Abstract
Duck plague virus (DPV) pUL48 is a homologous of herpes simplex virus VP16, and some studies have shown that VP16 is essential for viral replication and proliferation, but there are few studies on DPV pUL48. Therefore, in order to study the function of pUL48 protein, we constructed a UL48-deleted mutant (DPV-BAC-∆UL48) that completely reemoved the UL48 gene from the DPV BAC genome and the revertant virus (DPV-BAC-∆UL48R) by using the 2-step red recombination system. Compared with the parental virus (DPV-BAC) and the revertant virus, the titer of UL48-deleted mutant was reduced by more than 38.2%, and the efficiency of producing infectious virions was significantly reduced. In addition, the average size of plaques produced by UL48-deleted mutant was about 30% smaller than that of the parental and revertant viruses, suggesting that pUL48 protein affected the cell-to-cell transmission of DPV. Finally, pharmacological inhibition assay showed that pUL48 is a late protein of DPV. In this study, we found that UL48, as a late gene, plays an important role in viral replication by affecting the formation of DPV infectious virion, virus cell-to-cell transmission, and viral genome transcription, which may provide some help for the study of the function of DPV pUL48 protein and the prevention and control of DPV.
Key words: duck plague virus, pUL48 protein, late gene, viral replication
BACKGROUND
Duck plague virus (DPV), belonging to herpesvirales, alphaherpesvirinae, mardivirus, is a double-stranded linear DNA virus (Lefkowitz et al., 2018). DPV chinese virulent strain CHv (DPV CHv) genome has a typical alphaherpesvirus genome structure, which is composed of unique long region (UL), unique short region (US), the internal inverted repeat sequence (IRS) and terminal inverted repeat sequence (TRS) constitute the UL-IRS-US-TRS structure from 5′ to 3′. The DPV genome contains 78 open reading frames (ORFs), 68 in UL region, 11 in US region, and 2 ORFs located in IRS and TRS region (Wu et al., 2012).
UL48 gene, located in the UL region of herpesvirus genome, is responsible for encoding VP16 or Vmw65. VP16 not only regulates the assembly and maturation of virus, but also activates the transcription of viral gene inducing factor, so it is also called α gene transinducing factor (Fan et al., 2020). Studies have shown that the effcet of VP16 on viral replication varies among different alphaherpesviruses. For example, herpes simplex virus type 1 (HSV-1) deleted VP16 cannot detect progeny virus in non-complementary cells without VP16, and can only proliferate in complementary cell lines containing VP16 (Mossman et al., 2000). This indicates that VP16 is essential for replication and proliferation of HSV-1. Moreover, although the pseudorabies virus (PRV) with VP16 knocked out can still proliferated in host cells, the particle morphology was severely defective, with a large number of newly formed nucleocapsids remaining in the cytoplasm, while few mature extracellular enveloped virions (Fuchs et al., 2002). Therefore, the study of DPV pUL48 protein is very important to understand the pathogenesis of DPV.
In this study, we utilized a 2-step red recombination based on an infectious artificial chromosome (BAC) containing the DPV CHv genome system, the UL48-deleted mutant (DPV-BAC-∆UL48) with green flourescence and the revertant virus (DPV-BAC-∆UL48R) were successfully constructed and rescued.
MATERIALS AND METHODS
Virus, Cell, and Antibody
DPV-BAC (GenBank:JQ647509) strain was preserved by the Poultry Disease Control Research Center of Sichuan Agricultural University. In this study all the experiments were based on duck embryo fibroblasts (DEFs), which were prepared from 10-day-old duck embryos purchased from a duck farm in Pixian County, Sichuan Province. DEFs were grown in DMEM medium containing 10% newborn bovine serum (NBS; Gibco-BRL, Grand Island, NY) and the medium was changed to DMEM medium containing 2% NBS after transfection or infection.Taking 12-well plates as an example, each test used 5.86 × 105 cells. Rabbit anti-UL48 polyclonal antibody serum, rabbit anti-UL47 polyclonal antibody serum, rabbit anti-ICP4 polyclonal antibody serum, and rabbit anti-UL29 polyclonal antibody serum were prepared and preserved by the Research Center of Avian Disease Prevention and Control, Sichuan Agricultural University.Rabbit anti-beta (β)-actin antibody was obtained from Proteintech. HRP-conjugated goat anti-mouse IgG and HRP-conjugated goat anti-rabbit IgG were purchased from Beyotime Biotechnology (Shanghai, China).
Strains, Plasmids, and Cell Transfection
GS1783-pBAC-DPV strain, eukaryotic expression plasmids pCAGGS and pCAGGS-pUL48-HA were preserved and provided by the Poultry Disease Control Research Center of Sichuan Agricultural University. In this study, Lipofectamine 3000 reagent was used (Thermo Fisher Scientific, Waltham, MA). One μg of each plasmid DNA was mixed with volumes of 2 μL of each transfection reagent according to their related manufacturers` instructions (Rahimi et al., 2018).
Construction, Rescue, and Identification of Recombinant Viruses
The recombinant virus was constructed based on the 2-step red recombination system (Tischer et al., 2010). In a nutshell, targeted fragments were amplified by PCR, which targeted E.coli GS1783-pBAC-DPV receptive cells. Deletion of the Kan fragment was achieved by a second homologous recombination with the Kan resistance gene replacing the UL48 gene by the first homologous recombination, followed by cleavage of the I-SecI site. Recombinant pBAC plasmids were extracted using QIAGEN Plasmid Midi Kit (No.12143) and transfected into DEFs of 12-well plate by Lipofectamine 3000 Reagent (Thermo Fisher Scientific, Waltham, MA). Cultured until cells developed green fluorescent plaques representative of cytopathic, cells were collected and subcultured. The obtained viruses were identified by PCR, Western Blot, and IFA.
Western Blot
Samples lysed by RIPA lysis buffer (strong) (Beyotime Biotechnology, Shanghai, China) were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF, Millipore, Boston, MA). Subsequently, the PVDF was blocked with 5% skim milk at room temperature for 4 h and incubated overnight with primary antibody at 4℃. After that, it was incubated with secondary antibodies of HRP-conjugated goat anti-rabbit/mouse IgG (1:3,000) at room temperature for 1h. Finally, the protein signal was visualized with an enhanced chemiluminescence (ECL) Western blotting analysis system. The primary antibodies were used: rabbit anti-UL48 (1:800), rabbit anti-UL47(1: 500), rabbit anti-UL29 (1:500), and rabbit anti-ICP4 (1:400).
Real-Time PCR
Total RNA was extracted using RNAiso Plus (TaKaRa, Tokyo, Japan) and the PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa) was used to reverse transcribed the RNA into cDNA. A real-time PCR (RT-PCR) assay was used to detect mRNA levels of each gene by using the TB Green Premix Ex Taq II (TaKaRa) and corresponding quantitative primers. 18sRNA was used as an internal reference, and the relative transcription level of each gene was analyzed by the 2−ΔΔCt relative quantitative method. In addition, viral DNA was extracted using MiniBEST Viral RNA/DNA Extraction Kit Ver.5.0 (TaKaRa), and the UL30 probe and primers were used to detect the copy number of the viral genome.
Multistep Viral Growth Kinetics
DEFs in 24-well plates were infected with 0.01 MOI DPV-BAC, DPV-BAC-∆UL48, or DPV-BAC-∆UL48R, incubated at 37℃ for 2 h and then changed to 2% NBS DMEM. Samples of the infected cells and supernatants were collected at 24 h, 48 h, 72 h and 84 h postinfection,and the volume of each sample was increased to 500 μL. The titer of virus was assessed by determining the half tissue culture infectious dose (TCID50) of virus in cells and supernatant.
Plaque Morphology of the Recombinant Viruses
DEFs in 12-well plates were infected with 0.001MOI DPV-BAC, DPV-BAC-∆UL48, or DPV-BAC-∆UL48R, and then the supernatant was dicarded and 1% methylcellulose(Solarbio, Beijing, China) was added to cover the cells after incubation at 37 °C for 2 h. At 48 hpi, the green fluorescent plaques formation in cells was observed under a fluorescence microscope. Twelve fluorescent plaques were randomly selected for statistical analysis of their size, and the plaque size of the parental virus was set as 100%.
Adsorption, Invasion, Replication, and Release of Recombinant Viruses
To facilitate adsorption, invasion, and replication experiments, we adjusted the viral copy numbers of DPV-BAC, DPV-BAC-∆UL48 and DPV-BAC-∆UL48R to 1.0 × 108 copies/100 μL. Adsorption: DEFs were precooled at 4℃ for 1 h, then DPV-BAC, DPV-BAC-∆UL48, or DPV-BAC-∆UL48R were infected and incubated at 4℃ for 2 h (to permit binding, but to prevent viral internalization). The cell samples were washed with pre-cooled PBS for 5 times and added with DMEM. After freeze-thaw, the virus genome of the sample was extracted, and the virus copy number was detected. Invasion: DEFs were pre-cooled at 4℃ for 1 h and incubated at 4℃ for 2 h after inoculation with 3 viruses. The culture medium was then changed and the temperature was raised to 37℃ to allow entry of the virus. Three hours later, cell samples were collected and DNA was extracted for virus to detect copy number. Replication: DEFs were incubated with 0.05 MOI recombinant cirus at 37℃ for 6 h, and then the medium was changed to 2% NBS DMEM. Cell samples were collected for RT-qPCR at 6 h, 7 h, 8 h, 9 h and 10 h after medium was changed. Release: DEFs were incubated with 0.05 MOI recombinant cirus at 37℃ for 6 h, and then the medium was changed to 2% NBS DMEM. After 18 h, the culture medium was changed at 30, 60, 90, and 120 min after the change of medium to detect TCID50.
Pharmacological Inhibition Assay
Pharmacological inhibition assay was performed to confirm the DPV UL48 gene expression patterns. DEFs in 12-well plates were respectively added 350 μg/mL ganciclovir (GCV), a nucleic acid synthesis inhibitor, and 200 μg/mL cycloheximide (CHX), a protein synthesis inhibitor, and infected with 0.01MOI. DPV-BAC. Total RNA was isolated at 24 h postinfection and subsequently reverse transcribed into cDNA. The cDNA was used for subsequent PCR analysis, and the product was identified using a 1% agarose gel.
RESULTS AND DISCUSSION
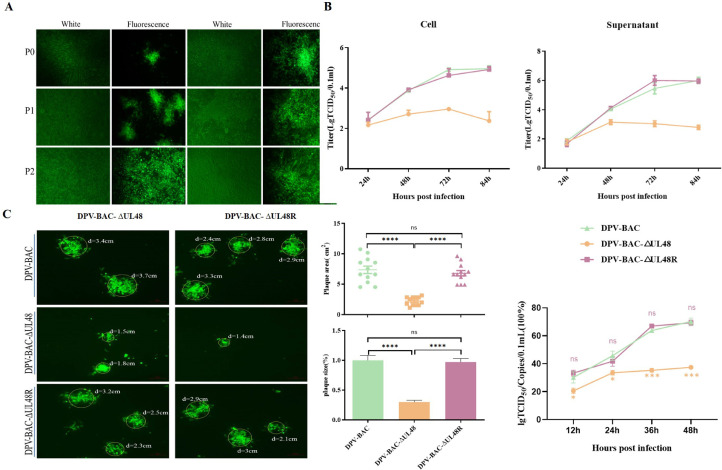
In this paper, the UL48-deleted mutant (DPV-BAC-∆UL48) and revertant virus (DPV-BAC-∆UL48R) were successfully constructed and rescued based on the lab-constructed bacterial artificial chromosome recombinant duck plague virus rescue platform (BAC-DPV) combined with red 2-step homologous recombination technology (Figure 1A). But it was found in the rescue process that exogenous expression of pUL48 was required to generate green fluorescent plaques formed by recombinant virus, which may be that the expression of exogenous pUL48 in transfected cells activated the expression of IE genes to further promote the formation of mature virions, and this also reflected that pUL48 had a great effect on the replication of DPV. The results of the multistep growth curve assay showed that UL48 deletion resulted in a several hundred fold decrease in viral titer due to the fact that the multistep growth curve traversed multiple replication cycles of the virus, which amplified the effect of UL48 deletion on viral replication. In addition, we also confirmed that the deletion of UL48 gene significantly reduced the efficiency of infectious virions formation by DPV (Figure 2B), indicating that pUL48 affects the ability of DPV to produce progeny virions.
Figure 1.
Determination of viral growth kinetics and cell-to-cell spread. (A) Rescue of DPV-BAC-∆UL48 and DPV-BAC-∆UL48R. (B) Intracellular and supernatant viral titers and the efficiency of infectious virion formation (Titers/copies). (C) Green fluorescent plaques produced by DPV-BAC, DPV-BAC-∆UL48, or DPV-BAC-∆UL48R. On the right are the corresponding statistics of area or size of green fluorescent plaque. t test was used to analyze the data differences between the two groups, and the significance was marked as follows: NS showed no difference, * * * * P < 0.0001. Error bars represent the SEMs.
Figure 2.
Identification of UL48 gene type. (A) Transcriptional phase of ICP4/UL29/UL47/UL48 was detected by RT-qPCR. (B) The expression phase of ICP4/UL29/UL47/pUL48 protein was detected by WB. (C) UL48 gene expression was inhibited by GCV/CHX.
The life cycle of herpesviruses is roughly divided into adsorption, invasion, replication of DNA, nucleocapsid assembly, release of mature virions, and cell-to-cell transmission (Owen et al., 2015). Adsorption is the first step of virus infection, and viral envelope glycoprotein adsorb to cell surface specific receptors, a process that tends to be completed in minutes to tens of minutes; The viral envelopment fuses with the cell membrane, forming a channel and thereby allowing the viral nucleocapsid and tegument proteins to enter the cytoplasm; Upon entry into the cell, the transcription system of the host nucleus is utilized to activate transcription and translation of the virus, synthesizing various components of the virus. Newly synthesized viral components are gradually packaged into complete virons within the infected cells; Finally, mature virions are eliminated from the cell through the exocytosis or endoplasmic reticulum system (Connolly et al., 2011; Hadigal and Shukla, 2013; Owen et al., 2015). Here, we found that pUL48 knockdown decreased virus adsorption, invasion, replication of viral DNA, and release of progeny virus to various degrees (Data not shown). But after the deletion of pUL48, there is a significant decrease in adsorption – the first step in virus-infected cells, and the subsequent decrease in invasion, replication, and release processes is likely because the altered adsorption capacity induces subsequent cascades. So, whether the DPV pUL48 only has an effect on the adsorption process of the virus or has an effect on various parts of the cycle follow-up needs to be studied in depth. We also found that after the deletion of pUL48, the number and the area of green fluorescent plaques produced by the recombinant virus decreased significantly (Figure 1C), indicating that UL48 not only affects the proliferation of DPV in DEFs, but also blocks the transmission of DPV between cells.
There is a strict cascade and timing in the transcription of genes during infection by herpesviruses and is controlled by viral proteins. In descending order of transcription, they are classified as IE, E, and L gene, which encode immediate early, early, and late protein, respectively .To determine the gene type of DPV UL48, we analyzed the trends of UL48 mRNA and protein levels at different time points. It was found that the trends of UL48 transcription and expression levels were generally consistent, and it could be detected as early as 12 h after virus infection, which was similar to the contrast gene UL47 (Figures 2A and 2B). GCV is a nucleotide analogue that, upon entry of drugs into virus-infected cells, is phosphorylated to an activated form of triphosphate and form an inhibition of viral replication by affecting the viral DNA polymerase as well as blocking DNA strand extension in 2 ways. CHX, a protein synthesis inhibitor produced by streptomyces griseus, effectively inhibits protein expression by interfering with the translocation of the protein synthesis process. Therefore, GCV and CHX are often used in studies of herpesvirus gene type determination. To further define the gene type of DPV UL48, we took advantage of both GCV and CHX inhibition and found that transcription of UL48 was sensitive to inhibition by both CHX and GCV, consistent with the results obtained transcription and expression phases, indicating that UL48 as a late gene of DPV (Figure 2C).
In summary, in the above studies, we found that the UL48 gene, as a late gene of DPV, plays an important role in the viral life and that complete deletion affects the viral replication, the formation of infectious virions, the ability of the virus to spread between cells. It is hoped that this study will provide basic information on the DPV UL48 gene and help in the study of pUL48 protein function and prevention and control of DPV.
ACKNOWLEDGMENTS
This work was supported by t grants from National Natural Science Foundation of China (31872476), the earmarked fund for China Agriculture Research System (CARS-42-17) and the Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System (SCCXTD-2020-18).
DISCLOSURES
The authors have no financial or non-financial competing interests to declare.
References
- Connolly S.A., Jackson J.O., Jardetzky T.S., Longnecker R. Fusing structure and function: a structural view of the herpesvirus entry machinery. Nat. Rev. Microbiol. 2011;9:369–381. doi: 10.1038/nrmicro2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan D., Wang M., Cheng A., Jia R., Yang Q., Wu Y., Zhu D., Zhao X., Chen S., Liu M., Zhang S., Ou X., Mao S., Gao Q., Sun D., Wen X., Liu Y., Yu Y., Zhang L., Tian B., Pan L., Chen X. The role of VP16 in the life cycle of alphaherpesviruses. Front. Microbiol. 2020;11:1910. doi: 10.3389/fmicb.2020.01910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs W., Granzow H., Klupp B.G., Kopp M., Mettenleiter T.C. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 2002;76:6729–6742. doi: 10.1128/JVI.76.13.6729-6742.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadigal S., Shukla D. Exploiting herpes simplex virus entry for novel therapeutics. Viruses. 2013;5:1447–1465. doi: 10.3390/v5061447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkowitz E.J., Dempsey D.M., Hendrickson R.C., Orton R.J., Siddell S.G., Smith D.B. Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV) Nucleic Acids Res. 2018;46 doi: 10.1093/nar/gkx932. D708-d17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossman K.L., Sherburne R., Lavery C., Duncan J., Smiley J.R. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 2000;74:6287–6299. doi: 10.1128/jvi.74.14.6287-6299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D.J., Crump C.M., Graham S.C. Tegument assembly and secondary envelopment of alphaherpesviruses. Viruses. 2015;7:5084–5114. doi: 10.3390/v7092861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi P., Mobarakeh V.I., Kamalzare S., SajadianFard F., Vahabpour R., Zabihollahi R. Comparison of transfection efficiency of polymer-based and lipid-based transfection reagents. Bratisl. Lek Listy. 2018;119:701–705. doi: 10.4149/BLL_2018_125. [DOI] [PubMed] [Google Scholar]
- Tischer B.K., Smith G.A., Osterrieder N. En passant mutagenesis: a two step markerless red recombination system. Methods Mol. Biol. 2010;634:421–430. doi: 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- Wu Y., Cheng A., Wang M., Yang Q., Zhu D., Jia R., Chen S., Zhou Y., Wang X., Chen X. Complete genomic sequence of Chinese virulent duck enteritis virus. J. Virol. 2012;86:5965. doi: 10.1128/JVI.00529-12. [DOI] [PMC free article] [PubMed] [Google Scholar]