Abstract
Introduction
The management of mid and low rectal cancer is based on neoadjuvant chemoradiotherapy (CRT) followed by standardised surgery. There is no biomarker in rectal cancer to aid clinicians in foreseeing treatment response. The determination of factors associated with treatment response might allow the identification of patients who require tailored strategies (eg, therapeutic de-escalation or intensification). Colibactin-producing Escherichia coli (CoPEC) has been associated with aggressive colorectal cancer and could be a poor prognostic factor. Currently, no study has evaluated the potential association between intestinal microbiota composition and tumour response to CRT in mid and low rectal cancer. The aim of this study is to assess the association between response to neoadjuvant CRT and faecal intestinal microbiota composition and/or CoPEC prevalence in patients with mid or low rectal cancer.
Methods and analysis
This is a non-randomised bicentric prospective clinical study with a recruitment capacity of 200 patients. Three stool samples will be collected from participants with histological-proven adenocarcinome of mid or low rectum who meet eligibility criteria of the study protocol: one before neoadjuvant treatment start, one in the period between CRT end and surgery and one the day before surgery. In each sample, CoPEC will be detected by culture in special media and molecular (PCR) approaches. The global microbiota composition will be also assessed by the bacterial 16S rRNA gene sequencing. Neoadjuvant CRT response and tumour regression grade will be described using the Dworak system at pathological examination. Clinical data and survival outcomes will also be collected and investigated.
Ethics and dissemination
MICARE was approved by the local ethics committee (Comité de Protection des Personnes Sud-Est II, 18 December 2019. Reference number 2019-A02493-54 and the institutional review board. Patients will be required to provide written informed consent. Results will be published in a peer reviewed journal.
Trial registration number
Keywords: ONCOLOGY, GASTROENTEROLOGY, RADIOTHERAPY, MICROBIOLOGY
Strengths and limitations of this study.
As far as we know, this is the first study to evaluate association between intestinal microbiota composition and tumour response to chemoradiotherapy in mid and low rectal cancer.
MICARE is a prospective clinical study including 200 patients.
This study is based on a non-invasive and reproductible faecal test.
Tumour response will be described at pathological examination after surgery.
The limitation of this study will include population stratification for delay between radiotherapy and surgery, and adjonction of neoadjuvant chemotherapy in tumour response evaluation.
Introduction
With more than 700 000 new cases and 300 000 deaths in 2018, rectal cancer is the eighth leading cause of cancer deaths worldwide.1 The initial management of mid and low rectal cancer is based on neoadjuvant chemoradiotherapy (CRT) for locally advanced tumours. This is associated with a significant decrease of the locoregional recurrence rate, but without survival improvement.2–4 Neoadjuvant treatment is followed by standardised surgery.5 Total mesorectal excision is crucial for reducing tumour recurrence,6 but its significant morbidity can affect the patients’ quality of life. Prognosis also depends on the tumour response to neoadjuvant CRT. Currently, the surgical strategy is adapted in function of the tumour response to neoadjuvant treatment, assessed by MRI after CRT end.7 Indeed, the objective is therapeutic deescalation with rectal preservation to decrease morbidity and functional disorders. For patients with complete response (up to 25% of patients), careful monitoring without surgery (‘watch and wait’ strategy) has been proposed.8 9 For small tumours with good response to CRT, transanal excision with rectal preservation seems to be feasible in terms of cancer prognosis.10 For patients with large tumours or a locally advanced disease, a tailored treatment strategy with total neoadjuvant therapy (TNT) is now a gold standard.11 12 After surgical excision, the tumour response is classified in five pathologic tumour response grades, according to the Dworak classification, on the basis of the pathology findings.13 Recent studies reported up to 30% of poor responders (grades 0 and 1).14 15 These data emphasise the importance of the initial tumour staging and response to neoadjuvant CRT for tailoring surgical strategies. MRI is an essential tool for these two assessments.16–18 These data highlight the need of response predictive models to adapt the TNT in mid and low rectal cancer.
Gut microbiota behaves as a real organ and participates in intestinal homeostasis. An imbalance in its composition (dysbiosis) could be involved in many pathologies, including colorectal cancer (CRC).19–21 Escherichia coli (E. coli) has been widely described as a bacteria which could be involved in CRC.22 23 E. coli is the predominant aero-anaerobic Gram-negative specie in human colon, but it is also a pathogen involved in various intestinal diseases.24 Indeed, some E. coli strains have acquired the capacity to produce toxins named cyclomodulins, including colibactin that is encoded by the pks island.25 Colibactin-producing E. coli (CoPEC) has genotoxic effects by inducing DNA damage and chromosomal instability.25–27 CoPEC implication in CRC has been demonstrated, particularly in aggressive forms.28–34 Specifically, higher E. coli colonisation rate and higher prevalence of CoPEC are found in patients with TNM stage III or IV tumours29 (UICC TNM Classification, 8th Edition, 2017).35 Moreover, CoPEC gut colonisation might contribute to modulate the immunotherapy efficacy.36 Recent clinical studies discussed the prognostic role of intestinal microbiota in the tumour response following surgery and chemotherapy or immunotherapy37 and suggested that it could be used as a biomarker to predict tumour response to neoadjuvant treatments. On the other hand, very few clinical studies have assessed the influence of gut microbiota on radiotherapy efficacy, especially in rectal cancer. Recently, a preclinical study showed that mice which survive a high dose of radiation, harboured gut microbiota enriched with Lachnospiraceae and Enterococcaceae.38 Yet, a description of the intestinal microbiota composition before neoadjuvant therapy could allow identifying predictive bacterial markers of tumour response in rectal cancer, and to adapt TNT.
Indeed, chronic exposure of the gastrointestinal tract to genotoxins could be a prognostic marker of radiotherapy response. CoPEC colonisation would start at the very beginning of life38 and might lead to exposure of the intestinal mucosa to chronic genotoxic stress. The resulting damage could give cells the ability to resist to other genotoxic stresses such as radiation therapy. One in vitro study already showed the decreased radiation sensitivity of cells incubated by colibactin.27 Therefore, developing a non-invasive method to analyse gut microbiota composition and to evaluate CoPEC implication in the response to CRT could help clinicians to tailor cancer management and to develop tools to control the pathologic microorganisms identified as new therapeutic targets.
Methods and analysis
This study protocol is written in accordance with the SPIRIT guidelines (online supplemental file 1).
bmjopen-2022-061527supp001.pdf (87.6KB, pdf)
Objectives
Primary objective
The study’s primary objective is to assess the correlation between response to neoadjuvant CRT and CoPEC presence in stool samples.
Secondary objectives
To analyse in a non-targeted manner the global microbiota composition before CRT and to evaluate the correlation between composition and response to treatment.
To study the modulation of the intestinal microbiota by CRT.
To describe the correlation between clinical data and microbiota composition modulation induced by CRT.
To determine microbiological prognostic factors of overall survival, disease-specific survival and relapse-free survival (locoregional and metastatic) in patients with low or mid-rectum cancer.
To create a microbiological database for future mechanistic analyses.
To study the modulation of CoPEC colonisation by CRT.
Study design
The study is a non-randomised bicentric prospective clinical study. Two surgical teams will be involved—Institut du Cancer de Montpellier and CHU de Clermont-Ferrand; and an INSERM Unit—M2iSH Clermont-Ferrand. The study actually started on January 2020 and the estimated study completion date is November 2027.
Patients’ selection
Inclusion criteria
Histologically-proven adenocarcinoma of low or mid rectum, of stage II or III (UICC TNM Classification, 8th Edition, 201735).
Patient eligible for neoadjuvant treatment (50 Gray radiation and capecitabine, CAP 50), according to the French national recommendations.5 39
Informed signed consent received.
Man or woman aged ≥18 years.
Appropriate contraceptive measures taken by men and pre-menopausal women before study entry and for at least 8 weeks after the last CRT cycle. Patients should be informed by the investigator on the contraceptive measures to use.
Exclusion criteria
Antibiotic treatment at the time of stool sampling or in the month before.
Presence of a derivative stoma.
Previous chemotherapy treatment for rectum cancer.
Patient not affiliated to the French social security system.
Patient with possible poor treatment compliance for psychologic, familial, social and geographic reasons.
Legal incapacity or limited legal capacity.
Pelvic radiotherapy or brachytherapy in the year before inclusion in the study.
History of other cancers in the 5 last years, except for cervical carcinoma in situ and skin carcinoma, but including melanoma under treatment.
Pregnant or breastfeeding woman.
Study sponsor
The sponsor (Montpellier Cancer Institute, ICM) is responsible for the study design and management, and for obtaining all study authorisations (Persons Protection Committee, National Agency for Medical Security). It will also declare to these authorities the inclusion period beginning and end, produce the final study report, inform the competent authorities of the trial results and store all study-related documents for at least 15 years after the study end.
Clinical study procedures
Inclusion in the study
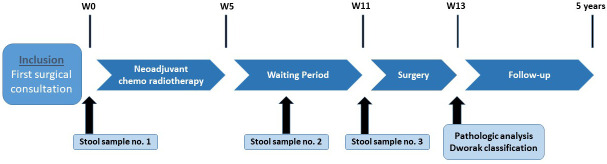
The study flow diagram is presented in figure 1.
Figure 1.
MICARE flow diagram.
Before study entry, all patients will receive exhaustive explanations on the study aims and procedures. A signed informed consent will be obtained from all patients before any study procedure (online supplemental file 2). At baseline, demographic (sex, age), clinical (performance status, weight, height, medical history, initial diagnosis date, tumour localisation, histologic type) and biological (complete blood count, carcinoembryonic antigen (CEA) level) data will be collected (table 1). Patients will undergo rectal examination and tumour staging by CT, rectal MRI and possibly rectal endoscopic ultrasound examination (depending on the centre decision).
Table 1.
Flowchart with the clinical and radiological evaluations
| Assessment | Baseline | Re-evaluation | Day before surgery | Follow-up every 6–8 months |
| Informed consent | X | |||
| Selection criteria validation | X | |||
| Demographic and clinical data | X | |||
| Physical examination | X | |||
| Patient inclusion | X | |||
| Stool sample | X | X | X | |
| Patient vital status | X | |||
| Tumour evaluation | ||||
| Rectal MRI | X | X | X | |
| CT | X | X | ||
| Rectal examination | X | X | X | |
bmjopen-2022-061527supp002.pdf (137.2KB, pdf)
During the surgical consultation, the first stool sample (stool sample N°1) may be collected during rectal examination (faeces left on the clinician’s glove), or by proctoscopy. Otherwise, the stool sample will be collected by the patient.
Neoadjuvant treatment
Patients will undergo neoadjuvant CRT in accordance with the French national guidelines.5 The recommended regimen is a concomitant oral chemotherapy (5-FU/CAPECITABINE) and 50 Gray radiotherapy. Despite PRODIGE 23 and RAPIDO trials, it is highly recommended to add a systemic chemotherapy (FOLFIRINOX or FOLFOX) to the CRT in locally advanced rectal cancer.12 CRT data (dose, possible dose modifications or interruptions) and CRT complications will be recorded.
Re-evaluation
During the consultation after CRT end and before surgery, a second stool sample (stool sample N°2) will be collected, as described for the baseline sample. If the patient has received antibiotics in the month before this consultation, stool sampling will not be performed.
This second consultation will include MRI examination as during the baseline visit. The tumour response will be described precisely with emphasis on the tumour regression grade according to the MERCURY study experience.7
Surgery
Surgical data (surgery type, digestive reconstruction or stoma and surgical outcomes), anatomopathological data (histologic type, ypTN grade, Dworak grade,13 Quirke classification,40 circumferential resection, distal margins and extramucosal vascular invasion) and biological data (RAS and BRAF mutational status, if available) will be collected. The day before surgery, before bowel mechanical preparation, the third stool sample (stool sample N°3) will be collected in hospital, as described for the previous samples. If the patient received antibiotics in the month before hospitalisation, stool sampling will not be performed.
Pathologic analysis
To meet the primary objective, the pathologic analysis of the surgical specimens will describe the tumour regression grade according to the Dworak classification13 (table 2). Patients with grade 0 and 1 tumours will be considered poor responders, in accordance with the literature.
Table 2.
Tumour regression grade (TRG), Dworak classification13
| TRG | Pathology |
| Grade 0 | No regression |
| Grade 1 | Dominant tumour mass with obvious fibrosis and/or vasculopathy |
| Grade 2 | Dominant fibrotic changes with few tumour cell groups (easy to find) |
| Grade 3 | Very few (difficult to find microscopically) tumour cells in fibrotic tissue with or without mucous substance |
| Grade 4 | No tumour cell, only fibrotic mass (total regression or response) |
Safety
All adverse events will be reported following the study sponsor’s pharmacovigilance procedures, and in accordance with the applicable regulation (online supplemental file 3).
bmjopen-2022-061527supp003.pdf (20KB, pdf)
Follow-up and study duration
Follow-up will last 5 years from the date of surgery. The frequency of follow-up visits will be decided at each centre. Every 6–8 months, the disease and survival status will be assessed. Recurrence will be investigated by clinical examination with rectal MRI and CT and a tumour marker test (CEA) (table 1). Locoregional or metastatic relapse will be reported in the case report form with the date of relapse diagnosis.
As the inclusion period will be of 36 months and the follow-up will last 5 years, the total study duration will be of 8 years.
Microbiological analyses
Sample handling
Three stool samples will be collected during the study (figure 1): (i) one at patient inclusion, before any treatment, to describe the baseline intestinal microbiota composition; (ii) one during the interval between the end of neoadjuvant CRT and surgery, at the surgical consultation for tumour reappraisal; and (iii) one just before bowel preparation (mechanical or antibiotics) for surgery.
Each sample will be divided into two cryotubes: one empty and one with 15% glycerol/DMEM to preserve cell integrity. Samples will be immediately stored at −80°C until transport to the M2iSH laboratory, Clermont-Ferrand, France, which will be in charge of the molecular analysis and storage of the samples.
E. coli strain identification and CoPEC detection
All microbiological analyses will be performed as previously described.28 After thawing, samples stored in DMEM/glycerol will be crushed and diluted in sterile phosphate buffered saline pH 7.4 before plating on TBX agar and chromogenic agar chromID CPS3 plates (bioMérieux) to allow the identification and quantitation of enterobacteria. Colonies (around 48 per sample) will be collected for molecular typing, and their identification will be confirmed with the automated Vitek II (bioMérieux) system. Enterobacterial Repetitive Intergenic Consensus PCR will be used as genotyping method to determine the number of E. coli strains per sample.28
E. coli harbouring the colibactin-encoding pks island will be identified by PCR analysis of each E.coli isolate.41 This will allow identifying the presence of CoPEC (primary objective).
Untargeted analysis of the local microbiota composition
Global microbiota modifications will be assessed by high-throughput sequencing of the bacterial 16S rRNA gene in DNA extracted from the three stool samples using the NucleoSpin DNA stool kit (Macherey-Nagel, Hoerdt, France), according to the manufacturer’s instructions. Quantitative PCR will be performed to quantify pro-carcinogenic bacterial species, such as Fusobacterium nucleatum, Enterococcus feacalis, bft-positive Bacteroides fragilis and CoPEC. In addition, the V4 region of the bacterial 16S rRNA gene will be amplified using the 515F/806R primer pair followed by Illumina high throughput sequencing on a MiSeq apparatus, according to the manufacturer’s guidelines. A global description of the intestinal microbiota could also be obtained by shotgun metagenomic sequencing to access the microbiota functional features after selection of the more informative samples.
Endpoints
Primary endpoint
The primary endpoint (associated with the primary objective) is the relative risk (RR) of poor response to neoadjuvant CRT in patients colonised by CoPEC (‘exposed’) compared with non-colonised patients (‘unexposed’).
Secondary endpoints
Prevalence and CoPEC colonisation rate before and after CRT.
Other bacterial strains present before CRT and relative risk of poor response to CRT in colonised and non-colonised patients.
Type, prevalence and colonisation rate of bacteria other than CoPEC in the microbiota, before and after CRT.
Percentage of colonised patients, depending on the bacterial type, according to the clinical parameters (age, sex, body mass index).
HR for overall survival, disease-specific survival and relapse-free survival (locoregional or metastatic) in colonised patients, for the different bacterial types, according to the overall bacterial composition (including CoPEC), and in non-colonised patients.
Data collection and management
The database will be managed by the sponsor, and data stored at the data processing centre, Biometrics Unit of the Montpellier Cancer Institute. Case report form design and clinical data management will be implemented using the Ennov Clinical software. Microbiological data will be collected in a database first stored at the M2iSH laboratory and then transferred to the sponsor database for analysis. Data and any trial documents will be made available on reasonable request and after signature of a data access agreement.
In accordance with the General Data Protection Regulation, a registration number will be used to identify each patient. The corresponding table will be encrypted and stored in a secure place. Special vigilance will be exercised throughout the study to maintain data anonymisation.
Study monitoring, quality control and audit
According to the sponsor’s risk-based monitoring plan (study participants, logistics, resources, impact), the collection of the patient informed consents and the respect of the study protocol and procedures will be monitored.
To guarantee the originality of all data and in accordance with the Good Clinical Practices, quality control will be performed by the sponsor. The study will be managed according to the sponsor procedures and in respect of the protocol, and the quality of the data included in the report forms will be checked.
The sponsor may wish to conduct an audit at some investigating centres. Audits may be conducted by the sponsor or any duly authorised person for at least 15 years after the trial.
Statistical considerations
Sample size
The recruitment capacity for this exploratory study will be around 200 patients. For a mean rate of 30% of poor responders to the neoadjuvant treatment among the patients not colonised by CoPEC (ie, a proportion of response P2=0.30 among unexposed patients), the study will be able to estimate a relative risk of 1.7 (RR=1.7) with a 30% precision and a CI at 95% (α=0.05). Patients in whom the CoPEC colonisation status cannot be determined at baseline, in whom CRT must be prematurely arrested, or who cannot undergo surgery will be considered non-evaluable.
Considering a 10% rate of potentially non-evaluable patients, a total of 220 patients (20 supplementary patients) will be included in the study.
Study population
Two populations will be defined for the analysis. The intention-to-treat population will be defined as all patients included in the study, treated (patients who received complete/partial neoadjuvant treatment) and not treated (patients who did not undergo CRT), eligible (ie, all patients who were included in the study without violation of a major inclusion or exclusion criterion) or not, and with/without baseline stool sample. The per-protocol population will include all eligible patients, treated (complete or partial CRT), and with baseline stool sample.
Statistical analyses
Qualitative variables will be described by frequencies and percentages, and quantitative variables with means, SD, medians and ranges. No imputation method will be used in case of missing data. Correlations between qualitative variables will be assessed using the χ2 or Fisher exact test. Quantitative variables will be compared using the Student’s t-test or the Kruskal-Wallis test. Comparison of quantitative variables at different times (before and after CRT) will be assessed using the Wilcoxon test for matched samples. The RR of poor response to neoadjuvant CRT in CoPEC-colonised patients (or colonised by other bacteria) compared with non-colonised patients will be estimated using a logistic regression (univariate analysis) and will be presented with the 95% CI. Survival analyses will be performed using the Kaplan-Meier method and survival distributions compared with the log rank test. HRs and their 95% CI will be estimated with a Cox proportional risk model. A detailed statistical analysis plan (SAP) will be written before the database is locked for analysis; supplementary subgroup analyses, if appropriate, will be specified in the SAP. All analyses will be performed using the Stata V.16 software (Stata, College Station, Texas).
Patient and public involvement
There was no patient or public involvement in the design of this study.
Discussion
The implication of intestinal microbiota in CRC has been widely demonstrated.42 Several recent studies suggest that different bacterial species, including CoPEC, could be used as biological biomarkers for CRC diagnosis and prognosis.29 36 41 43 44 The potential role of the gut microbiota in the modulation of the efficacy of anti-tumour treatments has been studied, with interesting results regarding chemotherapy and immunotherapy.37 However, these studies were focused on colon cancer dysbiosis and few data are available on rectal cancer and mucosa. Moreover, the correlation between gut microbiota homeostasis and radiation sensitivity remains unclear. Patients treated by pelvic radiation develop long-term complications that affect their quality of life, and have worse functional results than patients treated with surgery alone.45 46 It has been hypothesised that the intestinal microbiota has a significant impact on pelvic enteropathy47; however, pelvic irradiation is responsible for microbiota dysbiosis.48 49 To our knowledge, no previous study has assessed the local microbiota composition and its implication in the response to CRT in rectal cancer, although treatment response is one of the key points for prognosis estimation. Biomarkers to predict tumour response in rectal cancer are still crucially needed. Imaging techniques50 and biological markers51 52 have been evaluated, but they are often expensive and complicated to implement. Moreover, the results are still discussed. Currently, their use seems to be limited to research and expert centres. The present study will describe the intestinal microbiota composition in patients with rectal cancer receiving neoadjuvant CRT to show its potential correlation with the tumour response, focusing on CoPEC colonisation. In addition, the effect of radiotherapy on the local intestinal microbiota composition will be studied by comparing stool samples collected before and after CRT. Unlike studies on the intestinal microbiota in colon cancer in which tumour fragments are needed, in the case of mid or low rectal cancer, stool samples should be representative of the local microbiota.
One of the main hypotheses to explain CoPEC effect on CRT response is based on their capacity to induce DNA damage.25–27 Besides the direct effect on the cell, radiotherapy is also cytotoxic through the production of reactive oxygen species and reactive nitrogen species.53 Chronic genotoxic stress caused by CoPEC presence in gut mucosa could lead to an adaptation of the gut mucosa to genotoxic agents and consequently to reduce radiation sensitivity and resistance to therapy. For instance, in an in vitro study, Wilson et al observed less DNA damage in colibactin-positive epithelial cells infected by CoPEC.27 Moreover, radiation sensitivity is closely linked to autophagy regulation.54 55 Recent studies showed the involvement of gut microbiota in autophagy regulation, with a link to chemoresistance.56 Ionising radiation effects might be modified indirectly through autophagy deregulation induced by gut microbiota. In addition, radiotherapy cytotoxic effect could result in a modification of the local microenvironment with significant clinical consequences.57
The modulation of radiotherapy efficacy by the intestinal microbiota is an emerging concept in CRC, but its study faces many obstacles, especially sample availability. In this study, we want to develop a non-invasive reproducible faecal test that could become a key biomarker to predict tumour response to CRT. Our work will help clinicians to tailor neoadjuvant therapeutic strategies with the final goal of increasing tumour response, organ preservation, and reducing surgical morbidity, while maintaining oncological safety.
Ethics and dissemination
The study protocol (V.3.0, dated on 24 September 2019) was approved by the local ethics committee (Comité de Protection des Personnes Sud-Est II, 18 December 2019, Reference number 2019-A02493-54) and the institutional review board COMERE. The French National Drug Agency Authority (ANSM) was informed. The study was registered on Clinicaltrials.gov, identifier NCT04103567.
All patients will be informed of the study objectives and procedures by the investigators before enrolment. A signed informed consent will be obtained from all patients before their inclusion in the study and before any study procedure is performed. All patients may end their participation in the study at any time, for whatever reason, without any consequence or prejudice concerning their care. Study participants will be able to request global results from investigators as soon as study results become available.
In the event of substantial modification, the request will be sent by the sponsor to the ethics committee for an opinion. On receipt of the favourable opinion, the sponsor will send the amended version of the protocol to all investigators.
The study will be conducted in accordance with the current French and European Regulatory requirements, including regulations on biomedical research from the Public Health Code, the bioethics and data protection laws and decrees, the French Jardé’s law on research implicating human beings, the Good Clinical Practice and the Helsinki Declaration.
Supplementary Material
Acknowledgments
The authors thank the Clinical Research and Innovation Department of the Montpellier Cancer Institute for help with regulatory and administrative aspects of the study, Dr. Stéphanie Delaine for her help in obtaining funding for the study, Pierre Sauvanet and Michael Rodrigues for technical advice, and Maxime Tressol for the microbiological analyses.
Footnotes
Twitter: @matbonne
Contributors: GC, CT, JG, CF, MJ, GR, CC, P-EC, PR and MB wrote the protocol. MJ, GC, PR, CT and CF conducted statistical trial planning. PR, CT and CF handled ethics and regulatory affairs. GC, CT, HF, MB, CF, NB, PR and MJ wrote the paper draft. GC, MB, MJ, PR, CT and CF contributed to the trial design and modifications. All authors read and approved the final manuscript.
Funding: This work was supported by the SIRIC Montpellier Cancer, the Biocodex Microbiota Foundation and La Ligue Contre le Cancer (Herault committee). The funding body was not involved in the study design and will not be involved in data collection, data analysis and interpretation, and writing of the study report and publication.
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Ethics statements
Patient consent for publication
Consent obtained directly from patient(s)
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Sauer R, Becker H, Hohenberger W, et al. Preoperative versus postoperative chemoradiotherapy for rectal cancer. N Engl J Med 2004;351:1731–40. 10.1056/NEJMoa040694 [DOI] [PubMed] [Google Scholar]
- 3.Bosset J-F, Collette L, Calais G, et al. Chemotherapy with preoperative radiotherapy in rectal cancer. N Engl J Med 2006;355:1114–23. 10.1056/NEJMoa060829 [DOI] [PubMed] [Google Scholar]
- 4.van Gijn W, Marijnen CAM, Nagtegaal ID, et al. Preoperative radiotherapy combined with total mesorectal excision for resectable rectal cancer: 12-year follow-up of the multicentre, randomised controlled Tme trial. Lancet Oncol 2011;12:575–82. 10.1016/S1470-2045(11)70097-3 [DOI] [PubMed] [Google Scholar]
- 5.Lakkis Z, Manceau G, Bridoux V, et al. Management of rectal cancer: the 2016 French guidelines. Colorectal Dis 2017;19:115–22. 10.1111/codi.13550 [DOI] [PubMed] [Google Scholar]
- 6.Heald RJ, Husband EM, Ryall RD. The mesorectum in rectal cancer surgery--the clue to pelvic recurrence? Br J Surg 1982;69:613–6. 10.1002/bjs.1800691019 [DOI] [PubMed] [Google Scholar]
- 7.Patel UB, Taylor F, Blomqvist L, et al. Magnetic resonance imaging-detected tumor response for locally advanced rectal cancer predicts survival outcomes: mercury experience. J Clin Oncol 2011;29:3753–60. 10.1200/JCO.2011.34.9068 [DOI] [PubMed] [Google Scholar]
- 8.Habr-Gama A, Perez RO, Nadalin W, et al. Operative versus Nonoperative treatment for stage 0 distal rectal cancer following chemoradiation therapy: long-term results. Trans Meet Am Surg Assoc 2004;CXXII:309–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Renehan AG, Malcomson L, Emsley R, et al. Watch-and-wait approach versus surgical resection after chemoradiotherapy for patients with rectal cancer (the OnCoRe project): a propensity-score matched cohort analysis. Lancet Oncol 2016;17:174–83. 10.1016/S1470-2045(15)00467-2 [DOI] [PubMed] [Google Scholar]
- 10.Rullier E, Rouanet P, Tuech J-J, et al. Organ preservation for rectal cancer (GRECCAR 2): a prospective, randomised, open-label, multicentre, phase 3 trial. Lancet 2017;390:469–79. 10.1016/S0140-6736(17)31056-5 [DOI] [PubMed] [Google Scholar]
- 11.Rouanet P, Rullier E, Lelong B, et al. Tailored treatment strategy for locally advanced rectal carcinoma based on the tumor response to induction chemotherapy: preliminary results of the French phase II multicenter GRECCAR4 trial. Dis Colon Rectum 2017;60:653–63. 10.1097/DCR.0000000000000849 [DOI] [PubMed] [Google Scholar]
- 12.Conroy T, Bosset J-F, Etienne P-L, et al. Neoadjuvant chemotherapy with FOLFIRINOX and preoperative chemoradiotherapy for patients with locally advanced rectal cancer (UNICANCER-PRODIGE 23): a multicentre, randomised, open-label, phase 3 trial. Lancet Oncol 2021;22:702–15. 10.1016/S1470-2045(21)00079-6 [DOI] [PubMed] [Google Scholar]
- 13.Dworak O, Keilholz L, Hoffmann A. Pathological features of rectal cancer after preoperative radiochemotherapy. Int J Colorectal Dis 1997;12:19–23. 10.1007/s003840050072 [DOI] [PubMed] [Google Scholar]
- 14.Gérard J-P, Azria D, Gourgou-Bourgade S, et al. Comparison of two neoadjuvant chemoradiotherapy regimens for locally advanced rectal cancer: results of the phase III trial ACCORD 12/0405-Prodige 2. J Clin Oncol 2010;28:1638–44. 10.1200/JCO.2009.25.8376 [DOI] [PubMed] [Google Scholar]
- 15.Lefevre JH, Mineur L, Kotti S, et al. Effect of interval (7 or 11 weeks) between neoadjuvant radiochemotherapy and surgery on complete pathologic response in rectal cancer: a multicenter, randomized, controlled trial (GRECCAR-6). Journal of Clinical Oncology 2016;34:3773–80. 10.1200/JCO.2016.67.6049 [DOI] [PubMed] [Google Scholar]
- 16.Nougaret S, Reinhold C, Mikhael HW, et al. The use of MR imaging in treatment planning for patients with rectal carcinoma: have you checked the "DISTANCE"? Radiology 2013;268:330–44. 10.1148/radiol.13121361 [DOI] [PubMed] [Google Scholar]
- 17.Nougaret S, Rouanet P. Restaging rectal cancer after neoadjuvant treatment with multiparametric MRI: a landscape of new opportunities. Diagn Interv Imaging 2016;97:839–41. 10.1016/j.diii.2016.08.012 [DOI] [PubMed] [Google Scholar]
- 18.Kalisz KR, Enzerra MD, Paspulati RM. Mri evaluation of the response of rectal cancer to neoadjuvant chemoradiation therapy. Radiographics 2019;39:538–56. 10.1148/rg.2019180075 [DOI] [PubMed] [Google Scholar]
- 19.Boleij A, Tjalsma H. Gut bacteria in health and disease: a survey on the interface between intestinal microbiology and colorectal cancer. Biol Rev Camb Philos Soc 2012;87:701–30. 10.1111/j.1469-185X.2012.00218.x [DOI] [PubMed] [Google Scholar]
- 20.Ahn J, Sinha R, Pei Z, et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst 2013;105:1907–11. 10.1093/jnci/djt300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collins D, Hogan AM, Winter DC. Microbial and viral pathogens in colorectal cancer. Lancet Oncol 2011;12:504–12. 10.1016/S1470-2045(10)70186-8 [DOI] [PubMed] [Google Scholar]
- 22.Swidsinski A, Khilkin M, Kerjaschki D, et al. Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology 1998;115:281–6. 10.1016/S0016-5085(98)70194-5 [DOI] [PubMed] [Google Scholar]
- 23.Arthur JC, Jobin C. The complex interplay between inflammation, the microbiota and colorectal cancer. Gut Microbes 2013;4:253–8. 10.4161/gmic.24220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russo TA, Johnson JR. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect 2003;5:449–56. 10.1016/S1286-4579(03)00049-2 [DOI] [PubMed] [Google Scholar]
- 25.Nougayrède J-P, Homburg S, Taieb F, et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 2006;313:848–51. 10.1126/science.1127059 [DOI] [PubMed] [Google Scholar]
- 26.Cuevas-Ramos G, Petit CR, Marcq I, et al. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci U S A 2010;107:11537–42. 10.1073/pnas.1001261107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson MR, Jiang Y, Villalta PW, et al. The human gut bacterial genotoxin colibactin alkylates DNA. Science 2019;363:eaar7785. 10.1126/science.aar7785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buc E, Dubois D, Sauvanet P, et al. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS One 2013;8:e56964. 10.1371/journal.pone.0056964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonnet M, Buc E, Sauvanet P, et al. Colonization of the human gut by E. coli and colorectal cancer risk. Clin Cancer Res 2014;20:859–67. 10.1158/1078-0432.CCR-13-1343 [DOI] [PubMed] [Google Scholar]
- 30.Arthur JC, Gharaibeh RZ, Mühlbauer M, et al. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat Commun 2014;5:4724. 10.1038/ncomms5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cougnoux A, Dalmasso G, Martinez R, et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut 2014;63:1932–42. 10.1136/gutjnl-2013-305257 [DOI] [PubMed] [Google Scholar]
- 32.Dejea CM, Fathi P, Craig JM, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 2018;359:592–7. 10.1126/science.aah3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lucas C, Salesse L, Hoang MHT, et al. Autophagy of intestinal epithelial cells inhibits colorectal carcinogenesis induced by Colibactin-Producing Escherichia coli in ApcMin/+Mice. Gastroenterology 2020;158:1373–88. 10.1053/j.gastro.2019.12.026 [DOI] [PubMed] [Google Scholar]
- 34.Tomkovich S, Yang Y, Winglee K, et al. Locoregional effects of microbiota in a preclinical model of colon carcinogenesis. Cancer Res 2017;77:2620–32. 10.1158/0008-5472.CAN-16-3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edge SB, Compton CC. The American joint committee on cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol 2010;17:1471–4. 10.1245/s10434-010-0985-4 [DOI] [PubMed] [Google Scholar]
- 36.Lopès A, Billard E, Casse AH, et al. Colibactin-positive Escherichia coli induce a procarcinogenic immune environment leading to immunotherapy resistance in colorectal cancer. Int J Cancer 2020;146:3147–59. 10.1002/ijc.32920 [DOI] [PubMed] [Google Scholar]
- 37.Villéger R, Lopès A, Carrier G, et al. Intestinal microbiota: a novel target to improve anti-tumor treatment? Int J Mol Sci 2019;20:4584. 10.3390/ijms20184584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Payros D, Secher T, Boury M, et al. Maternally acquired genotoxic Escherichia coli alters offspring's intestinal homeostasis. Gut Microbes 2014;5:313–512. 10.4161/gmic.28932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gérard J-P, André T, Bibeau F, et al. Rectal cancer: French intergroup clinical practice guidelines for diagnosis, treatments and follow-up (SNFGE, FFCD, GERCOR, UNICANCER, SFCD, SFED, SFRO). Digestive and Liver Disease 2017;49:359–67. 10.1016/j.dld.2017.01.152 [DOI] [PubMed] [Google Scholar]
- 40.Nagtegaal ID, van de Velde CJH, van der Worp E, et al. Macroscopic evaluation of rectal cancer resection specimen: clinical significance of the pathologist in quality control. J Clin Oncol 2002;20:1729–34. 10.1200/JCO.2002.07.010 [DOI] [PubMed] [Google Scholar]
- 41.Gagnière J, Bonnin V, Jarrousse A-S, et al. Interactions between microsatellite instability and human gut colonization by Escherichia coli in colorectal cancer. Clin Sci 2017;131:471–85. 10.1042/CS20160876 [DOI] [PubMed] [Google Scholar]
- 42.Gagnière J, Raisch J, Veziant J, et al. Gut microbiota imbalance and colorectal cancer. World J Gastroenterol 2016;22:501. 10.3748/wjg.v22.i2.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang Q, Chiu J, Chen Y, et al. Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin Cancer Res 2017;23:2061–70. 10.1158/1078-0432.CCR-16-1599 [DOI] [PubMed] [Google Scholar]
- 44.Eklöf V, Löfgren-Burström A, Zingmark C, et al. Cancer-associated fecal microbial markers in colorectal cancer detection: fecal microbial markers in colorectal cancer detection. Int J Cancer 2017;141:2528–36. 10.1002/ijc.31011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bruheim K, Guren MG, Skovlund E, et al. Late side effects and quality of life after radiotherapy for rectal cancer. Int J Radiat Oncol Biol Phys 2010;76:1005–11. 10.1016/j.ijrobp.2009.03.010 [DOI] [PubMed] [Google Scholar]
- 46.Peeters KCMJ, van de Velde CJH, Leer JWH, et al. Late side effects of short-course preoperative radiotherapy combined with total mesorectal excision for rectal cancer: increased bowel dysfunction in irradiated patients--a Dutch colorectal cancer group study. J Clin Oncol 2005;23:6199–206. 10.1200/JCO.2005.14.779 [DOI] [PubMed] [Google Scholar]
- 47.Reis Ferreira M, Andreyev HJN, Mohammed K, et al. Microbiota- and radiotherapy-induced gastrointestinal side-effects (MARS) study: a large pilot study of the microbiome in acute and late radiation enteropathy. Clin Cancer Res 2019;25:clincanres.0960.2019:6487–500. 10.1158/1078-0432.CCR-19-0960 [DOI] [PubMed] [Google Scholar]
- 48.Gerassy-Vainberg S, Blatt A, Danin-Poleg Y, et al. Radiation induces proinflammatory dysbiosis: transmission of inflammatory susceptibility by host cytokine induction. Gut 2018;67:97–107. 10.1136/gutjnl-2017-313789 [DOI] [PubMed] [Google Scholar]
- 49.Nam Y-D, Kim HJ, Seo J-G, et al. Impact of pelvic radiotherapy on gut microbiota of gynecological cancer patients revealed by massive pyrosequencing. PLoS ONE 2013;8:e82659. 10.1371/journal.pone.0082659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pham TT, Liney G, Wong K, et al. Study protocol: multi-parametric magnetic resonance imaging for therapeutic response prediction in rectal cancer. BMC Cancer 2017;17:465. 10.1186/s12885-017-3449-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gonçalves-Ribeiro S, Sanz-Pamplona R, Vidal A, et al. Prediction of pathological response to neoadjuvant treatment in rectal cancer with a two-protein immunohistochemical score derived from stromal gene-profiling. Ann Oncol 2017;28:2160–8. 10.1093/annonc/mdx293 [DOI] [PubMed] [Google Scholar]
- 52.Zhang S, Bai W, Tong X, et al. Correlation between tumor microenvironment-associated factors and the efficacy and prognosis of neoadjuvant therapy for rectal cancer. Oncol Lett 2019;17:1062–70. 10.3892/ol.2018.9682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baskar R, Dai J, Wenlong N, et al. Biological response of cancer cells to radiation treatment. Front Mol Biosci 2014;1:24. 10.3389/fmolb.2014.00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuwahara Y, Oikawa T, Ochiai Y, et al. Enhancement of autophagy is a potential modality for tumors refractory to radiotherapy. Cell Death Dis 2011;2:e177. 10.1038/cddis.2011.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Digomann D, Linge A, Dubrovska A. SLC3A2/CD98hc, autophagy and tumor radioresistance: a link confirmed. Autophagy 2019;15:1850–1. 10.1080/15548627.2019.1639302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu T, Guo F, Yu Y, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 2017;170:e16:548–63. 10.1016/j.cell.2017.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barker HE, Paget JTE, Khan AA, et al. The tumour microenvironment after radiotherapy: mechanisms of resistance and recurrence. Nat Rev Cancer 2015;15:409–25. 10.1038/nrc3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2022-061527supp001.pdf (87.6KB, pdf)
bmjopen-2022-061527supp002.pdf (137.2KB, pdf)
bmjopen-2022-061527supp003.pdf (20KB, pdf)