Abstract
Background
Malaria rapid diagnostic tests (RDTs) remain the main point-of-care tests for diagnosis of symptomatic Plasmodium falciparum malaria in endemic areas. However, parasites with gene deletions in the most common RDT target, histidine rich protein 2 (pfhrp2/HRP2), can produce false-negative RDT results leading to inadequate case management. The objective of this study was to determine the prevalence of hrp2/3 deletions causing false-negative RDT results in Vietnam (Gia Lai and Dak Lak provinces).
Methods
Individuals presenting with malaria symptoms at health facilities were screened for P. falciparum infection using light microscopy and HRP2-RDT (SD Bioline Malaria Antigen Pf/Pv RDT, Abbott). Microscopically confirmed P. falciparum infections were analysed for parasite species by 18S rRNA qPCR, and pfhrp2 and pfhrp3 exon2 deletions were investigated by nested PCR. pfhrp2 amplicons were sequenced by the Sanger method and HRP2 plasma levels were determined by enzyme-linked immunosorbent assay (ELISA).
Results
The prevalence of false-negative RDT results among symptomatic cases was 5.6% (15/270). No pfhrp2 and pfhrp3 deletions were identified. False-negative RDT results were associated with lower parasite density (p = 0.005) and lower HRP2 plasma concentrations (p < 0.001), as compared to positive RDT.
Conclusions
The absence of hrp2/3 deletions detected in this survey suggests that HRP2-based malaria RDTs remain effective for the diagnosis of symptomatic P. falciparum malaria in Central Vietnam.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12936-022-04399-w.
Keywords: Malaria, Plasmodium falciparum, Diagnostics, Rapid Diagnostic Test, Histidine rich protein 2, Histidine rich protein 3, Gene deletion, Surveillance, Vietnam
Background
Malaria rapid diagnostic tests (RDT) play a key role in malaria case management, as they provide immediate diagnosis of malaria infections, especially in rural settings or in the absence of expert microscopy. RDT are lateral flow immunochromatographic tests designed to detect Plasmodium antigens present in human blood. The most commonly used RDT for Plasmodium falciparum targets histidine rich protein 2 (HRP2), an abundant secreted protein that is not present in other Plasmodium parasite species [1, 2]. These RDT are more sensitive and heat-stable than RDT detecting other antigens such as lactate-dehydrogenase (LDH) or aldolase. Importantly, antibodies in the test strip of HRP2-RDT may also cross-react with another antigen of the HRP family, namely HRP3, due to strong similarities in the amino acid repeat sequences [3].
Monitoring the accuracy of the RDT at point-of-care is critical for effective case management. Parasites with deletions in pfhrp2 and/or pfhrp3 genes can go undetected by HRP2-RDTs and are cause of concern in malaria endemic countries [4]. There are other common causes of false-negative RDT results, including product and storage quality, operator errors or parasite densities below the detection threshold of the device. However, some of these causes can be addressed with improved transport and storage of RDTs as well as end user training. The identification of P. falciparum parasites with hrp2/3 gene deletions was first reported in the Peruvian Amazon in 2010, where hrp2-deleted parasites now constitute more than 50% of P. falciparum infections [5, 6]. Besides South America, the prevalence of hrp2/3 deletions is highest in Eritrea and Djibouti (> 80%) [7, 8], prompting changes in national diagnostic guidelines towards LDH-based RDT. Data on hrp2/3 deletions through systematic surveillance studies is limited for other countries in Africa and Asia [6].
Given the relatively quick emergence of such parasite populations and the importance of HRP2-based detection for malaria control, the World Health Organization (WHO) is urging countries to assess the prevalence of hrp2/3 gene deletions causing false-negative tests. Prioritized areas include those with recognized discordance between HRP2-RDT and microscopy, with non-representative or sporadic reports of hrp2/3 deletions in the country, or those that neighbour an area where frequent hrp2/3 deletions have been identified [9]. If the prevalence of deletions known to cause false-negative RDT results among symptomatic individuals is > 5%, switching to non-HRP2-based RDT is recommended.
Vietnam has achieved a remarkable reduction in malaria cases in the past decade. Plasmodium falciparum and Plasmodium vivax remain endemic in Central and Southern provinces, with the former representing 60–70% of reported cases [10]. HRP2-RDT are the main diagnostic tool for P. falciparum at community health facilities, yet baseline information on prevalence of hrp2/3 deletions is very limited. Global genomic surveillance conducted by MalariaGen Pf3K project (Wellcome Sanger Institute) reported a 4.6% prevalence of pfhrp3-deletions but no pfhrp2-deletions among Vietnam samples (n = 216) [11]. The present survey was designed to determine the prevalence of deletions in hrp2/3 genes causing false-negative RDT results among symptomatic P. falciparum cases in Gia Lai and Dak Lak, the two provinces with the highest malaria burden in Vietnam.
Methods
Study design
A cross-sectional multi-site study was conducted on individuals clinically suspected of having P. falciparum malaria seeking medical care at health facilities. The study was designed considering WHO guidelines for surveillance of hrp2/3 deletions published in 2018 [9]. Screening for P. falciparum infection was conducted using routine HRP2-based RDT in accordance with National Malaria Control Programme malaria diagnosis policy, together with light microscopy (LM) as a confirmatory test.
Study site
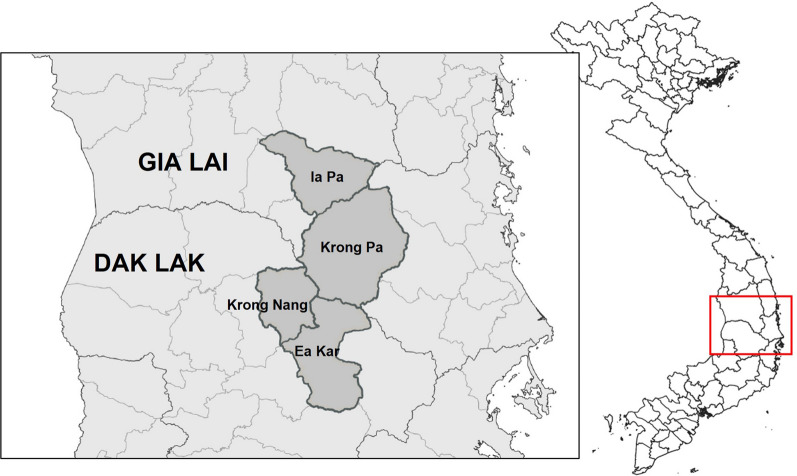
The study was conducted in four malaria endemic districts located along the border of Gia Lai (Krong Pa and Ia Pa districts) and Dak Lak (Ea Kar and Krong Nang districts) provinces, Central Highlands, Vietnam (Fig. 1). A total of 33 community health centres (CHC) were included: 21 in Gia Lai and 12 in Dak Lak. The area is characterized by a tropical climate and hilly geography with partially forested areas. The main occupation is agriculture in forest fields (maize, manioc, rice), which are often located far from villages of residence and require overnight stays in the forest. Malaria transmission mainly occurs in forested areas and is perennial with a peak between September and December and the lowest incidence from February to April. Both provinces have the highest burden of malaria in the country. In years 2018, 2019 and 2020, there were 946, 1804 and 530 malaria cases in Gia Lai and 733, 638 and 124 malaria cases in Dak Lak, respectively (National Institute of Malariology, Parasitology and Entomology, NIMPE). Plasmodium falciparum represented 70% of reported cases.
Fig. 1.
Study area. The study was conducted simultaneously in 33 CHC distributed across Ia Pa and Krong Pa districts (Gia Lai province) and Krong Nang and Ea Kar districts (Dak Lak). This map was developed for the purpose of this article
Sample size
Due to the absence of conclusive preliminary evidence on false-negative RDT prevalence or pfhrp2 deletions in the country, an initial sample size of 370 symptomatic P. falciparum patients per sampling domain was targeted as recommended in WHO guidelines [9]. This sample size would be adequate to demonstrate that a pfhrp2/3 deletion prevalence is below the 5% threshold for an expected population prevalence of 3.2% with 95% confidence. The four districts across the two provinces were considered as a single sampling domain.
Patients and enrolment procedures
Between June 2019 and June 2021 participants were recruited from individuals self-presenting at CHC with symptoms requiring a test for malaria, based on the criteria of the medical staff. Routine testing was conducted using SD Bioline Malaria Pf/Pv Antigen (Abbott) and LM, as further detailed below. Patients with confirmed P. falciparum diagnosis by LM and/or their parents/guardians were informed about the study objectives and invited to participate. Individuals with P. vivax malaria or other non-falciparum species by LM, including mixed infections, and those presenting signs or symptoms of severe malaria were excluded [12]. A clear explanation was provided in Vietnamese language and signed informed consent (or parental/guardian permission for those under 18 years of age and ≥ 12 months old and assent for those between 10 and 17 years of age) was obtained. For all consenting individuals, three dried blood spots (DBSs) containing 50 μl of blood each were obtained from a second finger prick, air-dried for 24 h and stored in silica gel containing bags, before being transferred to NIMPE (Hanoi, Vietnam) for molecular analysis. After sample collection, patients were treated for malaria as per Vietnam’s Ministry of Health (MoH) national guidelines, and asked to complete the short questionnaire on malaria symptoms and treatment, prevention habits and risk behaviour.
Light microscopy
All microscopy evaluations at point-of-care were conducted by expert microscopists (WHO Level 1 or equivalent). Blood slides were stained with Giemsa and examined at a magnification of 1000×. Parasite density was determined by counting the number of asexual parasites per 200 white blood cells (WBCs) with a hand tally counter. If more than 500 parasites were counted before reaching 200 WBCs, the count was stopped after completion of the field. Density was expressed as the number of asexual parasites per µl of blood, calculated by dividing the number of asexual parasites by the number of WBCs and corrected by the estimated WBCs density (typically 8000 per µl). A blood slide was considered negative when examination of 1000 WBCs revealed no asexual parasites. All blood films were stored in boxes and shipped to NIMPE (Hanoi, Vietnam) for double reading by WHO Level 1 microscopists to confirm results from the field and adjust the final diagnosis, if necessary. A third reading was conducted only in case of discrepancy (i.e., disagreement in species identification, in positive vs negative, or in parasite density estimation > 25%).
Quantitative PCR
DNA was extracted at NIMPE from 5 mm punches of DBS using QIAamp DNA 96 Blood kit (Qiagen), following manufacturer’s instructions and a final elution in 200 μl of water. A duplex quantitative real-time polymerase chain reaction (qPCR) targeting 18S ribosomal gene of P. falciparum and P. vivax was conducted to confirm species identified by LM [13]. Briefly, a 7.5 µl mix was prepared using primers and probes from QuantiNova Probe RT-PCR Kit (Qiagen) in a 7500 Real-Time PCR System (Applied Biosystems). Ct value > 40 was considered negative. Samples with no amplification in the Pf–Pv specific qPCR were checked for presence of other Plasmodium species using the generic QMAL qPCR assay [14].
Genotyping of pfhrp2 and pfhrp3
The regions covering exon 2 of both pfhrp2 and pfhrp3 genes were assessed by nested polymerase chain reaction (PCR) amplification using primers from Baker et al. [15] and HotStarTaq Plus Master Mix Kit (Qiagen), under the optimized cycling conditions described in Parr et al. [16]. DNA obtained from DBS of laboratory strains 3D7 (no deletions), Dd2 (pfhrp2-deletion; pfhrp3-wild type), HB3 (pfhrp2-wild type; pfhrp3-deletion) were used as positive controls (kindly provided by Pau Cisteró, ISGlobal, Barcelona). Nested PCR products were visualized in a 2% agarose gel and samples were considered positive if a band was observed in the 600-1000 bp range (in 3D7, expected products are 790 bp for pfhrp2 and 698 bp for pfhrp3). DNA samples testing negative for pfhrp2 and/or pfhrp3 were further analysed by nested PCR of P. falciparum glutamine-rich protein (pfglurp) [17]. Negative result in pfglurp confirmatory PCR was interpreted as low quality/insufficient DNA and invalidated pfhrp2 or pfhrp3 negative result as evidence of deletion. A random selection of ≈ 10% of the samples (n = 40) was shipped for external quality assessment of pfhrp2 and pfglurp nested PCRs at IMPE-Quy Nhon laboratory (Quy Nhon, Binh Dinh Province, Vietnam); results agreement was 100% (40/40) for both assays.
pfhrp2 sequencing
pfhrp2-exon2 PCR products were shipped to Genewiz (Suzhou, China) for DNA purification and bidirectional Sanger sequencing. Analysis was conducted in BioEdit 7.0.5. Quality of chromatograms of both forward and reverse sequences was inspected by eye, forward sequence and reverse complement of reverse sequence were aligned, and a consensus sequence was generated at overlapping regions. Nucleotide sequences were translated to amino acids and a multiple sequence alignment was performed using Clustal Omega in EMBL-EBI server (https://www.ebi.ac.uk/Tools/msa/clustalo/). pfhrp2sequence of the 3D7 strain (PF3D7_0831800) was included as reference.
The number and type of amino acid sequence repeats were identified based on the classification developed by Baker et al. [15]. The number of type 2 and type 7 repeats at each sequence was used to calculate type 2× type 7 score and classify sequences for their predicted sensitivity in RDT detection as type A (score > 100, “very sensitive”), type B (score 50–99; “sensitive”) or type C (< 50; “low or non-sensitive” group) [3, 15].
HRP2 levels by ELISA
The levels of HRP2 protein were measured in all RDT negative/LM positive samples and a random selection of RDT positive samples. Protein elution was conducted from 2 × 5 mm punch of DBS (equivalent to 11.2 μl of blood or 5.6 μl of plasma, [18]), with a final elution in 1120 μl of buffer (1/200) [19]. Quantitative ELISA-based detection was conducted using Quantimal™ CELISA Ultra-sensitive PfHRP2 Malaria (Cellabs, Australia) according to manufacturer’s instructions [20]. Samples were tested in duplicate. Optical density (OD) values were read in a spectrophotometer at 450 nm/620 nm, and cut-off was set at negative control OD + 0.1. HRP2 concentration in plasma was estimated as previously described [21]. HRP2 concentration was calculated by interpolating OD in the standard curve of recombinant HRP2 provided in the kit (0.01–10 ng/ml). Samples with positive OD but below the limit of quantification of final standards were given a value of 0.01 ng/ml. Results were adjusted for specimen pre-dilution and expressed as estimated concentration in plasma.
Definitions and statistical analysis
Prevalence of suspected false-negative HRP2-RDT results among symptomatic patients with P. falciparum malaria was determined from patient case report form data by dividing the number of patients testing RDT-negative and LM-positive by the total number of symptomatic P. falciparum cases. The prevalence of false-negative HRP2-RDT caused by hrp2/3 deletions was determined as the number of pfhrp2 or pfhrp3 deletions divided by the total number of symptomatic P. falciparum cases. Differences between point estimates across sociodemographic and parasitological variables were tested using chi-square or Fisher’s exact test. Mann–Whitney/Kruskal Wallis test was used to compare parasite densities between population groups.
Results
Baseline characteristics of study participants
A total of 9456 patients were screened at CHCs during the study period (5366 [56.7%] in Dak Lak and 4090 [43.3%] in Gia Lai). Microscopy screening identified 370 patients with P. falciparum infections (3.9%) who were invited to participate. RDTs from SD Bioline Malaria Antigen Pf/Pv (Abbott) were positive for 353 patients, resulting in a suspected false-negative RDT prevalence among symptomatic cases of 4.6% (17/370). Molecular characterization of Plasmodium infection was assessed in 270 out of the 370 DBS samples collected. Of note, there was no selection bias based on RDT result, as samples were processed as they were collected and RDT results were not available to the laboratory until initial qPCR screening was completed. According to WHO guidelines for the surveillance of hrp2/3 deletions, 270 cases should be sufficient to ensure the 95% confidence interval does not include the 5% pfhrp2-deletion warning threshold for an estimated pfhrp2-deletion population prevalence of 3.2% [9].
The baseline characteristics of 270 participants are provided in Table 1. Most individuals were adult males dedicated to farming and slash-and-burn agriculture as their main occupation. By district, Krong Pa accounted for 83.0% of the cases (224/270), followed by Krong Nang (24/270, 8.9%), Ea Kar (19/270, 7%) and Ia Pa (3/270, 1.1%). Headache and fever were the most common symptoms (264/265, 99.6%), followed by fatigue (245/265, 92.5%). Median parasite density was 6440 asexual parasites/μl (range 80–246,800; n = 264). Prevalence of false-negative RDT was 5.6% (15/270); 14 were from Krong Pa district and one from Ia Pa district.
Table 1.
Baseline characteristics of participants (N = 270)
| Variable | Gia Lai | Dak Lak | Overall |
|---|---|---|---|
| P. falciparum cases by LM | 227 | 43 | 270 |
| Number of CHC | 15 | 12 | 27 |
| Median age (range) | 27 (7–57) | 33 (21–58) | 28 (7–58) |
| Sex | |||
| Male | 222 (97.8%) | 43 (100%) | 265 (98.1%) |
| Female | 5 (2.2%) | 0 (0) | 5 (1.9%) |
| Occupationa | |||
| Farmer | 94 (42.0%) | 20 (48.8%) | 114 (43.0%) |
| Slash-and-burn cultivation | 110 (49.1%) | 13 (31.7%) | 123 (46.4%) |
| Other | 20 (8.9%) | 8 (19.5%) | 28 (10.6%) |
| Travel history in last 2 months | |||
| Visited forest fieldsa | 189 (84.4%) | 41 (100%) | 230 (86.8%) |
| Travelled outside village of residencea | 25 (11.2%) | 16 (39.0%) | 41 (15.5%) |
| Use of bednets | |||
| At homea | 199 (88.8%) | 37 (90.2%) | 236 (89.1%) |
| In forest fieldsa | 171 (76.3%) | 28 (68.3%) | 199 (75.1%) |
| Fevera | 223 (99.6%) | 41 (100%) | 264 (99.6%) |
| RDT result | |||
| P. falciparum | 212 (93.4%) | 43 (100%) | 255 (94.4%) |
| P. vivax or mixed | 0 (0) | 0 (0) | 0 (0) |
| False-negative for P. falciparum | 15 (6.6%) | 0 (0) | 15 (5.6%) |
an = 265 (Gia Lai, n = 224; Dak Lak, n = 41)
Molecular characterization of suspected false-negative RDT results
Duplex Pf-Pv qPCR was applied to all 270 samples. The screening confirmed 260 P. falciparum mono-infections, whereas 7 samples were P. falciparum-P. vivax co-infections (Table 2). Three samples were negative for both species and tested negative in the generic Plasmodium qPCR assay. Infections with false-negative RDT results were all P. falciparum mono-infections and individual details are provided in Additional file 1: Table S1. Parasite density by LM was lower in false-negative RDT result samples (2460 parasites/μl [interquartile range, IQR 1200–4800]; n = 14) as compared to RDT-positive cases (7160 parasites/μl [IQR 2280–20900]; n = 248; p = 0.005, Kruskal–Wallis). Similar results were found in qPCR, with higher Ct (i.e., lower parasite DNA concentration) in false-negative RDT cases as compared to RDT-positive cases (p = 0.028, Kruskal–Wallis; Table 2).
Table 2.
Molecular characterization of P. falciparum cases, stratified by RDT result
| Pf positive RDT result | False-negative RDT result | All | |
|---|---|---|---|
| DBS processed | 255 | 15 | 270 |
| Species qPCR | |||
| P. falciparum | 245 (96.1%) | 15 (100%) | 260 (96.3%) |
| Pf–Pv mixed infections | 7 (2.7%) | 0 (0) | 7 (2.6%) |
| Negative | 3 (1.2%) | 0 (0) | 3 (1.1%) |
| Ct (Pf), median [IQR] | 28.1 [26.6–29.9] | 30.0 [I27.5–31.4] | 28.2 [26.7–30.0] |
| hrp2/3 PCR positivity | |||
| pfhrp2 exon2 | 252/252 (100%) | 14/15 (93.3%) | 266/267 (99.6%) |
| pfhrp3 exon2 | 252/252 (100%) | 14/15 (93.3%) | 266/267 (99.6%) |
| HRP2 plasma ng/mL, median [IQR] | 54.5 [15.2–226.0]a | 11.8 [2–14.9] | 26.0 [11.9–209.7] |
an = 44
Fourteen out of the 15 suspected false-negative RDT results were successfully amplified at both pfhrp2 and pfhrp3 exon2 (Table 2 and Additional file 1: Table S1). The one sample which had no visible bands in both pfhrp2 and pfhrp3 was also negative when tested for pfglurp PCR amplification, suggestive of insufficient sensitivity of nested PCR methods (qPCR Ct of the sample = 39). Overall, the prevalence of false-negative HRP2-RDT caused by confirmed hrp2/3 deletions among symptomatic P. falciparum cases was 0% (0/266).
HRP2 expression was assessed in all false-negative RDT and in a subset of 44 positive RDT samples. All samples had OD values above the negative control cut-off, but levels were significantly lower in the false-negative RDT group (p < 0.001; Kruskal–Wallis; Table 2 and Additional file 1: Fig. S1). There was a positive correlation between HRP2 levels and parasite density by LM (Pearson’s coefficient: 0.450, p < 0.001; Additional file 1: Fig. S2A) or qPCR Ct values (Pearson’s coefficient: − 0.657, p < 0.001; Additional file 1: Fig. S2B).
HRP2 sequence variants
Genotyping of pfhrp2 and pfhrp3-exon2 was also conducted for the remaining P. falciparum RDT-positive samples (n = 255). Bands were observed for both genes in all the remaining cases. Sequencing of exon2 was successful in 235/266 pfhrp2 PCR products (Additional file 2: Data S1).
Three sequence variants were identified in the study population: VN1 (207/235, 88.1%), VN2 (24/235, 10.2%) and VN3 (4/235, 1.7%) (Table 3 and Additional file 1: Fig. S3). The type2 × type7 score and category is shown in Table 3. VN1 was classified as ‘C’ (predicted “low/no RDT sensitivity”), and VN2 and VN3 were classified as ‘B’ (predicted “sensitive”). The distribution of variants did not differ significantly by province (p = 0.465, Fisher’s exact). VN1 was found in all four study districts, VN2 in all except Ia Pa, and VN3 only in Krong Pa district. All false-negative RDT results with sequence data available (n = 13) corresponded to the predominant variant VN1 (Additional file 1: Table S1).
Table 3.
Amino acid repeats in HRP2 sequence variants identified in Vietnam
| Repeat id | Sequence | Sequence variant | ||
|---|---|---|---|---|
| VN1 (n = 207) | VN2 (n = 24) | VN3 (n = 4) | ||
| 1 | AHHAHHVAD | 1 | 2 | 3 |
| 2 | AHHAHHAAD | 15 | 12 | 11 |
| 3 | AHHAHHAAY | 1a | 2 | 1 |
| 4 | AHH | 0 | 0 | 1 |
| 5 | AHHAHHASD | 1 | 1 | 2 |
| 6 | AHHATD | 3 | 4 | 3 |
| 7 | AHHAAD | 2 | 6 | 5 |
| 8 | AHHAAY | 2 | 1 | 1 |
| 10 | AHHAAAHHATD | 1 | 2 | 2 |
| 12 | AHHAAAHHEAATH | 1 | 1 | 1 |
| type2 × type7 score | 30 | 72 | 55 | |
| type2 × type7 category | C | B | B | |
aFound in one sample only
Discussion
This survey was used to determine the contribution of hrp2/3 deletions to false-negative RDT results in Gia Lai and Dak Lak provinces between 2019 and 2021. Despite ≈ 5% of symptomatic cases confirmed by LM being negative by HRP2-RDT, there was no evidence of gene deletions in the surveyed population.
The rapid emergence of hrp2/3-deleted parasite populations in countries like Eritrea or Djibouti in recent years has raised alarms about the threat posed by parasite diagnostic escape to malaria control. In general, many malaria surveillance and/or clinical studies use HRP2-based RDT as a screening tool to identify malaria cases in human populations. This leads to an intrinsic selection bias of studied parasite population that has probably made difficult incidental and/or early detection of hrp2-deleted clones. In 2018, the WHO launched a reference protocol for the surveillance of hrp2/3-deletions to ensure elements were included in survey design [9]. All of the following were taken into account in the present survey: sampling in ≈ 10 health facilities across the sampling domain, quality-assured microscopy as a confirmatory test, confirmation of species and parasite density by qPCR, genotyping of both pfhrp2 and pfhrp3, amplification of a confirmatory single-copy gene in the event of hrp2/3 deletions and measurement of HRP2 concentration by serological analysis [6, 9]. In terms of sample size, molecular data was generated for 270 out of 370 participants initially recruited. The study was heavily affected in 2020 by both a marked reduction in malaria cases and the lockdowns caused by the SARS-CoV-2 pandemic in Vietnam, which made difficult the sample shipments and population movement to/from study sites. However, the final sample size had enough power for population prevalence of deletions up to 3.2%.
Molecular characterization of false-negative RDT results indicated that they were not caused by deletions in exon2 of pfhrp2. Although deletions can also occur around exon1 (exon1/2 region) [9], studies that genotyped this region show that almost all parasites lacking exon1/2 also lack exon2 [5, 22–24]. Moreover, previous molecular assays targeting exon1/2 can yield to spurious amplification of the paralogous gene [16]. The lower parasite densities found in false-negative RDT cases (further confirmed by lower HRP2 plasma levels) suggest that these infections could be below the test’s limit of detection (LOD). This hypothesis is not supported by LM parasitaemia—which was > 400 parasites/µl in all cases—but potential overestimation of parasitaemia by the LM-WBC count method cannot be excluded [25]. Interestingly, all false-negative RDT were caused by an HRP2 variant classified as category C—“low sensitivity”, according to predicted targeting of HRP2 epitope by RDT monoclonal antibodies [3, 15]. This variant was the most common overall and also present in most RDT positive samples, suggesting RDT escape properties are unlikely to explain negative test result. HRP2 sequence diversity was low with only three exon2 variants, probably due to focal transmission of a limited number of clones. Although this study targeted a large number of CHC within each district—to aim for maximum representativity of parasite population—this result highlights the risk of oversampling clonal populations when conducting hrp2/3 surveys in very low transmission/pre-elimination areas using the same study design as for high transmission areas.
The survey has other limitations. First, it was not possible to assess the contribution of causes other than gene deletions to false-negative RDT, such as adherence to manufacturer’s instructions, transportation conditions, or storage conditions. User interpretation errors were ruled out, as CHC staff took pictures from all RDTs to be double checked by the study team. Second, the detection of parasites with hrp2/3 gene deletions in polyclonal infections (i.e. infections with both hrp2/3-deleted and hrp2/3-wild type clones) may have been missed in the nested PCR approach.
In conclusion, there was no evidence of P. falciparum parasites carrying pfhrp2 or pfhrp3 gene deletions among symptomatic cases in Gia Lai and Dak Lak provinces, suggesting HRP2-RDTs remain effective for malaria case management. Given that parasites populations with hrp2/3-deletions have emerged rapidly in other countries [7], new surveys in these and other malaria-endemic provinces of Vietnam will be required in the future.
Supplementary Information
Additional file 1: Table S1. Parasitological characteristics of samples with false-negative RDT result (n = 15). Figure S1. HRP2 levels in plasma, by RDT result. Figure S2. Correlation between HRP2 levels in plasma and parasite density. Figure S3. HRP2 sequence variants in P. falciparum infections from Gia Lai and Dak Lak provinces, Vietnam.
Additional file 2: Data S1. HRP2 exon 2 sequences.
Acknowledgements
The authors would like to thank all the participants who volunteered their time to be in the study. Additionally, the authors are very grateful to the Centre for Disease Control and Prevention and the Health Services of Gia Lai and Dak Lak provinces.
Disclaimer
The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, the Department of Defense or the U.S. government and also those of the Australian Defence Force, Joint Health Command or any extant Australian Defence Force policy. For KAE (LCDR, MC, USN) and NJM (CDR, MC, USN): I am a military Service member. This work was prepared as part of my official duties. Title 17, U.S.C., §105 provides that copyright protection under this title is not available for any work of the U.S. Government. Title 17, U.S.C., §101 defines a U.S. Government work as a work prepared by a military Service member or employee of the U.S. Government as part of that person’s official duties.
Abbreviations
- CHC
Community health centre
- CI
Confidence interval
- ELISA
Enzyme-linked immunosorbent assay
- glurp
Glutamine-rich protein
- HRP
Histidine-rich protein
- IQR
Interquartile range
- LOD
Limit of detection
- NIMPE
National Institute of Malariology, Parasitology and Entomology
- OD
Optical density
- PCR
Polymerase chain reaction
- qPCR
Quantitative real-time PCR
- RDT
Rapid diagnostic test
- WBC
White blood cells
- WHO
World Health Organization
Author contributions
NDT conceived the study, obtained ethics approval and performed general oversight of the study. ERV developed the study protocols for field and laboratory work, analysed molecular data, performed statistical analysis, and wrote the main manuscript. NTHB performed the laboratory work and analysed molecular data. DVD coordinated field work, performed microscopy readings, and managed the study database. NTHN performed the laboratory work and analysed molecular data. TKL managed and curated study database and performed statistical analysis. TTD performed general oversight of the study. NJM conceived the study and obtained scientific and ethical approval. KAE conceived the study, obtained scientific and ethical approval, and contributed to draft the manuscript. All authors reviewed the final manuscript. All authors read and approved the final manuscript.
Funding
This work was funded by the US Defense Health Agency Research and Development Program (work unit number D1430).
Availability of data and materials
The authors confirm that the data supporting the findings of this study are available within the article and/or its supplementary materials. Any additional data that does not compromise the privacy of research participants is available from the corresponding author, ERV, upon reasonable request.
Declarations
Ethics approval and consent to participate.
Ethics approval was obtained from the Vietnam National Institute of Malaria, Parasitology and Entomology Institutional Review Board (Ministry of Health, Vietnam). Extra-mural review was conducted in accordance with U.S. Department of the Navy Human Research Protection Program guidelines in compliance with all applicable federal regulations governing the protection of human subjects. Written informed consent was obtained from participants and parents or guardians of minor participants, prior to enrolment. Children aged 10 to 17 years of age provided assent to participate in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Howard RJ, Uni S, Aikawa M, Aley SB, Leech JH, Lew AM, et al. Secretion of a malarial histidine-rich protein (Pf HRP II) from Plasmodium falciparum-infected erythrocytes. J Cell Biol. 1986;103:1269–1277. doi: 10.1083/jcb.103.4.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.UNITAID-World Health Organization. Malaria diagnostics landscape update. Geneva: World Health Organization; 2015. http://unitaid.org/assets/Malaria_Diagnostics_Landscape_Update_Fe_2015.pdf.
- 3.Baker J, Ho MF, Pelecanos A, Gatton M, Chen N, Abdullah S, et al. Global sequence variation in the histidine-rich proteins 2 and 3 of Plasmodium falciparum: implications for the performance of malaria rapid diagnostic tests. Malar J. 2010;9:129. doi: 10.1186/1475-2875-9-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gatton ML, Chaudhry A, Glenn J, Wilson S, Ah Y, Kong A, et al. Impact of Plasmodium falciparum gene deletions on malaria rapid diagnostic test performance. Malar J. 2020;19:392. doi: 10.1186/s12936-020-03460-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gamboa D, Ho M-F, Bendezu J, Torres K, Chiodini PL, Barnwell JW, et al. A large proportion of P. falciparum isolates in the Amazon Region of Peru Lack pfhrp2 and pfhrp3: implications for malaria rapid diagnostic tests. PLoS ONE. 2010;5:e8091. doi: 10.1371/journal.pone.0008091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomson R, Parr JB, Cheng Q, Chenet S, Perkins M, Cunningham J. Prevalence of Plasmodium falciparum lacking histidine-rich proteins 2 and 3: a systematic review. Bull World Health Organ. 2020;98:558–658. doi: 10.2471/BLT.20.250621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berhane A, Anderson K, Mihreteab S, Gresty K, Rogier E, Mohamed S, et al. Major threat to malaria control programs by Plasmodium falciparum lacking histidine-rich protein 2, Eritrea. Emerg Infect Dis. 2018;24:462–470. doi: 10.3201/eid2403.171723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iriart X, Menard S, Chauvin P, Mohamed HS, Charpentier E, Mohamed MA, et al. Misdiagnosis of imported falciparum malaria from African areas due to an increased prevalence of pfhrp2/pfhrp3 gene deletion: the Djibouti case. Emerg Microbes Infect. 2020;9:1984–1987. doi: 10.1080/22221751.2020.1815590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.WHO. Protocol for estimating the prevalence of pfhrp2/pfhrp3 gene deletions among symptomatic falciparum patients with false-negative RDT results. Geneva: World Health Organization; 2018. https://www.who.int/docs/default-source/malaria/mpac-documentation/mpac-oct2017-hrp2-deletion-protocol-session4.pdf?sfvrsn=2c9dfaf4_2.
- 10.Goldlust SM, Thuan PD, Giang DDH, Thang ND, Thwaites GE, Farrar J, et al. The decline of malaria in Vietnam, 1991–2014. Malar J. 2018;17:226. doi: 10.1186/s12936-018-2372-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunningham J. Update on Plasmodium falciparum hrp2/3 gene deletions. Malaria Policy Advisory Committee Meeting, World Health Organization; 2017. https://www.who.int/malaria/mpac/mpac-mar2017-hrp2-3-deletions-session7-presentation.pdf.
- 12.WHO . Management of severe malaria: a practical handbook. 3. Geneva: World Health Organization; 2012. [Google Scholar]
- 13.Kamau E, Alemayehu S, Feghali KC, Saunders D, Ockenhouse CF. Multiplex qPCR for detection and absolute quantification of malaria. PLoS ONE. 2013;8:e71539. doi: 10.1371/journal.pone.0071539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wampfler R, Mwingira F, Javati S, Robinson L, Betuela I, Siba P, et al. Strategies for detection of Plasmodium species gametocytes. PLoS ONE. 2013;8:e76316. doi: 10.1371/journal.pone.0076316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker J, McCarthy J, Gatton M, Kyle DE, Belizario V, Luchavez J, et al. Genetic diversity of Plasmodium falciparum histidine-rich protein 2 (PfHRP2) and its effect on the performance of PfHRP2-based rapid diagnostic tests. J Infect Dis. 2005;192:870–877. doi: 10.1086/432010. [DOI] [PubMed] [Google Scholar]
- 16.Parr JB, Anderson O, Juliano JJ, Meshnick SR. Streamlined, PCR-based testing for pfhrp2- and pfhrp3-negative Plasmodium falciparum. Malar J. 2018;17:137. doi: 10.1186/s12936-018-2287-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snounou G. Genotyping of Plasmodium spp. nested PCR. Methods Mol Med. 2002;72:103–116. doi: 10.1385/1-59259-271-6:103. [DOI] [PubMed] [Google Scholar]
- 18.Corran PH, Cook J, Lynch C, Leendertse H, Manjurano A, Griffin J, et al. Dried blood spots as a source of anti-malarial antibodies for epidemiological studies. Malar J. 2008;7:195. doi: 10.1186/1475-2875-7-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.San NN, Kien NX, Manh ND, Van Thanh N, Chavchich M, Binh NTH, et al. Cross-sectional study of asymptomatic malaria and seroepidemiological surveillance of seven districts in Gia Lai province, Vietnam. Malar J. 2022;21:40. doi: 10.1186/s12936-022-04060-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mihreteab S, Anderson K, Pasay C, Smith D, Gatton ML, Cunningham J, et al. Epidemiology of mutant Plasmodium falciparum parasites lacking histidine-rich protein 2/3 genes in Eritrea 2 years after switching from HRP2-based RDTs. Sci Rep. 2021;11:21082. doi: 10.1038/s41598-021-00714-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.WHO. Methods manual for laboratory quality control testing of malaria RDTs. Geneva: World Health Organization; 2020. https://www.who.int/publications/m/item/methods-manual-for-laboratory-quality-control-testing-of-malaria-rdts.
- 22.Bharti PK, Chandel HS, Ahmad A, Krishna S, Udhayakumar V, Singh N. Prevalence of pfhrp2 and/or pfhrp3 gene deletion in Plasmodium falciparum population in eight highly endemic States in India. PLoS ONE. 2016;11:e0157949. doi: 10.1371/journal.pone.0157949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta H, Matambisso G, Galatas B, Cisteró P, Nhamussua L, Simone W, et al. Molecular surveillance of pfhrp2 and pfhrp3 deletions in Plasmodium falciparum isolates from Mozambique. Malar J. 2017;16:416. doi: 10.1186/s12936-017-2061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar N, Pande V, Bhatt RM, Shah NK, Mishra N, Srivastava B, et al. Genetic deletion of HRP2 and HRP3 in Indian Plasmodium falciparum population and false negative malaria rapid diagnostic test. Acta Trop. 2013;125:119–121. doi: 10.1016/j.actatropica.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 25.Alves ER, Gomes LT, Ribatski-Silva D, Mendes CRJ, Leal-Santos FA, Simões LR, et al. Assumed white blood cell count of 8,000 cells/μL overestimates malaria parasite density in the Brazilian Amazon. PLoS ONE. 2014;9:e94193. doi: 10.1371/journal.pone.0094193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Parasitological characteristics of samples with false-negative RDT result (n = 15). Figure S1. HRP2 levels in plasma, by RDT result. Figure S2. Correlation between HRP2 levels in plasma and parasite density. Figure S3. HRP2 sequence variants in P. falciparum infections from Gia Lai and Dak Lak provinces, Vietnam.
Additional file 2: Data S1. HRP2 exon 2 sequences.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and/or its supplementary materials. Any additional data that does not compromise the privacy of research participants is available from the corresponding author, ERV, upon reasonable request.