Abstract
Background
Agents blocking programmed cell death protein 1/programmed death-ligand 1 (PD-1/PD-L1) have been approved for triple-negative breast cancer (TNBC). However, the response rate of anti-PD-1/PD-L1 is still unsatisfactory, partly due to immunosuppressive factors such as transforming growth factor-beta (TGF-β). In our previous pilot study, the bispecific antibody targeting TGF-β and murine PD-L1 (termed YM101) showed potent antitumor effect. In this work, we constructed a bispecific antibody targeting TGF-β and human PD-L1 (termed BiTP) and explored the antitumor effect of BiTP in TNBC.
Methods
BiTP was developed using Check-BODYTM bispecific platform. The binding affinity of BiTP was measured by surface plasmon resonance, ELISA, and flow cytometry. The bioactivity was assessed by Smad and NFAT luciferase reporter assays, immunofluorescence, western blotting, and superantigen stimulation assays. The antitumor activity of BiTP was explored in humanized epithelial-mesenchymal transition-6-hPDL1 and 4T1-hPDL1 murine TNBC models. Immunohistochemical staining, flow cytometry, and bulk RNA-seq were used to investigate the effect of BiTP on immune cell infiltration.
Results
BiTP exhibited high binding affinity to dual targets. In vitro experiments verified that BiTP effectively counteracted TGF-β-Smad and PD-L1-PD-1-NFAT signaling. In vivo animal experiments demonstrated that BiTP had superior antitumor activity relative to anti-PD-L1 and anti-TGF-β monotherapy. Mechanistically, BiTP decreased collagen deposition, enhanced CD8+ T cell penetration, and increased tumor-infiltrating lymphocytes. This improved tumor microenvironment contributed to the potent antitumor activity of BiTP.
Conclusion
BiTP retains parent antibodies’ binding affinity and bioactivity, with superior antitumor activity to parent antibodies in TNBC. Our data suggest that BiTP might be a promising agent for TNBC treatment.
Keywords: Immunotherapy; Breast Neoplasms; Tumor Microenvironment; Lymphocytes, Tumor-Infiltrating; Antibodies, Neoplasm
WHAT IS ALREADY KNOWN ON THIS TOPIC
Although anti-PD-1/PD-L1 therapy revolutionizes the treatment paradigm of triple-negative breast cancer (TNBC), the low response rate of anti-PD-1/PD-L1 therapy has not been well resolved.
For late-stage breast cancer, TGF-β promotes tumor progression by enhancing metastasis, treatment resistance, and immune escape.
WHAT THIS STUDY ADDS
We developed a bispecific antibody targeting TGF-β and human PD-L1 (termed BiTP) and explored the antitumor effect of BiTP in TNBC.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
BiTP exhibits bioactivities in vitro and potent antitumor activity in TNBC models.
Given the positive results of the preclinical study, BiTP is a promising candidate to enter further clinical study.
Background
Breast cancer is the leading threat to women’s health, causing nearly 68,500 deaths globally in 2020.1 2 According to the status of hormone receptors and human epidermal growth factor 2 (HER2), breast cancers are classified into four subtypes: luminal A/B, HER2-enriched, normal-like, triple-negative breast cancer (TNBC). TNBC is the most aggressive breast cancer subtype, lacking specific targets for effective therapeutic agents. Therefore, there is a critical need for innovative treatment strategies for TNBC. Immunotherapy, especially anti-programmed cell death protein 1/programmed death-ligand 1 (anti-PD-1/PD-L1), revolutionizes multiple cancers’ treatment paradigm.3–7 Currently, pembrolizumab combined with chemotherapy has been approved for TNBC.8 9 Nevertheless, the low response rate of anti-PD-1/PD-L1 therapy has not been well resolved.10 The therapeutic effect of anti-PD-1/PD-L1 is hampered by multiple immunosuppressive factors, including transforming growth factor-beta (TGF-β).11
As a multifunctional cytokine, the function of TGF-β is context-dependent. For normal tissue and early-stage breast cancer, TGF-β plays as a tumor suppressor by inducing cell-cycle arrest and apoptosis.12 However, for late-stage breast cancer, TGF-β promotes tumor progression by enhancing metastasis, radiotherapy or chemotherapy resistance, and the tumor microenvironment (TME) reprogramming.13–16 The immunosuppressive effects of TGF-β on the TME are multifaceted: impairing the activities of tumor-infiltrating lymphocytes (TILs), altering the polarization of macrophages, inducing regulatory T (Treg) cell differentiation, and limiting dendritic cell (DC) functions.17 18 Moreover, TGF-β undermines TIL penetration by increasing peritumoral collagen production of cancer-associated fibroblast (CAF).19
Accumulating data demonstrate high TGF-β is correlated with resistance to anti-PD-1/PD-L1 therapy,19 while neutralizing TGF-β improves the efficacy of anti-PD-1/PD-L1.19–21 In the previous study, we designed and constructed the bispecific antibody (BsAb) blocking TGF-β and murine PD-L1 (named YM101) based on Check-BODY platform. YM101 showed potent antitumor activity without observable toxicity in murine tumor models.22–24 Given the success of YM101 in the pilot study, we further developed the BsAb targeting TGF-β and human PD-L1 (termed BiTP) using Check-BODY platform. BiTP was constructed by modifying the structure of YM101, aiming to provide potential anti-TGF-β/PD-L1 BsAb for future clinical trials. Here, we measured the biochemistry characteristics and bioactivities of BiTP. In addition, we investigated the antitumor activity of BiTP in TNBC models and its underlying mechanisms.
Materials and methods
Cell lines and antibodies
Human peripheral blood mononuclear cells (hPBMCs), H358, NK92MI engineered to express CD16A (NK92MI-CD16A), A549, MCF7, BT474, 4T1-hPDL1, epithelial-mesenchymal transition (EMT)-6-hPDL1, and Jurkat-1-PD-1-NFAT-Luc were cultured in RPMI-1640 medium with 10% fetal bovine serum (FBS) (Excell). TF-1 was cultured in RPMI-1640 medium with 10% FBS and 2 ng/mL recombinant human granulocyte-macrophage colony stimulating factor (rhGM-CSF) (7954 GM, R&D). CHO-K1 cell engineered to express PD-L1 and anti-CD3 scFv (CHO-K1-PD-L1-CD3L) and HFL1 (human fetal lung fibroblast) were cultured with Ham’s F-12K medium with 10% FBS. Primary human fibroblast was maintained in DEME medium with 10% FBS. Therapeutic antibodies used in this work were purchased from Wuhan YZY Biopharma.
Molecular weight and purity test
The molecular weight and purity of BiTP were estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie Brilliant Blue staining, as previously described.25 The exact molecular weight was measured by liquid chromatograph-mass spectrometer, and dithiothreitol was added to detect the molecular weight of single-chain.26 According to the standard protocol, the exact purity was tested by capillary electrophoresis-SDS (CE-SDS).27 Iodoacetamide was used for non-reduced CE-SDS, while β-mercaptoethanol was used for reduced CE-SDS. Prepared mixture for CE-SDS was diluted in SDS-MW buffer, with a total volume of 101 µL. After incubation at 70℃ for 10 min, we performed CE-SDS separations in Beckman PA 800 plus platform.
Biacore surface plasmon resonance assay
The affinity of BiTP was measured by surface plasmon resonance (SPR) based on a Biacore T200 Biosensor instrument (GE Healthcare) with BiTP captured on Protein A chip. Different concentrations of TGF-β1 or PD-L1 (Wuhan YZY Biopharma) were injected to test the kinetical binding interaction. The results of SPR were further processed with Biacore T200 Biaevaluation V.2.0 software. Finally, we used a 1:1 binding model to calculate the association rate constant (Kon), dissociation rate constant (Koff), and equilibrium dissociation constant (Kd) in the assays.
ELISAs using plate-bound TGF-β
To detect the binding affinity of the anti-TGF-β moiety, we precoated plates with TGF-β (200 ng per well) at 37℃ for 2 hours. Next, the plates were washed using PBST and blocked using 5% bovine serum albumin (BOVOGEN) for 2 hours. Then, the plates were washed and probed with serially diluted BiTP for 1 hour. After washing with PBST, the secondary antibody (anti-hIgG-HRP, 1:5000, BOSTER) was added. In accordance with the standard ELISA protocol, the absorbance was detected at 450 nm.
To detect the simultaneous binding affinity of BiTP, we performed double-antigen sandwich ELISA. Plates were precoated with 100 ng TGF-β1 at 37℃ for 2 hours. After washing and blocking, plates were probed with serially diluted BiTP at 37°C for 1 hour. Following incubation and washing, PD-L1-HRP (5000 ng/mL, 100 µL per well, Wuhan YZY Biopharma) was added. The simultaneous binding of BiTP was quantified using absorbance at 450 nm.
ELISAs determining the binding of BiTP to human Fc gamma receptors (hFcγRs)
Plates were precoated with hFcγRI, hFcγRIIA, hFcγRIIB, hFcγRIIIA, and hFcγRIIIB (Acro) (100 ng per well) at 37°C for 2 hours. After washing and blocking using 5% BSA, the assay plates were probed with serially diluted BiTP for 1 hour. Anti-hFab-HRP (A0293, Sigma) was used as the secondary antibody. The absorbance value was detected at 450 nm.
In vitro antibody-dependent cellular cytotoxicity detection
H358 cells were stained with 2.5 µM carboxyfluorescein diacetate succinimidyl ester buffer (containing 1% FBS) at 37°C for 15 min. Then, H358 cells were washed by medium and seeded into 96 well plates (2×104 per well). The next day, NK92MI-CD16A (4×104 per well) and antibodies were added to plates. Cells were cultured at 37°C for 6 hours. After treatment with propidium iodide (1 µg/mL, P4170-25 mg, Sigma), the ratio of dead cells was measured by flow cytometry.
Cell-binding assay using flow cytometry
H358 cells were incubated with serially diluted BiTP at room temperature for 1 hour. After thoroughly washing, cells were incubated with APC-conjugated anti-hIgG Fc antibody (409306, BioLegend) for 30 min. The cell-binding activity was measured by mean fluorescence intensity.
BiTP competitively inhibited the binding of PD-1 and TGFBR2 to corresponding ligands
Plates were precoated with TGF-β1 (200 ng per well) at 37°C for 2 hours. After washing and blocking, the plates were added with TGFBR2-HIS (Wuhan YZY Biopharma) (5 ng/mL for TGF-β1, 10 µg/mL for TGF-β2, 0.14 µg/mL for TGF-β3) and serially diluted BiTP. Then, the plates were incubated with the secondary antibody (1:5000) (HRP-66005, Proteintech) for 1 hour. The absorbance value was detected at 450 nm. For the competitive inhibition of binding between PD-1 and PD-L1, plates were precoated with 200 ng PD-1-Fc (10377-H02H, Sino Biological). PD-L1-HIS (1 µg/mL, 10084-H08H, Sino Biological) and serially diluted BiTP were used in this assay.
Pharmacokinetics
CD-1 mice were used for the pharmacokinetic study. Mice received 3 mg/kg, 9 mg/kg, and 27 mg/kg BiTP treatment by intravenous injection. Then, 5 min, 2 hours, 8 hours, 24 hours, 3 days, 7 days, 14 days, 21 days, and 28 days later, 0.6 mL peripheral blood was collected. The concentration of BiTP in plasma was measured by ELISA assays. Goat anti-human IgG HRP Conjugated Min X Monkey (A80-319P, BETHYL) was used in this assay.
The influence of BiTP on the concentration of TGF-β in plasma
B-cell-depleted (μMT) mice were used to investigate the effect of BiTP treatment on TGF-β plasma concentration. Mice received 9 mg/kg BiTP and 6.6 mg/kg anti-PD-L1 by intravenous injection. Before treatment, as well as 2 hours, 6 hours, 24 hours, 48 hours, 72 hours, 120 hours, and 168 hours after treatment, peripheral blood was collected to measure the concentration of TGF-β. The level of TGF-β was detected using U-PLEX TGF-β Mouse SECTOR (K15242K-1, MSD) and MESO QuickPlex SQ 120 (MSD) according to the manufacturer’s recommendations.
CCK-8 assay
TF-1 proliferation is sensitive to TGF-β.28 We used CCK-8 reagent (Dojindo) to determine the capability of BiTP against the TGF-β-inhibited proliferation of TF-1 cells. 3×103 viable TF-1 cells were seeded in 96-well plates. Then, TGF-β (TGF-β1: 1 ng/mL; TGF-β2: 20 ng/mL; TGF-β3: 1 ng/mL) and antibodies were added. After culture for 6 days, CCK-8 reagent was added to assay plates. Following incubation for 3 hours, the absorbance values were measured by microplate reader (Molecular Devices).
Immunofluorescence
Before treatment, human primary fibroblast and HFL1 were cultured with the DMEM or Ham’s F-12K containing 1% FBS for 24 hours. Then, the medium was replaced with DMEM or Ham’s F-12K containing 1% FBS, 10 ng/mL TGF-β1, and 105 pM BiTP. After incubation for 72 hours, the cells were fixed with 4% paraformaldehyde for 30 min, permeabilized with 1% Triton-X 100 for 20 min, and incubated with 5% BSA for 1 hour. Then, the cells were incubated with primary antibodies (anti-α-SMA, 19245, CST, 1:300; anti-Collagen I, ab34710, Abcam, 1:300) at 4℃ overnight. The fluorescent secondary antibody (ab150077, Abcam, 1:300) was incubated at room temperature for 1 hour. The cell nuclei were counterstained with DAPI (4083, CST, 1 µg/mL).
Western blotting
Western blotting was performed as previously described.26 The following primary and secondary antibodies were used (all antibodies from CST): anti-E-cadherin (1:1000, 3195), anti-N-cadherin (1:1000, 13116), anti-vimentin (1:1000, 5741), anti-β-Actin (1:1000, 8457), and anti-rabbit-IgG-HRP antibody (1:3000, 7074).
Treg induction assay
Plates were coated with 10 µg/mL anti-human CD3 (Clone OKT3, BioLegend) at 37°C for 2 hours. The hPBMCs were maintained in RPMI-1640 with 10% FBS, 100 U/mL interleukin-2 (IL-2, Beijing Fourrings), 5 µg/mL anti-CD28 (Clone 28.2, BioLegend), 20 ng/mL TGF-β1, and BiTP for 6 days.
Cytokine detection assay
To measure the bioactivity of the anti-PD-L1 moiety of BiTP, we conducted a superantigen stimulation assay. Viable hPBMCs (5×104 /mL) were mixed with serially diluted antibodies. Staphylococcal enterotoxin A (500 ng/mL, Toxin Technology) was added to the hPBMC-antibody mixtures. Four days later, IL-2 concentration in the supernatants was detected by Multi-Analyte Flow Assay Kit (Human CD8/NK Panel, BioLegend).
Smad luciferase reporter assay
A549 cells were plated in 96-well plates (2×104 cells per well) on the day before transfection. Then, transient transfection with Smad luciferase reporter plasmid was performed by QIAGEN Effectene Transfection Reagent. Subsequently, cells were incubated with 20 ng/mL TGF-β1 and serially diluted antibodies for 24 hours. Luminescence was measured by Promega Bright-Glo Luciferase Assay System according to the manufacturer’s recommendations.
NFAT luciferase reporter assay in CHO-K1 cells
CHO-K1-PD-L1-CD3L cells (4×104 per well) were seeded in 96 well plates and cultured for 16 hours. Then, Jurkat-1-PD-1-NFAT-Luc cells (5×104 cells per well) and serially diluted antibodies were added to plates. After incubation for 6 hours, luminescence was measured by Promega Bright-Glo Luciferase Assay System.
Murine tumor models
Humanized TNBC models (EMT-6-hPDL1 and 4T1-hPDL1) were used to investigate the antitumor efficacy of BiTP. The endogenous mouse Cd274 of TNBC cells was replaced by human CD274. Due to the interaction between hPD-L1 and mouse PD-1 (mPD-1), this hPD-L1 expressing tumor model could be adopted to evaluate the efficacy of anti-hPD-L1 antibodies.29 Totally, we established two orthotopic breast cancer models (EMT-6-hPDL1 and 4T1-hPDL1) by inoculating 1×105 tumor cells into the right mammary fat pad of BALB/c mice. When tumor volume was nearly 100 mm3, tumor-bearing mice were randomized into four groups (n=8) and treatment started: Isotype (i.p., 6.6 mg/kg), anti-TGF-β (i.p., 6.6 mg/kg), anti-PD-L1 (i.p., 6.6 mg/kg), and BiTP (i.p., 9 mg/kg). All mice received six doses of treatment in the following 2 weeks. Tumor volume (length×width2×0.5) and mouse weight were recorded on alternate days. Mice were euthanized when the study ended or tumor volume exceeded 2500 mm3.
Flow cytometry for the TME
Tumors were collected, minced, and digested with collagenase B (0.5 mg/mL, Roche) and hyaluronidase (0.5 mg/mL, Absin) at 37°C for 1 hour. Suspensions were filtered through 40 µm cell strainers. Then, cells were suspended and stained with Fixable Viability Stain 700 (BD Biosciences). Cells were stained according to the standard protocol for flow cytometry. The following antibodies and buffers were used (all reagents from BD Biosciences unless otherwise indicated): anti-CD45 (560510), anti-CD3e (562600), anti-CD8α (563068), anti-CD25 (553075), anti-CD69 (566500), anti-Ki67 (556027), anti-TNF-α (563943), anti-IFN-γ (560660), anti-Perforin (ThermoFisher, 11-9392-82), anti-Granzyme-B (BioLegend, 372204), anti-CD11c (566504), anti-I-A/I-E (BioLegend, 107608), anti-CD80 (560016), anti-CD86 (561962), Brilliant Stain Buffer (563794), and FOXP3/Transcription Factor Staining Buffer Set (Invitrogen, 00-5523-00). Cells per 100 mg tumor were counted by Beckman Vi-Cell Auto. Flow cytometry was performed with Beckman CytoFLEX LX.
Immunohistochemistry and multiplex immunohistochemistry
Tumor tissues were fixed with 4% paraformaldehyde at room temperature for 2 days, dehydrated, and embedded in paraffin wax. Primary antibodies used in this study included antibodies recognizing CD8 (Abcam, ab217344), Vimentin (CST, 5741), E-cadherin (CST, 3195), α-SMA (CST, 19245), Cleaved-Caspase-3 (CST, 9661), Ki67 (Abcam, ab16667), and PCNA (CST, 13110). Direct Red 80 (Sigma, 365548) was used for picrosirius red staining. The fluorescent multiplex immunohistochemistry (IHC) staining with Tyramide Signal Amplification was conducted as previously described.30 IHC staining was performed following a two-step protocol.31 Two pathologists independently defined the regions of interest. IHC staining was scored using ImageJ software. The expression levels were quantified with integral optical density. T cell infiltration depth was measured by radius-scaled distance from CD8+ pixels to margin.19
Bulk RNA-seq assay
The bulk RNA-seq assay was performed by Seqhealth (Wuhan, China) (online supplemental table S1). Total RNA was extracted from the 4T1-hPDL1 tumor tissues by TRIzol reagent (Invitrogen, 15596026).31 After RNA extraction, DNA digestion was performed using DNaseI. RNA quality was assessed by A260/A280 with Nanodrop One Cspectrophotometer (Thermo Fisher). RNA integrity was measured with 1.5% agarose gel electrophoresis. Then, qualified RNA was quantified by Qubit3.0 with Qubit RNA Broad Range Assay kit (Life Technologies, Q10210).
jitc-2022-005543supp001.pdf (2.3MB, pdf)
Subsequently, 2 µg total RNA was used for stranded RNA sequencing library preparation with Ribo-off rRNA Depletion Kit (Illumina, MRZG12324) and KC-Digital Stranded mRNA Library Prep Kit (Wuhan Seqhealth, DR08502) following the manufacturer’s instruction. This kit labels the preamplified cDNA molecules with unique molecular identifier of 8 random bases, in order to eliminate duplication bias in PCR and sequencing steps. The library products corresponding to 200–500 bps were enriched, quantified, and sequenced on NovaSeq 6000 sequencer (Illumina) with PE150 model.
Deduplicated Reads were mapped to the reference genome of Mus_musculus.GRCm38 (https://ftp.ensembl.org/pub/release-87/fasta/mus_musculus/dna/) with the annotation from gff3 file (https://ftp.ensembl.org/pub/release-87/gff3/mus_musculus/) by STRA analysis tool (2.5.3a) using default parameters. Then, reads mapping to exons were counted using featureCounts tool (Subread-1.5.1; Bioconductor) and scaled with RPKM (Reads Per Kilobase per Million mapped reads) method. After assessing RNA-seq performance and quality (online supplemental table S2), a standard analysis pipeline was adopted to detect the biological signals of samples. Differentially expressed gene (DEG) was identified when the gene presented >2 fold expression difference and adjusted p<0.05. The DEG analysis was conducted by DESeq2 package (1.34.0).32 The results of DEG analysis were visualized by pheatmap package (1.0.12).33 The principal component analysis was performed using prcomp function of stats package. The functional enrichment was performed using the online tool The Database for Annotation, Visualization and Integrated Discovery. A pathway with adjusted p value (false discovery rate (FDR))<0.05 is regarded as statistically significant. According to public gene lists, immune signatures were constructed and scored.34 Briefly, signature scores were performed by calculating the mean value of scaled gene expression levels.
Statistical analyses
Statistical analyses were conducted by R software and GraphPad Prism V.8 software. Student’s t-test with or without Welch’s correction and Mann-Whitney test were adopted to compare two groups. Survival curves were compared by log-rank test. Data were presented as mean ±SE of mean (SEM) or SD. All tests were two sided, and p<0.05 was regarded as significant.
Results
The structure of BiTP
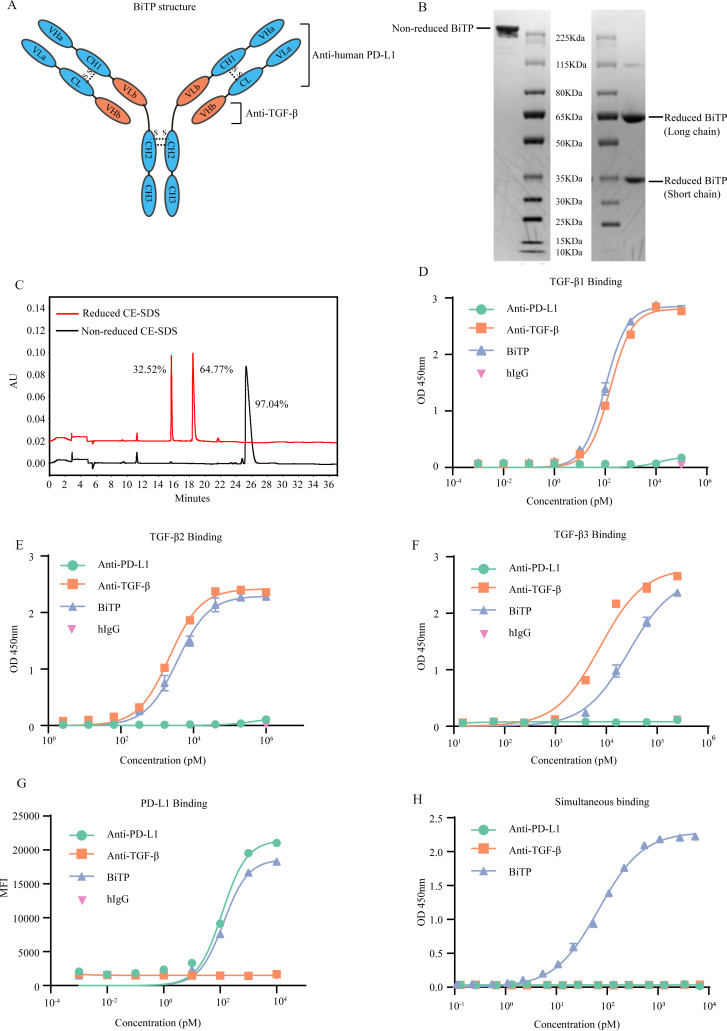
As an IgG1/IgG2 hybrid antibody, the Fc of BiTP contains IgG2-derived CH2 domain and IgG1-derived CH3 domain, with D270A modification to reduce the binding affinity to FcγR (online supplemental figure S1A–E). We did not observe significant antibody-dependent cell-mediated cytotoxicity for BiTP (online supplemental figure 1). The VLa, CL, VHa, and CH1 are derived from the corresponding domains of anti-human PD-L1. The VHb and VLb domains are derived from the corresponding domains of anti-TGF-β (figure 1A). Reduced SDS-PAGE showed BiTP consists of short chain and long chain (figure 1B). CE-SDS demonstrated the purity was more than 97% (figure 1c). The molecular weight of BiTP is about 201.5 KDa (short chain: 37.3 KDa; long chain: 63.4 KDa) (online supplemental figure S2A–C).
Figure 1.
The structure and binding affinity of BiTP. (A) The structure of BiTP. BiTP is an IgG1/IgG2 hybrid antibody, containing IgG2-derived CH2 domain and IgG1-derived CH3 domain. The VLa, CL, VHa, and CH1 are derived from the corresponding domains of anti-PD-L1. The VHb and VLb domains are derived from the corresponding domains of anti-TGF-β. (B) Non-reduced and reduced SDS-PAGE. (C) Non-reduced and reduced CE-SDS. (D–F) The binding affinity to TGF-β1. BiTP was captured by plate-coated TGF-β. The affinity was determined by ELISA. (G) The binding affinity to PD-L1. Antibodies were incubated with H358 cells. The binding affinity was measured by mean fluorescence intensity in flow cytometry assay. (H) The simultaneous binding to PD-L1 and TGF-β1. BiTP was captured by precoated TGF-β1. Then, PD-L1-HRP was added, and the simultaneous binding was detected by ELISA. CE-SDS, capillary electrophoresis-sodium dodecyl sulfate; MFI, mean fluorescence intensity; TGF-β, transforming growth factor-beta.
The binding affinity of BiTP
Compared with anti-TGF-β, BiTP bound to plate-bound TGF-β with similar or slightly weaker affinities (TGF-β1: Kd=101 pM for BiTP, 160 pM for anti-TGF-β; TGF-β2: Kd=3.59×103 pM for BiTP, 2.17×103 pM for anti-TGF-β; TGF-β3: Kd=2.76×105 pM for BiTP, 7.51×104 pM for anti-TGF-β) (figure 1D–F). In addition, BiTP bound to H358 cells (expressing a high level of PD-L1) with a similar affinity to anti-PD-L1 (Kd=133 pM for BiTP, 119 pM for anti-PD-L1) (figure 1G). Double-antigen sandwich ELISA showed that TGF-β1-captured BiTP simultaneously bound to PD-L1 (Kd=68 pM) (figure 1H).
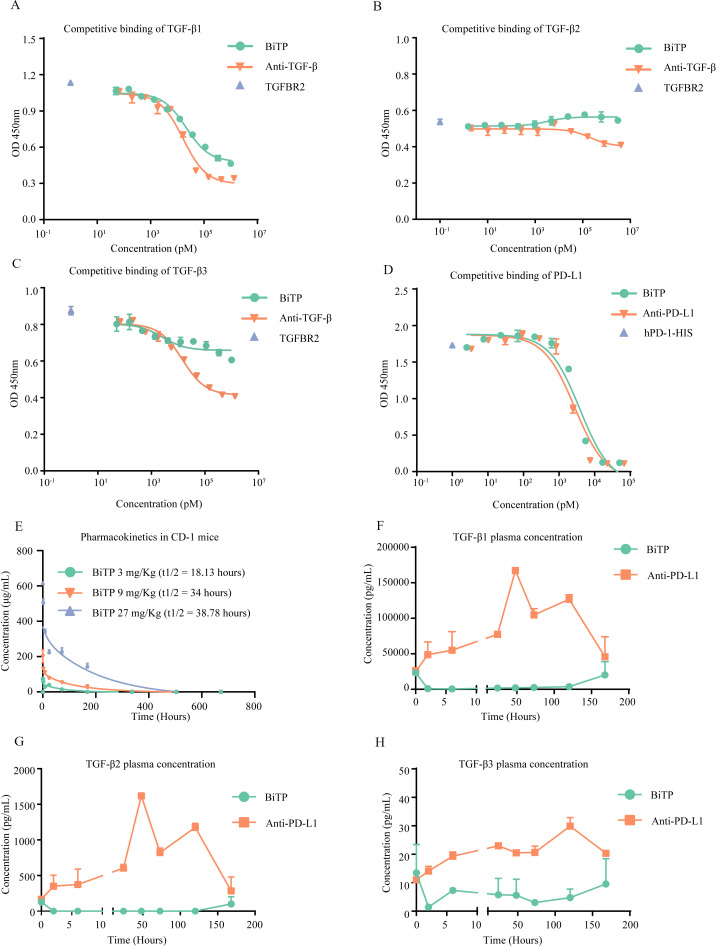
In addition, we used SPR Biacore assays to measure the kinetic binding of BiTP to TGF-β (Kon=3.20×106 M−1S−1, Koff=4.17×10−3 S−1, Kd=1.30×10−9 M) and PD-L1 (Kon=2.39×105 M−1S−1, Koff=3.48×10−4 S−1, Kd=1.45×10−9 M) (online supplemental figure S3A, B). The results of competitive binding assays showed that a high level of BiTP could competitively antagonize the binding between TGF-β1 and TGFBR2 (IC50=2.13×104 pM). However, BiTP had a modest effect on the binding of TGF-β2 or TGF-β3 to TGFBR2. Generally, the competitive binding property of BiTP was weaker than anti-TGF-β (figure 2A–C). Moreover, a high concentration of BiTP almost completely inhibited the binding of PD-1 to PD-L1 (IC50=3.84×103 pM) (figure 2D).
Figure 2.
Competitive binding experiments and pharmacokinetic study. (A–C) BiTP competitively inhibited the binding of TGFBR2 to TGF-β. TGFBR2-HIS and BiTP were incubated with precoated TGF-β. The competitive inhibition capability was evaluated with anti-HIS ELISA. (D) BiTP competitively inhibited the binding of PD-1 to PD-L1. PD-L1-HIS and BiTP were incubated with precoated PD-L1. The competitive inhibition capability was evaluated with anti-HIS ELISA. (E) CD-1 mice received 3 mg/kg, 9 mg/kg, 27 mg/kg BiTP treatment by intravenous injection. Then, 5 min, 2 hours, 8 hours, 24 hours, 3 days, 7 days, 14 days, 21 days, and 28 days later, 0.6 mL peripheral blood was collected to measure the concentration of BiTP in plasma. (F–H) The influence of BiTP on the concentration of TGF-β in plasma. B-cell-depleted (μMT) mice received 9 mg/kg BiTP and 6.6 mg/kg anti-PD-L1 treatment. Before treatment, as well as 2 hours, 6 hours, 24 hours, 24 hours, 72 hours, 120 hours, and 168 hours after treatment, peripheral blood was collected to measure the concentration of TGF-β. TGF-β, transforming growth factor-beta.
Pharmacokinetic study and TGF-β concentration in plasma
BiTP exhibited nonlinear pharmacokinetics in CD-1 mice. The half-life time (t1/2) of BiTP was 34 hours at the dose of 9 mg/kg (figure 2E). In μMT mice, after a single dose of BiTP, the TGF-β1 and TGF-β2 in plasma were almost completely depleted (from 2 to 120 hours) (figure 2F–G). In addition, after BiTP treatment, the concentration of TGF-β3 was maintained at a low level throughout all time points (figure 2H).
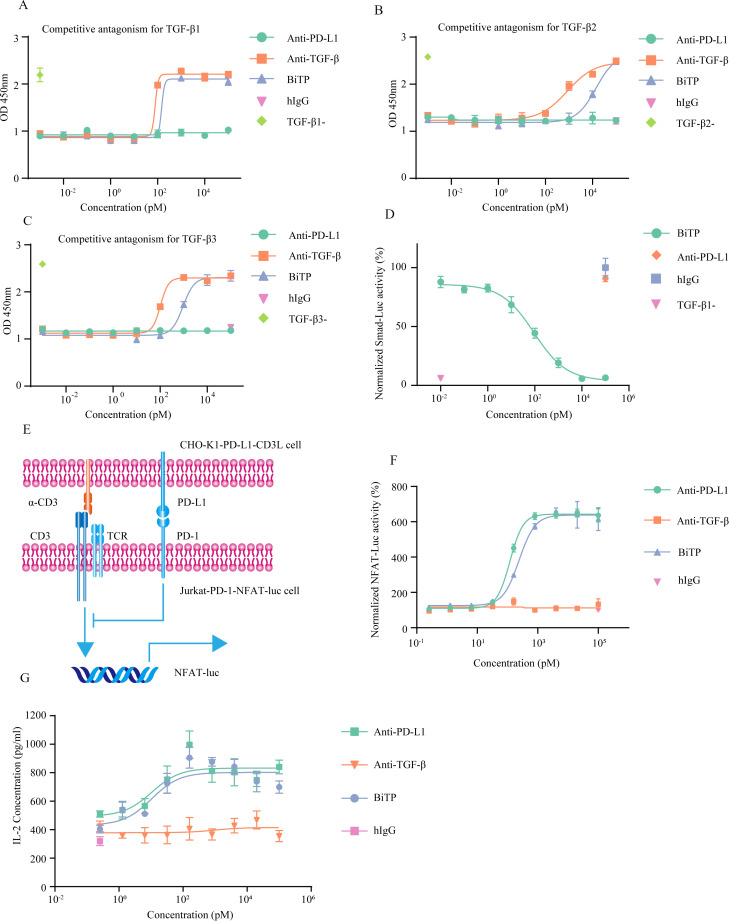
The bioactivity of BiTP’s anti-TGF-β moiety
The biological activity of TGF-β could be determined by TF-1 proliferation assay.35 TGF-β suppressed GM-CSF-dependent TF-1 proliferation in vitro. The TGF-β-caused impairment on TF-1 proliferation could be reversed by BiTP in a dose-dependent manner (EC50=148 pM for TGF-β1, 1.41×104 pM for TGF-β2, 939 pM for TGF-β3) (figure 3a–3c). In addition, BiTP effectively blocked canonical TGF-β-Smad signaling (EC50=92 pM) (figure 3d).
Figure 3.
The in vitro bioactivity of BiTP. (A–C) CCK-8 assays to detect the capability of BiTP against TGF-β-inhibited proliferation of TF-1 cells. (D) Luciferase reporter experiments to evaluate Smad-mediated transcriptional activity. Smad-Luc-transfected A549 cells were treated with 20 ng/mL TGF-β1 and antibodies for 24 hours. Then, luminescence was detected. (E) The diagram showing NFAT luciferase reporter system. In the system, Jurkat-1-PD-1-NFAT-Luc and CHO-K1-PD-L1-CD3L were used. The activity of NFAT-Luc could be hampered by PD-1-PD-L1 axis. (F) NFAT luciferase reporter experiments to detect PD-1 signaling. Jurkat-1-PD-1-NFAT-Luc and BiTP were incubated with CHO-K1-PD-L1-CD3L for 6 hours. Then, luminescence was detected. (G) Superantigen stimulation assay assessing the activity of the anti-PD-L1 moiety of BiTP. PBMCs were mixed with antibodies and staphylococcal enterotoxin A (SEA). Four days later, IL-2 concentration in the supernatants was detected. PBMCs, peripheral blood mononuclear cell; TGF-β, transforming growth factor-beta.
Then, we explored the antagonism of BiTP on TGF-β-mediated immune and nonimmune effects. Exogenous TGF-β1 promoted Treg differentiation in vitro. Different from isotype and anti-PD-L1, BiTP effectively inhibited TGF-β1-induced Treg differentiation (online supplemental figure S4A). In addition, in line with previous reports, TGF-β1 downregulated epithelial marker E-cadherin and increased mesenchymal markers N-cadherin and Vimentin. BiTP effectively reversed TGF-β1-induced EMT in breast cancer cells: increasing epithelial marker and decreasing mesenchymal markers (online supplemental figure S4B).
The bioactivity of BiTP’s anti-PD-L1 moiety
The bioactivity of BiTP’s anti-PD-L1 moiety was assessed by a luciferase reporter system containing Jurkat-1-PD-1-NFAT-Luc and CHO-K1-PD-L1-CD3L cells (figure 3E). BiTP counteracted PD-1/PD-L1 pathway-mediated suppression on NFAT signaling (EC50=240 pM) (figure 3F). In the superantigen stimulation assay, BiTP enhanced the secretion of IL-2 (figure 3G). This enhancement of T cell activity was related to the dose of BiTP (EC50=16.81 pM). The high dose of BiTP exhibited a stronger capability to promote T cell activation.
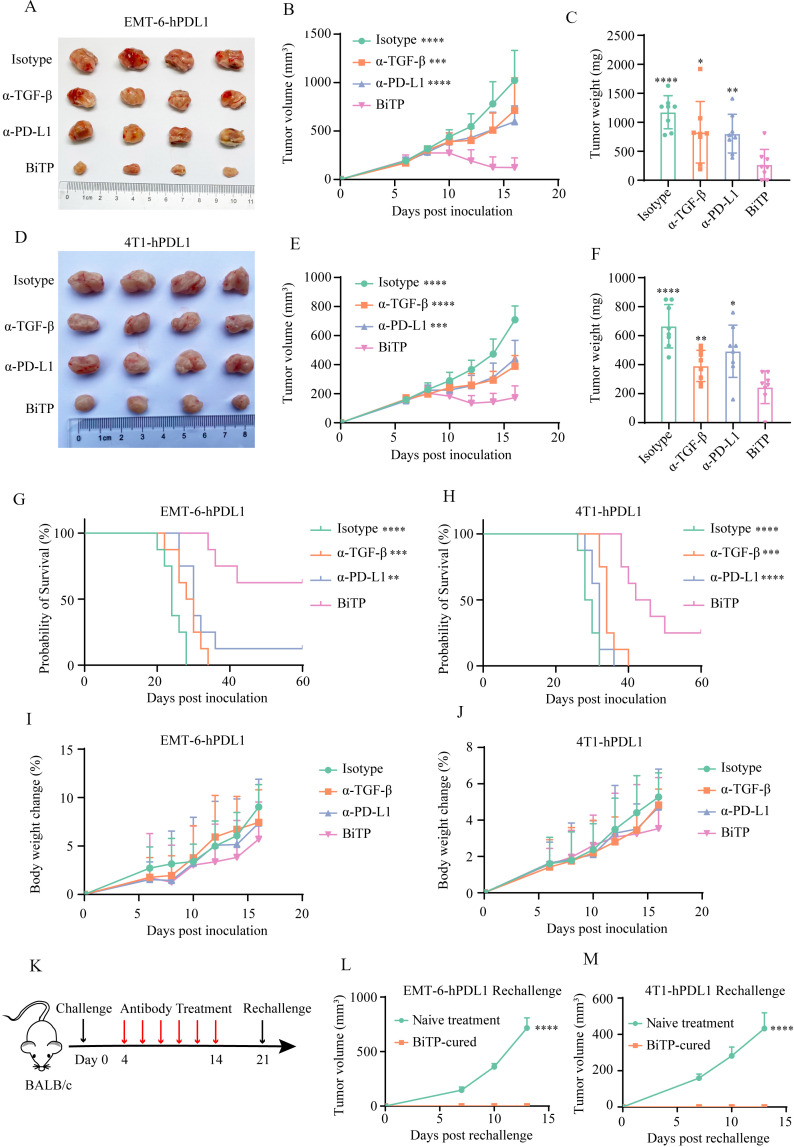
The antitumor activity of BiTP in murine models
The antitumor effects of BiTP were evaluated in the EMT-6-hPDL1 and 4T1-hPDL1 models. One week after tumor cell inoculation, mice were grouped and treated with six doses of antibodies. In two TNBC models, BiTP exhibited superior antitumor activity to parent antibodies. Compared with other three groups, BiTP also significantly slowed tumor growth and extended the survival of tumor-bearing mice (figure 4A–H). In addition, we did not observe overt toxicity and weight loss in mice receiving antibody treatment, indicating the acceptable safety and tolerability of BiTP (figure 4I, J). Rechallenge assays showed all BiTP-cured mice rejected rechallenged tumors, demonstrating BiTP established a potent antitumor immune memory (figure 4K–M).
Figure 4.
The antitumor activity of BiTP in murine TNBC models. (A) The representative images EMT-6-hPD-L1 implants in mice receiving antibody treatment. (B) The growth curves of EMT-6-hPD-L1 implants in mice receiving antibody treatment. (C) The weights of EMT-6-hPD-L1 implants in mice receiving antibody treatment. (D) The representative images 4T1-hPD-L1 implants in mice receiving antibody treatment. (E) The growth curves of 4T1-hPD-L1 implants in mice receiving antibody treatment. (F) The weights of 4T1-hPD-L1 implants in mice receiving antibody treatment. (G, H) Survival of tumor-bearing mice receiving antibody treatment. (I, J) Body weight changes of tumor-bearing mice following treatment. (K–M) Rechallenge assays. BALB/c mice were orthotopically inoculated with 105 tumor cells on day 0, and BiTP therapy was initiated on day 4. On day 21, BiTP-cured and treatment-naive mice were inoculated with 105 tumor cells for rechallenge assays. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 mean the significant difference compared with BiTP-treated samples. EMT, epithelial-mesenchymal transition; TNBC, triple-negative breast cancer.
BiTP suppressed collagen production and promoted T cell infiltration
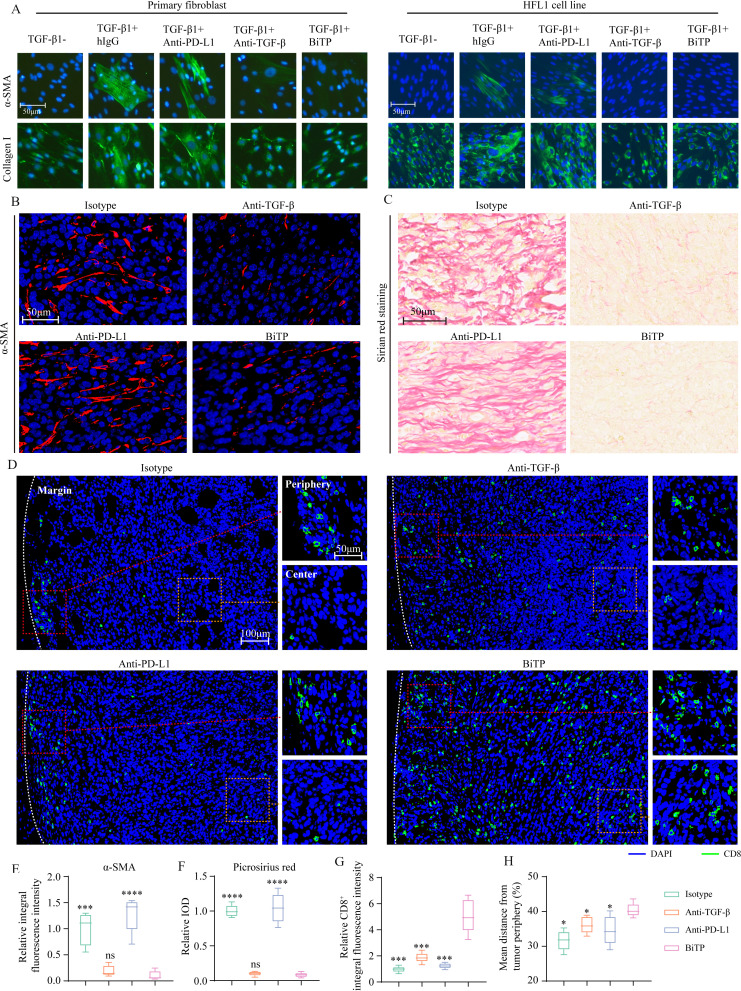
Our previous study showed that TGF-β blockade enhanced T cell infiltration by limiting CAF-derived collagen.22 Given that CAF-derived collagen undermines T cell penetration, we first explored the effects of BiTP on CAF activity in vitro. α-SMA is the marker of CAF.36 For primary fibroblast and HFL1, TGF-β1 increased α-SMA expression and collagen I generation (figure 5A). Meanwhile, BiTP treatment antagonized the upregulation of α-SMA and collagen I caused by TGF-β1.
Figure 5.
The effects of BiTP on collagen production and CD8+ T cell infiltration in vivo. (A) The effect of BiTP on TGF-β-mediated fibroblast activation. Primary human fibroblast from gastric tissue and HFL1 cells were cultured with TGF-β1 and antibodies. Immunofluorescent staining was performed to measure the levels of α-SMA and collagen I. (B) Multiplex IHC staining to evaluate the level of α-SMA in 4T1-hPD-L1 tumors. (C) Picrosirius red staining to measure collagen production in 4T1-hPD-L1 tumors. (D) Multiplex IHC staining to evaluate the infiltration of CD8+ T cell. The presentative images of infiltrating CD8+ T cells. (E, F) The quantification of anti-α-SMA and picrosirius red staining. (G, H) The quantification of CD8+ pixel and infiltration depth. *p < 0.05, ***p < 0.001, and ****p < 0.0001 mean the significant difference compared with BiTP-treated samples. HFL, human fetal lung fibroblast; IHC, immunohistochemistry; TGF-β, transforming growth factor-beta.
Next, we evaluated CAF activity and CD8+ T cell infiltration in 4T1-hPD-L1 tumors. Anti-α-SMA staining showed BiTP and anti-TGF-β downregulated α-SMA expression, indicating restrained CAF activity (figure 5B). Picrosirius red staining showed that collagen production was significantly decreased after BiTP and anti-TGF-β treatment (figure 5C). Then, we performed anti-CD8 staining to investigate the immune infiltration status. For mice treated with hIgG or anti-PD-L1, most CD8+ pixels were located in tumor margin. In contrast, for mice treated with anti-TGF-β or BiTP, more CD8+ pixels were in tumor center (figure 5D). Quantitative analysis showed that BiTP expanded T cell population and increased its infiltration depth. Collectively, BiTP suppressed CAF activity, reduced collagen deposition, and enhanced T cell infiltration (figure 5E–H).
Flow cytometry revealing alterations in the TME after BiTP treatment
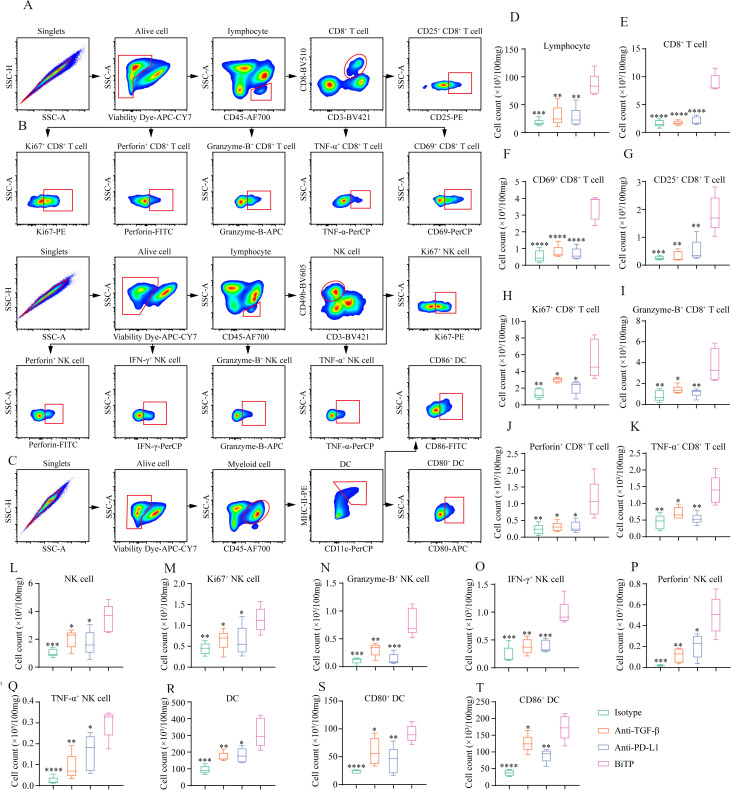
Subsequently, we collected 4T1-hPDL1 tissues to detect changes in immune populations by flow cytometry (figure 6A–C). Compared with the other three groups, BiTP significantly increased TIL, CD8+ T cell, activated (CD69+/CD25+) CD8+ T cell, proliferating (Ki67+) CD8+ T cell, cytotoxic (Granzyme-B+/Perforin+/TNF-α+) T cell, NK cell, proliferating (Ki67+) NK cell, and cytotoxic (granzyme-B+/IFN-γ+/Perforin+/TNF-α+) NK cell (figure 6D–Q). In addition to TIL, BiTP also expanded total DC and mature (CD80+/CD86+) DC (figure 6r–6t).
Figure 6.
Flow cytometry to explore the influence of BiTP on the TME in the 4T1-hPD-L1 tumors. (A–C) The gating strategies for CD8+ T cell, NK cell, and DC in flow cytometry assays. (D-T) The quantification of tumor-infiltrating lymphocyte, CD8+ T cell, CD69+ CD8+ T cell, CD25+ CD8+ T cell, Ki67+ CD8+ T cell, Granzyme-B+ CD8+ T cell, Perforin+ CD8+ T cell, TNF-α+ CD8+ T cell, NK cell, Ki67+ NK cell, Granzyme-B+ NK cell, IFN-γ+ NK cell, Perforin+ NK cell, TNF-α+ NK cell, DC, CD80+ DC, and CD86+ DC. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 mean the significant difference compared with BiTP-treated samples. DC, dendritic cell; TME, tumor microenvironment.
Bulk RNA-seq depicting the TME status
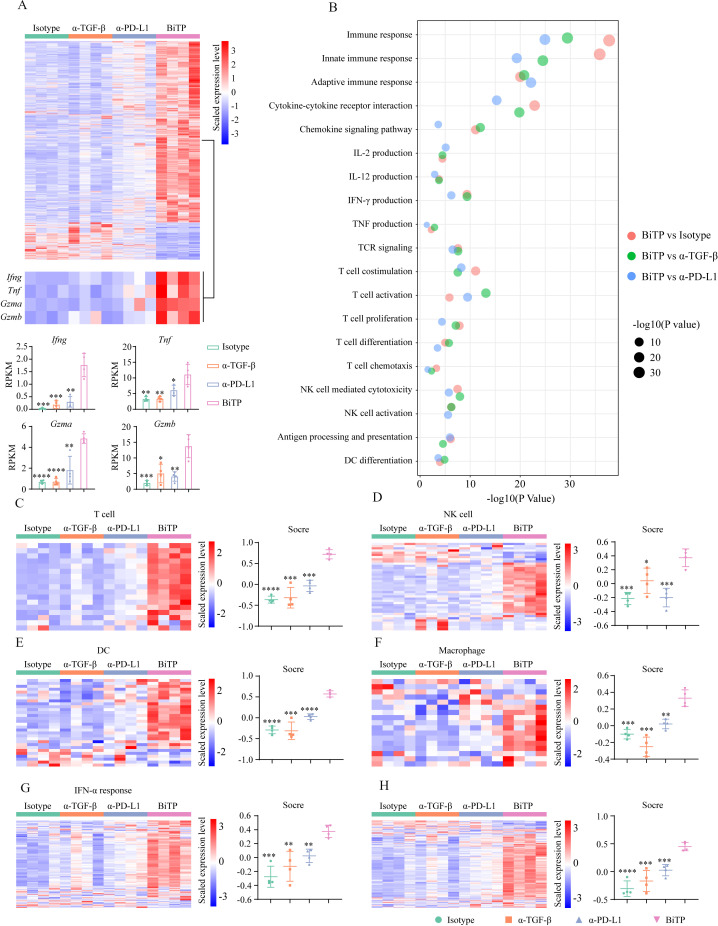
To explore the mechanisms of enhanced antitumor activity of BiTP, we performed bulk RNA-seq using 4T1-hPDL1 tissues. Theoretically, BiTP has the greatest ability among all antibodies to stimulate antitumor immune response. We used BiTP-treated tumors as the baseline to validate whether BiTP has superior immunostimulatory properties to others. Comparing BiTP vs other antibodies, nearly all DEGs were upregulated in BiTP-treated tumors. It is reasonable to assume that these upregulated DEGs might be mainly attributed to BiTP-boosted antitumor immunity. Here we focused on the overlapping DEGs among the three comparisons, which should converge toward immune activation in theory. BiTP-treated tumors had a distinguished transcriptomic profile. DEG analysis identified 2144 (BiTP vs hIgG), 2211 (BiTP vs anti-TGF-β), and 709 (BiTP vs anti-PD-L1) genes, respectively (figure 7A). In all DEGs identified in three comparisons, 598 genes overlapped (566 genes were upregulated and 32 genes were downregulated in BiTP-treated tumors). Enrichment analyses showed the overlapped DEGs were highly associated with immune activation (online supplemental figure S5A–D). As expected, some immune killing-associated genes such as Ifng, Tnf, Gzma, Gzmb were markedly upregulated in BiTP-treated samples. Then, KEGG and GO enrichment analyses based on the identified DEGs were performed. The results showed immune response, innate/adaptive immune response, cytokine-cytokine receptor interaction, chemokine signaling pathway, IL-2/IL-12/IFN-γ/TNF production, T cell functions (TCR signaling, T cell stimulation, activation, proliferation, differentiation, and chemotaxis), NK cell-mediated cytotoxicity, NK cell activation, antigen processing and presentation, DC differentiation were significantly enriched in BiTP-treated samples (figure 7b).
Figure 7.
Bulk RNA-seq to evaluate immune landscape in 4T1-hPD-L1 tumors. Tumor-bearing mice received six doses of antibodies. Then, tumor tissues were collected for bulk RNA-seq assay. (A) The heat map showing differentially expressed gene profiles. (B) The levels of Ifng, Tnf, Gzma, and Gzmb. (C–H) The heat maps representing the expression levels of genes belonging to immune signatures. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 mean the significant difference compared with BiTP-treated samples. TGF-β, transforming growth factor-beta.
Moreover, the effects of BiTP on the TME were comprehensively evaluated by six signatures. The scores of all immune signatures were highest in BiTP-treated samples (figure 7C–H). Also, the principal component analysis showed that tumors treated with hIgG, anti-TGF-β, and anti-PD-L1 shared similar mRNA profiles, while the gene expression pattern of BiTP-treated tumors was utterly different (online supplemental figure S5E). The results indicated that the immunostimulatory effects of anti-TGF-β and anti-PD-L1 monotherapies were weak in this model, and the suppressed antitumor immunity could not be fully restored. Generally, the transcriptomic data indicated that BiTP systemically boosted multiple immune components and enhanced antitumor immunity.
BiTP reversed EMT and inhibited tumor cell proliferation
We assessed EMT status in the 4T1-hPDL1 model after BiTP treatment. Anti-vimentin and anti-E-cadherin staining showed BiTP effectively reversed EMT in vivo (online supplemental figure S6A, B). In addition, we explored the influences of BiTP on tumor cell apoptosis and proliferation. The results indicated that BiTP increased cleaved-caspase-3 but decreased PCNA and Ki67 (online supplemental figure S7). Together, the IHC staining verified the EMT reversal and the enhanced tumor-killing activity of BiTP.
Discussion
Abundant extracellular matrix deposition in TNBC tissues forms a physical barrier, leading to poor immune cell infiltration and drug transport.37 Inhibiting the activation of CAF by neutralizing TGF-β signaling promotes the degradation of extracellular matrix and improves T cell penetration.38 As immune cell infiltration is improved by reprogramming extracellular matrix, the activity of TIL is commonly undermined in the immunosuppressive TME. How to rescue TIL from exhausted status is a great challenge to realize optimal immunotherapy effect. For some patients, PD-1/PD-L1 axis is the dominant rheostat in the cancer-immunity cycle, and PD-1/PD-L1 blockade effectively normalizes dysfunctional antitumor immunity.39 However, for most patients, PD-1/PD-L1 blockade monotherapy is insufficient to trigger the robust antitumor response. Apart from stromal cells, TGF-β also regulates the functions and activities of immune cells: suppressing the cytotoxicity of T cells, inducing Treg differentiation, recruiting myeloid-derived suppressor cells, and inhibiting the maturation of DCs.40 41 Collectively, remodeling extracellular matrix and counteracting immunoinhibitory regulators by TGF-β blockade are favorable to anti-PD-1/PD-L1 treatment. Hence, second-generation anti-PD-1/PD-L1 bifunctional agents such as M7824 and SHR1701 are developed to address this issue, which could simultaneously target TGF-β/TGF-βR and PD-1 signaling.
Both M7824 and SHR1701 are anti-PD-L1/TGF-βR bifunctional fusion proteins.42 43 The preclinical data of M7824 suppressed tumor growth and extended mouse survival more effectively than anti-PD-1.34 Explorations in the TME indicated M7824 significantly activated innate and adaptive immunity.34 Early clinical trial data also confirmed the potent antitumor effect of M7824 in patients.44 However, in the later phase 2/3 trials of bile duct cancer and non-small cell lung cancer, M7824 was not as effective as expected. Although reasons for clinical trial failures are still unclear, identifying predictors to select sensitive patients is valuable to improve the performance of these bifunctional antibodies. In our previous work, we constructed anti-TGF-β/PD-L1 BsAb YM101.22 Different from bifunctional fusion protein, YM101 was developed based on the symmetric tetravalency BsAb technology. The pilot study showed YM101 had an encouraging efficacy and tolerable safety profile.22 BiTP is the alternative analog of YM101, designed for further clinical trials. In this work, we found that BiTP had high binding affinities to targets and effectively blocked downstream signaling pathways. In vivo experiments demonstrated that BiTP significantly retarded tumor growth and prolonged survival of tumor-bearing mice. IHC staining showed BiTP downregulated α-SMA, reduced collagen production, and increased T cell infiltration depth in TNBC models. FACS and bulk RNA-seq data revealed BiTP reprogrammed the TME: increasing the number and improving the cytotoxicity of TILs, and elevating tumor-infiltrating DCs. Notably, we checked the levels of Tgfb1 and Cd274 with RNA-seq (online supplemental figure S8). We noticed Cd274 expression was significantly upregulated in BiTP-treated samples. Although murine Cd274 was silenced in 4T1-hPDL1 cells, non-tumor cell populations in the TME contributed to this Cd274 upregulation under immune pressure. However, no significant changes in Tgfb1 were observed. Actually, the BiTP treatment neutralized the TGF-β at protein level and had modest effects on Tgfb1 expression at transcriptional level. Together, BiTP achieved better tumor control by promoting the formation of hot tumors in TNBC models.
The molecular subtype of breast cancer is associated with tumor mutation burden (TMB) and immune cell infiltration. Relative to hormone receptor-positive breast cancer, TNBC commonly has more TMB and TILs, which might partly contribute to the benefits of anti-PD-1/PD-L1 in TNBC.45 46 However, the clinicopathological analysis indicated that TILs were more frequently dispersed in tumor stroma, having no direct interaction with carcinoma cells, raising the issue of whether stromal barriers undermine immune cell infiltration.47 48 Although the density of stromal TIL was also a favorable prognostic factor for TNBC,49 intratumoral TILs had direct contact with carcinoma cells which might be more critical to immunosurveillance in theory. In this work, BiTP provided an optimal position and distribution for T cell infiltration, converting immune-excluded to inflamed subtype in TNBC models. Considering the unsatisfactory response rate of anti-PD-1/PD-L1, BiTP would be a significant upgrade for current immunotherapy strategies. In addition, due to the role of TGF-β signaling in the plasticity, chemoresistance, and progression of TNBC,15 50 BiTP-involved combination therapies would be promising to change the treatment paradigm of TNBC in the future.
Conclusion
In this study, we design and construct the BsAb BiTP, targeting TGF-β and human PD-L1. After YM101, BiTP is the second BsAb developed using Check-BODYTM technology platform, which could be produced with high purity and quality. BiTP exhibits bioactivities in vitro and potent antitumor activity in TNBC models. Investigations in the TME show BiTP suppresses stromal collagen deposition, improves T cell infiltration, and decreases immunoinhibitory components. Given the positive results of the preclinical study, BiTP is a promising candidate to enter further clinical study.
Footnotes
MY, YW and MN contributed equally.
Contributors: MY, YW and MN conducted the experiments and drafted the manuscript. SZ, JZ, YY and PZ participated in the analysis and interpretation of data. KW and ZD designed and supervised the work, are responsible for the overall content of the publication and act as guarantors. All authors read and approved the final manuscript.
Funding: This work was supported by the National Natural Science Foundation of China (Nos. 82073370, 82272794, and 81874120).
Competing interests: JZ, YY and PZ were employees of Wuhan YZY Biopharma Co., Ltd.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request. All raw RNA-seq data are publicly available in National Center for Biotechnology Information Sequence Read Archive under accession number PRJNA867008.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
The animal operations in this study were evaluated and approved by the Institutional Animal Care and Use Committee of Tongji Hospital of Huazhong University of Sciences and Technology.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Yi M, Li T, Niu M, et al. Epidemiological trends of women's cancers from 1990 to 2019 at the global, regional, and national levels: a population-based study. Biomark Res 2021;9:55. 10.1186/s40364-021-00310-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Y, Gao M, Huang Z, et al. SBRT combined with PD-1/PD-L1 inhibitors in NSCLC treatment: a focus on the mechanisms, advances, and future challenges. J Hematol Oncol 2020;13:105. 10.1186/s13045-020-00940-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012;366:2443–54. 10.1056/NEJMoa1200690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marin-Acevedo JA, Kimbrough EO, Manochakian R, et al. Immunotherapies targeting stimulatory pathways and beyond. J Hematol Oncol 2021;14:78. 10.1186/s13045-021-01085-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larkin J, Chiarion-Sileni V, Gonzalez R, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med 2015;373:23–34. 10.1056/NEJMoa1504030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu L, Liu J, Chen J, et al. The developing landscape of combinatorial therapies of immune checkpoint blockade with DNA damage repair inhibitors for the treatment of breast and ovarian cancers. J Hematol Oncol 2021;14:206. 10.1186/s13045-021-01218-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmid P, Cortes J, Dent R, et al. Event-Free survival with pembrolizumab in early triple-negative breast cancer. N Engl J Med 2022;386:556–67. 10.1056/NEJMoa2112651 [DOI] [PubMed] [Google Scholar]
- 9.Yi M, Zheng X, Niu M, et al. Combination strategies with PD-1/PD-L1 blockade: current advances and future directions. Mol Cancer 2022;21:28. 10.1186/s12943-021-01489-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yi M, Jiao D, Xu H, et al. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol Cancer 2018;17:129. 10.1186/s12943-018-0864-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim B-G, Malek E, Choi SH, et al. Novel therapies emerging in oncology to target the TGF-β pathway. J Hematol Oncol 2021;14:55. 10.1186/s13045-021-01053-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moses H, Barcellos-Hoff MH. TGF-beta biology in mammary development and breast cancer. Cold Spring Harb Perspect Biol 2011;3:a003277. 10.1101/cshperspect.a003277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colak S, Ten Dijke P. Targeting TGF-β signaling in cancer. Trends Cancer 2017;3:56–71. 10.1016/j.trecan.2016.11.008 [DOI] [PubMed] [Google Scholar]
- 14.Yadav P, Shankar BS. Radio resistance in breast cancer cells is mediated through TGF-β signalling, hybrid epithelial-mesenchymal phenotype and cancer stem cells. Biomed Pharmacother 2019;111:119–30. 10.1016/j.biopha.2018.12.055 [DOI] [PubMed] [Google Scholar]
- 15.Xu X, Zhang L, He X, et al. TGF-β plays a vital role in triple-negative breast cancer (TNBC) drug-resistance through regulating stemness, EMT and apoptosis. Biochem Biophys Res Commun 2018;502:160–5. 10.1016/j.bbrc.2018.05.139 [DOI] [PubMed] [Google Scholar]
- 16.Bagati A, Kumar S, Jiang P, et al. Integrin αvβ6-TGFβ-SOX4 pathway drives immune evasion in triple-negative breast cancer. Cancer Cell 2021;39:54–67. 10.1016/j.ccell.2020.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bai X, Yi M, Jiao Y, et al. Blocking TGF-β signaling to enhance the efficacy of immune checkpoint inhibitor. Onco Targets Ther 2019;12:9527–38. 10.2147/OTT.S224013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Batlle E, Massagué J. Transforming growth factor-β signaling in immunity and cancer. Immunity 2019;50:924–40. 10.1016/j.immuni.2019.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mariathasan S, Turley SJ, Nickles D, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554:544–8. 10.1038/nature25501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tauriello DVF, Palomo-Ponce S, Stork D, et al. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 2018;554:538–43. 10.1038/nature25492 [DOI] [PubMed] [Google Scholar]
- 21.Terabe M, Robertson FC, Clark K, et al. Blockade of only TGF-β 1 and 2 is sufficient to enhance the efficacy of vaccine and PD-1 checkpoint blockade immunotherapy. Oncoimmunology 2017;6:e1308616. 10.1080/2162402X.2017.1308616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yi M, Zhang J, Li A, et al. The construction, expression, and enhanced anti-tumor activity of YM101: a bispecific antibody simultaneously targeting TGF-β and PD-L1. J Hematol Oncol 2021;14:27. 10.1186/s13045-021-01045-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yi M, Niu M, Zhang J, et al. Combine and conquer: manganese synergizing anti-TGF-β/PD-L1 bispecific antibody YM101 to overcome immunotherapy resistance in non-inflamed cancers. J Hematol Oncol 2021;14:146. 10.1186/s13045-021-01155-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yi M, Niu M, Wu Y, et al. Combination of oral STING agonist MSA-2 and anti-TGF-β/PD-L1 bispecific antibody YM101: a novel immune cocktail therapy for non-inflamed tumors. J Hematol Oncol 2022;15:142. 10.1186/s13045-022-01363-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan X, Zhou P, Fan T, et al. Immunoglobulin fragment F(ab')2 against RBD potently neutralizes SARS-CoV-2 in vitro. Antiviral Res 2020;182:104868. 10.1016/j.antiviral.2020.104868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu S, Zhang J, Yan Y, et al. A novel asymmetrical anti-HER2/CD3 bispecific antibody exhibits potent cytotoxicity for HER2-positive tumor cells. J Exp Clin Cancer Res 2019;38:355. 10.1186/s13046-019-1354-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dada OO, Rao R, Jones N, et al. Comparison of SEC and CE-SDS methods for monitoring hinge fragmentation in IgG1 monoclonal antibodies. J Pharm Biomed Anal 2017;145:91–7. 10.1016/j.jpba.2017.06.006 [DOI] [PubMed] [Google Scholar]
- 28.Tsang ML, Zhou L, Zheng BL, et al. Characterization of recombinant soluble human transforming growth factor-beta receptor type II (rhTGF-beta sRII). Cytokine 1995;7:389–97. 10.1006/cyto.1995.0054 [DOI] [PubMed] [Google Scholar]
- 29.Huang A, Peng D, Guo H, et al. A human programmed death-ligand 1-expressing mouse tumor model for evaluating the therapeutic efficacy of anti-human PD-L1 antibodies. Sci Rep 2017;7:42687. 10.1038/srep42687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang G, Achim CL, Hamilton RL, et al. Tyramide signal amplification method in multiple-label immunofluorescence confocal microscopy. Methods 1999;18:459–64. 10.1006/meth.1999.0813 [DOI] [PubMed] [Google Scholar]
- 31.Liu Q, Li A, Yu S, et al. DACH1 antagonizes CXCL8 to repress tumorigenesis of lung adenocarcinoma and improve prognosis. J Hematol Oncol 2018;11:53. 10.1186/s13045-018-0597-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016;32:2847–9. 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- 34.Lan Y, Zhang D, Xu C, et al. Enhanced preclinical antitumor activity of M7824, a bifunctional fusion protein simultaneously targeting PD-L1 and TGF-β. Sci Transl Med 2018;10:eaan5488. 10.1126/scitranslmed.aan5488 [DOI] [PubMed] [Google Scholar]
- 35.Kim G-H, Kang S-Y, Kwak H-J, et al. Transforming growth factor-β1 bioassay involving matrix metalloproteinase-2 induction. J Interferon Cytokine Res 2010;30:667–72. 10.1089/jir.2009.0058 [DOI] [PubMed] [Google Scholar]
- 36.De Wever O, Van Bockstal M, Mareel M, et al. Carcinoma-associated fibroblasts provide operational flexibility in metastasis. Semin Cancer Biol 2014;25:33–46. 10.1016/j.semcancer.2013.12.009 [DOI] [PubMed] [Google Scholar]
- 37.Zhang P, Qin C, Liu N, et al. The programmed site-specific delivery of LY3200882 and PD-L1 siRNA boosts immunotherapy for triple-negative breast cancer by remodeling tumor microenvironment. Biomaterials 2022;284:121518. 10.1016/j.biomaterials.2022.121518 [DOI] [PubMed] [Google Scholar]
- 38.Henke E, Nandigama R, Ergün S. Extracellular matrix in the tumor microenvironment and its impact on cancer therapy. Front Mol Biosci 2019;6:160. 10.3389/fmolb.2019.00160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanmamed MF, Chen L. A paradigm shift in cancer immunotherapy: from enhancement to normalization. Cell 2019;176:677. 10.1016/j.cell.2019.01.008 [DOI] [PubMed] [Google Scholar]
- 40.Thomas DA, Massagué J. TGF-beta directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell 2005;8:369–80. 10.1016/j.ccr.2005.10.012 [DOI] [PubMed] [Google Scholar]
- 41.Peng D, Fu M, Wang M, et al. Targeting TGF-β signal transduction for fibrosis and cancer therapy. Mol Cancer 2022;21:104. 10.1186/s12943-022-01569-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng B, Ding K, Chen P, et al. Anti-PD-L1/TGF-βR fusion protein (SHR-1701) overcomes disrupted lymphocyte recovery-induced resistance to PD-1/PD-L1 inhibitors in lung cancer. Cancer Commun 2022;42:17–36. 10.1002/cac2.12244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lind H, Gameiro SR, Jochems C, et al. Dual targeting of TGF-β and PD-L1 via a bifunctional anti-PD-L1/TGF-βRII agent: status of preclinical and clinical advances. J Immunother Cancer 2020;8:e000433. 10.1136/jitc-2019-000433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paz-Ares L, Kim TM, Vicente D, et al. Bintrafusp alfa, a bifunctional fusion protein targeting TGF-β and PD-L1, in second-line treatment of patients with NSCLC: results from an expansion cohort of a phase 1 trial. J Thorac Oncol 2020;15:1210–22. 10.1016/j.jtho.2020.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas R, Al-Khadairi G, Decock J. Immune checkpoint inhibitors in triple negative breast cancer treatment: promising future prospects. Front Oncol 2020;10:600573. 10.3389/fonc.2020.600573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yi M, Qin S, Zhao W, et al. The role of neoantigen in immune checkpoint blockade therapy. Exp Hematol Oncol 2018;7:28. 10.1186/s40164-018-0120-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salgado R, Denkert C, Demaria S, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an international TILs Working group 2014. Ann Oncol 2015;26:259–71. 10.1093/annonc/mdu450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.El Bairi K, Haynes HR, Blackley E, et al. The tale of TILs in breast cancer: a report from the International Immuno-Oncology biomarker Working group. NPJ Breast Cancer 2021;7:150. 10.1038/s41523-021-00346-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kos Z, Roblin E, Kim RS, et al. Pitfalls in assessing stromal tumor infiltrating lymphocytes (sTILs) in breast cancer. NPJ Breast Cancer 2020;6:17. 10.1038/s41523-020-0156-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jalalirad M, Haddad TC, Salisbury JL, et al. Aurora-A kinase oncogenic signaling mediates TGF-β-induced triple-negative breast cancer plasticity and chemoresistance. Oncogene 2021;40:2509–23. 10.1038/s41388-021-01711-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-005543supp001.pdf (2.3MB, pdf)
Data Availability Statement
Data are available upon reasonable request. All raw RNA-seq data are publicly available in National Center for Biotechnology Information Sequence Read Archive under accession number PRJNA867008.