Abstract
Salmonella serovars are associated with human diseases that range from mild gastroenteritis to host-disseminated enteric fever. Human infections by Salmonella enterica serovar Typhi can lead to typhoid fever, but this serovar does not typically cause disease in mice or other animals. In contrast, S. enterica serovar Typhimurium and S. enterica serovar Enteritidis, which are usually linked to localized gastroenteritis in humans and some animal species, elicit a systemic infection in mice. To better understand these observations, multiple strains of each of several chosen serovars of Salmonella were tested for the ability in the nonopsonized state to enter, survive, and replicate within human macrophage cells (U937 and elutriated primary cells) compared with murine macrophage cells (J774A.1 and primary peritoneal cells); in addition, death of the infected macrophages was monitored. The serovar Typhimurium strains all demonstrated enhanced survival within J774A.1 cells and murine peritoneal macrophages, compared with the significant, almost 100-fold declines in viable counts noted for serovar Typhi strains. Viable counts for serovar Enteritidis either matched the level of serovar Typhi (J774A.1 macrophages) or were comparable to counts for serovar Typhimurium (murine peritoneal macrophages). Apoptosis was significantly higher in J774A.1 cells infected with serovar Typhimurium strain LT2 compared to serovar Typhi strain Ty2. On the other hand, serovar Typhi survived at a level up to 100-fold higher in elutriated human macrophages and 2- to 3-fold higher in U937 cells compared to the serovar Typhimurium and Enteritidis strains tested. Despite the differential multiplication of serovar Typhi during infection of U937 cells, serovar Typhi caused significantly less apoptosis than infections with serovar Typhimurium. These observations indicate variability in intramacrophage survival and host cytotoxicity among the various serovars and are the first to show differences in the apoptotic response of distinct Salmonella serovars residing in human macrophage cells. These studies suggest that nonopsonized serovar Typhimurium enters, multiplies within, and causes considerable, acute death of macrophages, leading to a highly virulent infection in mice (resulting in death within 14 days). In striking contrast, nonopsonized serovar Typhi survives silently and chronically within human macrophages, causing little cell death, which allows for intrahost dissemination and typhoid fever (low host mortality). The type of disease associated with any particular serovar of Salmonella is linked to the ability of that serovar both to persist within and to elicit damage in a specific host's macrophage cells.
Salmonella serovars are responsible for human diseases that range from localized gastroenteritis to systemic infections (6). The virulence of specific strains in humans and other animals is frequently serovar specific. Salmonella enterica serovar Typhi causes typhoid fever in humans, but no disease is associated with experimental infections of mice (8). On the other hand, serovar Typhimurium and serovar Enteritidis possess a broad host specificity, causing disease in a variety of animals (14). Strains of serovar Typhimurium are usually associated with localized gastroenteritis in humans; however, mice infected with this serovar display a systemic infection (17) that serves as an experimental model for typhoid fever. The capacity of Salmonella serovars to enter, survive and replicate within, and cause cytotoxicity of macrophage cells may play a major role in their ability to cause disease in particular animal species (2, 6, 17, 26). For example, both opsonized and nonopsonized, invasive serovar Typhimurium can enter murine macrophage cells in organelles called spacious phagosomes (1), eventually replicate within these macrophage cells (2, 20, 24), and apparently utilize these host vehicles to disseminate via the lymphatic system. This serovar is adapted to growth in the murine macrophage, yet it does not survive as well in macrophage cells of human origin (2, 3). Conversely, nonopsonized serovar Typhi strains can enter and thrive in human macrophage cells (11, 26), but these same bacteria are killed more readily in murine macrophage cells (2, 3).
Once salmonellae invade macrophage cells, they replicate and produce cytotoxins within the these host cells. Previous studies have demonstrated that Salmonella spp. are capable of causing cytotoxic effects in macrophage cells of murine origin (4, 5), and these effects have been shown to be at least in part due to the induction of an apoptotic response by the salmonellae (9, 21). Both serovar Typhimurium (9, 21) and serovar Typhi (9) cause cell death in murine tissue culture-derived macrophage cells. This cell death has been linked to the type III secretion system of serovar Typhimurium (9) which probably secretes the inducing factor into the phagosomal compartment.
The precise molecular basis for host specific intramacrophage survival by various Salmonella serovars is still unresolved, although a number of bacterial genetic loci have been implicated (1, 2, 5a, 12). Further, no study has been conducted to examine apoptosis in human macrophages infected with salmonellae and to determine whether there is a difference in the extent of apoptosis caused by different serovars. To address the question of intracellular survival and host cytotoxicity caused by intracellular, nonopsonized salmonellae and determine the frequency of apoptosis among various Salmonella serovars, we have tested multiple strains of each of several Salmonella serovars for the ability to enter, survive, and replicate within human versus murine macrophage cells (tissue culture and primary cells). Moreover, we show that there is a significant difference in intracellular bacterial survival and in cytotoxicity to murine and human macrophages infected with serovar Typhimurium compared to infections with serovar Typhi.
MATERIALS AND METHODS
Bacteria and culture conditions.
Strains of the different Salmonella serovars used in this study are listed in Table 1. All serovar Typhimurium strains except the highly avirulent M206 strain are relatively virulent for mice. All serovar Typhi strains were considered virulent. Wild-type serovar Typhi strain ISP1820 was obtained from David Hone (University of Maryland), and the serovar Enteritidis clinical isolates from patients with gastroenteritis were obtained from B. Swaminathan (Centers for Disease Control and Prevention, Atlanta, Ga.). All Salmonella strains were grown to mid-logarithmic phase in Luria (L) broth (19) containing 10 g of NaCl per liter.
TABLE 1.
Salmonella strains used in this study
| Serovar | Strain | Source |
|---|---|---|
| Typhimurium | LT2 | LESTDa |
| C5 | LESTD | |
| TML | LESTD | |
| W118-2 | LESTD | |
| M206 | LESTD | |
| Typhi | Ty2 | LESTD |
| 643 | LESTD | |
| ISP1820 | D. Hone | |
| Enteritidis | 48-86 | B. Swaminathan |
| 464-86 | B. Swaminathan |
LESTD, Laboratory of Enteric and Sexually Transmitted Diseases Culture Collection, Center for Biologics Evaluation and Research, Food and Drug Administration.
Escherichia coli parent strain MP180 (HfrH thi-1) and derivative strains UM120, UM122, and UM202, containing Tn10 in katE, katF, and katG, respectively (22a; kindly provided by J. L. Rosner, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, Md.) served as experimental controls to monitor rpoS phenotype. To test for survival, stationary-phase L-broth cultures of bacteria were exposed for 30 and 60 min to hydrogen peroxide added to a final concentration of 42 mM as described previously (27a). This assay provided a rapid assessment of the rpoS gene status of the chosen Salmonella serovars.
Eukaryotic cells.
Tissue culture cell lines J774A.1 (murine macrophage-like) and U937 (human macrophage-like), obtained from the American Type Culture Collection, were grown in RPMI 1640 medium (BioWhittaker) supplemented with glutamine (Gibco/BRL, Gaithersburg, Md.) and heat-treated (56°C, 30 min) 10% fetal calf serum (Gibco/BRL) (this medium will henceforth be called RPMI complete). All eukaryotic cells were incubated at 37°C under an atmosphere of 95% air–5% CO2. The U937 cells were activated with phorbol 12-myristate 13-acetate (Sigma Chemical Co., St. Louis, Mo.) at a concentration of 10−8 M (25) for 12 to 14 h before being harvested and seeded into 24-well plates; this treatment caused the U937 cells to become adherent and activated.
Elutriated human macrophage cells, obtained from peripheral blood and suspended in phosphate-buffered saline (PBS, pH 7.2; BioWhittaker) were provided by K. Faust and K. Clouse (Center for Biologics Evaluation and Research, Food and Drug Administration). These cells were incubated in RPMI complete supplemented with heat-treated, 5% human serum (Sigma) to provide macrophage colony-stimulating factor. Eighteen to twenty-four hours before invasion assays, the medium was switched to RPMI complete without human serum.
Murine peritoneal macrophages were harvested as previously described (13). Briefly, BALB/c mice (Jackson Laboratories) were injected intraperitoneally with 1 ml of NIH thioglycolate broth (Difco). Animal care was according to National Institutes of Health regulations. After 3 to 6 days, the mice were killed by cervical dislocation, and 6 to 8 ml of RPMI (Gibco/BRL) was injected into the peritoneal cavity of each mouse. The abdomen of each mouse was massaged vigorously, and peritoneal macrophages were withdrawn using a syringe. Cells were pelleted by centrifugation (250 × g, 10 min), suspended in RPMI complete, and distributed in 24-well plates to allow the macrophages to adhere to the plastic. This incubation was for 30 min, and nonadherent (i.e., nonmacrophage) cells were washed away with RPMI complete. These enriched populations of macrophages were then cultured in RPMI complete.
Macrophage invasion assays.
The invasion assays were performed as previously described (27). Nonopsonized, mid-log-phase grown cultures of different serovars of Salmonella were added to macrophage cell monolayers (i.e., 4 × 105 to 5 × 105 cells/well) in 24-well tissue culture plates at a multiplicity of infection (MOI) generally of 10 bacteria per eukaryotic cell for most assays, although MOIs of 1:1 and 100:1 were also used. After incubation at 37°C for 45 min (0-h time point), infected cell monolayers were washed three times with PBS; then RPMI complete containing 50 μg of gentamicin (Gibco/BRL) per ml was added to kill any remaining extracellular bacteria. After 2 h of further incubation at 37°C, the medium in the 24-well plates was replaced again with RPMI complete containing 5 μg of gentamicin per ml. Host cells remained in this medium for the remainder of the infection to prevent extracellular growth of any released bacteria. Note that all assays were conducted in triplicate and repeated at least three times on different days. The results are presented as the mean ± standard error of the mean.
At different time points following infection with salmonellae, the cell monolayers were processed in one of three ways. For viable count determinations, the infected macrophage monolayers were washed three times with PBS, and salmonellae were harvested by adding 300 μl of 0.1% Triton X-100 in distilled water to each well. After 3 min, cell lysates were collected and serially diluted 10-fold in PBS, and aliquots were plated onto L agar to assess bacterial CFU. A second processing involved Giemsa staining of the cells, as described below. A third treatment involved a trypan blue exclusion protocol, also described below. Statistical analyses were conducted using Student's t test.
Giemsa staining.
Giemsa staining of macrophage cells was carried out as previously reported (27). Briefly, eukaryotic cells were seeded (4 × 105 to 5 × 105) onto circular coverslips lying in 24-well tissue culture plates. Monolayers were infected and processed as described above. At specific times after infection with the bacteria, the coverslips were washed two times with PBS and fixed for 5 to 7 min with methanol at room temperature. The coverslips were air dried and stained for 15 to 60 min with Giemsa stain (Sigma) prepared as instructed by the manufacturer. After the coverslips were washed three times with distilled water, they were air dried and observed microscopically under oil immersion. Time points at 2 and 8 h postinfection were examined.
Eukaryotic cell viability assay.
To assess the overall viability of macrophage cells following bacterial infection, monolayers were infected as described above. At particular time points after infection with bacteria, the monolayers were washed two times with PBS and treated with a 0.4% solution of trypan blue for 2 to 3 min. Those eukaryotic cells that are still intact will exclude trypan blue and be visualized as clear cells under the microscope. Eukaryotic cells that are no longer viable, which have damaged membranes that allow entry of the dye, stain blue. Assays were performed in triplicate and repeated at least three times. The number of intact viable cells was expressed as a percentage of total cells and was assessed at different times postinfection.
Measurement of apoptosis.
To detect and quantify the level of apoptosis at the single-cell level, U937 and J774A.1 cells were seeded into 96-well tissue culture plates at 7 × 104 cells/well. After 24 h, the monolayers were infected as noted above with an MOI of 10:1. At specific time points postinfection, apoptosis was detected in individual macrophage cells, via the TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) reaction, using a fluorescence in situ apoptosis detection kit (Boehringer Mannheim) according to the manufacturer's instructions. The percentages of macrophage cells undergoing programmed cell death were counted using a Zeiss Axioplan fluorescence microscope with halogen illumination and an Omega triple-band filter (Molecular Probes, Inc.). Under these conditions, macrophage cells that were apoptotic stained bright green. A total of 300 macrophage cells were counted to obtain the final percentage. Probabilities were calculated by the chi-square test.
RESULTS
Differential survival of Salmonella serovars in tissue culture-derived macrophage cells.
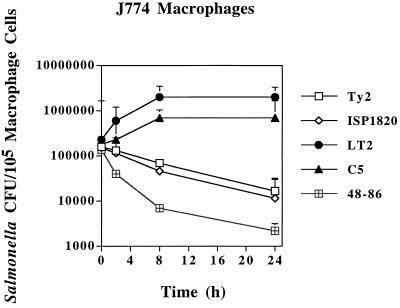
To verify and expand on previous reports of differences in intramacrophage survival abilities of serovar Typhimurium and serovar Typhi (2, 3), we tested several virulent strains of serovar Typhimurium and serovar Typhi as well as two strains of serovar Enteritidis in both murine J774A.1 and human U937 macrophage cell lines. In murine J774A.1 cells, virulent nonopsonized serovar Typhimurium strains LT2 and C5 showed increased viable counts over a 24-h infection time frame (Fig. 1), resulting in up to a 1-log increase in intracellular bacteria. Serovar Typhimurium strains TML and W118-2 behaved similarly (data not shown). On the other hand, each of three strains of serovar Typhi demonstrated at least a 1-log decline in viable counts during the same time frame, as exemplified in Fig. 1. Even more pronounced declines in bacterial viable counts were seen for J774A.1 cells infected with serovar Enteriditis strain 48-86 (Fig. 1). Virtually identical results were seen for serovar Enteritidis strain 464-86. Besides the viable count numbers, Giemsa staining of serovar Typhimurium LT2-infected and serovar Typhi Ty2-infected J774A.1 macrophages visually confirmed the numerical differences between the serovars. Avirulent serovar Typhimurium strain M206 showed a drop in intracellular viable counts of 1.5 logs after 24 h in J774A.1 cells (data not shown).
FIG. 1.
Survival of Salmonella strains in murine J774A.1 tissue-cultured macrophages. Bacterial CFU per 105 macrophage cells (y axis) and time after addition of gentamicin (x axis) are indicated. All values are the means ± standard deviations of at least three experiments done in triplicate.
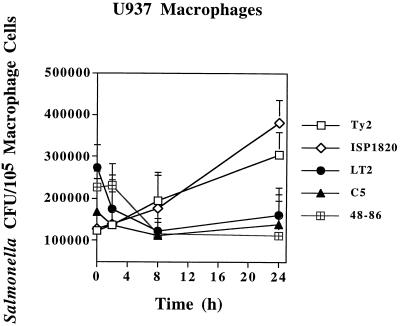
An examination of salmonellae survival within human U937 macrophages cells was also undertaken with multiple strains of each serovar as discussed above. Viable counts of virulent strains of serovar Typhimurium uniformly decreased initially at 2 h postinfection and then either stabilized or increased slightly, but not to the level of the 0-h (45-min real time) postinfection numbers, as exemplified in Fig. 2. The serovar Enteriditis counts within U937 cells also declined twofold and stayed at that level through the time course. Conversely, virulent serovar Typhi levels never declined and slowly increased to a point where the numbers doubled. Giemsa staining of infected U937 cells showed more bacteria in the serovar Typhi-infected macrophages by 8 h postinfection but no increases in the numbers of serovar Typhimurium LT2 (data not shown). These observations suggested a host-specific difference with regard to survival of salmonellae in the intracellular environments of each mammalian cell line.
FIG. 2.
Survival of Salmonella strains in human U937 tissue-cultured macrophages. Bacterial CFU per 105 macrophage cells (y axis) and time after addition of gentamicin (x axis) are indicated. All values are the means ± standard deviations of at least three experiments done in triplicate.
Differential survival of Salmonella serovars in primary macrophage populations of murine or human origin.
The analyses with the immortalized macrophage cell lines (J774A.1 and U937) suggested differences in intramacrophage survival ability of the Salmonella serovars tested. Survival inside tissue culture cells offers some useful information on pathogenesis but may not always reflect what occurs in host primary macrophage cells. To assess whether similar trends occur in primary macrophages, murine peritoneal macrophages and elutriated human macrophages were isolated and infected with a subset of the Salmonella strains used in the above studies.
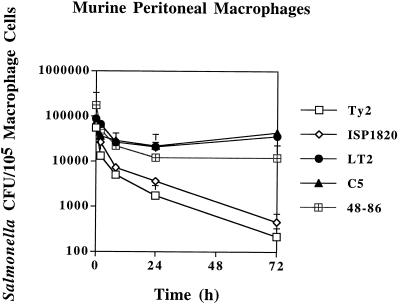
Infections of murine peritoneal macrophages by Salmonella spp. produced CFU decreases for all tested strains over a 72-h time period (Fig. 3). These professional phagocytic cells appeared to be much more bacteriocidal than their cell line counterparts. Strains of virulent serovar Typhimurium (C5 and LT2) and serovar Enteritidis showed the least reduction in bacterial numbers. For serovar Typhimurium, viable counts declined for 8 h and then increased slightly. On the other hand, both virulent strains of serovar Typhi (Ty2 and ISP1820) showed substantial reductions of >2 logs in viability over 72 h that were statistically significant compared to the other strains (P < 0.001 and P < 0.01, respectively).
FIG. 3.
Survival of Salmonella strains in murine peritoneal macrophages. Bacterial CFU per 105 macrophage cells (y axis) and time after addition of gentamicin (x axis) are indicated. All values are the means ± standard deviations of at least three experiments done in triplicate.
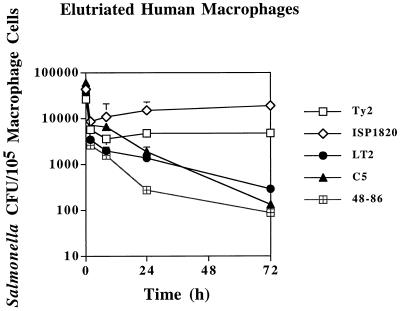
Survival in elutriated human macrophages was tested next. As shown for murine peritoneal macrophages, the primary human macrophages were more effective at eliminating salmonellae than the U937 tissue culture cells. Both serovar Typhi strains used to infect the elutriated macrophages displayed initial declines in viability at 2 h postinfection (Fig. 4). Past this time point, both strains showed slight increases in bacterial numbers through 72 h postinfection. Even by 7 days postinfection, serovar Typhi Ty2 CFU were still as high as 103 per 105 macrophage cells, and viable bacteria persisted inside the elutriated human macrophages after 21 days (data not shown). In striking contrast, both virulent serovar Typhimurium strains showed continued sharp (i.e., >2-log) reductions in viable counts through 72 h (Fig. 4). Viable counts of strain LT2 were statistically lower at 24 h than counts of both serovar Typhi strains (Ty2 and 1820; P < 0.01 and P < 0.002, respectively), and strain C5 was also statistically less numerous than serovar Typhi strain ISP1820 (P < 0.01). Levels of viable serovar Enteritidis strain 48-86 declined to 5 × 102 CFU per 105 macrophage at 24 h and were significantly lower than the levels of serovar Typhi (P < 0.001). By 72 h postinfection, the differences in viable counts between serovar Typhi and either serovar Typhimurium or serovar Enteritidis strains had increased substantially. After 7 days postinfection with serovar Typhimurium or serovar Enteritidis strains, no viable bacteria were detected in the primary human macrophages.
FIG. 4.
Survival of Salmonella strains in elutriated human macrophages. Bacterial CFU per 105 macrophage cells (y axis) and time after addition of gentamicin (x axis) are indicated. All values are the means ± standard deviations of at least three experiments done in triplicate.
Effects of Salmonella serovar MOI on macrophage viability.
It has been known for quite a while that strains of serovar Typhimurium can have a cytotoxic effect on host cells (4, 5). Our Giemsa staining of J774A.1 macrophage cells infected with serovar Typhimurium strain LT2 also suggested a cytotoxic effect by the bacteria on infected macrophage cells (data not shown). A more thorough examination was carried out to determine the effects of infection with various Salmonella serovars on host cell viability.
Since the initial infections of J774A.1 macrophage cells were performed at an MOI of 10:1, the host cell viability analyses were also conducted initially at this same MOI and later at MOIs 10-fold below or above this number. The virulent serovar Typhimurium strain LT2 caused a substantial drop in J774A.1 cell viability (Table 2) as measured by trypan blue exclusion over 24 h of infection (93% down to 57% at MOI = 10). Similar cytotoxicity to J774A.1 cells was observed following infection with other virulent serovar Typhimurium strains (i.e., C5, TML, and W118-2). The total number of J774A.1 cells, measured microscopically by Giemsa stain, also decreased over the infection time period. At 8 h following infection with serovar Typhimurium there was a 5 to 10% reduction in the total number of plate-attached J774A.1 cells. This loss of cells increased to 25 to 35% after 24 h of infection, adding to the damage assessed by dye exclusion. The serovar Typhi strain Ty2 (or ISP1820 [data not shown]) had a nominal effect on J774A.1 viability at 24 h postinfection (91% viability, compared to 97% for the noninfected macrophage control) and did not cause J774A.1 cell detachment. The serovar Enteritidis strains, like the serovar Typhimurium strains, caused significant reductions in host cell viability after 24 h of incubation with an MOI of 10:1. At an MOI of 1:1, very little difference was observed between the serovars after 8 h of infection, but the viability of J774A.1 cells declined by 13 to 25% after 24 h of infection with serovar Typhimurium or Enteritidis (Table 2). The percentage of viable macrophages changed considerably when the MOIs were increased to 100:1. By 24 h, viability of the remaining 65 to 75% of plate-attached J774A.1 cells was reduced in half for serovar Typhimurium or for serovar Enteritidis. In marked contrast, serovar Typhi strain Ty2 showed only a marginal decrease in J774A.1 viability even after 24 h at an MOI of 100:1 (97% down to 92%). Preliminary cytotoxicity studies with murine, peritoneal macrophage cells indicated a differential adverse effect caused by infection with serovar Typhimurium or Enteritidis strains compared to serovar Typhi, but overall macrophage killing was reduced (Table 3). These results suggested that serovars Typhimurium and Enteritidis have a more toxic effect than serovar Typhi on macrophage cells of murine origin.
TABLE 2.
Percent murine macrophage (J774A.1) viability over 24 h based on MOI
| Infectious agent | Viabilitya
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 hb
|
8 h
|
24 h
|
|||||||
| 1c | 10 | 100 | 1 | 10 | 100 | 1 | 10 | 100 | |
| Serovar Typhimurium LT2 | 96.9 ± 0.57 | 92.8 ± 2.63 | 90.9 ± 0.99 | 94.9 ± 0.73 | 76.4 ± 4.14 | 60.2 ± 2.44 | 75.6 ± 5.40 | 57.0 ± 7.48 | 48.4 ± 2.66 |
| Serovar Typhi Ty2 | 97.9 ± 1.20 | 97.2 ± 0.79 | 96.8 ± 0.79 | 98.2 ± 0.63 | 96.8 ± 1.03 | 94.2 ± 2.39 | 96.8 ± 0.79 | 93.2 ± 3.01 | 91.6 ± 4.57 |
| Serovar Enteritidis 48-86 | 97.4 ± 0.96 | 92.2 ± 2.20 | 87.1 ± 3.14 | 95.8 ± 0.63 | 81.6 ± 2.95 | 65.8 ± 6.96 | 87.8 ± 4.05 | 67.1 ± 3.51 | 50.3 ± 3.56 |
| None | 97.8 ± 1.13 | 97.8 ± 0.63 | 96.8 ± 0.63 | ||||||
Mean percentage of viable, trypan-negative, eukaryotic cells ± standard deviation based on at least three separate experiments done in triplicate.
Time after infection with the salmonellae.
MOI.
TABLE 3.
Murine peritoneal macrophage viability after infection with selected Salmonella serovars
| Infectious agent | Viabilitya
|
||
|---|---|---|---|
| 0 hb | 8 h | 24 h | |
| Serovar Typhimurium LT2 | 93.5 ± 0.71 | 89.0 ± 1.41 | 83.0 ± 0.00 |
| Serovar Typhi Ty2 | 95.0 ± 0.00 | 94.5 ± 0.71 | 92.5 ± 0.71 |
| Serovar Enteritidis 49-86 | 94.0 ± 0.00 | 89.0 ± 0.00 | 85.0 ± 0.00 |
| None | 96.0 ± 0.00 | 95.5 ± 0.71 | 95.0 ± 0.00 |
Mean percentage of viable, trypan-negative, eukaryotic cells ± standard deviation based on at least three separate experiments done in triplicate.
Time after infection with the salmonellae.
Human U937 macrophage cell viability following infection with the same Salmonella serovars was also examined. The level of viability for noninfected U937 cells was approximately 10% lower (Table 4) than that observed for J774A.1 cells (Table 2). Through 24 h of infection, virulent serovar Typhimurium strains reduced U937 cell viability by 10 to 20% at an MOI of 10:1 (Table 4). Invasion by serovar Typhi led to only a ∼5% reduction in viability, although serovar Enteritidis infections led to substantial U937 cell leakiness (i.e., ∼65% viability). An MOI of 1:1 led to little difference in macrophage viability when infections were conducted with serovar Typhimurium or Typhi, but serovar Enteritidis-infected U937 cells showed a 10% reduction in viability after 24 h. Once the MOI was increased to 100:1, host cell damage became detectable in as little as 45 min (0 h) of infection with serovar Typhimurium or Enteritidis. By 24 h of infection, more than half of the U937 macrophage population was injured by these same serovars (i.e., 43 or 45% remaining U937 viability, respectively). At this high MOI, however, serovar Typhi-infected U937 cells were only slightly affected following 24 h (∼7% reduction in viability). Roughly similar amounts of macrophage cell detachment occurred after infection with serovars Typhimurium and Enteritidis as seen over time with J774A.1 cells. These results show that human macrophage viability is affected by the specific Salmonella serovar and the concentration of salmonellae used in the infection. Preliminary viability analyses of primary human macrophages also show less overall cytotoxicity than with immortalized cell lines but more damage mediated by serovars Typhimurium and Enteritidis compared to little cytotoxicity caused by serovar Typhi (Table 5). Therefore, these results suggest that serovars Typhimurium and Enteritidis damage mammalian macrophage cells more severely than serovar Typhi and that higher MOIs result in more cell damage.
TABLE 4.
Percent human macrophage (U937) viability over 24 h based on MOI
| Infectious agent | Viabilitya
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 hb
|
8 h
|
24 h
|
|||||||
| 1c | 10 | 100 | 1 | 10 | 100 | 1 | 10 | 100 | |
| Serovar Typhimurium LT2 | 84.9 ± 0.74 | 84.4 ± 0.96 | 80.7 ± 1.05 | 87.7 ± 0.67 | 75.8 ± 2.20 | 67.6 ± 1.94 | 82.0 ± 1.41 | 66.8 ± 3.22 | 42.6 ± 4.32 |
| Serovar Typhi Ty2 | 86.6 ± 0.83 | 86.1 ± 0.73 | 85.2 ± 0.79 | 88.6 ± 0.68 | 87.2 ± 0.63 | 85.8 ± 1.03 | 84.7 ± 0.94 | 82.8 ± 4.49 | 79.2 ± 1.47 |
| Serovar Enteritidis 48-86 | 87.8 ± 0.79 | 85.9 ± 1.10 | 78.3 ± 1.15 | 87.7 ± 0.94 | 77.6 ± 2.41 | 72.8 ± 2.74 | 77.7 ± 2.31 | 64.6 ± 3.77 | 44.8 ± 2.70 |
| None | 86.8 ± 0.92 | 88.3 ± 0.94 | 88.0 ± 1.33 | ||||||
Mean percentage of viable, trypan-negative, eukaryotic cells ± standard deviation based on at least three separate experiments done in triplicate.
Time after infection with the salmonellae.
MOI.
TABLE 5.
Elutriated human macrophage viability after infection with selected Salmonella serovars
| Infectious agent | Viabilitya
|
||
|---|---|---|---|
| 0 hb | 8 h | 24 h | |
| Serovar Typhimurium LT2 | 89.0 ± 0.00 | 84.3 ± 0.58 | 81.0 ± 1.00 |
| Serovar Typhi Ty2 | 89.0 ± 0.00 | 88.7 ± 0.58 | 87.7 ± 0.58 |
| Serovar Enteritidis 49-86 | 87.7 ± 0.58 | 82.0 ± 1.00 | 80.7 ± 1.53 |
| None | 89.7 ± 0.58 | 89.0 ± 0.00 | 88.7 ± 0.58 |
Mean percentage of viable, trypan-negative, eukaryotic cells ± standard deviation based on at least three separate experiments done in triplicate.
Time after infection with the salmonellae.
Testing the extent of apoptosis among tissue culture-derived macrophage cells infected with different Salmonella serovars.
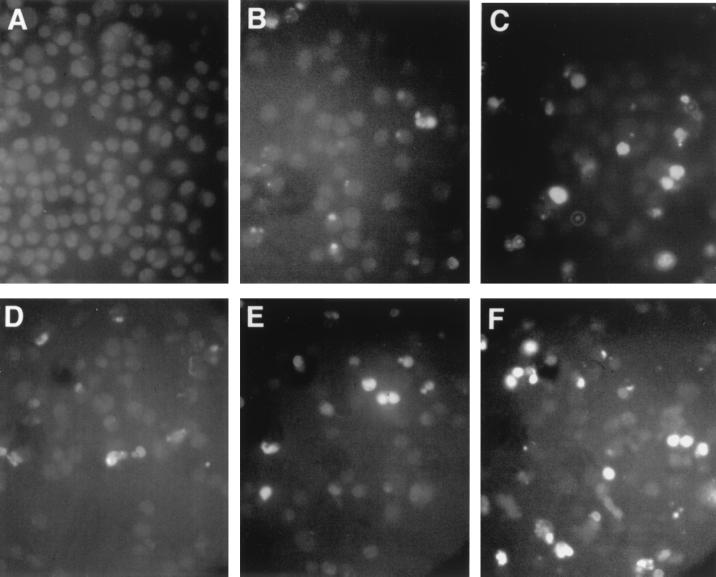
Previous studies have shown that Salmonella spp. are capable of inducing apoptosis in murine macrophages (9, 21), but no studies have examined this phenomenon in human macrophages. To repeat the prior studies and expand the observations to include human macrophage cells, an in situ apoptosis detection assay was used to observe and quantify the amount of apoptosis in infected murine and human macrophage cells. Murine J774A.1 macrophage cells infected with serovar Typhi at an MOI of 10:1 showed very little (i.e., 0.4%) apoptosis after a 2-h incubation time that increased only to 4.2% after 24 h (Table 6). Infections with serovar Typhimurium caused an increase in the level of apoptosis from 0.8% at 2 h to 10.9% at 24 h. Figure 5A to C shows a typical field of view for the J774A.1 cells that were uninfected or infected with serovar Typhi or Typhimurium. Human U937 cells already exhibited a measurable amount of apoptosis (1.6 to 2.0% [Table 6]) before infection. Even after a 2-h infection with either serovar Typhi Ty2 or serovar Typhimurium LT2, there was little change in the percentage of apoptotic macrophage cells. This level of programmed cell death increased to 6.4% in Ty2-infected macrophages and up to 14.9% in LT2-infected U937 cells. Representative fields of view for noninfected cells versus U937 cells infected with serovar Typhi Ty2 or serovar Typhimurium LT2 are shown in Fig. 5D to F. The results demonstrate that significantly more apoptosis is induced by serovar Typhimurium than by serovar Typhi.
TABLE 6.
Percentage of apoptotic J774A.1 and U937 macrophage cells following infection with serovar Typhimurium or Typhi
| Macrophage | Time (h) after infection | % of apoptotic cellsa infected with:
|
||
|---|---|---|---|---|
| LT2 | Ty2 | No bacteria (control) | ||
| J774A.1 | 2 | 0.8 ± 0.3 | 0.4 ± 0.2 | 0.0 |
| 24 | 10.9 ± 2.1 | 4.2 ± 2.6 | 0.0 | |
| U937 | 2 | 1.2 ± 0.3 | 2.0 ± 0.3 | 1.6 ± 0.4 |
| 24 | 14.9 ± 1.7 | 6.4 ± 0.9 | 2.1 ± 0.3 | |
Mean percentage of apoptotic macrophage cells ± standard deviation based on at least 250 cells done at least two times.
FIG. 5.
Fluorescent TUNEL reaction examination of macrophage cells undergoing programmed cell death following a 24-h infection with Salmonella. (A to C) Murine J774A.1 macrophage cells; (D to F) human U937 macrophage cells; (A and D) noninfected cells; (B and E) serovar Typhi strain Ty2-infected macrophages; (C and F) serovar Typhimurium LT2-infected macrophage cells. Weak, diffuse backround fluorescence staining of all cells was enhanced with Adobe Photoshop to allow differentiation between densely stained areas in apoptotic cells and total cells in the field.
DISCUSSION
Comparative studies of intramacrophage survival of salmonellae have previously indicated that serovar Typhimurium has a survival advantage in murine macrophage cells; however, in human macrophages, serovar Typhi seems to be favored (2, 3). These past studies were limited in scope and were based on one strain of each Salmonella serovar. Our previous findings also suggested such a difference, but these were only preliminary in nature (26). In this study, we have expanded on these initial observations and have also investigated for the first time apoptosis in human macrophage cells infected with Salmonella. Through the use of multiple strains of nonopsonized virulent serovars Typhimurium and Typhi, we confirm that serovar Typhi has a distinct survival advantage over serovar Typhimurium in human macrophage cells, whereas the converse appears to be true in murine macrophages. Furthermore, we report that strains of serovars Typhimurium and Enteritidis appear to damage mammalian macrophage cells much more extensively than do strains of serovar Typhi or related human-specific serovars (e.g., serovar Paratyphi [W. R. Schwan and D. J. Kopecko, unpublished data]).
These intramacrophage survival data correlate well with previous animal studies in which certain Salmonella serovars cause systemic disease. Virulent serovar Typhimurium strains cause a systemic infection in mice (17), growing in the splenic macrophages of the infected animals (7, 16). Bacterial persistence and growth in the host's macrophage populations plus the ability of certain Salmonella serovars to initiate death of infected macrophages likely contribute to the type and severity of disease caused by Salmonella strains. Pathogenic serovar Typhimurium strains that were tested in this report either maintained stable cell numbers or grew within murine J774A.1 macrophages, mimicking findings of earlier studies (7, 12, 20). These high numbers of serovar Typhimurium in murine macrophages take on enhanced significance when one takes into account that after 24 h of infection, ∼30% of J774A.1 cells have detached (and were not assessed here) and ∼50% of the remaining attached cells are dead (i.e., trypan blue positive). These same serovar Typhimurium strains were more readily killed in resident murine peritoneal macrophages, similar to what has been previously described (2, 7, 12), but still maintained high intracellular, viable counts over 24 h.
Serovar Enteritidis, another broad-host-range serovar, can also grow and cause disease in mice (10), but serovar Typhi and related human-specific serovars, serovar Paratyphi A and B, do not grow or cause disease in mice (8, 23). Poor survival within murine macrophages could largely explain why serovar Typhi infections of mice do not lead to disease. As shown in Fig. 1, all serovars invaded J774A.1 cells equally well after the 45-min infection period, resulting in a time zero average of ∼2 bacteria internalized per host macrophage. Despite starting with equal numbers of internalized serovar Typhi versus serovar Typhimurium and the fact that much less cytotoxicity occurred in serovar Typhi-infected cells, all three virulent serovar Typhi strains exhibited substantial drops in viable counts inside J774A.1 or peritoneally derived murine macrophage cells, coinciding with results of previous reports (2, 3). The serovar Enteritidis strain was cleared from J774A.1 cells at levels similar to those for the virulent serovar Typhi strains. However, in murine peritoneal macrophages, serovar Enteritidis levels dropped, but only to the levels noted for serovar Typhimurium strains at 24 h postinfection. These findings demonstrate that macrophage killing of salmonellae is complicated and can vary depending not only on the Salmonella serovar or strain (and possibly on the completeness of opsonization) but also on the source and state of activation of the macrophage cells. Additionally, some other bacterial factors may affect survival of serovar Enteritidis within immortalized murine macrophages compared to primary cell macrophage populations. The changes in intracellular bacterial number over time reflect serovar-specific bacterial ability to survive and multiply within that macrophage type and are also affected by Salmonella-induced cytotoxicity and macrophage cell detachment. J774A.1 macrophage cell death increased with an increase in the number of intracellular serovar Typhimurium, but equal macrophage death occurred with decreasing and much lower numbers of serovar Enteritidis. Thus, cell death did not specifically correlate with the number of viable, intracellular Salmonella.
Similar to Salmonella infection of J774A.1 cells, invasion of U937 cells by nonopsonized salmonellae resulted in an average of one to three bacteria internalized per macrophage cell for all serovars. However, Salmonella serovar survival abilities in human macrophages were in stark contrast with the results in murine macrophages. Instead of the prolific growth observed for serovar Typhimurium in murine macrophages, virulent serovar Typhimurium strains were initially killed and only later were able to grow slightly in U937 cells, but nevertheless caused equivalent cytotoxicity to that observed in J774A.1 cells. However, serovar Typhi strains were able to undergo from one to three rounds of replication in the tissue culture-derived U937 cells and, despite high intracellular bacterial numbers, caused much less cytotoxicity than the other serovars. Differential serovar persistence was perhaps demonstrated more dramatically in elutriated human macrophages; serovar Typhi strains were shown to multiply after an initial killing phase, whereas serovar Typhimurium and serovar Enteritidis viable numbers continued to decline dramatically throughout 72 h of infection. These differences between serovar Typhi and serovar Typhimurium survival have been observed before with a single strain of each serovar (2, 3) or in a more limited study (26), and our findings here further substantiate that serovar Typhi has a host-specific survival advantage over serovar Typhimurium in human macrophages. Thus, host specificity of the serovar would appear to play an important role in observed cytotoxicity and ability of intracellular bacterial to survive and multiply. In humans, serovars Typhi and Paratyphi survive and multiply within macrophages, which likely serve as the conduits for deep tissue infection. However, some Salmonella serovars (e.g., Enteritidis and Typhimurium) do not typically survive for extended periods of time in human macrophages or cause a disseminated infection in humans but do cause mild gastroenteritis (6). Presumably, long-term (i.e., many days) survival within macrophage populations enable salmonellae to spread from a localized infection in the intestine to a systemic one.
In addition to the observed differences in bacterial intramacrophage survival associated with specific Salmonella serovars, we also discovered variability in the degree of cytotoxicity that these serovars exert toward the host cell, as well as the prevalence of apoptosis initiated by these serovars. Cytotoxicity has been reported previously for Salmonella spp. (4, 5, 9, 21). Although all of the limited Salmonella serovars studied herein displayed some degree of cytotoxicity, some serovars were much more destructive than others. Virulent serovar Typhimurium and serovar Enteritidis strains significantly reduced the viability and total number of plate-attached, infected J774A.1 or U937 cells, whereas virulent serovar Typhi minimally affected viability (and caused no detachment) of these host cells even at later times postinfection. These differences in cytotoxic properties could be a reflection of either restrictions in host range of the toxin(s) or the amount of cytotoxin secreted. Ashkenazi et al. (4) have reported that serovar Typhi secretes 2.5-fold less cytotoxin than strains of serovar Enteritidis and 5.5-fold less than serovar Choleraesuis, suggesting that certain serovars secrete less cytotoxin than do others.
Virulent serovar Typhimurium strains survived intracellularly and multiplied rapidly in J774A.1 cells during the first few hours (this study and references 7, 12, and 20), but these bacteria were very destructive by 8 to 24 h postinfection, killing and/or detaching about two-thirds of the macrophages. Serovar Typhimurium did not survive or multiply as well in U937 cells but still caused similar levels of host cell death. As the MOI rose, more host cells were also damaged, presumably because more cytotoxin was produced. Two recent studies have shed some light on the basis for the cytotoxic effect of salmonellae on murine macrophage cells (9, 21). Both reports demonstrate that many Salmonella serovars can trigger cytotoxicity in murine macrophages and that apoptosis is responsible for some of the observed overall host cell death. Furthermore, apoptosis requires a functional Salmonella type III secretion system (i.e., the cytotoxin must be secreted [9]) and actively replicating salmonellae (21). Thus, higher amounts of proteins secreted into the host cells at higher MOIs may lead to increased host cell death even if the number of bacteria internalized did not change significantly. Our study demonstrated that macrophage infections with equal numbers of internalized salmonellae of both serovars Typhimurium and Typhi initiated apoptosis in J774A.1 cells, but the level of apoptosis observed was much less than the level noted by Chen et al. (9). Moreover, we saw a significant difference between serovars Typhimurium and Typhi that was not observed in this previous study. Interesting with regard to the findings of Ashkenazi et al. (4) noted above, serovar Typhimurium caused 2.6-fold more apoptosis than serovar Typhi in J774A.1 cells; the overall levels of apoptosis caused by serovar Typhimurium were closer to the data reported by Monack et al. (21). It seems plausible that both methodological and strain differences could in part explain these discrepancies. Nevertheless, trypan blue exclusion analysis of J774A.1 cells infected with serovar Typhi strain ISP1820 reflected the minimal level of cytotoxicity observed for Ty2-infected cells (data not shown). In human U937 cells, the same trend was seen. Serovar Typhimurium-infected macrophages were significantly more apoptotic than serovar Typhi-infected U937 cells (14.9 and 6.4%, respectively). These are the first results to show apoptosis in human macrophage cells infected with salmonellae, and they also demonstrate that serovar Typhimurium strains are more cytotoxic than serovar Typhi strains toward human macrophage cells.
Swords et al. (29) showed that a defective Salmonella rpoS gene results in avirulence in the mouse infection model. Since the rpoS phenotype of Salmonella can change during passage and affects catalase gene expression, we wondered whether this phenotype in our tested strains might affect intramacrophage survival. Using a stationary-phase bacterial survival assay in H2O2 (27a), we determined that our LT2 and TML strains were likely wild type with respect to rpoS, but that C5 was rpoS defective. Both serovar Enteritidis strains and serovar Typhi ISP1820 appeared wild type for rpoS, but Ty2 appeared to be rpoS defective. Regardless of the apparent rpoS phenotype differences, all serovar Typhi strains behaved virtually identically in macrophages. Similarly, all pathogenic serovar Typhimurium strains showed equal intracellular survival and cytotoxic behaviors. Thus, we do not believe that the rpoS phenotype affects our interpretations.
How might these differences in observed Salmonella serovar-specific intramacrophage survival and host cell cytotoxicity relate to pathogenicity in humans versus mice? Serovars Typhimurium and Enteritidis, which do not survive well in vitro in human macrophages, generally produce in humans a more localized infection which results in mild gastroenteritis. During infections of mice, these and other mouse-pathogenic salmonellae are presumed to enter M cells in the ileum and destroy these modified epithelial cells (15, 24), after which they are engulfed by underlying macrophages, which may act as vehicles for widespread dissemination within the host. The ability of these serovars to replicate within and kill macrophages leads to an acute systemic disease in mice that is associated with high mortality. In this circumstance, bacterial multiplication may overwhelm the macrophage and cause a majority of nonprogrammed cell demise. In contrast, serovar Typhi, which can infect mice via the oral route, does not survive well in murine macrophages and cannot cause disease in mice. On the other hand, serovar Typhi survives well in human macrophages while causing only slight macrophage cell death, almost all of which is orderly and controlled by apoptosis. The ability of this serovar (or related serovar Paratyphi strains) to translocate asymptomatically across the human intestine, trigger uptake into and survival for long periods (i.e., many days) within human macrophages, and multiply within these cells without inducing much host cell death may allow this serovar to cross the ileal epithelium silently, moving stealthily within macrophage vehicles to deep tissues, where acute multiplication in selected tissues (e.g., liver) ultimately results in typhoid fever.
Finally, as recently proposed by Godfrey (13a), some infectious diseases might involve simultaneous interrelated cycles of chronic and acute infections among a limited number of host cell types, rather than overt infection of all cells in an organism. For serovar Typhi, the chronic phase of infection may occur in the human macrophage, whereas disease symptoms may occur via more acute multiplication in selected tissues. Although both opsonized and nonopsonized Salmonella strains have been reported to be internalized by mouse and human macrophages (1, 2, 21), long-term intramacrophage survival of bacteria taken up by different mechanisms has not yet been studied systematically. The uptake into macrophage of nonopsonized Salmonella, perhaps via a bacterium-induced mechanism(s), may enhance Salmonella's ability to enter a privileged niche (e.g., spacious phagosome [1, 2]) where it is protected from killing. Further studies comparing nonopsonized versus opsonized Salmonella survival in macrophages and clarification of the mechanism(s) by which nonopsonized salmonellae enter macrophage cells should aid our understanding of host specificity and intramacrophage persistence by salmonellae.
ACKNOWLEDGMENTS
We thank K. Elkins and F. Collins for critical reading of the manuscript, D. Hone, J. L. Rosner, and B. Swaminathan for bacterial strains, K. Elkins and A. Jerse for mice, and K. Faust and K. Clouse for elutriated human macrophages.
This work was performed in part while W. R. Schwan, X.-Z. Huang, and L. Hu were postdoctoral fellows of the National Research Council and the Fogarty International Center of the National Institutes of Health.
REFERENCES
- 1.Alpuche-Aranda C M, Racoosin E L, Swanson J A, Miller S I. Salmonella stimulate macrophage macropinocytosis and persist within spacious phagosomes. J Exp Med. 1994;179:601–608. doi: 10.1084/jem.179.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alpuche-Aranda C M, Berthiaume E P, Mock B, Swanson J A, Miller S I. Spacious phagosome formation within mouse macrophages correlates with Salmonella serotype, pathogenicity, and host susceptibility. Infect Immun. 1995;63:4456–4462. doi: 10.1128/iai.63.11.4456-4462.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arai T, Ishibashi Y, Matsui K. Mechanism of selective pathogenesis of Salmonella serovars and suggestions for chemotherapy and development of safe vaccines for typhoid fever. In: Pang T, Koh C L, Putchucheary S D, editors. Typhoid fever strategies for the 90's. River Edge, N.J: World Scientific; 1992. pp. 140–147. [Google Scholar]
- 4.Ashkenazi S, Cleary T G, Murray B E, Wanger A, Pickering L K. Quantitative analysis and partial characterization of cytotoxin production by Salmonella strains. Infect Immun. 1988;56:3089–3094. doi: 10.1128/iai.56.12.3089-3094.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baloda S B, Faris A, Krovacek K, Wadstrom T. Cytotoxic enterotoxins and cytotoxic factors produced by S. enteritidis and S. typhimurium. Toxicon. 1983;21:785–790. doi: 10.1016/0041-0101(83)90067-3. [DOI] [PubMed] [Google Scholar]
- 5a.Blanc-Potard A-B, Groisman E A. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 1997;16:5376–5385. doi: 10.1093/emboj/16.17.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaser M J, Newman L S. A review of human salmonellosis. I. Infective dose. Rev Infect Dis. 1982;4(Suppl. 6):1096–1106. doi: 10.1093/clinids/4.6.1096. [DOI] [PubMed] [Google Scholar]
- 7.Buchmeier N A, Heffron F. Intracellular survival of wild-type Salmonella typhimurium and macrophage-sensitive mutants in diverse populations of macrophages. Infect Immun. 1989;57:1–7. doi: 10.1128/iai.57.1.1-7.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter P B, Collins F M. Growth of typhoid and paratyphoid bacilli in intravenously infected mice. Infect Immun. 1974;10:816–822. doi: 10.1128/iai.10.4.816-822.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L M, Kaniga K, Galan J E. Salmonella spp. are cytotoxic for cultured macrophages. Mol Microbiol. 1996;21:1101–1115. doi: 10.1046/j.1365-2958.1996.471410.x. [DOI] [PubMed] [Google Scholar]
- 10.Collins F M. Salmonellosis in orally infected specific pathogen-free C57BL mice. Infect Immun. 1972;5:191–198. doi: 10.1128/iai.5.2.191-198.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dragunsky E M, Rivera E, Hochstein H D, Levenbook I S. In vitro characterization of Salmonella typhi mutants strains for live oral vaccines. Vaccine. 1990;8:263–268. doi: 10.1016/0264-410x(90)90056-r. [DOI] [PubMed] [Google Scholar]
- 12.Fields P I, Swanson R V, Haidaris C G, Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fortier A, Falk L A. Isolation of murine macrophages. In: Coligan J E, Kruisbeek A M, Margulies D H, Shevach E M, Strober W, editors. Current protocols in immunology. New York, N.Y: John Wiley & Sons, Inc.; 1994. pp. 14.1.1–14.1.6. [Google Scholar]
- 13a.Godfrey H. Pathogenesis of chronic bacterial infections. Trends Microbiol. 1998;6:303. doi: 10.1016/s0966-842x(98)01323-7. [DOI] [PubMed] [Google Scholar]
- 14.Hook E W. Salmonella species (including typhoid fever) In: Mandell G L, Douglas R G, Bennett J E, editors. Principles and practice of infectious diseases. 3rd ed. New York, N.Y: Churchill Livingstone; 1990. pp. 1700–1716. [Google Scholar]
- 15.Jones B D, Ghori N, Falkow S. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J Exp Med. 1994;180:15–23. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lissner C R, Swanson R L, O'Brien A D. Genetic control of the innate resistance of mice to Salmonella typhimurium: expression of the ity gene in peritoneal and splenic macrophages isolated in vitro. J Immunol. 1983;131:3006–3013. [PubMed] [Google Scholar]
- 17.Mackaness G B, Blanden R V, Collins F M. Host-parasite relations in mouse typhoid. J Exp Med. 1966;124:573–583. doi: 10.1084/jem.124.4.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCormick B A, Miller S I, Carnes D, Madar J L. Transepithelial signaling to neutrophils by salmonellae: a novel virulence mechanism for gastroenteritis. Infect Immun. 1995;63:2302–2309. doi: 10.1128/iai.63.6.2302-2309.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 20.Mills S D, Finlay B B. Isolation and characterization of Salmonella typhimurium and Yersinia pseudotuberculosis-containing phagosomes from infected mouse macrophages: Y. pseudotuberculosis traffics to terminal lysosomes where they are degraded. Eur J Cell Biol. 1998;77:35–47. doi: 10.1016/S0171-9335(98)80100-3. [DOI] [PubMed] [Google Scholar]
- 21.Monack D M, Raupach B, Hromockyj A E, Falkow S. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc Natl Acad Sci USA. 1996;93:9833–9838. doi: 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morello J A, Baker E E. Interaction of Salmonella with phagocytes in vitro. J Infect Dis. 1965;115:131–141. doi: 10.1093/infdis/115.2.131. [DOI] [PubMed] [Google Scholar]
- 22a.Mulvey M R, Switala J, Borys A, Loewen P C. Regulation of transcription of katE and katF in Escherichia coli. J Bacteriol. 1990;172:6713–6720. doi: 10.1128/jb.172.12.6713-6720.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Brien A D. Innate resistance of mice to Salmonella typhi infection. Infect Immun. 1982;38:948–952. doi: 10.1128/iai.38.3.948-952.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pascopella L, Raupach B, Ghori N, Monack D, Falkow S, Small P L C. Host restriction phenotypes of Salmonella typhi and Salmonella gallinarum. Infect Immun. 1995;63:4329–4335. doi: 10.1128/iai.63.11.4329-4335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pearlman E, Jiwa A H, Engleberg N C, Eisenstein B I. Growth of Legionella pneumophila in a human macrophage-like (U937) cell line. Microb Pathog. 1988;5:87–95. doi: 10.1016/0882-4010(88)90011-3. [DOI] [PubMed] [Google Scholar]
- 26.Schwan W R, Kopecko D J. Serovar specific differences in Salmonella survival within macrophage cells. Adv Exp Med Biol. 1997;412:277–278. doi: 10.1007/978-1-4899-1828-4_46. [DOI] [PubMed] [Google Scholar]
- 27.Schwan W R, Demuth A, Kuhn M, Goebel W. Phosphatidylinositol-specific phospholipase C from Listeria monocytogenes contributes to intracellular survival and growth of Listeria innocua. Infect Immun. 1994;62:4795–4803. doi: 10.1128/iai.62.11.4795-4803.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.Shiba T, Tsutsumi K, Yano H, Ihara Y, Kameda A, Tanaka K, Takahashi H, Munekata M, Rao N N, Kornberg A. Inorganic polyphosphate and the induction of rpoS expression. Proc Natl Acad Sci USA. 1997;94:11210–11215. doi: 10.1073/pnas.94.21.11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sizemore D R, Elsinghorst E A, Eck L C, Branstrom A A, Hoover D L, Warren R L, Rubin F A. Interaction of Salmonella typhi strains with cultured human monocyte-derived macrophages. Infect Immun. 1997;65:309–312. doi: 10.1128/iai.65.1.309-312.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swords W E, Cannon B M, Benjamin W H., Jr A virulence of LT2 strains of Salmonella typhimurium results from a defective rpoS gene. Infect Immun. 1997;65:2451–2453. doi: 10.1128/iai.65.6.2451-2453.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]