Abstract
The fate of oceanic carbon and nutrients depends on interactions between viruses, prokaryotes, and unicellular eukaryotes (protists) in a highly interconnected planktonic food web. To date, few controlled mechanistic studies of these interactions exist, and where they do, they are largely pairwise, focusing either on viral infection (i.e., virocells) or protist predation. Here we studied population-level responses of Synechococcus cyanobacterial virocells (i.e., cyanovirocells) to the protist Oxyrrhis marina using transcriptomics, endo- and exo-metabolomics, photosynthetic efficiency measurements, and microscopy. Protist presence had no measurable impact on Synechococcus transcripts or endometabolites. The cyanovirocells alone had a smaller intracellular transcriptional and metabolic response than cyanovirocells co-cultured with protists, displaying known patterns of virus-mediated metabolic reprogramming while releasing diverse exometabolites during infection. When protists were added, several exometabolites disappeared, suggesting microbial consumption. In addition, the intracellular cyanovirocell impact was largest, with 4.5- and 10-fold more host transcripts and endometabolites, respectively, responding to protists, especially those involved in resource and energy production. Physiologically, photosynthetic efficiency also increased, and together with the transcriptomics and metabolomics findings suggest that cyanovirocell metabolic demand is highest when protists are present. These data illustrate cyanovirocell responses to protist presence that are not yet considered when linking microbial physiology to global-scale biogeochemical processes.
Subject terms: Bacteriophages, Water microbiology
Introduction
Microbes drive planetary nutrient and energy cycles, but they do so under bottom-up (resources) and top-down (predation and infection) constraints within intricate food webs. In the oceans, resource availability was historically thought to affect microbial biomass and composition more strongly than predation [1]. However, recent global ocean species interaction networks suggest that biotic factors are stronger predictors of microbial community structure than abiotic factors [2]. Predation rates are commonly on par with microbial growth in low-nutrient surface ocean waters. However, the fate of nutrients differs by predator [3–6], with the paradigm being that viruses “shunt” carbon (C) and nutrients toward the dissolved phase [7–10] or “pump” C to the deep ocean [11], and heterotrophic protists serve as a critical link between lower and higher trophic levels [12–14]. Yet, viral and protistan interplay in controlling prokaryote populations and nutrient cycling [15–17] remains poorly characterized, despite indications of potential synergism of viral and protist impacts on prey populations [16, 18–20] and viral influence on protist predation and growth [21, 22].
In nature, Synechococcus, which together with Prochlorococcus contributes about one-quarter of total global oceanic primary productivity and is predicted to become more abundant with continued climate change [23], is both preyed upon by protists and infected by viruses (phages). For protists, field data show that they reduce Synechococcus abundances via direct ingestion (grazing) [24], while laboratory experiments demonstrate that protist predation alters freshwater Synechococcus cell morphology [25–27] in ways that could impact photosynthesis or nutrient cycling. For viruses, virus-infected cells (virocells) are physiologically and metabolically distinct from uninfected cells and display unique metabolite footprints that could impact the surrounding ecosystem [28–33]. For cyanobacteria virocells (cyanovirocells) specifically, while the physiological response of Synechococcus to infection has been characterized via genome-wide transcriptomics [34, 35] and photophysiology [36–38], the broader cellular response (e.g., metabolomics) and how the presence of a protist impacts cyanovirocell metabolic reprogramming and ecosystem footprints have not been studied. Ecosystems are comprised of communities of simultaneously interacting organisms, and yet phage-protist-cyanobacteria interactions remain understudied.
Here we assess phage-protist-cyanobacteria interactions by examining how population-level marine Synechococcus virocell physiology is impacted by the presence of a protist. We use a systems biology approach including time-resolved genome-wide transcriptomics, endo- and exo-metabolomics, photosynthetic efficiency measurements, and microscopy in a culture-based model system. Together these efforts provide mechanistic grounding to interactions among T4-like phages, Synechococcus, and dinoflagellates implicated in microbial interaction networks that are highly predictive of global ocean carbon flux [39].
Materials and methods
Culture conditions
Using semi-batch culturing, axenic Synechococcus strain WH8102 was grown in SN media prepared with sterile 0.1 μm-filtered, autoclaved seawater obtained from Scripps Pier in January 2014. Cultures were maintained at 20 ± 1 °C on a light: dark cycle of 14 h:10 h at 60 μmol photons m−2 s−1. The protist Oxyrrhis marina (CCMP3375; NCMA), a dinoflagellate of size 10–20 μm, was grown in Scripps Pier (January 2014) seawater with 1 × 105 cells mL−1 Isochrysis galbana (CCMP1323; NCMA) as prey. Prey levels were depleted just prior to the experiment (see below).
The T4-like Myophage S-SSM5 (dsDNA genome of 176.18 Kbp) was propagated on axenic exponentially growing WH8102 within 14 days of beginning the combined phage infection and protist co-culture experiment, filtered through a 0.45-μm Supor filter (Pall Corp, cat. no. PN4614) and stored in the dark at 4 °C. Phage titer was determined by the most probable number assay [40, 41] using three independent dilution series on WH8102 grown in 96-well plates under the growth conditions described above. Preliminary viral infection dynamics were assessed using exponentially growing WH8102 at 60 μmol photons m−2 s−1 with a light: dark cycle of 14 h:10 h.
Sampling
Twenty liters of exponentially growing Synechococcus strain WH8102 were aseptically pooled and split into four treatments: cyanobacteria only, cyanobacteria with phage, cyanobacteria with protist, and cyanobacteria with both phage and protist, with four biological replicates of each (each 1.2 L volume). Phage infection was initiated at the onset of the light cycle in eight separate flasks, each with 3 × 107 pfu mL−1 (phage) incubated with 2.8 × 107 host cells mL−1 (+phage treatment average) and 2.9 × 107 host cells mL−1 (+phage and protist treatment average) for a multiplicity of infection of 1.1. Encounter theory estimates based on the infectious phage to host cell numbers present in the infection incubation predict that 67% of WH8102 cells would be phage-infected. After 1 h, cells were centrifuged and the supernatant containing free phages was removed, cells were washed and resuspended in spent culture medium (same for the cyanobacterium-only and cyanobacteria with protist treatments), and these samples were counted via flow cytometry. Synechococcus sp. starting concentrations were 1.9 ± 0.3 × 107, 1.7 ± 0.1 × 107, 2.2 ± 0.1 × 107, and 1.7 ± 0.3 × 107 cells mL−1 in the untreated control cyanobacteria only, phage infection, protist co-culture, and combined phage and protist treatments, respectively (Ave ± SD). In addition, two 1.2 L replicates of a 0.45-μm-filtered medium served as a control for exometabolite analyses.
Protist Oxyrrhis marina (CCMP3375) cells were allowed to reduce Isochrysis sp. CCMP1323 maintenance prey concentrations over 3 days. CCMP3375 was concentrated 1 h prior to the experiment via gentle gravity filtration over 10 μm pore size polyethersulfone membrane filters (PES, Sterlitech Corp. cat. no. PES 8025100), with continual flushing of sterile (0.22-μm-filtered and autoclaved) seawater. O. marina CCMP3375 was added to both cyanobacteria and cyanobacteria with phage treatments at a final concentration of 43 cells mL−1 and final protest: Synechococcus ratio of 1:512,000 and 1:395,000, respectively, which marked the start of the experiment. All treatments were maintained at 20 ± 1 °C under 60 μmol photons m−2 s−1.
Samples (transcriptomics, metabolomics, flow cytometry, microscopy, photosynthetic efficiency) were collected between 2 and 12 h every ~2 h during the daylight portion of the light: dark cycle, by gently mixing and pouring 100 mL into a sterile 250 mL polycarbonate flask. Cell and phage abundance samples were preserved with a final concentration of 0.1% EM-grade glutaraldehyde (Acros Organics, cat. no. 233280250), stored at 4 °C for 15–30 min, then flash-frozen in liquid nitrogen and stored at −80 °C until counted via flow cytometry (Synechococcus and phage) or fluorescence microscopy (O. marina). Active fluorescence samples were stored in darkened tubes for 15 min at 20 °C, then immediately analyzed (see below). For transcriptomics, ~2 × 108 Synechococcus cells were centrifuged at 10,000 g for 10 min, flash-frozen in liquid nitrogen, and stored at −80 °C until RNA extraction. For exo-metabolomics, ~2 × 108 Synechococcus cells were filtered onto 0.22-μm pore size PES filters (Sterlitech, cat. no PES0225100) and the filtrate was stored at −20 °C until analysis. For endo-metabolomics, the ~2 × 108 Synechococcus cells retained on 0.22-μm pore size PES filters were washed twice with 5 mL of sterile phosphate-buffered saline, and filters were transferred to Eppendorf tubes, flash-frozen in liquid nitrogen, and stored at −80 °C until extraction. All downstream analyses were performed with a minimum of three biological replicates.
Photosynthetic measurements
From at least three biological replicates, the maximum photosynthetic efficiency of photosystem II (PSII) expressed as Fv/Fm was measured using the FastAct Fast Repetition Rate fluorometer (FRRf; Chelsea Technologies Group, Surrey, UK). The minimum (Fo) and maximum (Fm) fluoresence yields, Fv/Fm were calculated as ((Fm–Fo)/Fm) using cells that were dark-adapted at 20 °C for 15 min prior to analysis. Prior to experimental analyses, optimum excitation parameters were determined experimentally using cell cultures as recommended by the instrument manufacturer (Chelsea Technologies Ltd., FastPro Manual, pp. 33). Based on these data, the instrument was programmed to generate single turnover saturation using 100 excitation flashlets on a 2-μs pitch with a combination of excitation wavelengths (450, 530, and 624 nm) and included a relaxation phase of 40 flashlets on a 50-μs pitch. Acquisition sequences were repeated five times with 100–120 ms between sequences, as recommended. Cell-free controls (0.2-µm-filtered cultures; Pall Supor Acrodisk, Cat. No. 4612) were used to determine blank values.
Statistics
Besides the method-specific statistics employed for transcriptomics and metabolomics, which are explained in their respective sub-sections, all instances where “significance” is mentioned in the results and discussion section refer to a two-tailed t-test with a p value <0.05.
Transcriptomics
Transcriptomes were generated for six timepoints from each condition and RNA was extracted from frozen pellets using Qiagen RNEasy Mini Kit, total RNA was quantified and quality-assessed using the 2100 Expert Prokaryote Total RNA Pico kit and Agilent Bioanalyzer, rRNA sequences were depleted using an Illumina Ribo-Zero Plus rRNA Depletion Kit, libraries were created using the Illumina TruSeq Stranded Total RNA Kit, and sequenced on a HiSeq-2500 1 TB device, yielding paired-end 2 × 101 bp reads. Between 13 and 29 million reads were obtained per sample (median = 18.5 million reads). BBtools v35.84 following JGI’s default pipeline was used on raw reads to remove those containing two or more “N” bases with an average quality score across the read <10, with a length ≤51 bp, containing known Illumina artifacts, or mapping to PhiX, human, cat, dog, and mouse genomes with the identity of ≥93%. BBtools v35.84 trimmed reads to remove known Illumina artifacts in 5’ and 3’ ends, or when with a base quality score under 6 on 3’. Differential expression analyses were conducted with a custom-made R script used in numerous prior phage-host ‘omics studies [30, 42, 43]. Treatment samples were compared to their corresponding untreated control at each time point (e.g., T2 cyanovirocells versus T2 cyanobacteria only; T2 cyanovirocells with protist versus T2 cyanobacteria only).
Metabolomics
For differential analyses, peak height was log10-transformed using a custom R script and used as a proxy for metabolite abundance. Metabolites detected (raw peak height ≥10,000) in <2 biological replicates were excluded. Differential abundance between cyanobacteria only (control) and each infection and/or protist co-culture treatment was computed using a fold change (FC) between log10-transformed peak heights (averaged across replicates). False discovery rate (FDR) was estimated using a t-test with Benjamini–Hochberg-corrected p values. A metabolite was considered significantly different between control and treatment if log10FC ≥ 0.1 (an approximate change of 25% between treatment and control) and FDR ≤ 0.05 (5% false positives).
Results and discussion
Experimental design and infection dynamics
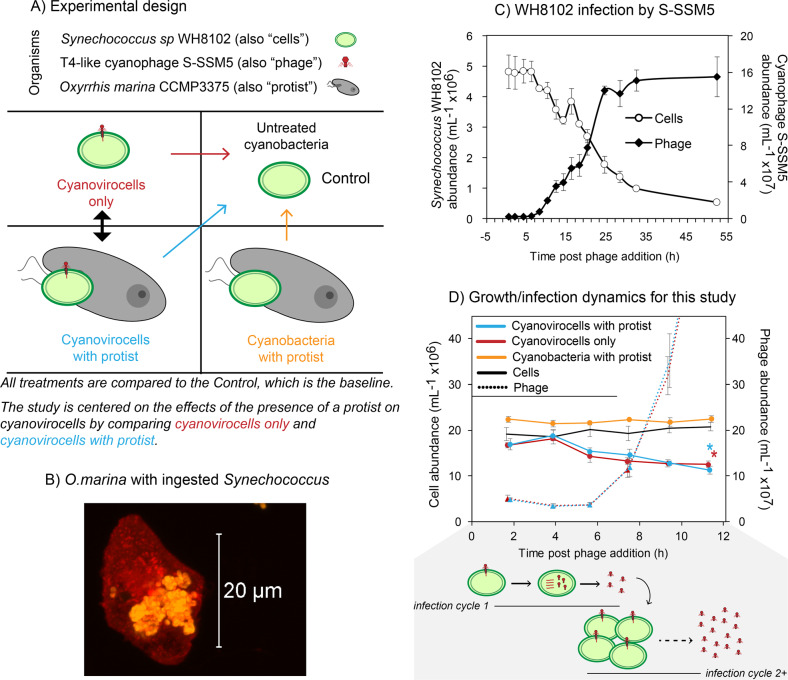
We sought to understand the effect that the presence of a protist, regardless of significant predation, would have on a population of phage-infected marine Synechococcus (i.e., virocells), through a time-resolved systems biology approach. To this end, we generated Synechococcus virocells (i.e., cyanovirocells) by adding the T4-like Myovirus S-SSM5, and we compared these cyanovirocells to cyanovirocells co-cultured with the protist O. marina. These cyanovirocells were compared to uninfected Synechococcus co-cultured with the protist, and the baseline for all treatments was a Synechococcus control lacking both phage and protist (Fig. 1A). Microscopy measurements confirmed that approximately half the protist population were actively feeding upon Synechococcus (Fig. 1B), even though they did not significantly alter their overall abundance due to the low protist: prey ratio. Furthermore, we calculated an encounter rate of 444 prey cells mL−1 s−1 following conservative approaches [44, 45], which represents 8–97% of the total prey population directly encountered by the protist by 2 and 10 h, respectively. O. marina contacting prey would presumably induce prey stress signals, as previously found [46, 47], but neither direct prey ingestion nor prey contact is necessary for the protist to elicit prey responses [48–52]. Here we investigate Synechococcus virocell physiological responses to the presence of the protist, rather than responses to predation.
Fig. 1. Experimental design and impacts of a protist on cyanobacteria and cyanobacteria virocells (cyanovirocells).
A Experimental design. Synechococcus WH8102 cells were infected with the T4-like myophage S-SSM5 to generate cyanovirocells. Cyanovirocells were studied over the course of infection either alone (“Cyanovirocells only” treatment) or in the presence of the protist Oxyrrhis marina (“Cyanovirocells with protist”). Arrows denote the comparisons: all treatments are compared against the Control to detect significant changes to the transcripts, metabolites, and photosynthetic efficiency. In addition, the focus of this study is the cyanovirocell responses to the presence of a protist, which is the contrast between cyanovirocells only and cyanovirocells with protist. B Fluorescent microscopic image of O. marina with ingested Synechococcus cells (protists were stained red with WGA - Alexa 488 and Synechococcus were detected with phycoerythrin pigment autofluorescence). Image was acquired using Zeiss LSM 710 confocal microscope with a 63× (1.4 N.A.) objective. C General infection dynamics of Synechococcus infected with S-SSM5, followed over 55 h post phage addition. This time course captures multiple cycles of infection. D Cell and phage abundances over the course of this study. Samples for genome-wide transcriptomics, endo-metabolomics, exo-metabolomics, and microscopy and photosynthetic efficiency measurements were taken every 2 h between 2 and 12 h post phage and/or protist addition. This time course captures a population-level view of infection, whereby more than one infection cycle is taking place (shaded in gray): the phages released from “infection cycle 1” infect new cells and undergo “infection cycle 2+” (i.e., two or more asynchronized infection cycles may be happening at this time). Both the burst size (here depicted as 4 phages released per infected cell), and the number of infection cycles are simplified. Cell abundance decreases significantly from start to end of ‘omics sampling during phage infection (t-test, p value <0.05), with or without protist (indicated by an asterisk for each treatment), and it does not significantly change in the presence of the protist alone. All experiments included a minimum of three biological replicates and the average with standard error is shown.
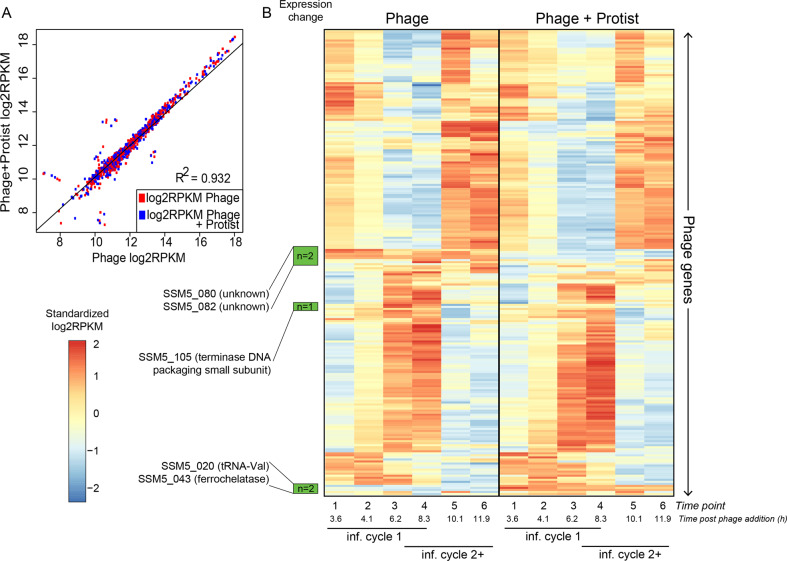
As for the phage, we examined infection dynamics over the course of multiple infection cycles, expecting the first infection cycle to be similar to that of the closely related T4-like phage Syn9 (77% average nucleotide identity; Supplementary Fig. S1) infecting our same host [34, 53]. Results indicated that, indeed, the phage had a ~6–8-h latent period and significantly reduced Synechococcus population abundances (Fig. 1C). These dynamics were maintained when the protist was added (Fig. 1C), confirming that protist addition did not catastrophically interfere with phage infection. Similarly, phage infection did not alter protist feeding upon Synechococcus (Supplementary text). Finally, phage transcription was also mostly unaltered when the protist was added (Fig. 2), as the phage maintained the temporal transcriptional dynamics expected for T4-like myoviruses [34, 54] with or without protist, and only five phage genes significantly changed expression with the protist (Supplementary Figs. S2 and S3 and Supplementary text). Overall, protists were unaffected by phages and phage infection was unaffected by protists. The experimental design thus enables us to examine the ways in which cyanovirocells respond to external protist presence, not necessarily predation, from a transcriptional, metabolic, and ecological perspective.
Fig. 2. Phage expression in the presence and absence of the protist.
A Pearson correlation of the phage gene expression values (as log2RPKM) with and without protist shows that phage expression levels are largely the same in both treatments. B Heatmap representing the expression (as standardized log2RPKM) of all phage genes (vertical) in the treatment with (right) or without (left) protists. Genes that change temporal dynamics in the presence of protists are marked in green (5 in total) and their annotations are displayed. The known transcriptional categories for this and generally the T4-like myophages (early, middle and late-expressed genes) are unaltered with protist for all except 5 genes (see Supplementary text). The time course for the infection captures more than one infection cycle: the first one with the phages added at the start of the experiment (“inf. cycle 1”) and the ones that follow once the phages are released from “inf. cycle 1” and infect new cells (“inf. cycle 2+”).
Protist impact on cyanovirocell endometabolites
To date, virocell studies have demonstrated that while phage transcription may largely be invariant, the host has a unique transcriptional response depending on the phage and the environment [34, 42, 43, 55, 56]. However, the transcriptional and metabolic responses of virocells during co-culture with protists are unknown.
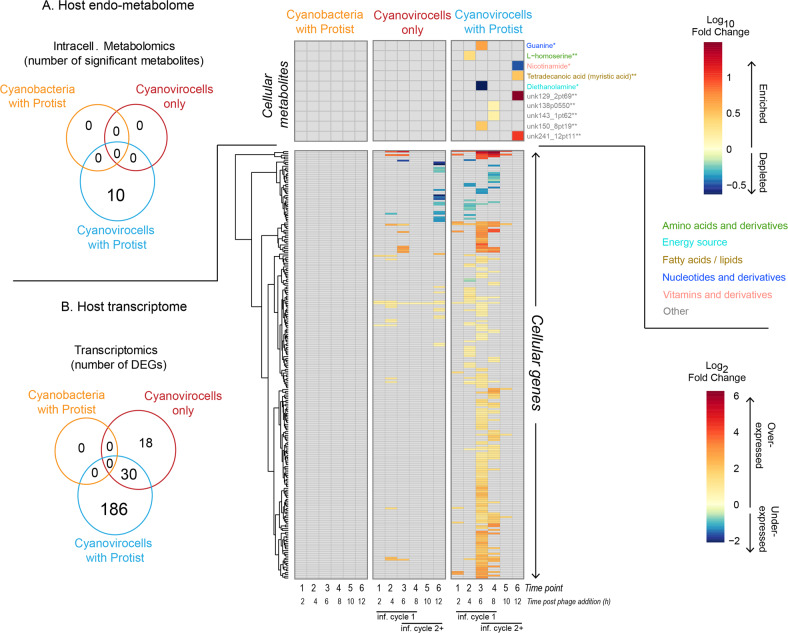
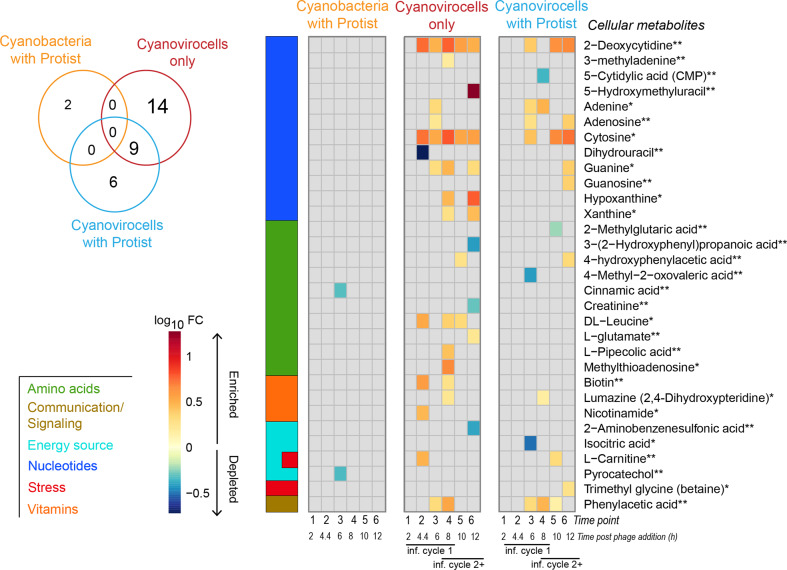
We investigated whether the presence of a protist would alter cyanovirocell transcriptional and metabolic profiles given the stress of protist–prey contact, handling, and ingestion/rejection [46–52, 57, 58]. We first investigated population-level intracellular metabolite profiles, which has not been explored previously. For this, we compared each treatment (cyanobacteria with protist, cyanovirocells without protist and cyanovirocells with protists) to the control (cyanobacteria only) lacking both phages and protists. We found that 0, 0, and 10 intracellular metabolites changed in each of those treatments relative to the control, respectively (Fig. 3A and Supplementary Table S1). The ten metabolites significantly changing in the cyanovirocells with protists are fewer than those reported in the three other prior temporally resolved virocell endometabolomic studies [59–61]. A detailed comparison with those studies (Supplementary Table S2) revealed that we used more stringent criteria for significance and that, had we used the prior, more permissive analytical procedures, those would have identified up to 17× more metabolites (Supplementary Table S3 and Supplementary text). As marine endo- and exo-metabolomics analyses are not yet standardized across the field, we opted for a more conservative approach for our metabolomics analyses.
Fig. 3. Host genes and metabolites altered in each treatment relative to the untreated control.
A Number of intracellular metabolites detected as significantly changing in each of the treatments (Venn diagram) and their log10FC value in the treatment relative to the untreated cells (heatmap). Compound names are colored by the class to which they belong, and the confidence in their detection is indicated by an asterisk next to the name. Confidence of level 1 (1 asterisk) indicates the compound had three identification criteria in positive or negative mode (i.e., retention peak, mz, and msms), whereas level 2 (2 asterisks) denotes two of the three criteria were met. Compounds denoted as “Other” represent detected features that were not identified. B Number of host genes differentially expressed in response to protist only (“cyanobacteria with protist”), phage only (“cyanovirocells only”), or phage plus protist (“cyanovirocells with protist”) pictured in a Venn diagram as well as their heatmap, which denotes gene expression as log2fold change (FC) values in the treatment compared to the untreated control over the experiment time course. Gray heatmap cells denote genes or metabolites that did not significantly change their expression or abundance, respectively, in the treatment relative to the control. All represented data derive from the average of a minimum of three biological replicates in which the treatment was compared to the uninfected control (cells only, no phage, no protist). The time course for the cyanovirocells and cyanovirocells with protist captures more than one infection cycle: the first one with the phages added at the start of the experiment (“inf. cycle 1”) and the ones that follow once the phages are released from “inf. cycle 1” and infect new cells (“inf. cycle 2+”).
Of the ten endometabolites significantly changing in the cyanovirocells with protist, the majority (60%, n = 6) changed in the population undergoing the first infection cycle (ending at 6–8 h), whereas the rest changed at 12 h post phage addition; in the population undergoing subsequent infections (Fig. 3A). Of the six metabolites changing within the first infection cycle, five were enriched (guanine, L-homoserine and three others with no database hits, thus termed “unknown”) and one was depleted (diethanolamine) in the protist-treated cyanovirocells relative to the control lacking phages or protists, and all between 4 and 8 h. Metabolites could be enriched because cyanovirocells either increase production intracellularly or incorporate them from the environment. Both guanine (a nucleotide) and L-homoserine (an amino acid) are building blocks that could be resources for phage reproduction, as it has previously been posited from transcriptome [55, 60, 62–65] and metabolome [59–61] studies of other phage infections. Especially in cyanobacteria, cyanophages are thought to be highly proficient at deviating cellular resources toward phage replication [31, 34, 66], which in this case would fuel the middle and late stages of infection. Alternatively, homoserine is also known to increase in cells responding to stress, likely as a consequence of protein degradation [67]. Therefore, the enrichment of these metabolites in the protist-treated cyanovirocells could reflect both stress and a need for additional resources for phage replication that are not as needed in the protist-free cyanovirocells.
Finally, the remaining four endometabolites that changed after the first infection cycle included three enriched (the lipid myristic acid and two more “unknown”) and one depleted (nicotinamide, a vitamin B3 derivative) compound. Here the signal derives from an asynchronous population composed of an uninfected minority, as well as a majority of cells undergoing different stages of infection. The lipid increase is consistent with viral infection [59, 68–70] and cyanophage-mediated restructuring of cellular membranes [71], as well as with microbial responses to protists (without phages) [72]. The decrease in intracellular nicotinamide may reflect cyanovirocells utilizing it for survival, given its critical biological role in myriad enzymatic reactions [73].
Protist impact on cyanovirocell transcription
Focusing next on gene expression, we again compared all treatments (cyanobacteria with protist, cyanovirocells only, and cyanovirocells with protist) to the untreated control. Each of those had 0, 48, and 216 differentially expressed (DE) genes (Fig. 3B and Supplementary Table S1). The 48 DE genes in the cyanovirocells only treatment represent a smaller host response than that reported for this same host during infection by myovirus Syn9 [34] which we attribute to differences in both the phage used and the experimental design as explained in the Supplementary text. The lack of DE genes in the protist-only treatment suggests that the transcriptional differences across the two cyanovirocell treatments (the 216 vs 48 DE genes) were due to protist presence, with a 4.5-fold increase in the number of host genes changing expression in the cyanovirocells in response to the protist. Both the direction (increased or decreased expression relative to the control; here “over-expressed” and “under-expressed”, respectively) and timing of cyanovirocell gene expression also varied with and without protist, as follows. Host genes were mostly over-expressed—65 and 89% in the absence and presence of the protist, respectively (Supplementary Fig. S5A)—thus suggesting that a larger number of host genes enhanced their expression in the cyanovirocells co-cultured with the protist than without it. In addition, when protist was added, the cellular transcriptional response was greater toward the end of the infection cycle, as the majority (52%, n = 153) of the genes were DE at 6 h post phage addition, compared with only 18% at that same time without the protist (Supplementary Fig. S5B). Thus, when protist was added, host transcriptional responses were greatest toward the end of the first infection cycle. Again, the majority (97%, n = 149) of those 153 genes were over-expressed.
We next examined the functions of these 153 host transcripts that responded at 6 h in the cyanovirocells with protists by grouping them into functional categories. The category with the most genes was the “unknown” (not functionally annotated, n = 74 genes) and its average fold change (aFC) of expression in the cyanovirocells treated with the protist relative to the cyanobacteria only control was 3.7 (Supplementary Fig. S5C), which highlights the need for further experimental work to characterize unknown genes even in relatively well-studied marine cyanobacteria.
The three most highly expressed categories with known functions were “Phosphate Metabolism” (aFC = 5.8, n = 3 genes), “Central Carbon Metabolism” (aFC = 5.4, n = 5 genes) and “Protein Metabolism” (aFC = 4.9, n = 15 genes) (Supplementary Fig. S5C). The “Phosphate Metabolism” category included the inorganic phosphate (Pi) transporter pstS and the transcriptional regulator phoB that respectively transport Pi into the cell and activate downstream cellular metabolic pathways under P-limitation [74]. Expression of these genes suggests that the cyanovirocells were P-limited, which is expected from cyanobacteria-cyanophage gene expression studies lacking protists [34, 75–78]. As phoB was not over-expressed without the protist, only pstS (Fig. 4A), assuming the PhoB protein is also highly produced, these data suggest that the presence of the protist induces a low-P-mediated cell-wide metabolic change in the cyanovirocells.
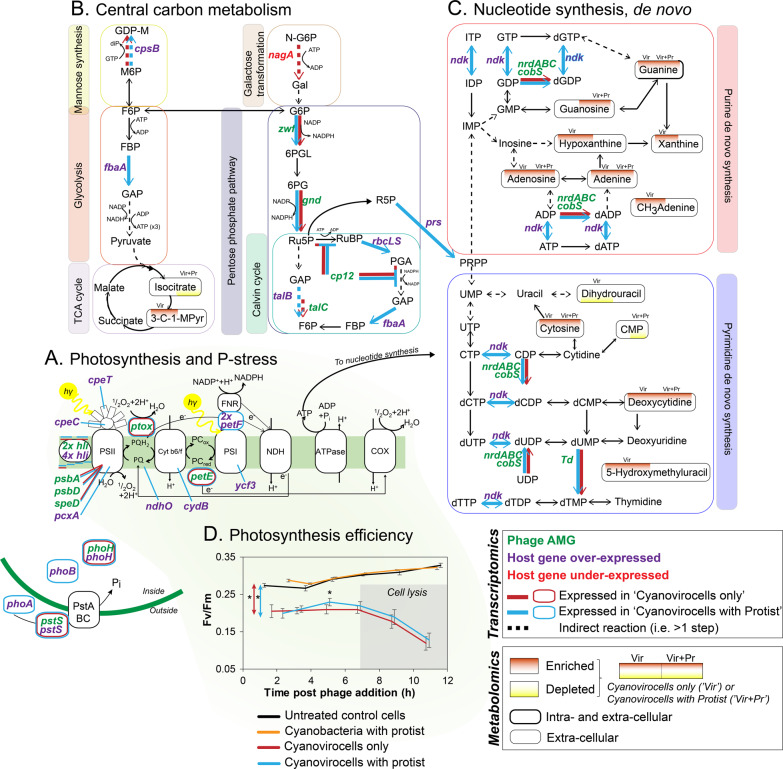
Fig. 4. Cyanovirocell energy and resource metabolic pathways responding to the protist.
A Photosynthesis and phosphate (P) stress. B Central C metabolism, including mannose synthesis, glycolysis, galactose metabolism pentose phosphate pathway (PPP), Calvin cycle, and the Tricarboxylic acid (TCA) cycle. C De novo purine and pyrimidine nucleotide synthesis. Phage auxiliary metabolic genes (AMGs) are in green text. Host over-expressed genes are in purple, while the under-expressed one is in red. The color of the arrow or the border (orange or blue) denotes if the gene is expressed in response to phage only or phage and protist. Metabolites that change significantly have a background shade colored red (if enriched) or yellow (if depleted) and the position of the shade denotes under which treatment they are altered: in the cyanovirocells only or in the cyanovirocells co-cultured with protists. D Photosynthetic efficiency of Synechococcus cells under each condition. Asterisks indicate when treatments are significantly different (t-test, p value <0.05). Cyanovirocells and cyanovirocells with protist both have significantly lower photosynthetic efficiency than uninfected cells (red and blue arrows with asterisks, respectively), but cells with just the protist do not (t-test, p value >0.05). Cyanovirocells with protist significantly increase in photosynthetic efficiency from ~2 to ~5 h post infection relative to cyanovirocells alone (t-test, p value <0.05). Abbreviations can be found in the Supplementary text.
Next, the “Central Carbon Metabolism” category included genes involved in glycolysis, pentose phosphate pathway (PPP), and the Calvin cycle. These genes have been found to be expressed during cyanophage infection [34, 78, 79] and are assumed to shift cellular energy production toward phage replication. Given that they were only over-expressed in the protist-treated cyanovirocells, it suggests that protist presence changes the energy needs of the cyanovirocells, especially toward the end of infection (see below).
Finally, the “Protein Metabolism” category mainly included genes for chaperones such as Hsp20, GroES, and GroEL, which were not DE in the cyanovirocells when the protist was absent. These chaperones were found over-expressed in Polynucleobacter asymbioticus responding to a protist [47], and are known to respond to various stresses in cyanobacteria, including phage infection [80], heat-shock, salinity, and oxidative stress [81, 82], but can also protect the photosystem II from photoinhibition [81], or help assemble key proteins, including Rubisco [83]. Since the host Rubisco genes as well as phage structural and lysis genes (Supplementary Fig. S2) were also over-expressed at this time, these host chaperones may help assemble key host and phage proteins during the final stages of infection.
We next asked if the presence of protists enhanced cellular stress. While only one gene in the “Stress” category was DE (and under-expressed, relative to the control) in the cyanovirocells without protist, 14 genes were DE (and over-expressed, relative to the control) throughout the infection when the protist was added, with the majority (n = 8 genes) over-expressed with an aFC = 3.4 at 6 h post phage addition, including the above chaperones (Hsp20, GroES, and GroEL), which contribute to stress as mentioned above, and Rubisco (Supplementary Fig. S5D). These data provide further evidence that the stress was higher in the cyanovirocells when the protist was present than when it was absent. We posit this stress could increase the energy and metabolic demand of phage infection in the protist-exposed cyanovirocells, even without significant predation. This could happen either directly through metabolic changes [84, 85], or indirectly through enhancing antipredator strategies that are known to occur in aquatic bacteria (e.g., increased motility, metabolite release), which are energetically costly [3, 86, 87].
Altogether, the transcriptomics and endo-metabolomics data suggest that the presence of the protist, even without significant predation, (i) augments the transcriptional and endometabolomic response of cyanovirocells, (ii) shifts cyanovirocell intracellular physiological responses largely toward the end of an infection cycle, and (iii) induces a larger stress, energy and resource demand in the cyanovirocells than in its absence.
Protist impact on cyanovirocell resource and energy production
The above results suggested that cyanovirocells nearing the end of the infection cycle required more cellular energy and resources in the presence of protists than in their absence. Cyanophages often encode auxiliary metabolic genes (AMGs) that alter (a) host photosynthesis and central C metabolism pathways (e.g., PPP and Calvin cycle) for obtaining energy (ATP), reducing power (NADPH), and nucleotide precursors (ribose-5-phosphate), and (b) P acquisition and nucleotide synthesis pathways for deviating cellular metabolism toward building new phages [33–37, 66, 77–79, 88–94]. Specifically, photosynthetic reaction center proteins, proteins that stabilize such reaction centers, and proteins that enhance light harvesting during infection are among the cyanophage AMGs that “boost” host machinery to ensure photosynthesis during infection. In addition, AMGs from central C metabolism pathways inhibit the Calvin cycle to prevent ATP consumption and to redirect C flow toward the PPP, from which NADPH and ribose-5-phosphate are obtained. With these resources, as well as with acquired P (with P-stress transporters), cyanophages can replicate their genomes (reviewed in [31, 78]; Fig. 4A–C).
We next examined the expression of the phage AMGs and host genes involved in those metabolic pathways. Phage S-SSM5 encodes AMGs involved in all of these pathways: (i) P-stress (pstS, phoH), (ii) photosynthesis (petE, ptox, hli (n = 2), psbA, psbD, speD), (iii) central C metabolism (zwf, gnd, talC, cp12), and (iv) nucleotide synthesis (nrdA, nrdB, nrdC, td, cobS) (Fig. 4A–C) [31, 32, 78], and were all expressed regardless of protist presence (Supplementary Table S4 and Supplementary Fig. S6). Contrastingly, the cyanovirocells over-expressed 4 and 23 (the same 4 plus 19 more) host genes in the absence and presence of the protist, respectively, relative to the control (Supplementary Table S5 and Supplementary Fig. S7). These genes are for (i) P-stress (phoA, phoB, pstS and phoH), (ii) photosynthesis (hli, cpeT, pcxA, cpeC, ndhO, cydB, petF, ycf3), (iii) central C metabolism (cpsB, fbaA, talB, rbcL and rbcS), and (iv) de novo nucleotide synthesis (prs and ndk). These were involved in different steps of the reprogrammed metabolic pathways than the phage AMGs (Fig. 4), with transaldolase being the exception, as both the phage (talC) and bacterial (talB) homologs were expressed in the same step of PPP. Presumably, this is because an enzymatic “double dose” is helpful for that step, given that it may be a bottleneck of PPP [79], whereas the rest of the steps can be driven by just the phage or the host enzyme. Finally, most genes were expressed at the end of the infection (6–8 h, n = 21 genes) in the protist-treated cyanovirocells; Supplementary Fig. S7). These data suggest that the phage specifically directed over-expression of host genes complementary to its AMGs to boost metabolic pathways necessary for the last stages of phage reproduction with and without the protist, but more so in the presence of the protist given the larger number of DE genes.
In summary, protist-treated cyanovirocells had a greater number of over-expressed host genes involved in (a) P-stress, which suggests that cyanovirocells co-cultured with protists have higher P demands than cyanovirocells without protists (b) nucleotide synthesis, which comes into play after recycling host DNA [95] and is an energetically costly process, and (c) photosynthesis, which suggest a larger energetic demand on Synechococcus during phage infection under protist predation (Fig. 4A–C). Overall, these data suggest that cyanovirocells have higher resource and energy demand when exposed to protists, regardless of significant predation.
Given these findings, we sought to assess whether the protist imposed a larger energetic demand on cyanovirocells through measuring the photosynthetic efficiency Fv/Fm. While the overall initial Fv/Fm of the Synechococcus cells were lower (0.20–0.29) than those observed in other photosynthesis studies of Synechococcus phage infection (0.6–0.7; [36–38]), the discrepancy is likely due the differences in irradiance and the bacterial strain used. Specifically, our cells were grown at 20 °C and 60 μmol photons m−2s−1 temperature and irradiance, respectively, and under these same conditions, three Synechococcus strains displayed Fv/Fm values of 0.26–0.34 [96]. In addition, work with our same strain grown at 10 and 150 μmol photons m−2s−1 obtained Fv/Fm values of 0.36 and 0.15, respectively [97].
The photosynthetic efficiency of cyanobacteria cells co-cultured with protists (Fv/Fm values of 0.28–0.32) was not significantly different from control cells (Fv/Fm values of 0.27–0.33) at any time point. This may be due to the low abundance of protists relative to the cyanobacteria. In contrast, the cyanovirocells, with or without protists, displayed significantly lower Synechococcus photosynthetic efficiency compared to the untreated control—by 35% as early as 2 h post-infection, and 64% by 12 h post-infection (Fig. 4D and Supplementary Fig. S8), which coincides with the first and subsequent infection cycles, respectively. The reduction in Synechococcus photosynthetic efficiency in response to phage infection is similar to that observed in virus-infected eukaryotic phytoplankton [98, 99], but contrasts with prior findings in Synechococcus or Prochlorococcus phage infection experiments [36, 37, 100], which demonstrated no change in Fv/Fm prior to host cell lysis. This discrepancy is likely due to instrumental differences as we used a single instead of multiple turnover flash fluorometer, and this methodological difference enabled us to observe phage-mediated plastoquinol quenching, indicative of deviation of light energy away from C fixation toward ATP production, which is not detectable using multiple turnover flash fluorometers [101–103]. Consistent with this physiological measurement, phage plastoquinol terminal oxidase (ptox) was expressed during early infection with or without protist (Supplementary Fig. S6A), and ptox has been hypothesized to facilitate alternative electron flows that prevent damage to PSII [89, 104].
Finally, while cyanovirocell photosynthetic efficiency was overall lower than the cyanobacteria only control as mentioned above, protist-treated cyanovirocells had a significant increase of ~15% (±5%) just before the first infection cycle cell lysis (~5 h post-infection), relative to the protist-free cyanovirocells (Fig. 4D and Supplementary Fig. S8). These results show that the energy required for phage infection was higher with protist presence toward the end of infection. Together with the genome-wide transcriptomics and endo-metabolomics findings, these results suggest that cyanovirocells co-cultured with a protist require greater resources (reflected in the transcriptional and metabolic changes) and energy (reflected in increased photosynthetic efficiency and metabolic reprogramming) to support phage reproduction, regardless of whether protists were actively feeding upon Synechococcus.
Protist impacts on cyanovirocell extracellular metabolites
We next profiled the exometabolome of all treatments (cyanobacteria with protist, cyanovirocells only, and cyanovirocells with protist) for dissolved chemical cues relative to the untreated control [6, 86, 87], to evaluate whether the presence of a protist changes ecosystem footprints. A total of 2, 23, and 15 exometabolites significantly changed in each respective treatment (Fig. 5 and Supplementary Table S1). In the protist-only treatment, the two metabolites were cinnamic acid and pyrocatechol, which were significantly depleted. Their depletion could mean either that the protist induced Synechococcus to release fewer compounds, or that one of the organisms, the protist or Synechococcus, were utilizing these metabolites, thus leading to decreasing their extracellular abundance. As these two exometabolites were absent from the cyanovirocell samples, we interpret that the protist alone had a unique impact on Synechococcus’s exometabolome in that it was different from the impact induced by viral infection. We next focused on the exometabolite changes induced by viral infection with and without the presence of the protist.
Fig. 5. Changes in the extracellular metabolome of cyanobacteria across three treatments.
Venn diagram: number of significantly changing metabolites detected extracellularly in each treatment. Heatmap: abundance of all significantly changing metabolites detected extracellularly with each treatment, in log10-fold change (FC) of the values in the treatment relative to the uninfected control. The colored bar on the left represents the class to which the metabolites belong. The confidence in metabolite detection is indicated by an asterisk next to the name. Confidence of level 1 (1 asterisk) indicates that the compound met all three identification criteria in positive or negative mode (i.e., retention peak, mz, and msms), whereas level 2 (2 asterisks) denotes two of the three criteria were met. The time course for cyanovirocells and cyanovirocells with protist captures more than one infection cycle: the first one with the phages added at the start of the experiment (“inf. cycle 1”) and the ones that follow once the phages are released from “inf. cycle 1” and infect new cells (“inf. cycle 2+”).
In the cyanovirocells only treatment, we evaluated the timing and functional annotation of the 23 exometabolites that changed relative to the cyanobacteria only control (Fig. 5). Within the first 6 h of infection, 10 metabolites were enriched extracellularly, indicating pre-lysis phage-induced metabolite release from intact cells. Functional annotation suggested that the exometabolites were nucleotides (n = 5), amino acids (n = 1), vitamins (n = 2) and potential communication (n = 1) and stress (n = 1) molecules. These could be resources for cellular or phage growth (e.g., the nucleotides, amino acids, and vitamins [59, 61, 105]), or cell-to-cell communication molecules [106]. Such virus-induced changes could have consequences for the microbial food web. For example, viruses can induce biochemical changes in the coccolithophore Emiliania huxleyi that decrease copepod and protist grazing [22, 107]. These findings suggest that phages shape both the intra- and the extracellular landscape, and that cyanovirocells release nutrients prior to cell lysis. Alternatively, as this treatment was composed of ~67% cyanovirocells and ~33% uninfected cells (Supplementary Dataset), it is possible that the enriched exometabolites derive from uninfected cells responding to cyanovirocells. However, considering that the majority of cells were infected and phage-mediated metabolite restructuring and release has been previously reported [59, 61, 108, 109], we favor the hypothesis that the cyanovirocells are the exometabolite source. Regardless, the organic matter release undoubtedly would benefit the larger microbial community by providing resources [110].
The eight unique exometabolites that were enriched in the subsequent infection cycles (≥8 h) of the protist-free cyanovirocells were our next focus. We reasoned that these could derive either from a minority of cyanovirocells undergoing different infection stages or a majority of lysed cells. Given that they were not detected in prior timepoints, during the infection cycle, and before lysis, we presume that the latter scenario is most likely. These were amino acids (n = 4), nucleotides (n = 3) and vitamins (n = 1), thus supporting prior findings that viral lysis releases organic matter [59, 109, 111]. Together, the exometabolite data from the protist-free cyanovirocell treatment suggests that (i) stress and communication molecules were unique to intact cells, as they were found only in the first infection cycle, and (ii) cyanovirocell-derived amino acids, nucleotides, and vitamins are organic resources available to a microbial community pre- and post-cell lysis.
Finally, we evaluated the 15 exometabolites differentially detected in the cyanovirocells co-cultured with protists relative to the cyanobacteria only control, 9 of which were shared with the cyanovirocells only treatment (and were also enriched), and 6 of which were new (2 enriched, 4 depleted). The decrease in the number of enriched exometabolites in the cyanovirocells with protist treatment relative to the cyanovirocells only treatment suggests that either in response to the protist the cyanovirocells were decreasing export/diffusion, or that one of the microbes (cyanobacterial cells, cyanovirocells, or protists) were utilizing the metabolites. For example, Sulfitobacter virocells [59] have both been shown to uptake exometabolites, and both Synechococcus and the protist are capable of directly importing dissolved molecules [110, 112]. The 4 exometabolites were derivatives of a nucleotide, two amino acids, and a TCA cycle metabolite intermediate from which bacteria obtain energy. All but one were depleted during the first infection cycle (Fig. 5), suggesting consumption by the cyanovirocells to supply extra resources for phage infection in the presence of the protist.
Ecological implications of protist-cyanovirocell interactions
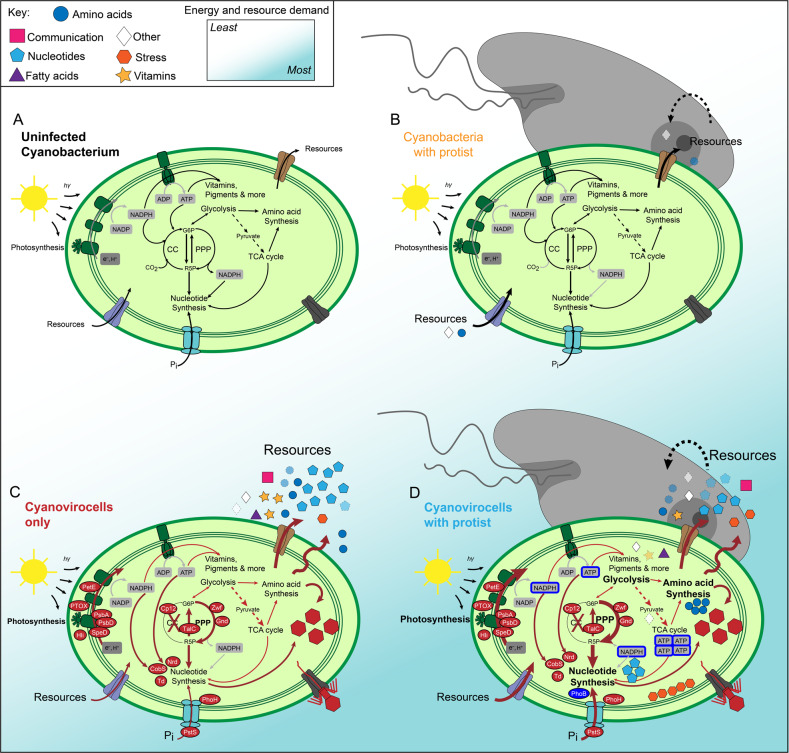
In this work we sought to move beyond the focus on the biogeochemical impacts of viral lysis and metabolic reprogramming of viral-infected cyanobacteria [31] toward building a conceptual model for the impacts of how protist presence impacts cyanovirocell infection dynamics and physiology (Fig. 6). To this end, we assessed cross-kingdom interactions between viruses, protists and cyanobacteria from the cyanovirocell perspective and its intra- and extracellular responses to the presence of a protist, with the untreated cell (Fig. 6A) as a baseline for multi-omics analyses. First, protist presence alone induced no measurable impact on Synechococcus endometabolites or transcripts, which could be due to the low relative abundance of the protist. However, protist presence did alter metabolite output, thus suggesting even low protist numbers have a measurable ecosystem impact (Fig. 6B).
Fig. 6. Metabolic reprogramming and ecosystem impact of protist- and/or phage-treated Synechococcus.
A Uninfected cyanobacterium. B Cyanobacteria with protist. The presence of the protist alters the abundance of some exometabolites. C Cyanovirocells only. Cyanophages reprogram P-acquisition, photosynthesis, energy pathways, and nucleotide metabolism toward building new phages. D Cyanovirocells with protist. The energy (e.g., ATP) and resource (e.g., reducing power, phosphate, nucleotides, and amino acids) demand of phage infection is highest for the cyanovirocells co-cultured with a protist. Cyanovirocells have the largest changes in the exometabolites as seen from the release of nutrients, either via diffusion or active transport across the membrane (C, D). This nutrient pool is available for the ecosystem, including uptake by protists (D).
Second, without the protist, Synechococcus had a small intracellular response to phage infection, with (i) very few transcriptional changes, (ii) expected phage AMG-driven metabolic reprogramming (reviewed in [31, 78]), and (iii) no significant impacts on detected endometabolites (Fig. 6C). Such a small intracellular impact suggests that this phage can replicate in this host using existing cellular resources, as previously observed for heterotroph-infecting phages that are well adapted to their host [30]. However, the phage-mediated cyanovirocell extracellular footprint was larger than the intracellular one, with multiple enriched exometabolites that spanned various nutrient classes (e.g., nucleotides, amino acids, vitamins, and communication/stress molecules) and which reinforce that cyanovirocells, similar to other known, marine organisms [113], can be a source of diverse “public goods” to feed and communicate with other organisms.
Third, when cyanovirocells were co-cultured with protists (Fig. 6D), even without significant population reduction from grazing, the cyanovirocell intracellular response increased, both at the transcriptional and metabolic levels, and was greatest toward the end of the infection cycle. Expression of phage structural genes at that time suggests that phage S-SSM5 was utilizing such host resources for building virions. Functionally, the protist-treated cyanovirocells were more stressed, more P-limited, and enhanced their protein folding and energy acquisition capabilities—observed in the over-expression of central C metabolism genes and increased photosynthetic efficiency. Metabolically, cyanovirocells increased their nucleotides and amino acids, again suggesting protist presence places a higher metabolic demand on infected cyanobacteria. The extracellular metabolome may also reflect such stress as well as prokaryote-eukaryote competition for nutrients given that when the protist was present the resources decreased.
Though only a first step toward mechanism, these findings highlight key areas for future hypothesis-driven experimental work as follows. To analyze the impact of the exometabolites on protist–prey interactions, microfluidic chemotaxis assays (e.g., [114]) combined with live cell video microscopy could quantify the response of diverse bacteria and protists to virocell-released exometabolites and provide an alternative to bulk population-based approaches [21, 22, 107]. In addition, to examine which microbes benefit from the exometabolites and to quantify nutrient utilization, generation, and exchange, stable isotope labeling of virocells, followed by a suite of methodological approaches such as nanoSIMS for spatially resolved compound flux visualization [115], stable isotope probing [116], or quantitative fluxomics [117] could be employed. Finally, the metabolites presented here offer a catalog of specific molecules to target via rapidly advancing mass spectrometry approaches [118], which, along with Bioorthogonal Noncanonical Amino Acid Tagging [119–121], would enable the discovery of virocell-derived signaling molecules, chemoattractants, and chemorepellents—all of which could have profound implications for cell–cell interactions and food web dynamics. As virocell studies mature, a grand challenge for the field will be to incorporate virocells into synthetic microbial communities [122] and ecosystems [123] along with applying advanced mathematical modeling [124]. These added efforts will enable the community to bridge the gap between model organism studies and observational accounts of complex natural communities.
Conclusions
A decade has passed since the marine scientific community was “dared” to examine interactions between viral infection and protists [17]. Using Synechococcus and their viruses, identified as key predictors of biological carbon flux [39], we mechanistically explored interactions among cyanobacteria, cyanovirocells, and protists. We found that cyanovirocells are not the same with and without the protist, such that even low protist abundance increases the energy (e.g., ATP) and resource (e.g., reducing power, phosphate, nucleotides, and amino acids) demand of cyanovirocells relative to protist-free cyanovirocells, especially toward the end of the infection cycle. Here we take a first approach to understanding how the presence of a protist impacts cyanovirocells, not how cyanovirocells respond to protist predation given that the low protist abundance would be a limitation for the latter endeavor. Future approaches with higher protist numbers may investigate how cyanovirocells respond to predation or even how different virocells respond to different predators. While ecosystems biology modeling is in its infancy [124–127], experimental findings like those here are critical to provide baseline data needed to help incorporate viruses and other important top-down predators into our understanding of ecosystem processes.
Supplementary information
Acknowledgements
We thank Benjamin Bolduc, Katherine Hargreaves, Lacey Orsini, and Bonnie Poulos for assistance with experimental sampling; Natalie Solonenko for assistance with RNA extraction, and Jordan Mullens, Ryan Marie Kelly, and Glen Marrs for assistance with confocal microscopy method development and analysis. We also thank Steve Archer for lending us the Chelsea Technologies FRRf instrument. This work was supported by NSF (OCE#1536989, 1829831, 2019589) and Moore Foundation Investigator Award (#3790) to MBS, and The Arnold and Mabel Beckman Foundation Postdoctoral Fellowship and NSF (OCE#1829905) to SAF. This is the NSF Center for Chemical Currencies of a Microbial Planet (C-CoMP) publication #010. The work (proposal: 10.46936/10.25585/60001407) conducted by the U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337), a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy operated under Contract No. DE-AC02-05CH11231.
Author contributions
SAF and MBS obtained funding for and designed the study. SAF and SS performed the culture experiments, generated all samples, and generated and analyzed flow cytometry data. SAF generated and analyzed fluorescence data. SS performed confocal microscopy and analyzed data with SAF. TW generated and CH-V and SR analyzed transcriptomic data. CH-V curated the genomes, curated the metabolisms, integrated the transcriptomics and metabolomics data, and generated all figures. LPS and RL generated and SAF, CH-V, BPB, SR, and TN analyzed the metabolomics data. SAF, CH-V, and MBS wrote the manuscript with contributions from all authors.
Data availability
Raw data, calculations, and statistical tests are available in the Supplementary Dataset, which has been deposited, along with the scripts used in the ‘omics analyses, the metabolomics quality control data, and the genomes used, in the public repositories Cyverse (https://datacommons.cyverse.org/browse/iplant/home/shared/iVirus/Cyano_phage_grazer_omics/) or GitHub (https://github.com/simroux/Metabolomics_CyanophageGrazer). Supplementary files also include expanded methods and analyses for culture conditions, photosynthetic measurements, transcriptomic analyses, LC-MS metabolomics procedures and analyses, choice of protist, protist–prey contact calculations, enumeration of organisms, phage titer determination, and further results and discussion on transcriptomics and metabolomics findings.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Matthew B. Sullivan, Email: sullivan.948@osu.edu
Sheri A. Floge, Email: floges@wfu.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s43705-022-00169-6.
References
- 1.Verity PG, Smetacek V, Smayda TJ. Status, trends and the future of the marine pelagic ecosystem. Environ Conserv. 2002;29:207–37. doi: 10.1017/S0376892902000139. [DOI] [Google Scholar]
- 2.Lima-Mendez G, Faust K, Henry N, Decelle J, Colin S, Carcillo F, et al. Determinants of community structure in the global plankton interactome. Science. 2015;348:1262073. doi: 10.1126/science.1262073. [DOI] [PubMed] [Google Scholar]
- 3.Pernthaler J. Predation on prokaryotes in the water column and its ecological implications. Nat Rev Microbiol. 2005;3:537–46. doi: 10.1038/nrmicro1180. [DOI] [PubMed] [Google Scholar]
- 4.Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Microbiol. 2007;5:782–91. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- 5.Brum JR, Morris JJ, Décima M, Stukel MR. Mortality in the oceans: causes and consequences. In: Kemp PF, editor. Eco-DAS IX symposium proceedings. Waco, TX, USA: ASLO; 2014. p. 16–48.
- 6.Worden AZ, Follows MJ, Giovannoni SJ, Wilken S, Zimmerman AE, Keeling PJ. Environmental science. Rethinking the marine carbon cycle: factoring in the multifarious lifestyles of microbes. Science. 2015;347:1257594. doi: 10.1126/science.1257594. [DOI] [PubMed] [Google Scholar]
- 7.Fuhrman J. Bacterioplankton roles in cycling of organic matter: the microbial food web BT. In: Falkowski PG, Woodhead AD, Vivirito K, editors. Primary productivity and biogeochemical cycles in the sea. Boston, MA: Springer US; 1992. p. 361–83. 10.1007/978-1-4899-0762-2_20.
- 8.Fuhrman JA, Suttle CA. Viruses in marine planktonic systems. Oceanography. 1993;6:51–63. doi: 10.5670/oceanog.1993.14. [DOI] [Google Scholar]
- 9.Fuhrman JA. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399:541–8. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- 10.Wilhelm SW, Suttle CA. Viruses and nutrient cycles in the sea. Bioscience. 1999;49:781–8. doi: 10.2307/1313569. [DOI] [Google Scholar]
- 11.Suttle CA. Marine viruses-major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–12. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 12.Putland JN. Microzooplankton herbivory and bacterivory in Newfoundland coastal waters during spring, summer and winter. J Plankton Res. 2000;22:253–77. doi: 10.1093/plankt/22.2.253. [DOI] [Google Scholar]
- 13.Schmoker C, Hernández-León S, Calbet A. Microzooplankton grazing in the oceans: impacts, data variability, knowledge gaps and future directions. J Plankton Res. 2013;35:691–706. doi: 10.1093/plankt/fbt023. [DOI] [Google Scholar]
- 14.Armengol L, Calbet A, Franchy G, Rodríguez-Santos A, Hernández-León S. Planktonic food web structure and trophic transfer efficiency along a productivity gradient in the tropical and subtropical Atlantic Ocean. Sci Rep. 2019;9:1–19. doi: 10.1038/s41598-019-38507-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinbauer MG, Hornák K, Jezbera J, Nedoma J, Dolan JR, Šimek K. Synergistic and antagonistic effects of viral lysis and protistan grazing on bacterial biomass, production and diversity. Environ Microbiol. 2007;9:777–88. doi: 10.1111/j.1462-2920.2006.01200.x. [DOI] [PubMed] [Google Scholar]
- 16.Miki T, Jacquet S. Complex interactions in the microbial world: underexplored key links between viruses, bacteria and protozoan grazers in aquatic environments. Aquat Microb Ecol. 2008;51:195–208. doi: 10.3354/ame01190. [DOI] [Google Scholar]
- 17.Breitbart M. Marine viruses: truth or dare. Ann Rev Mar Sci. 2012;4:425–48. doi: 10.1146/annurev-marine-120709-142805. [DOI] [PubMed] [Google Scholar]
- 18.Bonilla-Findji O, Herndl GJ, Gattuso JP, Weinbauer MG. Viral and flagellate control of prokaryotic production and community structure in offshore Mediterranean waters. Appl Env Microbiol. 2009;75:4801–12. doi: 10.1128/AEM.01376-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ory P, Hartmann HJ, Jude F, Dupuy C, Del Amo Y, Catala P, et al. Pelagic food web patterns: do they modulate virus and nanoflagellate effects on picoplankton during the phytoplankton spring bloom? Environ Microbiol. 2010;12:2755–72. doi: 10.1111/j.1462-2920.2010.02243.x. [DOI] [PubMed] [Google Scholar]
- 20.Longnecker K, Wilson MJ, Sherr EB, Sherr B. Effect of top-down control on cell-specific activity and diversity of active marine bacterioplankton. Aquat Microb Ecol. 2010;58:153–65. doi: 10.3354/ame01366. [DOI] [Google Scholar]
- 21.Evans C, Wilson WH. Preferential grazing of Oxyrrhis marina on virus infected Emiliania huxleyi. Limnol Oceanogr. 2008;53:2035–40. doi: 10.4319/lo.2008.53.5.2035. [DOI] [Google Scholar]
- 22.Goode AG, Fields DM, Archer SD, Martínez Martínez J. Physiological responses of Oxyrrhis marina to a diet of virally infected Emiliania huxleyi. PeerJ. 2019;7:e6722. doi: 10.7717/peerj.6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flombaum P, Gallegos JL, Gordillo RA, Rincón J, Zabala LL, Jiao N, et al. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc Natl Acad Sci USA. 2013;110:9824–9. doi: 10.1073/pnas.1307701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chow C-ET, Kim DY, Sachdeva R, Caron DA, Fuhrman JA. Top-down controls on bacterial community structure: microbial network analysis of bacteria, T4-like viruses and protists. ISME J. 2014;8:816–29. doi: 10.1038/ismej.2013.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hahn MW, Höfle MG. Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol Ecol. 2001;35:113–21. doi: 10.1111/j.1574-6941.2001.tb00794.x. [DOI] [PubMed] [Google Scholar]
- 26.Callieri C, Amalfitano S, Corno G, Bertoni R. Grazing-induced Synechococcus microcolony formation: experimental insights from two freshwater phylotypes. FEMS Microbiol Ecol. 2016;92:fiw154. [DOI] [PubMed]
- 27.Parnasa R, Nagar E, Sendersky E, Reich Z, Simkovsky R, Golden S, et al. Small secreted proteins enable biofilm development in the cyanobacterium Synechococcus elongatus. Sci Rep. 2016;6:32209. doi: 10.1038/srep32209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forterre P. Manipulation of cellular syntheses and the nature of viruses: the virocell concept. Comptes Rendus Chim. 2011;14:392–9. doi: 10.1016/j.crci.2010.06.007. [DOI] [Google Scholar]
- 29.Rosenwasser S, Ziv C, Creveld SG, Vardi A. Virocell metabolism: metabolic innovations during host-virus interactions in the ocean. Trends Microbiol. 2016;24:821–32. doi: 10.1016/j.tim.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 30.Howard-Varona C, Lindback MM, Bastien GE, Solonenko N, Zayed AA, Jang HB, et al. Phage-specific metabolic reprogramming of virocells. ISME J. 2020;14:881–95. https://www.nature.com/articles/s41396-019-0580-z. [DOI] [PMC free article] [PubMed]
- 31.Zimmerman AE, Howard-Varona C, Needham DM, John SG, Worden AZ, Sullivan MB, et al. Metabolic and biogeochemical consequences of viral infection in aquatic ecosystems. Nat Rev Microbiol. 2019 doi: 10.1038/s41579-019-0270-x. [DOI] [PubMed] [Google Scholar]
- 32.Warwick-Dugdale J, Buchholz HH, Allen MJ, Temperton B. Host-hijacking and planktonic piracy: how phages command the microbial high seas. Virol J. 2019;16:15. doi: 10.1186/s12985-019-1120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobson TB, Callaghan MM, Amador-Noguez D. Hostile takeover: how viruses reprogram prokaryotic metabolism. Annu Rev Microbiol. 2021;75:515–39. 10.1146/annurev-micro-060621-043448. [DOI] [PubMed]
- 34.Doron S, Fedida A, Hernndez-Prieto MA, Sabehi G, Karunker I, Stazic D, et al. Transcriptome dynamics of a broad host-range cyanophage and its hosts. ISME J. 2016;10:1437–55. doi: 10.1038/ismej.2015.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang S, Sun Y, Zhang S, Long L. Temporal transcriptomes of a marine cyanopodovirus and its Synechococcus host during infection. Microbiologyopen. 2020;10:e1150. https://pubmed.ncbi.nlm.nih.gov/33377630/. [DOI] [PMC free article] [PubMed]
- 36.Lindell D, Jaffe JD, Johnson ZI, Church GM, Chisholm SW. Photosynthesis genes in marine viruses yield proteins during host infection. Nature. 2005;438:86–9. doi: 10.1038/nature04111. [DOI] [PubMed] [Google Scholar]
- 37.Puxty RJ, Millard AD, Evans DJ, Scanlan DJ. Viruses inhibit CO2 fixation in the most abundant phototrophs on Earth. Curr Biol. 2016;26:1585–9. doi: 10.1016/j.cub.2016.04.036. [DOI] [PubMed] [Google Scholar]
- 38.Puxty RJ, Evans DJ, Millard AD, Scanlan DJ. Energy limitation of cyanophage development: implications for marine carbon cycling. ISME J. 2018;12:1273–86. doi: 10.1038/s41396-017-0043-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guidi L, Chaffron S, Bittner L, Eveillard D, Larhlimi A, Roux S, et al. Plankton networks driving carbon export in the oligotrophic ocean. Nature. 2016;532:465–70. doi: 10.1038/nature16942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zimmerman AE. MPN (Most Probable Number) assay for infectivity of algal viruses. protocols.io. 1–4. 10.17504/protocols.io.gunbwve.
- 41.TAYLOR J. The estimation of numbers of bacteria by tenfold dilution series. J Appl Bacteriol. 1962;25:54–61. doi: 10.1111/j.1365-2672.1962.tb01119.x. [DOI] [Google Scholar]
- 42.Howard-Varona C, Roux S, Dore H, Solonenko NE, Holmfeldt K, Markillie LM, et al. Regulation of infection efficiency in a globally abundant marine Bacteriodetes virus. ISME J. 2017;11:284–95. doi: 10.1038/ismej.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howard-Varona C, Hargreaves KR, Solonenko NE, Markillie LM, White RA, Brewer HM, et al. Multiple mechanisms drive phage infection efficiency in nearly identical hosts. ISME J. 2018;12:1605–18. doi: 10.1038/s41396-018-0099-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evans GT. The encounter speed of moving predator and prey. J Plankton Res. 1989;11:415–7. doi: 10.1093/plankt/11.2.415. [DOI] [Google Scholar]
- 45.Kiørboe T. A mechanistic approach to plankton ecology. Princeton University Press; 2008. https://books.google.com/books?id=H6fLBGvKg6AC.
- 46.Harke MJ, Jankowiak JG, Morrell BK, Gobler CJ. Transcriptomic responses in the bloom-forming Cyanobacterium Microcystis induced during exposure to Zooplankton. Appl Environ Microbiol. 2017;83:e02832–16. doi: 10.1128/AEM.02832-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beisser D, Bock C, Hahn MW, Vos M, Sures B, Rahmann S, et al. Interaction-specific changes in the transcriptome of polynucleobacter asymbioticuscaused by varying protistan communities. Front Microbiol. 2019;10:1–14. doi: 10.3389/fmicb.2019.01498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Long JD, Smalley GW, Barsby T, Anderson JT, Hay ME. Chemical cues induce consumer-specific defenses in a bloom-forming marine phytoplankton. Proc Natl Acad Sci USA. 2007;104:10512–7. doi: 10.1073/pnas.0611600104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Selander E, Thor P, Toth G, Pavia H. Copepods induce paralytic shellfish toxin production in marine dinoflagellates. Proceedings Biol Sci. 2006;273:1673–80. doi: 10.1098/rspb.2006.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Selander E, Jakobsen HH, Lombard F, Kiørboe T. Grazer cues induce stealth behavior in marine dinoflagellates. Proc Natl Acad Sci USA. 2011;108:4030–4. doi: 10.1073/pnas.1011870108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harvey EL, Menden-Deuer S. Predator-induced fleeing behaviors in phytoplankton: a new mechanism for harmful algal bloom formation? PLoS One. 2012;7:e46438. doi: 10.1371/journal.pone.0046438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harvey EL, Jeong HJ, Menden-Deuer S. Avoidance and attraction: chemical cues influence predator-prey interactions of planktonic protists. Limnol Oceanogr. 2013;58:1176–84. doi: 10.4319/lo.2013.58.4.1176. [DOI] [Google Scholar]
- 53.Fedida A, Lindell D. Two Synechococcus genes, two different effects on cyanophage infection. Viruses. 2017;9:136. doi: 10.3390/v9060136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kutter E, Bryan D, Ray G, Brewster E, Blasdel B, Guttman B. From host to phage metabolism: hot tales of phage T4’s takeover of E. coli. Viruses. 2018;10:387. https://www.ncbi.nlm.nih.gov/pubmed/30037085. [DOI] [PMC free article] [PubMed]
- 55.Blasdel BG, Chevallereau A, Monot M, Lavigne R, Debarbieux L. Comparative transcriptomics analyses reveal the conservation of an ancestral infectious strategy in two bacteriophage genera. ISME J. 2017;11:1988–96. doi: 10.1038/ismej.2017.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brandão A, Pires DP, Coppens L, Voet M, Lavigne R, Azeredo J. Differential transcription profiling of the phage LUZ19 infection process in different growth media. RNA Biol. 2021;18:1778–90. [DOI] [PMC free article] [PubMed]
- 57.Fenchel T, Blackburn N. Motile chemosensory behaviour of phagotrophic protists: mechanisms for and efficiency in congregating at food patches. Protist. 1999;150:325–36. doi: 10.1016/S1434-4610(99)70033-7. [DOI] [PubMed] [Google Scholar]
- 58.Wolfe GV. The chemical defense ecology of marine unicellular plankton: constraints, mechanisms, and impacts. Biol Bull. 2000;198:225–44. doi: 10.2307/1542526. [DOI] [PubMed] [Google Scholar]
- 59.Ankrah NYD, May AL, Middleton JL, Jones DR, Hadden MK, Gooding JR, et al. Phage infection of an environmentally relevant marine bacterium alters host metabolism and lysate composition. ISME J. 2013;8:1089–100. doi: 10.1038/ismej.2013.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chevallereau A, Blasdel BG, De Smet J, Monot M, Zimmermann M, Kogadeeva M, et al. Next-generation “-omics” approaches reveal a massive alteration of host RNA metabolism during bacteriophage infection of Pseudomonas aeruginosa. PLoS Genet. 2016;12:e1006134. doi: 10.1371/journal.pgen.1006134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Smet J, Zimmermann M, Kogadeeva M, Ceyssens P-J, Vermaelen W, Blasdel B, et al. High coverage metabolomics analysis reveals phage-specific alterations to Pseudomonas aeruginosa physiology during infection. ISME J. 2016;10:1823–35. doi: 10.1038/ismej.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ainsworth S, Zomer A, Mahony J, van Sinderen D. Lytic infection of Lactococcus lactis by bacteriophages Tuc2009 and c2 triggers alternative transcriptional host responses. Appl Env Microbiol. 2013;79:4786–98. doi: 10.1128/AEM.01197-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mojardín L, Salas M. Global transcriptional analysis of virus-host interactions between phage φ29 and Bacillus subtilis. J Virol. 2016;90:9293–304. doi: 10.1128/JVI.01245-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sacher JC, Flint A, Butcher J, Blasdel B, Reynolds HM, Lavigne R, et al. Transcriptomic analysis of the campylobacter Jejuni response to T4-like phage NCTC 12673 infection. Viruses. 2018;10:332. https://www.ncbi.nlm.nih.gov/pubmed/29914170. [DOI] [PMC free article] [PubMed]
- 65.Yang Z, Yin S, Li G, Wang J, Huang G, Jiang B, et al. Global transcriptomic analysis of the interactions between phage φAbp1 and extensively drug-resistant Acinetobacter baumannii. mSystems. 2019;4:e00068–19. doi: 10.1128/mSystems.00068-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thompson LR, Zeng Q, Chisholm SW. Gene expression patterns during light and dark infection of Prochlorococcus by Cyanophage. PLoS One. 2016;11:e0165375. doi: 10.1371/journal.pone.0165375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jozefczuk S, Klie S, Catchpole G, Szymanski J, Cuadros-Inostroza A, Steinhauser D, et al. Metabolomic and transcriptomic stress response of Escherichia coli. Mol Syst Biol. 2010;6:364. doi: 10.1038/msb.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fulton JM, Fredricks HF, Bidle KD, Vardi A, Kendrick BJ, Ditullio GR, et al. Novel molecular determinants of viral susceptibility and resistance in the lipidome of Emiliania huxleyi. Environ Microbiol. 2014;16:1137–49. doi: 10.1111/1462-2920.12358. [DOI] [PubMed] [Google Scholar]
- 69.Rose SL, Fulton JM, Brown CM, Natale F, Van Mooy BA, Bidle KD. Isolation and characterization of lipid rafts in Emiliania huxleyi: a role for membrane microdomains in host-virus interactions. Env Microbiol. 2014;16:1150–66. doi: 10.1111/1462-2920.12357. [DOI] [PubMed] [Google Scholar]
- 70.Zeng J, Liu S, Cai W, Jiang H, Lu X, Li G, et al. Emerging lipidome patterns associated with marine Emiliania huxleyi-virus model system. Sci Total Environ. 2019;688:521–8. doi: 10.1016/j.scitotenv.2019.06.284. [DOI] [PubMed] [Google Scholar]
- 71.Roitman S, Hornung E, Flores-Uribe J, Sharon I, Feussner I, Béjà O. Cyanophage-encoded lipid desaturases: oceanic distribution, diversity and function. ISME J. 2018;12:343–55. doi: 10.1038/ismej.2017.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Amato A, Sabatino V, Nylund GM, Bergkvist J, Basu S, Andersson MX, et al. Grazer-induced transcriptomic and metabolomic response of the chain-forming diatom Skeletonema marinoi. ISME J. 2018;12:1594–604. doi: 10.1038/s41396-018-0094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Monteverde DR, Gómez-Consarnau L, Suffridge C, Sañudo-Wilhelmy SA. Life’s utilization of B vitamins on early Earth. Geobiology. 2017;15:3–18. doi: 10.1111/gbi.12202. [DOI] [PubMed] [Google Scholar]
- 74.Santos-Beneit F. The Pho regulon: a huge regulatory network in bacteria. Front Microbiol. 2015;6:402. doi: 10.3389/fmicb.2015.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thompson AW, Huang K, Saito MA, Chisholm SW. Transcriptome response of high- and low-light-adapted Prochlorococcus strains to changing iron availability. ISME J. 2011;5:1580–94. doi: 10.1038/ismej.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zeng Q, Chisholm SW. Marine viruses exploit their host’s two-component regulatory system in response to resource limitation. Curr Biol. 2012;22:124–8. doi: 10.1016/j.cub.2011.11.055. [DOI] [PubMed] [Google Scholar]
- 77.Lin X, Ding H, Zeng Q. Transcriptomic response during phage infection of a marine cyanobacterium under phosphorus-limited conditions. Environ Microbiol. 2016;18:450–60. doi: 10.1111/1462-2920.13104. [DOI] [PubMed] [Google Scholar]
- 78.Gao E-B, Huang Y, Ning D. Metabolic genes within cyanophage genomes: implications for diversity and evolution. Genes (Basel) 2016;7:80. doi: 10.3390/genes7100080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thompson LR, Zeng Q, Kelly L, Huang KH, Singer AU, Stubbe J, et al. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc Natl Acad Sci USA. 2011;108:E757–64. doi: 10.1073/pnas.1102164108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morimoto D, Kimura S, Sako Y, Yoshida T. Transcriptome analysis of a bloom-forming Cyanobacterium Microcystis aeruginosa during Ma-LMM01 phage infection. Front Microbiol. 2018;9:2. doi: 10.3389/fmicb.2018.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hossain MM, Nakamoto H. Role for the cyanobacterial HtpG in protection from oxidative stress. Curr Microbiol. 2003;46:70–6. doi: 10.1007/s00284-002-3831-5. [DOI] [PubMed] [Google Scholar]
- 82.Chaurasia AK, Apte SK. Overexpression of the groESL operon enhances the heat and salinity stress tolerance of the nitrogen-fixing Cyanobacterium Anabaena sp. Strain PCC7120. Appl Environ Microbiol. 2009;75:6008–12. doi: 10.1128/AEM.00838-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Webb R, Sherman LA. The cyanobacterial heat-shock response and the molecular chaperones BT. In: Bryant DA, editor. The molecular biology of Cyanobacteria. Dordrecht: Springer Netherlands; 1994. p. 751–67. 10.1007/978-94-011-0227-8_26.
- 84.Sokolova IM. Energy-limited tolerance to stress as a conceptual framework to integrate the effects of multiple stressors. Integr Comp Biol. 2013;53:597–608. doi: 10.1093/icb/ict028. [DOI] [PubMed] [Google Scholar]
- 85.Guo MS, Gross CA. Stress-induced remodeling of the bacterial proteome. Curr Biol. 2014;24:R424–34. doi: 10.1016/j.cub.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pohnert G, Steinke M, Tollrian R. Chemical cues, defence metabolites and the shaping of pelagic interspecific interactions. Trends Ecol Evol. 2007;22:198–204. doi: 10.1016/j.tree.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 87.Roberts EC, Legrand C, Steinke M, Wootton EC. Mechanisms underlying chemical interactions between predatory planktonic protists and their prey. J Plankton Res. 2011;33:833–41. doi: 10.1093/plankt/fbr005. [DOI] [Google Scholar]
- 88.Sullivan MB, Huang KH, Ignacio-Espinoza JC, Berlin AM, Kelly L, Weigele PR, et al. Genomic analysis of oceanic cyanobacterial myoviruses compared with T4-like myoviruses from diverse hosts and environments. Env Microbiol. 2010;12:3035–56. doi: 10.1111/j.1462-2920.2010.02280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Clokie MRJ, Millard AD, Mann NH. T4 genes in the marine ecosystem: studies of the T4-like cyanophages and their role in marine ecology. Virol J. 2010;7:291. doi: 10.1186/1743-422X-7-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Crummett LT, Puxty RJ, Weihe C, Marston MF, Martiny JBH. The genomic content and context of auxiliary metabolic genes in marine cyanomyoviruses. Virology. 2016;499:219–29. doi: 10.1016/j.virol.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 91.Clokie MR, Shan J, Bailey S, Jia Y, Krisch HM, West S, et al. Transcription of a “photosynthetic” T4-type phage during infection of a marine cyanoacterium. Env Microbiol. 2006;8:827–35. doi: 10.1111/j.1462-2920.2005.00969.x. [DOI] [PubMed] [Google Scholar]
- 92.Dammeyer T, Bagby SC, Sullivan MB, Chisholm SW, Frankenberg-Dinkel N. Efficient phage-mediated pigment biosynthesis in oceanic cyanobacteria. Curr Biol. 2008;18:442–8. doi: 10.1016/j.cub.2008.02.067. [DOI] [PubMed] [Google Scholar]
- 93.Lindell D, Jaffe JD, Coleman ML, Futschik ME, Axmann IM, Rector T, et al. Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature. 2007;449:83–6. doi: 10.1038/nature06130. [DOI] [PubMed] [Google Scholar]
- 94.Sullivan MB, Lindell D, Lee JA, Thompson LR, Bielawski JP, Chisholm SW. Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. PLoS Biol. 2006;4:e234. doi: 10.1371/journal.pbio.0040234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Friedman S, Gots JS. The purine and pyrimidine metabolism of normal and phage-infected Escherichia coli. J Biol Chem. 1953;201:125–35. doi: 10.1016/S0021-9258(18)71354-9. [DOI] [PubMed] [Google Scholar]
- 96.Jodłowska S, Śliwińska S. Effects of light intensity and temperature on the photosynthetic irradiance response curves and chlorophyll fluorescence in three picocyanobacterial strains of Synechococcus. Photosynthetica. 2014;52:223–32. doi: 10.1007/s11099-014-0024-y. [DOI] [Google Scholar]
- 97.Kolodny Y, Zer H, Propper M, Yochelis S, Paltiel Y, Keren N. Marine cyanobacteria tune energy transfer efficiency in their light-harvesting antennae by modifying pigment coupling. FEBS J. 2021;288:980–94. doi: 10.1111/febs.15371. [DOI] [PubMed] [Google Scholar]
- 98.Kimmance S, Allen M, Pagarete A, Martínez Martínez J, Wilson WH. Reduction in photosystem II efficiency during a virus-controlled Emiliania huxleyi bloom. Mar Ecol Prog Ser. 2014;495:65–76. doi: 10.3354/meps10527. [DOI] [Google Scholar]
- 99.Gilg I, Archer S, Floge S, Fields D, Vermont A, Leavitt A, et al. Differential gene expression is tied to photochemical efficiency reduction in virally infected Emiliania huxleyi. Mar Ecol Prog Ser. 2016;555:13–27. doi: 10.3354/meps11805. [DOI] [Google Scholar]
- 100.Puxty RJ, Millard AD, Evans DJ, Scanlan DJ. Shedding new light on viral photosynthesis. Photosynth Res. 2015;126:71–97. doi: 10.1007/s11120-014-0057-x. [DOI] [PubMed] [Google Scholar]
- 101.Kolber ZS, Prasil O, Falkowski PG. Measurements of variable chlorophyll fluorescence using fast repetition rate techniques: defining methodology and experimental protocols. Biochim Biophys Acta. 1998;1367:88–106. doi: 10.1016/S0005-2728(98)00135-2. [DOI] [PubMed] [Google Scholar]
- 102.Kromkamp JC, Forster RM. The use of variable fluorescence measurements in aquatic ecosystems: differences between multiple and single turnover measuring protocols and suggested terminology. Eur J Phycol. 2003;38:103–12. doi: 10.1080/0967026031000094094. [DOI] [Google Scholar]
- 103.Suggett DJ, Moore CM, Oxborough K, Geider RJ. Fast repetition rate (FRR) chlorophyll a fluorescence induction measurements. Chelsea Technol Gr. 2006;53. http://www.psi.cz/ftp/publications/kautsky/FRRFmethodsManual.pdf%5Cnpapers://64c875cb-35e9-49cc-86a7-69f7bda703f6/Paper/p289.
- 104.Philosof A, Battchikova N, Aro E-M, Béjà O. Marine cyanophages: tinkering with the electron transport chain. ISME J. 2011;5:1568–70. doi: 10.1038/ismej.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Waldbauer JR, Coleman ML, Rizzo AI, Campbell KL, Lotus J, Zhang L. Nitrogen sourcing during viral infection of marine cyanobacteria. Proc Natl Acad Sci USA. 2019;116:15590–5. doi: 10.1073/pnas.1901856116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fontaine BM, Duggal Y, Weinert EE. Exploring the links between nucleotide signaling and quorum sensing pathways in regulating bacterial virulence. ACS Infect Dis. 2018;4:1645–55. doi: 10.1021/acsinfecdis.8b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vermont AI, Martínez Martínez J, Waller JD, Gilg IC, Leavitt AH, Floge SA, et al. Virus infection of Emiliania huxleyi deters grazing by the copepod Acartia tonsa. J Plankton Res. 2016;38:1194–205. doi: 10.1093/plankt/fbw064. [DOI] [Google Scholar]
- 108.Sheik AR, Brussaard CPD, Lavik G, Lam P, Musat N, Krupke A, et al. Responses of the coastal bacterial community to viral infection of the algae Phaeocystis globosa. ISME J. 2014;8:212–25. doi: 10.1038/ismej.2013.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ma X, Coleman ML, Waldbauer JR. Distinct molecular signatures in dissolved organic matter produced by viral lysis of marine cyanobacteria. Environ Microbiol. 2018;20:3001–11. doi: 10.1111/1462-2920.14338. [DOI] [PubMed] [Google Scholar]
- 110.Baran R, Bowen BP, Northen TR. Untargeted metabolic footprinting reveals a surprising breadth of metabolite uptake and release by Synechococcus sp. PCC 7002. Mol Biosyst. 2011;7:3200–6. doi: 10.1039/c1mb05196b. [DOI] [PubMed] [Google Scholar]
- 111.Middelboe M, Jørgensen NOG. Viral lysis of bacteria: an important source of dissolved amino acids and cell wall compounds. J Mar Biol Assoc United Kingdom. 2006;86:605–12. doi: 10.1017/S0025315406013518. [DOI] [Google Scholar]
- 112.Baran R, Lau R, Bowen BP, Diamond S, Jose N, Garcia-Pichel F, et al. Extensive turnover of compatible solutes in cyanobacteria revealed by deuterium oxide (D2O) stable isotope probing. ACS Chem Biol. 2017;12:674–81. doi: 10.1021/acschembio.6b00890. [DOI] [PubMed] [Google Scholar]
- 113.Moran MA, Kujawinski EB, Schroer WF, Amin SA, Bates NR, Bertrand EM, et al. Microbial metabolites in the marine carbon cycle. Nat Microbiol. 2022;7:508–23. doi: 10.1038/s41564-022-01090-3. [DOI] [PubMed] [Google Scholar]
- 114.Seymour JR, Simó R, Ahmed T, Stocker R. Chemoattraction to dimethylsulfoniopropionate throughout the marine microbial food web. Science. 2010;329:342–5. doi: 10.1126/science.1188418. [DOI] [PubMed] [Google Scholar]
- 115.Nuñez J, Renslow R, Cliff JB, III, Anderton CR. NanoSIMS for biological applications: current practices and analyses. Biointerphases. 2017;13:03B301. doi: 10.1116/1.4993628. [DOI] [PubMed] [Google Scholar]
- 116.Neufeld JD, Vohra J, Dumont MG, Lueders T, Manefield M, Friedrich MW, et al. DNA stable-isotope probing. Nat Protoc. 2007;2:860–6. doi: 10.1038/nprot.2007.109. [DOI] [PubMed] [Google Scholar]
- 117.Hui S, Cowan AJ, Zeng X, Yang L, TeSlaa T, Li X, et al. Quantitative fluxomics of circulating metabolites. Cell Metab. 2020;32:676–88.e4. doi: 10.1016/j.cmet.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Steen AD, Kusch S, Abdulla HA, Cakić N, Coffinet S, Dittmar T, et al. Analytical and computational advances, opportunities, and challenges in marine organic biogeochemistry in an era of “omics”. Front Mar Sci. 2020;7:718. doi: 10.3389/fmars.2020.00718. [DOI] [Google Scholar]
- 119.Hatzenpichler R, Scheller S, Tavormina PL, Babin BM, Tirrell DA, Orphan VJ. In situ visualization of newly synthesized proteins in environmental microbes using amino acid tagging and click chemistry. Env Microbiol. 2014;16:2568–90. [DOI] [PMC free article] [PubMed]
- 120.Hatzenpichler R, Connon SA, Goudeau D, Malmstrom RR, Woyke T, Orphan VJ. Visualizing in situ translational activity for identifying and sorting slow-growing archaeal-bacterial consortia. Proc Natl Acad Sci USA. 2016;113:E4069–78. doi: 10.1073/pnas.1603757113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hatzenpichler R, Krukenberg V, Spietz RL, Jay ZJ. Next-generation physiology approaches to study microbiome function at single cell level. Nat Rev Microbiol. 2020;18:241–56. doi: 10.1038/s41579-020-0323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Datta MS, Sliwerska E, Gore J, Polz MF, Cordero OX. Microbial interactions lead to rapid micro-scale successions on model marine particles. Nat Commun. 2016;7:11965. doi: 10.1038/ncomms11965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zengler K, Hofmockel K, Baliga NS, Behie SW, Bernstein HC, Brown JB, et al. EcoFABs: advancing microbiome science through standardized fabricated ecosystems. Nat Methods. 2019;16:567–71. doi: 10.1038/s41592-019-0465-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zomorrodi AR, Segrè D. Synthetic ecology of microbes: mathematical models and applications. J Mol Biol. 2016;428:837–61. doi: 10.1016/j.jmb.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wigington CH, Sonderegger D, Brussaard CPD, Buchan A, Finke JF, Fuhrman JA, et al. Re-examination of the relationship between marine virus and microbial cell abundances. Nat Microbiol. 2016;1:15024. doi: 10.1038/nmicrobiol.2015.24. [DOI] [PubMed] [Google Scholar]
- 126.Macklin DN, Ahn-Horst TA, Choi H, Ruggero NA, Carrera J, Mason JC, et al. Simultaneous cross-evaluation of heterogeneous E. coli datasets via mechanistic simulation. Science. 2020;369:eaav3751. https://science.sciencemag.org/content/369/6502/eaav3751. [DOI] [PMC free article] [PubMed]
- 127.Dukovski I, Bajić D, Chacón J, Quintin M, Vila J, Sulheim S, et al. Computation Of Microbial Ecosystems in Time and Space (COMETS): an open source collaborative platform for modeling ecosystems metabolism. Nat Protoc. 2021;16:5030–82. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data, calculations, and statistical tests are available in the Supplementary Dataset, which has been deposited, along with the scripts used in the ‘omics analyses, the metabolomics quality control data, and the genomes used, in the public repositories Cyverse (https://datacommons.cyverse.org/browse/iplant/home/shared/iVirus/Cyano_phage_grazer_omics/) or GitHub (https://github.com/simroux/Metabolomics_CyanophageGrazer). Supplementary files also include expanded methods and analyses for culture conditions, photosynthetic measurements, transcriptomic analyses, LC-MS metabolomics procedures and analyses, choice of protist, protist–prey contact calculations, enumeration of organisms, phage titer determination, and further results and discussion on transcriptomics and metabolomics findings.