Abstract
Phytoplankton-derived metabolites fuel a large fraction of heterotrophic bacterial production in the global ocean, yet methodological challenges have limited our understanding of the organic molecules transferred between these microbial groups. In an experimental bloom study consisting of three heterotrophic marine bacteria growing together with the diatom Thalassiosira pseudonana, we concurrently measured diatom endometabolites (i.e., potential exometabolite supply) by nuclear magnetic resonance (NMR) spectroscopy and bacterial gene expression (i.e., potential exometabolite uptake) by metatranscriptomic sequencing. Twenty-two diatom endometabolites were annotated, with nine increasing in internal concentration in the late stage of the bloom, eight decreasing, and five showing no variation through the bloom progression. Some metabolite changes could be linked to shifts in diatom gene expression, as well as to shifts in bacterial community composition and their expression of substrate uptake and catabolism genes. Yet an overall low match indicated that endometabolome concentration was not a good predictor of exometabolite availability, and that complex physiological and ecological interactions underlie metabolite exchange. Six diatom endometabolites accumulated to higher concentrations in the bacterial co-cultures compared to axenic cultures, suggesting a bacterial influence on rates of synthesis or release of glutamate, arginine, leucine, 2,3-dihydroxypropane-1-sulfonate, glucose, and glycerol-3-phosphate. Better understanding of phytoplankton metabolite production, release, and transfer to assembled bacterial communities is key to untangling this nearly invisible yet pivotal step in ocean carbon cycling.
Subject terms: Biogeochemistry, Ecology
Introduction
Phytoplankton bloom development and senescence are closely entangled with heterotrophic bacterial community activities, mediated through phytoplankton-derived dissolved organic carbon [1–3]. Newly released phytoplankton metabolites can be rapidly consumed by bacterial assemblages, often within minutes to days [4]. As marine phytoplankton are responsible for half of Earth’s photosynthesis and much of the fixed carbon is passed on to heterotrophic bacteria, quantifying this step is important for modeling global carbon flux [5–7].
Metabolite pools derived from phytoplankton consist of hundreds of unique organic compounds, most of which accumulate in only trace amounts [8]. This diversity coupled to high turnover rates poses a challenge for chemical identification, with only 1–5% of compounds identified thus far [8–10]. Transfer of these metabolites to bacteria has been even more difficult to quantify. Recent studies have made headway by analysis of bacterial gene expression as an indication of uptake and catabolism of substrates [11–14], yet information is lacking on the diversity of roles of bacterial community members in determining exometabolite flux and fate.
The extent and composition of direct release of phytoplankton photosynthate is influenced by the physiological state of the cell [15–17]. Healthy phytoplankton cells release labile compounds such as sugars, sugar alcohols, amino acids, and carboxylic acids [2, 18, 19], which may dominate during early phases of a bloom. As blooms progress towards senescence, the amount of metabolites released increases, and larger molecules such as polysaccharides take on more importance [2, 20, 21]. This release of labile carbon from phytoplankton to surrounding organisms can occur by multiple mechanisms [22]. The simplest is diffusion between intracellular pools and external seawater [23], although this is constrained to molecules of relatively small size [24]. Alternatively, molecules can be actively released via overflow pathways when rates of photosynthesis exceed their needs for growth, for example, carbohydrates are released in response to photorespiration [18, 23, 25]. Active release of metabolites can also occur in response to associated microbes [3, 26], such as through the release of molecules that serve as bacterial chemoattractants [27, 28].
Substrate release sets the stage for bacterial heterotrophy, with different substrate preferences governing the succession of taxa during a bloom [19]. Among heterotrophic bacterial taxa consistently found associated with phytoplankton there is evidence for specialization on certain components of extracellular release [2, 19], even when the extracellular release originates from a single phytoplankton species [14]. The ability to use common but distinct substrate sets is likely a benefit to these bacterial groups, contributing to their success in surface ocean communities [2, 29]. Detailed understanding of endometabolite composition, release, and utilization by heterotrophic bacteria under different stages of phytoplankton growth is still limited, however, particularly when there are multiple bacterial species that can compete and interact.
Globally, primary production is strongly influenced by annual spring blooms in temperate regions, commonly dominated by fast-growing diatoms [19, 30]. To mimic a diatom bloom under controlled laboratory conditions, Thalassiosira pseudonana was co-cultured with a synthetic community consisting of three heterotrophic bacteria (Ruegeria pomeroyi, Stenotrophomonas sp., and Polaribacter dokdonensis) representing taxa typically associated with phytoplankton blooms [19]. We used nuclear magnetic resonance (NMR) spectroscopy to identify the endometabolites of the diatom and transcriptomics to trace their potential transfer to bacteria during early and late bloom stages. Coupling diatom endometabolite quantification with bacterial gene expression analysis also enabled us to observe temporal patterns, either matched or mismatched, that are potentially informative of extracellular release mechanisms.
Methods
Co-culture conditions
During this synthetic bloom experiment, axenic cultures of the diatom T. pseudonana CCMP1335 (National Center for Marine Algae) were inoculated with equal cell numbers (~3 x 104 cells ml−1) of the heterotrophic bacteria R. pomeroyi DSS-3 (Rhodobacterales; ATCC 700808; isolated from southeastern US seawater [31]), Stenotrophomonas sp. SKA14 (Xanthomonadales; provided by J. Pinhassi, Linnaeus University Sweden; isolated from the Skagerrak Sea [32]) and P. dokdonensis MED152 (Flavobacteriales; provided by J. Pinhassi, Linnaeus University Sweden; isolated from the Mediterranean Sea [33]). The strains have high 16S rRNA gene identity to bacteria associated with phytoplankton cultures or flow-sorted with phytoplankton cells, with percent similarities up to 99.6% for R. pomeroyi [3, 34–36], 98.8% for Stenotrophomonas sp. SKA14 [34], and 97.2% for P. dokdonensis [35, 37]. The diatom was grown in organic carbon-free L1 medium [38] prepared in acid-washed glass containers at a salinity of 35 [39] for one week prior to the start of the experiment. Cultures were grown with a 16:8 h light:dark cycle under 160 µmol photons m−2 s−1 at 18 °C and checked for bacterial contamination by plating on rich medium (YTSS). On day 0 of the experiment, diatoms were transferred into 1.9 L culture flasks containing 1 L of medium to a final concentration of ~2 × 103 cells ml-1. The medium was made with 13C-bicarbonate (Cambridge Isotope Libraries, Inc., Tewksbury, MA, USA) to enhance NMR signals. One flask was kept as L1 medium without organisms for use as a background control for NMR analyses.
The three strains of heterotrophic bacteria were grown overnight in either YTSS at 30 °C (R. pomeroyi and Stenotrophomonas) or 1/5YTSS at 25 °C (P. dokdonensis) made with salinity 20 artificial seawater. Cells were harvested in exponential growth phase and washed five times in the same artificial seawater used for preparing the L1 medium (1 ml wash volume). The bacteria were inoculated in equal proportions of OD600 into 15 flasks containing diatoms, with a final combined concentration of ~1 × 105 cells ml−1, which is comparable to pre-bloom conditions during a natural bloom [19]. One set of three flasks remained axenic. Three co-culture flasks were sacrificed after 8 h of light (day 0), and then on days 3, 8, 15, 20. The axenic flasks were only sampled on day 15.
To trace diatom and bacterial growth, 1 ml subsamples were fixed with glutaraldehyde (1% final concentration), stored overnight at 4 °C, and thereafter at −80 °C until flow cytometric analysis. The samples were stained with SYBR® Green I (final concentration 1×; Life Technologies, Carlsbad, CA, USA) and analyzed on a CyAn ADP flow cytometer (Beckman Coulter, Hialeah, Florida) using 5-µm fluorescent particles (Spherotech, Lake Forest, IL, USA) for enumeration. The diatom specific growth rate (μ d−1) was calculated as (ln DE − ln DS)/(tE − tS), where DE is cell number at the end of a period and DS at the start of the experiment, and tE is the end day and tS the start day. The three morphologically distinct bacteria (P. dokdonensis by orange color; Stenotrophomonas sp. by fast growth and large colonies; R. pomeroyi by slower growth and small colonies; Fig. S1) were individually quantified as colony-forming units (CFUs) by dilution plating. Subsamples of 100 µl were diluted 10−1 to 10−7 times and spread onto both YTSS and 1/5YTSS agar plates. The plates were incubated at 30 °C (YTSS) or 25 °C (1/5YTSS) and counted after three and four days, respectively. Total bacterial cell numbers measured by flow cytometry correlated well with the sum of the species-specific CFUs (Fig. S1).
For sampling diatoms, subsamples (100–200 ml for RNA and 700–1000 ml for endometabolites) were gently filtered using a peristaltic pump onto 2.0 µm pore-size polycarbonate IsoporeTM filters (Millipore, Burlington, MA, USA). For sampling bacteria, the filtrate from the endometabolite samples was re-filtered onto 0.2 µm pore-size Supor® PES filters (PALL, Port Washington, NY, USA). Filters collected for diatom and bacterial RNA were immediately flash-frozen in liquid nitrogen, and along with endometabolite filters transferred to −80 °C until processing. Subsamples of the final filtrate were collected for dissolved inorganic nutrient analysis (10 ml stored at −20 °C).
Diatom endometabolite analysis
For endometabolite analysis (day 3 and 15), filters in tubes were sonicated for 7 min while submerged in ice-water to remove diatom cells (50 s on and 10 s off sequence) using an SLPe sonifier (Branson, Brookfield, CT, USA) after adding 15 ml of ultrapure water, and the liquid fraction was collected in fresh tubes as described in Uchimiya et al. [40]. This process was repeated three times and combined fractions stored at −80 °C until processing. Samples were lyophilized (Labconco, Kansas City, MO, USA) and pellets were mixed with 600 µL of phosphate buffer (30 mM phosphate in deuterated water, pH 7.4) and 1 mM internal standard (2, 2-dimethyl-2-silapentane-5-sulfonate). Samples were vortexed for 5 min, centrifuged at 20 800 RCF for 10 min, and supernatants were transferred to 5 mm NMR tubes (Bruker, Billerica, MA, USA). One pooled quality control sample was prepared by combining aliquots of all the samples and used for annotation. All sample processing was carried out at 4 °C. Metabolites were analyzed by NMR spectroscopy using a 600 MHz AVANCE III HD instrument (Bruker) equipped with a 5 mm TXI probe and pulse programs of 1H-13C heteronuclear single quantum correlation (HSQC, hsqcetgpprsisp2.2 by Bruker nomenclature) and 1H-13C HSQC-total correlation spectroscopy (HSQC-TOCSY, hsqcdietgpsisp.2). Data were processed by TopSpin version 4.0 (Bruker). Peak intensity was extracted by rNMR version 1.11 [41], normalized by cell number and auto-scaled. Metabolites were annotated based on chemical shift (HSQC) and spin network information (HSQC-TOCSY) (Fig. S2). Chemical shift values for candidate peaks were obtained from Biological Magnetic Resonance Data Bank [42], and raw HSQC spectra for validation from Human Metabolome Databases [43]. Four compounds of interest that are not in these databases were annotated using literature values (homarine, [44]; 2,3-dihydroxypropane-1-sulfonate (DHPS), dimethylsulfoniopropionate (DMSP), and β-1,3-glucan, [17]). A confidence level of annotation was assigned to each metabolite, where 1 = putative compounds with functional group information; 2 = partially matched to HSQC chemical shift information in the databases or literature; 3 = fully matched to HSQC chemical shift; 4 = fully matched to HSQC chemical shift and validated by HSQC-TOCSY; 5 = validated by a spiking experiment. All the data, sample preparation protocols, and NMR analysis and processing parameters are deposited in Metabolomics Workbench under Project ID 001231 (10.21228/M8KT3K). Data were converted to Z-scores (value – mean/standard deviation). Statistical analysis of day 3 vs day 15 co-cultures, and day 15 co-cultures versus day 15 axenic cultures was conducted by using unpaired T-tests (p ≤ 0.05, n = 3).
RNA extraction
RNA was extracted using the ZymoBIOMICS RNA Miniprep Kit (Zymo Research, Irvine, CA, USA) according to the manufacturer’s protocol with 20 min beating and on-column DNase treatment. Following extraction, an additional DNA removal was performed using the TURBO DNA-free Kit (Invitrogen, ThermoFisher Scientific, Vilnius, Lithuania) following standard kit procedures. Stranded RNA libraries were prepared using the Zymo-Seq RiboFree Total RNA Library Kit (ZymoBIOMICS) with rRNA depletion. Libraries and rRNA depletion for samples with low RNA concentration were prepared at HudsonAlpha Discovery (Huntsville, AL, USA) and all libraries were sequenced on the Illumina NextSeq platform (SE, 75 bp).
RNA-Seq and differential gene expression analyses
The TrimGalore toolkit was used for sequence trimming and quality control, imposing a minimum quality score of 20. Reads aligning to the rRNA sequences of the microbial taxa were removed using SortMeRNA. STAR aligner was used to map remaining reads to the genome of each of the species and HTSeq to count reads mapped to each gene. Genes with differential expression between co-culture time points (day 3 and 15) and between co-culture and axenic conditions (day 15; diatoms only) were identified using DESeq2 in R (Version 4.0.0). Biosynthesis pathways were identified based on Biocyc [45] and PhyloDB [46]. Heatmaps were created using the pheatmap package in R.
RNA-Seq data from a previous study [14] that was collected for each bacterial strain when in individual co-culture with the diatom was compared to the day 3 RNA-Seq data from this study. The datasets were analyzed using DESeq2 as described above, with each co-culture compared to the same reference dataset [14], and significantly enriched genes emerging from the analyses were compared. The reference dataset was established by growing bacteria in the same medium as used for both co-cultures, except that T. pseudonana was not inoculated and 2.5 mM glucose was added [14]; this provided transcriptomes of actively growing bacteria on a defined carbon source against which the two co-culture datasets were analyzed.
Dissolved inorganic nutrients
Dissolved inorganic nutrient concentrations were measured at day 15 in both axenic and co-cultures as well as in the media-only control (Table S1). Nitrate and nitrite were measured by the automated cadmium reduction method, phosphate by the automated ascorbic acid reduction method, and ammonium by the automated phenate method [47]. Pure water was used as a blank and 2 ml samples were analyzed using standard curves of 0, 50, 100, 200 ppb on a Alpkem RFA 300 (UniGreenScheme, UK). Silicate was analyzed spectrophotometrically [48, 49] (1 ml sample + 4 ml water; Spectronic 301; Milton Roy, Ivyland, PA, USA) using a standard curve composed of 0, 5, 10, 20, and 40 μM solutions and a water blank processed through same chemistry as the samples.
Results and discussion
Co-culture dynamics
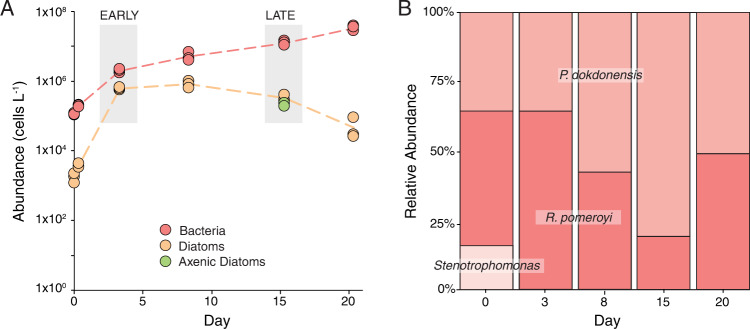
This study was designed to enhance understanding of metabolite release and utilization across bloom stages in a simple community of phytoplankton and heterotrophic bacteria. The synthetic community was established with the diatom T. pseudonana and the bacterial strains R. pomeroyi DSS-3, Stenotrophomonas sp. SKA14, and P. dokdonensis MED152. These bacterial strains have high genetic similarity to isolates from phytoplankton cultures [14] and represent taxa that are common in phytoplankton blooms. Metabolites derived from the diatom were the sole source of carbon available for the bacteria, since no organic substrates were added. In addition, none of the bacteria can assimilate nitrate, and usable nitrogen was only available as diatom or bacterial extracellular products. The diatom had its highest specific growth rate of 1.65 d−1 during days 0–3, after which the rate declined (Fig. 1A). The total abundance of heterotrophic bacteria increased steadily but there was a succession that favored P. dokdonensis through day 15, and then R. pomeroyi by day 20; Stenotrophomonas disappeared from the model system by day 3 (Fig. 1B). The presence of bacteria did not affect the growth of diatoms based on comparisons of abundance in co-cultures versus axenic cultures at day 15 (Fig. 1A), as has been found previously [14, 26]. Inorganic nutrients were not limiting (>5 μM at day 15; Table S1).
Fig. 1. Time course of microbial abundances.
A Cell abundance based on flow cytometric analysis for co-cultures (5 time points) and axenic cultures (day 15 only) (n = 3). The intensive sampling dates for the early and late bloom comparisons are marked with gray boxes. B Mean relative abundance of bacterial species is based on CFUs (n = 3). The day 0 samples were collected 8 h after inoculation.
Diatom endometabolite shifts
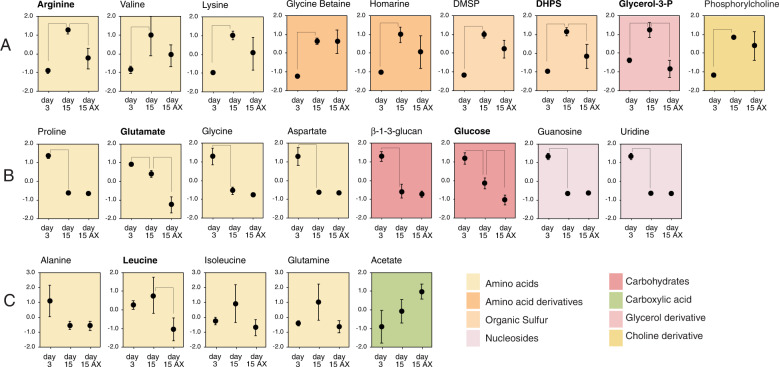
Analyses focused on the day 3 (early bloom) and day 15 (late bloom) co-culture time points, for which a complete set of metabolomic and transcriptomic data were collected. Twenty-two diatom endometabolites that were annotated with high confidence by NMR analysis (Table S2) and quantified after normalizing to diatom cell number revealed that endometabolome composition differed substantially between bloom stages. Metabolites with significantly different cellular concentrations included nine compounds that were higher in intracellular concentration during the late bloom; these were arginine, valine, lysine, DHPS, glycerol-3-phosphate, phosphorylcholine, DMSP, glycine betaine, and homarine (T-test; P < 0.05, n = 3; Fig. 2A; Table S3), of which the last three are known to function as osmolytes [50, 51]. Elevated internal DMSP concentrations in phytoplankton cells have been linked to nitrogen, CO2, silicate, and phosphate limitation, increasing during stationary growth phase and potentially replacing nitrogen-rich osmolytes or serving as an antioxidant under stressful conditions such as CO2 limitation and low temperature [52]. All three osmolytes have also been identified in endometabolomes of natural plankton communities from surface seawater [53]. Eight metabolites were significantly lower in intracellular concentration in the late bloom co-cultures; these were proline, glutamate, glycine, β-1,3-glucan, aspartate, glucose, guanosine, and uridine (Fig. 2B). Five metabolites did not change between early and late bloom stages; these were alanine, leucine, isoleucine, glutamine, and acetate (Fig. 2C).
Fig. 2. Relative endometabolite abundance in diatom cells.
Abundance is expressed as mean Z-score of per cell concentration in early bloom co-cultures (day 3), late bloom co-cultures (day 15), and axenic late bloom cultures (day 15 AX). Metabolites present in significantly different per cell concentrations are linked by brackets (T-test, p ≤ 0.05, n = 3); no statistical comparisons were made between day 3 and day 15 AX. Row A Endometabolites with significantly higher concentration in day 15 co-cultures compared to day 3 co-cultures; Row B Endometabolites with significantly higher concentration in day 3 co-cultures compared to day 15 co-cultures; Row C Endometabolites not significantly different between day 3 and day 15. Bold font highlights the metabolites accumulating to higher concentrations in 15 d co-cultures compared to 15 d axenic cultures. Plots are colored according to metabolite class. Error bars represent standard deviations. See Table S3 for metabolite intensity per cell.
Differences in diatom endometabolome composition were also evident in comparisons of the 15 d cultures with and without a bacterial community. Six diatom endometabolites had accumulated to higher concentrations in the presence of bacteria compared to axenic cultures by day 15; these were glutamate, arginine, leucine, DHPS, glucose, and glycerol-3-phosphate; none accumulated to higher concentrations in axenic cultures (Fig. 2), similar to previous analyses [17]. Alteration of phytoplankton physiology in the presence of bacteria has been reported previously and attributed to processes such as bacterial remineralization of ammonium from dissolved organic matter [54] and release of vitamins [13] or hormones [3]. In this study, phytoplankton were both nutrient and vitamin replete and there was no evidence that co-culturing with bacteria enhanced growth. Nonetheless, the diatom cells accumulated endometabolites differently depending on the presence or absence of bacteria.
Diatom gene expression
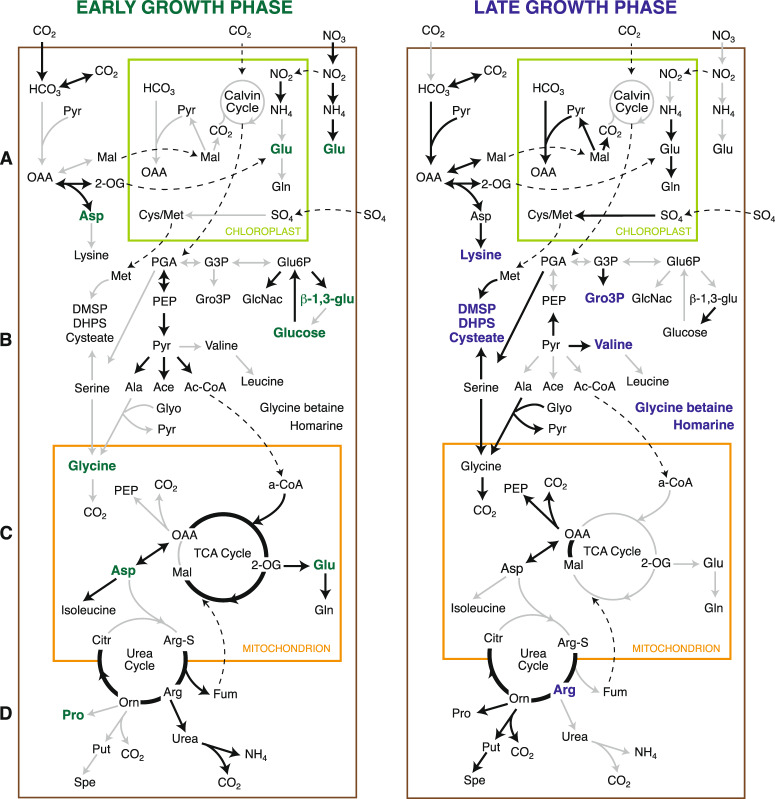
Diatom gene expression provided insights into physiological changes associated with growth stage. Relative gene expression fell into two distinct expression clusters consisting of early (days 0, 3) and late (days 8, 15, 20) sample times (Fig. S3). Comparing the day 3 and day 15 time points, 6 637 of the 11 675 predicted genes in the T. pseudonana genome (59%) had significantly different relative contributions to the transcriptome. The diatom’s early bloom transcriptome was highly enriched in transcripts for CO2 acquisition via carbonic anhydrases (Fig. 3A). Transcripts for synthesis of glycolysis products pyruvate, acetate, and acetyl-CoA (Fig. 3B) and for channeling acetyl-CoA into the tricarboxylic acid (TCA) cycle were also enriched, as were transcripts for multiple central TCA cycle steps from oxaloacetate to succinate (Fig. 3C). Transcription patterns indicated that recently fixed carbon was directed toward chitin precursor N-acetyl-D-glucosamine (GlcNAc) and chrysolaminarin backbone β-1,3-glucan (Table S4). These transcriptome features were consistent with the higher concentrations of glucose and β-1,3-glucan in the diatom endometabolome on day 3. Overall, this gene expression pattern indicated an emphasis on carbon fixation and biomass building in the early bloom (Fig. 2) and is consistent with the diatom’s highest specific growth rate occurring during days 0–3 (Fig. 1).
Fig. 3. Integration of diatom endometabolite and gene expression data for early-stage (left panel) and late-stage (right panel) bloom phases.
Black lines indicate relative gene expression that is significantly higher in one growth stage compared to the other, gray lines indicate expression that is not significantly higher. Green font indicates significantly higher metabolite concentration in early-stage cells, and blue font indicates higher concentration in late-stage cells (see Fig. 2). A Carbon and nitrogen assimilation. B Glycolysis/Gluconeogenesis. C TCA cycle. D Urea cycle. Metabolite abbreviations are as follows: Ala alanine, Ace acetate, Ac-CoA acetyl-CoA, Arg arginine, β-1,3-glu β-1,3-glucan, Citr citrulline, Cyst cysteate, Arg-S Arginine-succinate, Fum fumarate, GlcNac N-acetyl-D-glucosamine, Glu glutamate, Gln glutamine, Glyo glyoxylate, Mal malate, G3P glyceraldehyde-3-phosphate, Gro3P glycerol-3-phosphate, 2-OG oxoglutarate, OAA oxaloacetate, PEP phosphoenolpyruvate, Orn ornithine, Pyr pyruvate, PGA phosphoglycerate, Put putrescine, Spe spermidine.
Early-stage transcript enrichment was also observed for genes mediating nitrate assimilation and conversion to ammonium. Glutamate is a central component of biosynthesis pathways for amino acids and the assimilation pathway for ammonium, which serves as the nitrogen homeostasis mechanism. Compared to late phase T. pseudonana cells, glutamate synthesis genes were enriched in both the cytosol and mitochondrion (Fig. 3A, B and Table S4). Indeed, higher internal endometabolome concentrations of glutamate as well as other nitrogen-rich metabolites (amino acids proline, glycine, and aspartate; and nucleosides guanosine and uridine) were evident in the early growth stage diatoms (Fig. 2). T. pseudonana’s urea cycle coordinates cellular nitrogen and carbon status [55], and several genes in this cycle were enriched during early growth (Fig. 3D). These patterns are consistent with peak concentrations of nitrogen-containing amino acids and nucleosides in the early bloom, and together indicate higher nitrogen requirements by T. pseudonana during the biomass building phase.
Diatom transcripts in the late bloom phase were enriched instead with genes that synthesize malate and oxaloacetate, two metabolites of the C4 delivery pathway for CO2 that performs well under low CO2 concentrations (Fig. 3A). Whether C4 metabolism is functional in diatoms is controversial (e.g., refs [56–58]) but if so, it would be beneficial in cases where inorganic carbon concentrations decrease in late phase blooms. Late-phase enriched transcripts were also found in pathways for putrescine and spermidine synthesis. These polyamines have several known functions in diatom cells, among them a role in stress response [59]. In field studies, seawater concentrations of polyamines increased during diatom bloom decline [60]. Evidence for physiological stress on late bloom diatoms also included increased expression of genes for synthesis of the osmolyte DMSP (Fig. 3); the diatom pathways for synthesizing osmolytes glycine betaine and homarine are unknown.
Genes related to sulfate assimilation and subsequent synthesis of the organic sulfur compounds cysteate, sulfopyruvate, sulfolactate, and DHPS, in addition to DMSP mentioned above, were highly enriched in the later stage cultures, with up to 100-fold increases in relative expression (Fig. 3B and Table S4). These data are congruent with higher DMSP and DHPS endometabolite concentrations in late- compared to early-stage diatom cells (Fig. 2). The temporal switch from greater investments in synthesis of nitrogen-rich compounds in the early-stage bloom to organic sulfur compounds in the late stage could reflect the higher energy costs for nitrate assimilation compared to sulfate assimilation (41 versus 33 ATP per mol assimilated; [61]). Similar to nitrate transformation to nitrite and ammonium, sulfate transformation to cysteate also consumes NADPH, and thus removes excess reductants produced during photosynthesis [62]. Genes for cysteate catabolism, believed to be an intermediate in DHPS formation [50], were significantly enriched during the late growth phase. Relatively higher sulfate usage as compared to nitrogen in aging diatom cultures or during bloom senescence is generally associated with nitrate limitation [62], but here we observed this even under nitrate replete conditions, suggesting a physiological response by the cells as their growth rate slowed. Giordano and Raven [61] suggest that low early ocean sulfate concentrations may have influenced the evolution of marine phytoplankton, manifested in divergent present-day strategies for regulation of nitrogen versus sulfur metabolism.
Bacterial gene expression
The early versus late bloom differences evident in the diatom metabolome and transcriptome set the stage for responses by associated heterotrophic bacteria in their substrate acquisition patterns. The substrates available to the co-cultured bacteria were inferred from significant differences in relative expression of genes diagnostic for uptake of organic matter (i.e., those mediating transport or initial catabolism of exogenous molecules). We focus on R. pomeroyi and P. dokdonensis in this analysis as Stenotrophomonas sp. was not present in the co-cultures at day 15 (Fig. 1B).
Expression patterns of R. pomeroyi genes suggested higher relative availability of taurine, glycerol-3-phosphate, lactate, DHPS, putrescine, alanine, and glycine betaine in the early bloom; and of N-acetyltaurine, glycine, choline, spermidine, glycolate, trimethylamine, and sugars during late bloom (Tables 1 and S5). Transcription patterns suggested that DMSP was available at both time points but degraded by different pathways in the early (demethylation pathway) versus late (cleavage pathway) bloom [63], a temporal switch in pathway dominance consistent with previous studies in cultures [26, 64] and in a natural phytoplankton bloom [65], and potentially driven by differences in reactive oxygen stress between pathways [65]. Enriched bacterial gene expression for spermidine transport coincided with enriched diatom gene expression for its biosynthesis. Overall, however, there were few positive relationships between shifts in R. pomeroyi gene expression for uptake of a metabolite and shifts in phytoplankton intracellular concentration or biosynthetic gene expression for that same metabolite. For example, of eleven metabolites found in the diatom endometabolome and for which the R. pomeroyi transporters are known, we found that glycine betaine, DHPS, and glycerol-3-phosphate had higher concentrations in phytoplankton cells on day 15 but their transporters were enriched in the bacterial transcriptome on day 3; glycine had higher concentrations in phytoplankton cells on day 3 but the transporter was enriched on day 15; alanine and acetate did not differ in concentration between early versus late bloom phytoplankton cells but their transporters were enriched on day 3 and 15, respectively; and proline, aspartate, glucose, glutamate, and phosphorylcholine were either more abundant in day 3 or day 15 phytoplankton cells but their transporter expression in R. pomeroyi was not different. Assuming bacterial transporter expression is induced primarily by substrate availability [66], these mismatches argue for a minor role for passive leakage in metabolite release and instead support active release mechanisms that cannot be predicted from endometabolite concentrations. An additional ten metabolites detected in the phytoplankton metabolome do not have confirmed transporters in the R. pomeroyi genome: lysine, glutamine, arginine, valine, leucine, isoleucine, homarine, guanosine, uridine, and β-1,3-glucan.
Table 1.
Genes from R. pomeroyi (top) and P. dokdonensis (bottom) with relevance to diatom endometabolome composition and with significantly higher relative expression (p ≤ 0.05) during early (left) or late (right) diatom growth phase.
| Early growth phase | Late growth phase | ||
|---|---|---|---|
| Function | Locus tag | Function | Locus tag |
| Ruegeria pomeroyi | |||
| Glycine betaine transporter | SPO2441 | N-acetyltaurine transporter | SPO0661 |
| DMSP – demethylation | SPO0677, 3804 3805 | DMSP – cleavage | SPO1703 |
| Taurine transporter | SPO0674, 0676 | Glycine | SPOA0310 |
| Putrescine transporter | SPO3473, 3474 | Choline | SPO1083, 0084 |
| DHPS degradation | SPO0158 | Spermidine transporter | SPO3467 |
| Lactic acid transporter | SPO1017-1021 | Glycolate | SPOA0143 |
| Glycerol-3-phosphate transporter | SPO0239, 0240 | Glycerol transporter | SPO0608-0612 |
| Alanine symporter | SPO2370 | Trimethylamine transporter | SPO1551, 1552, 1562 |
| Amino acid transporter | SPO1516 | Sugar transporter | SPO0608-0612 |
| Acetate | SPO2963 | ||
| Polaribacter dokdonensis | |||
| Hypothetical protein PUL2 | MED152_00445 | MFS transporter (fucose) PUL3 | MED152_05095 |
| Glycosyl hydrolase family 17 PUL2 | MED152_00450 | MFS transporter (fucose) PUL3 | MED152_05115 |
| O-glycosyl hydrolase family 30 PUL2 | MED152_00455 | MFS transporter (maltose) PUL3 | MED152_05120 |
| MFS transporter (glycoside etc. family) PUL2 | MED152_00460 | AraC family transcriptional regulator PUL3 | MED152_05185 |
| Choline dehydrogenase PUL3 | MED152_05190 | Sugar kinase PUL3 | MED152_05080 |
| Sugar transporter (glucose/galactose) PUL5 | MED152_08460 | Alginate lyase | MED152_06195 |
| DNA-binding response regulator PUL5 | MED152_08465 | Sugar transporter | MED152_09090 |
| Histidine kinase PUL5 | MED152_08470 | Polysaccharide transporter | MED152_03130 |
| SusC/RagA family TonB-linked outer membrane protein PUL5 | MED152_08475 | Glyceraldehyde-3-P dehydrogenase | MED152_13444 |
| SusD/RagB family nutrient binding outer membrane lipoprotein PUL5 | MED152_08480 | ||
| Glucosamine-6-phosphate deaminase PUL5 | MED152_08485 | ||
| CAZyme; beta-N-acetylhexosaminidase (GH20) PUL5 | MED152_08490 | ||
| N-acetylglucosaminase kinase (GlcNac) PUL5 | MED152_08495 | ||
| Glycosyl hydrolase 81 (GH81) | MED152_00755 | ||
| Glycogen synthase (GT4) | MED152_05855 | ||
| Glycine cleavage | MED152_06700 | ||
| 1,4-α-glucan branching enzyme | MED152_05875 | ||
| Glyceraldehyde-3-P dehydrogenase | MED152_03295 | ||
PUL2Chrysolaminarin uptake; PUL3Unknown/multiple function; PUL5N-acetyl-D-glucosamine uptake.
Flavobacteriia member P. dokdonensis increased in abundance through time in the co-cultures, as has been observed for marine flavobacteria in natural diatom bloom progression [19]. This pattern has been attributed to specialization by this taxon for glycan utilization, release of which increases in aging phytoplankton cells [67]. Gene expression patterns by P. dokdonensis were also consistent with increasing glycan importance in the late bloom, particularly for genes active in polysaccharide utilization loci (PULs) [68]. These genomic regions enable hydrolysis of polysaccharides to monomers, which are transported into the periplasm and subsequently transported across the cell membrane. Several genes within PUL3 (as designated in the CAZy database; Taxonomy ID 313598 [14, 69]) were significantly enriched during the late growth phase, including two annotated as fucose permeases (Table 1 and S6). Polaribacter species were shown to peak concurrently with fucosidase activity in a natural bloom [19], indicating an association with fucose degradation within the genus. Genes in PUL5 hypothesized to transport and degrade chitin subunits [14] exhibited up to 40-fold enrichment in late bloom P. dokdonensis transcriptomes. Chitin is a common organic molecule in the ocean [70–72], and serves as component of cell walls and a locus for bacterial attachment in diatoms [73, 74].
In addition to utilization of structural polysaccharides, gene expression in P. dokdonensis PUL2 suggested utilization of the storage polysaccharide chrysolaminarin ([14]; Table 1). This parallels the high transcriptional signal and endometabolite concentration for the β-1,3-glucan building block of chrysolaminarin in early-stage T. pseudonana (Fig. 3B and Table S4) [75, 76]. Phytoplankton were recently shown to exhibit daily cycles of internal chrysolaminarin concentrations synchronized with diel light patterns [17, 76], with this storage polysaccharide comprising up to 80% of diatom carbon under certain conditions [75]. The utilization of both structural and storage polysaccharides represented a distinct niche for P. dokdonensis in the T. pseudonana co-cultures. As found previously [14], R. pomeroyi and P. dokdonensis transcriptomes indicated little overlap in resource use, with each species responding to different suites of organic compounds in early and late bloom stages.
There was temporal matching of peaks in diatom endometabolite concentrations and P. dokdonensis gene expression for early bloom peaks of proline, glucose, and β-1,3-glucan. Of these, proline is a candidate for passive diffusion because of its small size, but the higher molecular weight metabolites glucose and β-1,3-glucan are less likely to diffuse through the diatom membrane, suggesting instead a link to export for physiological balance [18, 23, 25, 77]. Previous studies have found glucose release from diatoms when carbon fixation rates are high [78], likely due to photosynthetic overflow pathways. Yet the poor match between peaks in diatom endometabolites and peaks in both P. dokdonensis and R. pomeroyi transcription indicate that endometabolite abundance cannot predict the release of metabolites into surrounding seawater (Table S2), and argues for a key role for active (i.e., non-diffusive) mechanisms of diatom metabolite release. Also of interest are the six metabolites (arginine, DHPS, glycerol-3-phosphate, glutamate, glucose, and leucine) with higher concentrations in phytoplankton cells in co-culture compared to axenic culture (Fig. 2). Although the mechanism is unclear, bacteria appeared to directly or indirectly affect the internal concentrations of these metabolites.
Community vs. individual bacterial gene expression
These same three bacterial species were co-cultured individually with T. pseudonana in a prior study [14], providing the opportunity to explore gene expression as a member of a bacterial community versus when alone. Several conditions differed between the studies; in Ferrer-González et al. [14], exometabolites built up in axenic T. pseudonana cultures for 7 days prior to inoculation of the bacteria, followed by sampling after 8 h. In the present study, the diatom and bacteria were inoculated at the same time, with sampling after 3 d. In both studies, however, the diatom was in exponential growth at the time of bacterial sampling. Significantly enriched bacterial genes were identified based on a reference dataset collected from bacteria growing in a glucose medium. Of the 24 enriched transporters in R. pomeroyi, 12 were enriched only in the individual co-cultures, including those with experimentally-verified substrates N-acetyltaurine and urea; and four were enriched only in the community co-cultures, including those with experimentally-verified substrates choline, acetate, and DMSP (Table S7). There was no difference in distribution of the three dominant compound classes represented by the transporters (organic acids, amino acids, and organic sulfur compounds), nor in the distribution of nitrogenous compound transporters (Table S7). For P. dokdonensis, the enrichment of genes in PULs 2, 3, 5, 6, and 7 seen in this study matched the Ferrer-González study [14]. However, PUL4 (no annotation available) had only one enriched gene matching and PUL8 (annotated for siderophore uptake and potentially colicin toxins [79]) had none (Table S8). Because the study designs differed in factors other than just the number of bacterial species, these comparisons are viewed as preliminary. Nonetheless, they suggest differences in uptake when bacteria are members of a community, with potential mechanisms including altered metabolite release by the diatom, or antagonism or competition for resources by the bacteria.
Conclusions
The interaction between phytoplankton and bacteria in the surface ocean represents a central biogeochemical relationship that is relevant at the global scale. In a model bloom experiment with the marine diatom T. pseudonana providing the only source of organic matter to a heterotrophic bacterial community, we characterized potential substrates and considered their mechanisms of release. The diatom’s endometabolome was dynamic, differed with bloom stage, and was affected by the presence of bacteria. The diatom’s transcriptome similarly differed between bloom stages, changing from a composition enabling carbon fixation and nitrogen metabolism regulation to one focused on organic sulfur compound synthesis. Bacterial transporter expression suggested that metabolite availability differed between early and late bloom, and that the bacterial species maintained distinct resource niches through the bloom. The dynamics of bacterial uptake system expression matched the dynamics of endometabolite concentrations in just a few cases, specifically for proline, glucose, and β-1,3-glucan. The majority of molecules, however, did not have synchronous patterns, suggesting complex interaction scenarios that reflect both phytoplankton physiology and bacterial influence.
Supplementary information
Acknowledgements
The authors thank S. Sharma and B. Nowinski for bioinformatic advice, B. Durham for sharing expertise on the T. pseudonana genome, and J. Nelson at the UGA CTEGD Cytometry Resource for flow cytometry expertise. This work was supported by the Swedish Research Council grant 2018-06571 to MO, National Science Foundation grants OCE-1948104 and OCE-2019589 to MAM and ASE, and by the Simons Foundation grant 542391 within the Principles of Microbial Ecosystems (PriME) to MAM. This is publication #004 of the NSF Center for Chemical Currencies of a Microbial Planet.
Author contributions
MO, MAM, and ASE conceptualized the study; MO, FXF-G, JES, and CBS conducted the experiment; MU and NRH carried out metabolite analysis; and MO and MAM wrote the manuscript with input from all authors.
Data availability
Transcriptome data and associated metadata are deposited at NCBI under BioProject ID PRJNA758094. Metabolomics data and associated sample preparation protocols and NMR analysis and processing parameters are deposited at the Metabolomics Workbench Data Repository under Project ID 001231 (10.21228/M8KT3K). Additional data products and metadata are available at GitHub (10.5281/zenodo.6344452).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s43705-022-00116-5.
References
- 1.Martin A. The seasonal smorgasbord of the seas. Science. 2012;337:46–47. doi: 10.1126/science.1223881. [DOI] [PubMed] [Google Scholar]
- 2.Buchan A, LeCleir GR, Gulvik CA, González JM. Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat Rev Microbiol. 2014;12:686–98. doi: 10.1038/nrmicro3326. [DOI] [PubMed] [Google Scholar]
- 3.Amin SA, Hmelo LR, van Tol HM, Durham BP, Carlson LT, Heal KR, et al. Interaction and signaling between a cosmopolitan phytoplankton and associated bacteria. Nature. 2015;522:98–101. doi: 10.1038/nature14488. [DOI] [PubMed] [Google Scholar]
- 4.Fuhrman JA, Ferguson RL. Nanomolar concentrations and rapid turnover of dissolved free amino acids in seawater: agreement between chemical and microbiological measurements. Mar Ecol Prog Ser. 1986;33:237–42. doi: 10.3354/meps033237. [DOI] [Google Scholar]
- 5.Azam F, Fenchel T, Field JG, Gray JS, Meyer-Reil LA, Thingstad F. The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser. 1983;10:257–63. doi: 10.3354/meps010257. [DOI] [Google Scholar]
- 6.Cole JJ, Findlay S, Pace ML. Bacterial production in fresh and saltwater ecosystems: a cross-system overview. Mar Ecol Prog Ser. 1988;43:1–10. doi: 10.3354/meps043001. [DOI] [Google Scholar]
- 7.Moran MA, Kujawinski EB, Stubbins A, Fatland R, Aluwhihare LI, Buchan A, et al. Deciphering ocean carbon in a changing world. Proc Natl Acad Sci USA. 2016;113:3143–51. doi: 10.1073/pnas.1514645113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker JW, Berube PM, Follett CL, Waterbury JB, Chisholm SW, DeLong EF, et al. Closely related phytoplankton species produce similar suites of dissolved organic matter. Front Microbiol. 2014;5:111. doi: 10.3389/fmicb.2014.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouslimani A, Sanchez LM, Garg N, Dorrestein PC. Mass spectrometry of natural products: current, emerging and future technologies. Nat Prod Rep. 2014;31:718–29. doi: 10.1039/c4np00044g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson WM, Longnecker K, Kido Soule MC, Arnold WA, Bhatia MP, Hallam SJ, et al. Metabolite composition of sinking particles differs from surface suspended particles across a latitudinal transect in the South Atlantic. Limnol Oceanogr. 2019;65:111–27. doi: 10.1002/lno.11255. [DOI] [Google Scholar]
- 11.McCarren J, Becker JW, Repeta DJ, Shi Y, Young CR, Malmstrom RR, et al. Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc Natl Acad Sci USA. 2010;107:16420–7. doi: 10.1073/pnas.1010732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi Y, Tyson GW, Eppley JM, DeLong EF. Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J. 2011;5:999–1013. doi: 10.1038/ismej.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durham BP, Sharma S, Luo H, Smith CB, Amin SA, Bender SJ, et al. Cryptic carbon and sulfur cycling between surface ocean plankton. Proc Natl Acad Sci USA. 2015;12:453–7. doi: 10.1073/pnas.1413137112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrer-González FX, Widner B, Holderman NR, Glushka J, Edison AS, Kujawinski EB, et al. Resource partitioning of phytoplankton metabolites that support bacterial heterotrophy. ISME J. 2021;15:762–73. doi: 10.1038/s41396-020-00811-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Obernosterer I, Herndl GJ. Phytoplankton extracellular release and bacterial growth: dependence on the inorganic N:P ratio. Mar Ecol Progr Ser. 1995;116:247–57. doi: 10.3354/meps116247. [DOI] [Google Scholar]
- 16.Mühlenbruch M, Grossart HP, Eigemann F, Voss M. Mini‐review: Phytoplankton‐derived polysaccharides in the marine environment and their interactions with heterotrophic bacteria. Environ Microbiol. 2018;20:2671–85. doi: 10.1111/1462-2920.14302. [DOI] [PubMed] [Google Scholar]
- 17.Uchimiya M, Schroer W, Olofsson, M, Edison AS, Moran, MA. Diel investments in metabolite production and consumption in a model microbial system. ISME J. 2021. 10.1038/s41396-021-01172-w. [DOI] [PMC free article] [PubMed]
- 18.Myklestad S, Holm-Hansen O, Vårum KM, Volcani BE. Rate of release of extracellular amino acids and carbohydrates from the marine diatom Chaetoceros affinis. J Plankton Res. 1989;11:763–73. doi: 10.1093/plankt/11.4.763. [DOI] [Google Scholar]
- 19.Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM, et al. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science. 2012;336:608–11. doi: 10.1126/science.1218344. [DOI] [PubMed] [Google Scholar]
- 20.Biddanda B, Benner R. Carbon, nitrogen, and carbohydrate fluxes during the production of particulate and dissolved organic matter by marine phytoplankton. Limnol Oceanogr. 1997;42:506–18. doi: 10.4319/lo.1997.42.3.0506. [DOI] [Google Scholar]
- 21.Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Microbiol. 2007;5:782–91. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- 22.Thornton DC. Dissolved organic matter (DOM) release by phytoplankton in the contemporary and future ocean. Eur J Phycol. 2014;49:20–46. doi: 10.1080/09670262.2013.875596. [DOI] [Google Scholar]
- 23.Bjørnsen PK. Phytoplankton exudation of organic matter: why do healthy cells do it? Limnol Oceanogr. 1988;33:151–4. doi: 10.4319/lo.1988.33.1.0151. [DOI] [Google Scholar]
- 24.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol R. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fogg GE. The ecological significance of extracellular products of phytoplankton photosynthesis. Bot Mar. 1983;26:3–14.. doi: 10.1515/botm.1983.26.1.3. [DOI] [Google Scholar]
- 26.Durham BP, Dearth SP, Sharma S, Amin SA, Smith CB, Campagna SR, et al. Recognition cascade and metabolite transfer in a marine bacteria-phytoplankton model system. Environ Microbiol. 2017;19:3500–13. doi: 10.1111/1462-2920.13834. [DOI] [PubMed] [Google Scholar]
- 27.Miller TR, Hnilicka K, Dziedzic A, Desplats P, Belas R. Chemotaxis of Silicibacter sp. strain TM1040 toward dinoflagellate products. Appl Environ Microbiol. 2004;70:4692–701. doi: 10.1128/AEM.70.8.4692-4701.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seymour JR, Simó R, Ahmed T, Stocker R. Chemoattraction to dimethylsulfoniopropionate thorughout the marine microbial food web. Science. 2010;329:342–5. doi: 10.1126/science.1188418. [DOI] [PubMed] [Google Scholar]
- 29.Luo H, Moran MA. How do divergent ecological strategies emerge among marine bacterioplankton lineages? Trends Microbiol. 2015;23:577–84. doi: 10.1016/j.tim.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 30.Bergkvist J, Klawonn I, Whitehouse MJ, Lavik G, Brüchert V, Ploug H. Turbulence simultaneously stimulates small- and large-scale CO2 sequestration by chain-forming diatoms in the sea. Nat Commun. 2018;9:3046. doi: 10.1038/s41467-018-05149-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalez JM, Covert JS, Whitman WB, Henriksen JR, Mayer F, Scharf B, et al. Silicibacter pomeroyi sp. nov. and Roseovarius nubinhibens sp. nov., dimethylsulfoniopropionate-demethylating bacteria from marine environments. Int J Syst Evol Micr. 2003;53:1261–9. doi: 10.1099/ijs.0.02491-0. [DOI] [PubMed] [Google Scholar]
- 32.Hagström Å, Pinhassi J, Zweifel UL. Biogeographical diversity among marine bacterioplankton. Aquat Microb Ecol. 2000;21:231–44. doi: 10.3354/ame021231. [DOI] [Google Scholar]
- 33.González JM, Fernández-Gómez B, Fernández-Guerra A, Gómez-Consarnau L, Sánchez O, Coll-Lladó M, et al. Genome analyses of the proteorhodopsin-containing marine bacterium Polaribacter sp. MED152 (Flavobacteria) Proc Natl Acad Sci USA. 2008;105:872–9. doi: 10.1073/pnas.0712027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alavi M, Miller T, Erlandson K, Schneider R, Belas R. Bacterial community associated with Pfiesteria‐like dinoflagellate cultures. Environ Microbiol. 2001;3:380–96. doi: 10.1046/j.1462-2920.2001.00207.x. [DOI] [PubMed] [Google Scholar]
- 35.Behringer G, Ochsenkühn MA, Fei C, Fanning J, Koester JA, Amin SA. Bacterial communities of diatoms display strong conservation across strains and time. Front Microbiol. 2018;9:659. doi: 10.3389/fmicb.2018.00659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Green DH, Llewellyn LE, Negri AP, Blackburn SI, Bolch CJ. Phylogenetic and functional diversity of the cultivable bacterial community associated with the paralytic shellfish poisoning dinoflagellate Gymnodinium catenatum. FEMS Microbiol Ecol. 2004;47:345–57. doi: 10.1016/S0168-6496(03)00298-8. [DOI] [PubMed] [Google Scholar]
- 37.Hold GL, Smith EA, Rappë MS, Maas EW, Moore ER, Stroempl C, et al. Characterisation of bacterial communities associated with toxic and non-toxic dinoflagellates: Alexandrium spp. and Scrippsiella trochoidea. FEMS Microbiol Ecol. 2001;37:161–73. doi: 10.1111/j.1574-6941.2001.tb00864.x. [DOI] [Google Scholar]
- 38.Guillard RRL, Hargraves PE. Stichochrysis immobilis is a diatom, not a chrysophyte. Phycologia. 1993;32:234–6. doi: 10.2216/i0031-8884-32-3-234.1. [DOI] [Google Scholar]
- 39.Harrison PJ, Waters RE, Taylor FJR. A broad spectrum artificial sea water medium for coastal and open ocean phytoplankton. J Phycol. 1980;16:28–35. [Google Scholar]
- 40.Uchimiya M, Tsuboi Y, Ito K, Date Y, Kikuchi J. Bacterial substrate transformation tracked by stable-isotope-guided NMR metabolomics: application in a natural aquatic microbial community. Metabolites. 2017;7:52. doi: 10.3390/metabo7040052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis IA, Schommer SC, Markley JL. rNMR: open source software for identifying and quantifying metabolites in NMR spectra. Magn Reson Chem. 2009;47:S123–S126. doi: 10.1002/mrc.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, et al. BioMagResBank. Nucleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vázquez-Fresno R, et al. HMDB 4.0 — the human metabolome database for 2018. Nucleic Acids Res. 2018;4:D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boroujerdi AFB, Lee PA, DiTullio GR, Janech MG, Vied SB, Bearden DW. Identification of isethionic acid and other small molecule metabolites of Fragilariopsis cylindrus with nuclear magnetic resonance. Anal Bioanal Chem. 201;404:777-84. [DOI] [PubMed]
- 45.Karp PD, Billington R, Caspi R, Fulcher CA, Latendresse M, Kothari A, et al. The BioCyc collection of microbial genomes and metabolic pathways. Brief Bioinf. 2019;20:1085–93. doi: 10.1093/bib/bbx085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.PhyloDB 1.076. https://drive.google.com/drive/u/0/folders/0B-BsLZUMHrDQfldGeDRIUHNZMEREY0g3ekpEZFhrTDlQSjQtbm5heC1QX2V6TUxBeFlOejQ?resourcekey=0-UEs0-cDRRm46O7ExECdI_g. Assessed September 2021.
- 47.APHA. Standard methods for the examination of water and wastewater. 21st edn. Washington DC: American Public Health Association; 2005).
- 48.APHA. Standard methods for the examination of water and wastewater. 15th edn. Washington DC: American Public Health Association; 1981.
- 49.Koroleff F. Determination of silicon. In: Grasshoff K, Ehrhardt M, Kremling K, editors. Methods of seawater analysis. Weinheim, Germany: Verlag Chemie; 1983. p. 174–87.
- 50.Durham BP, Boysen AK, Carlson LT, Groussman RD, Heal KR, Cain KR, et al. Sulfonate-based networks between eukaryotic phytoplankton and heterotrophic bacteria in the surface ocean. Nat Microbiol. 2019;4:1706–15. doi: 10.1038/s41564-019-0507-5. [DOI] [PubMed] [Google Scholar]
- 51.Torstensson A, Young JN, Carlson LT, Ingalls AE, Deming JW. Use of exogenous glycine betaine and its precursor choline as osmoprotectants in Antarctic sea-ice diatoms. J Phycol. 2019;55:663–75. doi: 10.1111/jpy.12839. [DOI] [PubMed] [Google Scholar]
- 52.Bucciarelli E, Sunda WG. Influence of CO2, nitrate, phosphate, and silicate limitation on intracellular dimethylsulfoniopropionate in batch cultures of the coastal diatom Thalassiosira pseudonana. Limnol Oceanogr. 2003;48:2256–65. doi: 10.4319/lo.2003.48.6.2256. [DOI] [Google Scholar]
- 53.Heal KR, Durham BP, Boysen AK, Carlson LT, Qin W, Ribalet F, et al. Marine community metabolomes carry fingerprints of phytoplankton community composition. mSystems. 2021;6:e01334–20. doi: 10.1128/mSystems.01334-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olofsson M, Robertson EK, Edler L, Arneborg L, Whitehouse MJ, Ploug H. Nitrate and ammonium fluxes to diatoms and dinoflagellates at a single cell level in mixed field communities in the sea. Sci Rep. 2019;9:1424. doi: 10.1038/s41598-018-38059-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Allen AE, Dupont CL, Oborník M, Horák A, Nunes-Nesi A, McCrow JP, et al. Evolution and metabolic significance of the urea cycle in photosynthetic diatoms. Nature. 2011;473:203–7. doi: 10.1038/nature10074. [DOI] [PubMed] [Google Scholar]
- 56.Riebesell U. Carbon fix for a diatom. Nature. 2000;407:959–60. doi: 10.1038/35039665. [DOI] [PubMed] [Google Scholar]
- 57.Kutska AB, Milligan AJ, Zheng H, New AM, Gates C, Bidle KD, et al. Low CO2 results in a rearrangement of carbon metabolism to support C4 photosynthetic carbon assimilation in Thalassiosira pseudonana. New Phytol. 2014;204:507–20. doi: 10.1111/nph.12926. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka R, Kikutani S, Mahardika A, Matsuda Y. Localization of enzymes relating to C4 organic acid metabolisms in the marine diatom, Thalassiosira pseudonana. Photosynth Res. 2014;121:251–63. doi: 10.1007/s11120-014-9968-9. [DOI] [PubMed] [Google Scholar]
- 59.Lin H-Y, Lin H-J. Polyamines in microalgae: something borrowed, something new. Mar Drugs. 2018;17:1. doi: 10.3390/md17010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu Y, Zhao W, Li C, Miao H. Free polyamine content during algal bloom succession in the East China Sea in spring 2010. Chin J Ocean Limnol. 2017;35:215–23. doi: 10.1007/s00343-016-5089-y. [DOI] [Google Scholar]
- 61.Giordano M, Raven JA. Nitrogen and sulfur assimilation in plants and algae. Aquat Bot. 2014;118:45–61. doi: 10.1016/j.aquabot.2014.06.012. [DOI] [Google Scholar]
- 62.van Tol HM, Armbrust EV. Genome-scale metabolic model of the diatom Thalassiosira pseudonana highlights the importance of nitrogen and sulfur metabolism in redox balance. PLoS ONE. 2021;16:e0241960. doi: 10.1371/journal.pone.0241960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moran MA, Reisch CR, Kiene RP, Whitman WB. Genomic insights into bacterial DMSP transformations. Ann Rev Mar Sci. 2012;4:523–42. doi: 10.1146/annurev-marine-120710-100827. [DOI] [PubMed] [Google Scholar]
- 64.Landa M, Burns AS, Roth SJ, Moran MA. Bacterial transcriptome remodeling during sequential co-culture with a marine dinoflagellate and diatom. ISME J. 2017;11:2677–90. doi: 10.1038/ismej.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Varaljay VA, Robidart J, Preston CM, Gifford SM, Durham BP, Burns AS, et al. Single-taxon field measurements of bacterial gene regulation controlling DMSP fate. ISME J. 2015;9:1677–86. doi: 10.1038/ismej.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nowinski B, Moran MA. Niche dimensions of a marine bacterium are identified using invasion studies in coastal seawater. Nat Microbiol. 2021;6:524–32. doi: 10.1038/s41564-020-00851-2. [DOI] [PubMed] [Google Scholar]
- 67.Passow U. Transparent exopolymer particles (TEP) in aquatic environments. Prog Oceanogr. 2002;55:287–333. doi: 10.1016/S0079-6611(02)00138-6. [DOI] [Google Scholar]
- 68.Grondin JM, Tamura K, Déjean G, Abbott DW, Brumer H. Polysaccharide utilization loci: fueling microbial communities. J Bacteriol. 2017;199:e00860–16. doi: 10.1128/JB.00860-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–5. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aluwihare LI, Repeta DJ, Pantoja S, Johnson CG. Two chemically distinct pools of organic nitrogen accumulate in the ocean. Science. 2005;308:1007–10. doi: 10.1126/science.1108925. [DOI] [PubMed] [Google Scholar]
- 71.Gooday GW The ecology of chitin degradation. In: Marshall KC, editor. Advances in microbial ecology, vol. 11. New York: Plenum Press, Inc.; 1990). p. 387–429.
- 72.Jeuniaux C, Voss-Foucart MF. Chitin biomass and production in the marine environment. Biochem Syst Ecol. 1991;19:347–56. doi: 10.1016/0305-1978(91)90051-Z. [DOI] [Google Scholar]
- 73.Frischkorn KR, Stojanovski A, Paranjpye R. Vibrio parahaemolyticus type IV pili mediate interactions with diatom-derived chitin and point to an unexplored mechanism of environmental persistence. Environ Microbiol. 2013;15:1416–27. doi: 10.1111/1462-2920.12093. [DOI] [PubMed] [Google Scholar]
- 74.Li Y, Lei X, Zhu H, Zhang H, Guan C, Chen Z, et al. Chitinase producing bacteria with direct algicidal activity on marine diatoms. Sci Rep. 2016;6:21984. doi: 10.1038/srep21984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hildebrand M, Manandhar-Shrestha K, Abbriano R. Effects of chrysolaminarin synthase knockdown in the diatom Thalassiosira pseudonana: Implications of reduced carbohydrate storage relative to green algae. Algal Res. 2017;23:66–77. doi: 10.1016/j.algal.2017.01.010. [DOI] [Google Scholar]
- 76.Becker S, Tebben J, Coffinet S, Wiltshire K, Iversen MH, Harder T, et al. Laminarin is a major molecule in the marine carbon cycle. Proc Natl Acad Sci USA. 2020;117:6599–607. doi: 10.1073/pnas.1917001117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carlson CA, Hansell DA. DOM sources, sinks, reactivity, and budgets. In: Hansell DA, Carlson CA, editors. Biogeochemistry of marine dissolved organic matter. 3rd edn. Boston: Academic Press; 2015. p. 65–126.
- 78.Urbani R, Magaletti E, Sist P, Cicero AM. Extracellular carbohydrates releeased by the marine diatoms Cylindrotheca closerium, Thalassiosira pseudonana and Skeletonema costatum: effect of P-depletion and growth status. Sci Total Environ. 2005;353:300–6. doi: 10.1016/j.scitotenv.2005.09.026. [DOI] [PubMed] [Google Scholar]
- 79.Buchanan SK, Lukacik P, Grizot S, Ghirlando R, Ali MM, Barnard TJ. Structure of colicin I receptor bound to the R‐domain of colicin Ia: implications for protein import. EMBO J. 2007;26:2594–604. doi: 10.1038/sj.emboj.7601693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Transcriptome data and associated metadata are deposited at NCBI under BioProject ID PRJNA758094. Metabolomics data and associated sample preparation protocols and NMR analysis and processing parameters are deposited at the Metabolomics Workbench Data Repository under Project ID 001231 (10.21228/M8KT3K). Additional data products and metadata are available at GitHub (10.5281/zenodo.6344452).