeen K P, Waldor M K, Beattie D T, Spriggs D R, Kenner J R, Trofa A, Sadoff J C, Mekalanos J J, Taylor D N. Safety, immunogenicity, and efficacy of live attenuated Vibrio cholerae O139 vaccine prototype. Lancet. 1995;345:949–952. doi: 10.1016/s0140-6736(95)90698-3. [DOI] [PubMed] [Google Scholar]
- 3.Dietrich G, Bubert A, Gentschev I, Sokolovic Z, Simm A, Catic A, Kaufmann S H, Hess J, Szalay A A, Goebel W. Delivery of antigen-encoding plasmid DNA into the cytosol of macrophages by attenuated suicide Listeria monocytogenes. Nat Biotechnol. 1998;16:181–185. doi: 10.1038/nbt0298-181. [DOI] [PubMed] [Google Scholar]
- 4.Donnelly J J, Ulmer J B, Shiver J W, Liu M A. DNA vaccines. Annu Rev Immunol. 1997;15:617–648. doi: 10.1146/annurev.immunol.15.1.617. [DOI] [PubMed] [Google Scholar]
- 5.Eko F O, Szostak M P, Wanner G, Lubitz W. Production of Vibrio cholerae ghosts (VCG) by expression of a cloned phage lysis gene: potential for vaccine development. Vaccine. 1994;12:1231–1237. doi: 10.1016/0264-410x(94)90249-6. [DOI] [PubMed] [Google Scholar]
- 6.Focareta T, Manning P A. Distinguishing between the extracellular DNases of Vibrio cholerae and development of a transformation system. Mol Microbiol. 1991;5:2547–2555. doi: 10.1111/j.1365-2958.1991.tb02101.x. [DOI] [PubMed] [Google Scholar]
- 7.Focareta T, Manning P A. Extracellular proteins of Vibrio cholerae: molecular cloning, nucleotide sequence and characterization of the deoxyribonuclease (DNase) together with its periplasmic localization in Escherichia coli K-12. Gene. 1987;53:31–40. doi: 10.1016/0378-1119(87)90090-4. [DOI] [PubMed] [Google Scholar]
- 8.Garrett J, Bruno C, Young R. Lysis protein S of phage lambda functions in Saccharomyces cerevisiae. J Bacteriol. 1990;172:7275–7277. doi: 10.1128/jb.172.12.7275-7277.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrett J, Fusselman R, Hise J, Chiou L, Smith-Grillo D, Schulz J, Young R. Cell lysis by induction of cloned lambda lysis genes. Mol Gen Genet. 1981;182:326–331. doi: 10.1007/BF00269678. [DOI] [PubMed] [Google Scholar]
- 10.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hohmann E L, Oletta C A, Killeen K P, Miller S I. phoP/phoQ-deleted Salmonella typhi (Ty800) is a safe and immunogenic single-dose typhoid fever vaccine in volunteers. J Infect Dis. 1996;173:1408–1414. doi: 10.1093/infdis/173.6.1408. [DOI] [PubMed] [Google Scholar]
- 12.Kenner J R, Coster T S, Taylor D N, Trofa A F, Barrera-Oro M, Hyman T, Adams J M, Beattie D T, Killeen K P, Spriggs D R, et al. Peru-15, an improved live attenuated oral vaccine candidate for Vibrio cholerae O1. J Infect Dis. 1995;172:1126–1129. doi: 10.1093/infdis/172.4.1126. [DOI] [PubMed] [Google Scholar]
- 13.Kotloff K L, Noriega F, Losonsky G A, Sztein M B, Wasserman S S, Nataro J P, Levine M M. Safety, immunogenicity, and transmissibility in humans of CVD 1203, a live oral Shigella flexneri 2a vaccine candidate attenuated by deletions in aroA and virG. Infect Immun. 1996;64:4542–4548. doi: 10.1128/iai.64.11.4542-4548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahan M J, Slauch J M, Mekalanos J J. Selection of bacterial virulence genes that are specifically induced in host tissues. Science. 1993;259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- 15.Neutra M R, Frey A, Kraehenbuhl J P. Epithelial M cells: gateways for mucosal infection and immunization. Cell. 1996;86:345–348. doi: 10.1016/s0092-8674(00)80106-3. [DOI] [PubMed] [Google Scholar]
- 16.Newland J W, Green B A, Foulds J, Holmes R K. Cloning of extracellular DNase and construction of a DNase-negative strain of Vibrio cholerae. Infect Immun. 1985;47:691–696. doi: 10.1128/iai.47.3.691-696.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rekkedal G. WHO world health report 1997. Tidsskr Sykepl. 1997;85:37–39. [PubMed] [Google Scholar]
- 18.Sizemore D R, Branstrom A A, Sadoff J C. Attenuated Shigella as a DNA delivery vehicle for DNA-mediated immunization. Science. 1995;270:299–302. doi: 10.1126/science.270.5234.299. [DOI] [PubMed] [Google Scholar]
- 19.Valdivia R H, Falkow S. Fluorescence-based isolation of bacterial genes expressed within host cells. Science. 1997;277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- 20.Young R, Blasi U. Holins: form and function in bacteriophage lysis. FEMS Microbiol Rev. 1995;17:191–205. doi: 10.1111/j.1574-6976.1995.tb00202.x. [DOI] [PubMed] [Google Scholar]