Abstract
Eight patients from five families with undiagnosed dominant distal myopathy underwent clinical, neurophysiological and muscle biopsy examinations. Molecular genetic studies were performed using targeted sequencing of all known myopathy genes followed by segregation of the identified mutations in the affected families using Sanger sequencing. Two novel mutations in DNAJB6 J domain, c.149C>T (p.A50V) and c.161A>C (p.E54A), were identified as the cause of disease. The muscle involvement with p.A50V was distal calf-predominant, and the p.E54A was more proximo-distal. Histological findings were similar to those previously reported in DNAJB6 myopathy. In line with reported pathogenic mutations in the glycine/phenylalanine (G/F) domain of DNAJB6, both the novel mutations showed reduced anti-aggregation capacity by filter trap assay and TDP-43 disaggregation assays. Modeling of the protein showed close proximity of the mutated residues with the G/F domain. Myopathy-causing mutations in DNAJB6 are not only located in the G/F domain, but also in the J domain. The identified mutations in the J domain cause dominant distal and proximo-distal myopathy, confirming that mutations in DNAJB6 should be considered in distal myopathy cases.
Keywords: DNAJB6 gene, Limb-girdle muscular dystrophy, Chaperonopathy
1. Introduction
Mutations in DNAJB6 were identified in 2012 to cause autosomal dominant limb-girdle muscular dystrophy type 1D (LGMD1D, OMIM 603511; current proposed nomenclature LGMD D1 DNAJB6-related) [1,2]. Later, several other DNAJB6 mutations have been identified in Asian, European and North American populations [3-11]. Interestingly, all reported mutations have affected the glycine/phenylalaninerich (G/F) domain of DNAJB6. LGMD1D was originally described as late-onset, slowly progressive disease, most patients remained ambulant even at high age. However, some mutations cause an earlier, more severe or distal-onset disease [2,5-8,12,13].
Muscle pathology in LGMD1D includes protein accumulations followed by autophagic rimmed vacuoles. The protein accumulations commonly contain proteins of the sarcomeric Z-disk, whereas the rimmed vacuoles are positive for LC3, SQSTM1 and TARDBP [2,5,14].
DNAJB6 is a member of the HSP40 (J-protein) family of co-chaperones. These co-chaperones interact with the HSPA (Hsp70) chaperones through their J domains, delivering client proteins to HSPAs and stimulating their ATPase activity [15]. DNAJB6 has been shown to bind and stimulate HSPA8 (Hsc70) [16,17], but it can also counteract protein aggregation in an HSPA-independent manner [18]. The protein accumulations in DNAJB6-mutated muscle indicate a reduced ability to handle normally occurring misfolded proteins. Indeed, all reported disease-causing DNAJB6 mutations have shown reduced anti-aggregation capacity in vitro [1,5,9,19].
By using targeted sequencing of muscle disease genes [20,21] we studied families with distal myopathies. Here we present evidence of two novel mutations in the J domain of DNAJB6 in five separate families as the cause of dominant adult-onset distal and proximo-distal myopathy.
2. Patients and methods
2.1. Study subjects and clinical examinations
Five families with eight affected patients with unknown distal myopathy were investigated. The families were from Spain (Family 1), Norway (Family 2), Italy (Family 3), Finland (Family 4) and Japan (Family 5). There were affected family members in two or three generations (except for Family 2) showing autosomal dominant inheritance pattern (Fig. 1). The patients had been clinically examined, and medical and family histories reviewed by neurologists. Electrophysiological examinations consisting of nerve conduction studies and needle electromyogram (EMG), creatine kinase (CK) measurements, and in a subset of patients pulmonary or cardiac evaluations, were performed. Muscle imaging was available from seven and muscle biopsy from six patients.
Fig. 1.
Pedigrees of the five families. DNAJB6 variants segregated with the disease in families 1–5. Individuals for whom DNA was available are marked with *. The genetically tested family members are indicated as negative (−/−) or heterozygous (+/−) for the DNAJB6 variants c.149C>T (p.A50V, in F1–4) or c.161A>C (p.E54A, in F5).
In Family 2 the proband was the only affected member; her parents were deceased and not available for the study. Her two daughters and one granddaughter were analysed as part of the segregation studies. The daughter (F2 III:1) and granddaughter (IV:1) were diagnosed with chronic fatigue syndrome. No muscle weakness or atrophy was observed on their clinical examination.
2.2. Standard protocol approvals, registrations and patient consents
The study was approved by the IRB of Tampere University Hospital. All participants provided appropriate consent and the study was conducted according to the Helsinki Declaration.
2.3. Genetics
Genomic DNA was isolated from peripheral blood lymphocytes using standard techniques. DNA samples from the probands of Families 1–4 were sequenced using the targeted high-throughput sequencing panel MYOcap as previously described [20]. The DNA sample from the proband of Family 5 was sequenced using the targeted high-throughput sequencing panel “myopathy with protein aggregations/rimmed vacuoles” as previously described [21]. The identified DNAJB6 variants and the segregation in available family members were verified by Sanger sequencing. Primer sequences are available on request. Sequence variants in DNAJB6 are described according to the coding DNA reference sequence (NG_032573.1), covering DNAJB6 transcript variant-1 (NM_058246.3). The minor allele frequency (MAF) reported refers to the frequency in gnomAD database (http://gnomad.broadinstitute.org)
2.4. Muscle pathology and antibodies
Snap-frozen patient muscle biopsies were cryo-sectioned and stained for endogenous proteins using antibodies against DNAJB6 (2C11-C1, Novus Biologicals; ab96539, Abcam); TIA1 (ab61700, Abcam); TARDBP (TDP-43) (WH0023435M1-1, Sigma-Aldrich); SQSTM1 (p62) (P0067, MilliporeSigma), and myotilin (10731-1-A, Proteintech). Histological stainings and electron microscopy were performed using standard methods.
2.5. Structure modeling
The 3D-structure of wild-type DNAJB6b was modelled using I-TASSER [22-24] and visualized using SwissPdbViewer v4.1.0 [25] and POV-Ray v3.7.
2.6. Protein sequence alignment
Protein sequences of vertebrate DNAJB6 orthologs were retrieved from UniProt (https://www.uniprot.org). Multiple sequence alignment of the N-terminal parts of the proteins, covering the J and G/F domains, was generated with Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/).
2.7. Plasmid constructs
The inducible DNAJB6a wild-type, DNAJB6b wild-type, and DNAJB6b F93L constructs in pCDNA5/TO, and 120Q-HTT in pEGFP-N1 have been described previously [1,26]. The c.149C>T (p.A50V), c.161A>C (p.E54A), c.170C>T (p.S57L), and c.93T>A (p.H31Q) mutations and double mutations were introduced by site-directed mutagenesis. To create GFP-DNAJB6b constructs, inserts encoding wild-type and variant DNAJB6b were ligated into the pcDNA3.1 vector containing a GFP tag. To create mCherry-TDP-43, full length TDP-43 cDNA was cloned into the pCherry-C1 vector. All constructs were verified by Sanger sequencing.
2.8. Filter-trap assay
The ability of DNAJB6 to prevent aggregation of polyQ-huntingtin was tested using filter-trap assay as described earlier [1,5,9] with minor modifications. In brief, T-REx 293 cells (Invitrogen) were grown at 37 °C, 5% CO2, in DMEM supplemented with 10% fetal bovine serum, GlutaMax, penicillin/streptomycin, and blasticidin S. The cells were transfected with GFP-tagged 120Q-HTT and inducible V5-tagged DNAJB6-constructs, expression of DNAJB6 induced after 5 h (or 4 h in experiments involving p.H31Q constructs) and the cells harvested 48 h post transfection. The soluble fraction of 120Q-HTT was measured using western blotting and the aggregates trapped in a 0.2 μm cellulose acetate filter (Whatman GmbH). The membranes and filters were stained for GFP (A-11122, Invitrogen) and V5 (R960-25, Invitrogen), scanned on an Odyssey scanner (LI-COR) and quantified using ImageStudio v. 3.1.4 (LI-COR) or ImageJ. The aggregation score was calculated from the intensities of aggregated and soluble GFP-HTT as (aggregated/soluble)induced/(aggregated/soluble)uninduced and statistical significance calculated using a two-tailed Mann–Whitney U test without multiple testing correction.
2.9. TDP-43 aggregation assay
The TDP-43 aggregation assay has been described previously [19,27]. In brief, U2OS cells were transfected with wild-type or variant GFP-DNAJB6b constructs together with mCherry-TDP-43. After 24 h, the cells were heat-shocked at 42 °C for 1 h prior to fixing. The proportion of cells containing mCherry-TDP-43 aggregates was determined by fluorescence microscopy. For each condition at least 300 cells were counted by a blinded assessor and the experiment repeated three times. Statistical significance was calculated using a two-tailed t-test without multiple testing correction.
2.10. Data availability statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
3. Results
3.1. Clinical features
The probands of Families 1–4 had distal lower leg weakness and findings compatible with distal myopathy. Two patients (F3 I:2 and III:2) carrying the DNAJB6 mutation were subjectively asymptomatic. However, objectively F3 III:2 had mild but clear typical findings on muscle MRI and slightly limping gait. F3 I:2 was not available for investigation. The mean age of onset was 48 years. The patients had difficulties in walking and especially walking on toes as a first sign. The symptoms were slowly progressive. There was muscle atrophy in the feet and calf muscles, and later in the hands with mild upper limb weakness. The probands of Family 1 (II:1) and 3 (II:1) also had proximal lower limb weakness resulting in difficulties in climbing stairs or rising from chair. One patient had mild dysphagia, others had no bulbar symptoms or facial muscle weakness.
The proband of Family 5 had more pronounced lower limb weakness causing difficulties in walking, climbing stairs and tendency to fall. On examination he had both proximal and distal lower limb weakness. He had also Achilles tendon contractures and decreased tendon reflexes. His mother had mild proximal and distal muscle weakness and dysarthria but was not available for further examination.
CK levels were normal or slightly elevated in all patients and EMG showed myopathic changes especially in the distal limb muscles. Clinical details of the patients are presented in Table 1 and the pedigrees in Fig. 1.
Table 1.
Clinical data for the patients.
| Patient | Sex/ Age |
Age at onset |
First symptoms |
Muscle weakness | CK level | EMG | Muscle biopsy | Muscle imaging | Other |
|---|---|---|---|---|---|---|---|---|---|
|
F1 II:1 |
M/75 | 69 | Distal weakness | Distal LL (anterior and posterior compartment), mild in proximal LL, very mild in UL | Normal | Myopathy | RV, myofibrillar aggregates | Severe: Gmed, Glat, S, PL (left), hamstrings Moderate: vastii | COPD, no cardiac symptoms |
| III:2 | F/50 | n.a. | Distal weakness | No detectable weakness | 206 | Myopathy | Size variation, internal nuclei | Moderate: Gmed, Glat | |
|
F2 II:1 |
F/65 | 40 | Distal lower leg weakness | Distal LL, distal UL with muscle atrophy | Normal to slightly elevated | Myopathy | Atrophic fibers | Severe: Gmed, Glat, S | |
|
F3 II:1 |
M/70 | 40 | Waddling gait | Distal and proximal LL, distal UL, mild dysphagia; ambulant 500 m with/without cane | 655 | n.a. | RV, myofibrillar aggregates | Severe: Gmed, Glat, S, PL, hamstrings | VC 97% Echo hypertrophy (hypertension) |
| III:2 | M/39 | n.a. | n.a. | No clear weakness but limping gait | 239 | n.a. | n.a. | Moderate: Gmed (right>>left) | |
|
F4 II:1 |
M/52 | 47 | Difficulty walking on toes | Distal LL with muscle atrophy in EDB and calf muscles | Normal | Mild myopathy | RV | Severe: Gmed, Glat, S Moderate: hamstrings | FEV1 78%, FVC 64%, no dyspnea Echo normal |
|
F5 II:2 |
F/81* | n.a. | Lower limb muscle weakness | Mild distal and proximal, dysarthria | n.a. | n.a. | n.a. | n.a. | |
| III:2 | M/51 | 20 | Difficulty walking | Distal and proximal with diffuse muscle atrophy, Achilles tendon contractures | 213 | Small motor units, fibrillation, fasciculations | RV | Diffuse in thigh and leg muscles (prox>dist) | ECG normal, no cardiac symptoms |
COPD, chronic obstructive pulmonary disease; ECG, electrocardiography; EDB, extensor digitorum brevis; FEV1, forced expiratory volume in one second; FVC, forced vital capacity; Glat, gastrocnemius lateralis; Gmed, gastrocnemius medialis; n.a., not available; LL, lower limbs; PL, peroneus longus; RV, rimmed vacuoles; S, soleus; UL, upper limbs; VC, vital capacity.
Age at death.
3.2. Muscle imaging
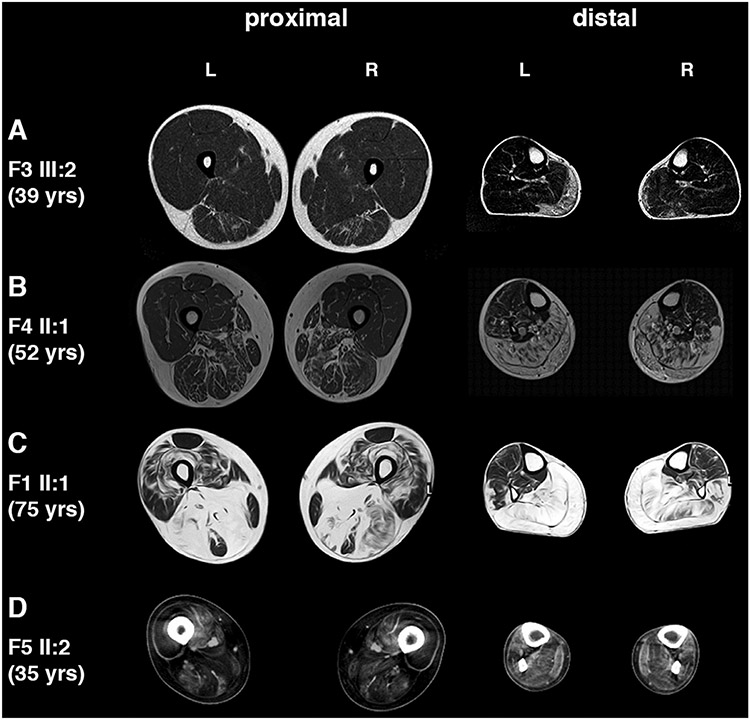
Muscle imaging showed consistent involvement of calf muscles in all examined patients in Families 1–4. Younger patients with the mildest symptoms had fatty degenerative changes in gastrocnemius medialis and no other or only very mild involvement of soleus and semitendinosus muscles. The most severe changes were observed in F1 II:1 (Fig. 2) and F3 II:1 (not shown) who had degenerative fatty replacement in almost all posterior thigh and calf muscles.
Fig. 2.
Muscle imaging. (A) Patient F3 III:2 at age 39. Moderate fatty degenerate changes are seen in gastrocnemius medialis muscle more on the right and mild in semitendinosus muscle. (B) Patient F4 II:1 at age 52. Severe involvement in all calf muscles and milder in hamstrings. (C) Patient F1 II:1 at age 75. Severe changes are seen in all calf muscles symmetrically and peroneus longus on the left. Hamstring muscles are also severely affected with less involvement in vastii muscles. Rectus femoris is best spared muscle. (D) Patient F5 II:2 at age 35. Severe involvement in anterior and posterior thigh muscles and diffuse fatty degenerative changes in all lower leg muscles, most prominent in peroneus longus and tibialis posterior.
Muscle CT of the proband in Family 5 revealed fatty degenerative changes in almost all thigh muscles. Lower leg muscles showed diffuse involvement, most prominent in peroneus longus and tibialis posterior (Fig. 2).
3.3. Muscle histology
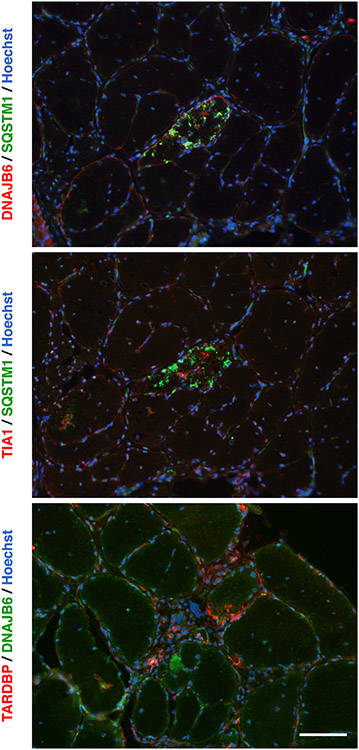
Muscle biopsies from the patients generally showed myofibrillar and rimmed-vacuolar pathology (Table 1 and Fig. 3(A)-(B)). In electron microscopy, myofibrillar disruptions and autophagic rimmed vacuolar changes were evident in a biopsy from F1 II:1 (Fig. 3(C)-(D)). The biopsy from his daughter (F1 III:2) mostly showed mild fiber size variation and some internal nuclei (not shown). Immunofluorescence staining of the biopsy from F1 II:1 showed rimmed vacuoles (Fig. 4) and myofibrillar protein accumulations staining positive for myotilin (not shown). Accumulation of DNAJB6, SQSTM1 (p62), TARDBP (TDP-43), and TIA1 was observed in the rimmed vacuolar fibers, as shown previously in patients with G/F domain DNAJB6 mutations [1,5,14,28].
Fig. 3.
Muscle pathology. (A) and (B) Serial sections of a muscle biopsy from patient F3 II:1 in hematoxylin/eosin (A) and modified Gomori trichrome stainings (B), showing myofibrillar and rimmed-vacuolar pathology. (C) and (D) Electron micrographs of a muscle biopsy from patient F1 II:1 showing Z-disk disruption and protein accumulations (C), and vacuolar pathology (D). Scale bar = 2 μm.
Fig. 4.
Immunofluorescence microscopy. Immunofluorescence images of a muscle biopsy from patient F1 II:1 showing DNAJB6, SQSTM1, TIA1, and TARDBP in protein accumulations as seen also with G/F-domain mutations in DNAJB6. Scale bar = 100 μm.
3.4. Molecular genetics
Targeted sequencing identified a DNAJB6 c.149C>T (p.A50V) mutation (Fig. 1) in all of the probands in families 1–4. In family 5, a DNAJB6 c.161A>C (p.E54A) variant was identified. The relatives in families 1, 2, 3, and 5 for whom DNA was available were Sanger sequenced, and the correct segregation of the disease mutations was confirmed in all four families. In the proband of F2 (II:1), also a c.410C>T variant (rs149027078, p.T137M, MAF=0.00009 in non-Finnish Europeans) in DNAJB6 was identified. This variant was also present in the healthy relatives F2 III:1 and III:2 and was therefore in trans with the c.149C>T change.
3.5. DNAJB6 structure
A homology model of the J and G/F domains of DNAJB6 (Fig. 5(A)) showed that the mutated residues A50 and E54 are located in the region of the J domain facing the G/F domain and are in close proximity with the previously identified F91 mutation site in the G/F domain. In contrast, the S57 residue, affected by the previously reported p.S57L variant [29] (see below) is not located in this interface.
Fig. 5.
Structural analysis of DNAJB6 mutations. (A) Protein homology model showing predicted location of residues A50, E54, S57, and F91. The region coded for by exon 5 (G/F domain) is shown in blue and the J domain in orange. A50 and E54 are located close to F91 in the interface between the J- and G/F-domains, whereas S57 is not in this area. (B) Protein sequence alignment of selected vertebrate DNAJB6 orthologs, showing conservation of the A50 and E54 residues. For the full alignment, see online supplementary file 1.
Protein sequence analysis indicated that the mutated residues are highly conserved in evolution. A50 was present in all the tetrapod DNAJB6 orthologs found from UniProt, and E54 in all vertebrates (Fig. 5(B) and online Supplementary file 1). Overall, the C-terminal half of the J domain showed a high degree of conservation, and the same was true for the 12-residue stretch containing all the published G/F domain mutations.
3.6. Functional studies
To test the effects of the novel mutations on DNAJB6b function, we investigated the mutant proteins in vitro. The original Finnish G/F-domain mutation p.F93L was included for comparison. We also analyzed the J-domain variant p.S57L, which has been previously reported in a single myopathic patient [29] but not studied functionally.
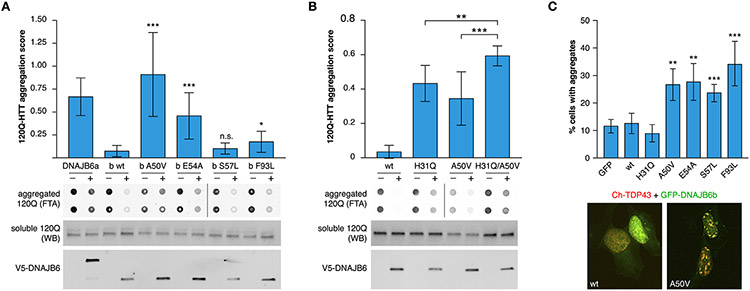
In filter trap assay, the p.A50V and p.E54A mutations caused a significant loss of anti-aggregation capacity towards 120Q-HTT, whereas p.S57L did not appear to affect this capacity (Fig. 6(A) and online Supplementary file 2). The anti-aggregation activity of DNAJB6b towards polyglutamine clients has been shown to be a combination of J-domain/HSPA-dependent and -independent functions [18]. To understand the effects of the disease-causing variants on these pathways, we studied the p.A50V and p.F93L variants in combination with p.H31Q, an HPD triad mutation that abolishes the interaction of the J domain with HSPA [30]. In 120Q-HTT filter trap assay, DNAJB6b harboring p.H31Q with either of the myopathy mutations showed significantly impaired anti-aggregation activity compared to any of the mutations alone (Fig. 6(B) and online Supplementary file 2).
Fig. 6.
Impaired function of mutant DNAJB6b. (A) Filter-trap assay showed significant loss of 120Q-HTT anti-aggregation activity for p.A50V and p.E54A, but not p.S57L, compared to wild-type DNAJB6b. The previously described p.F93L variant is included as reference and wild-type DNAJB6a (inactive towards cytoplasmic aggregation) as a negative control. Induction delay 5 h. (B) The p.H31Q mutation showed an additive impairing effect on DNAJB6b anti-aggregation activity towards 120Q-HTT when combined with p.A50V in filter-trap assay. Induction delay 4 h. Bars show mean ± SD of aggregation score (ratio of aggregated/soluble 120Q-HTT in induced versus uninduced cells) of 9 (A) or 12 (B) replicates. Representative FTA membranes show aggregated 120Q-HTT (in technical duplicates), whereas western blots show soluble 120Q-HTT and V5-DNAJB6 in uninduced (−) and induced (+) cells. For all FTA and WB images, see online supplementary file 2. (C) All investigated DNAJB6b variants (p.A50V, p.E54A, p.S57L, and p.F93L) increased nuclear TDP-43 aggregation after heat shock, whereas wild-type and p.H31Q DNAJB6b and GFP had no such effect. Bars show the percentage of cells with TDP-43 accumulations (mean ± SD) in 8 replicates. Representative cell images of DNAJB6b wild-type and p.A50V are shown under the graph; see online supplementary file 3 for the full series of representative images. *p<0.05, **p<0.01, ***p<0.001 compared to wild-type DNAJB6b (A) and (C) or for the indicated pairs (B).
As a complementary approach, we studied the effects of DNAJB6 variants on TDP-43 aggregation in heat-shocked cells. In this assay, all the J-domain variants as well as p.F93L similarly increased the proportion of cells showing nuclear TDP-43 accumulations (Fig. 6(C) and online supplementary file 3). DNAJB6b p.F93L performed more efficiently than the J-domain mutants in 120Q-HTT filter trap assay but comparably in the TDP-43 assay, in line with the idea that anti-aggregation activity towards different client proteins involves different molecular mechanisms [18,31].
4. Discussion
Mutations in the G/F domain of DNAJB6 are by now well-known causes of LGMD1D, and several mutations have been identified in populations from around the Northern hemisphere. Even though most mutations give rise to an adult-onset LGMD, some mutations cause distal onset myopathy [2,8,12,13]. We here describe novel disease-causing mutations in the J domain of DNAJB6, thus expanding the region of myopathy mutations outside of the G/F domain encoded by exon 5. The two novel mutations result in a predominantly distal phenotype, with distal myopathy seen in the p.A50V patients, and more proximo-distal in the p.E54A family.
The phenotype of p.A50V patients was very mild and late-onset, with symptoms starting in the calf muscles, and not all patients being even aware of the disease. With advanced age, however, the symptoms involved also proximal muscles, causing more disability. In the proband with the p.E54A mutation, the disease was more severe at younger age, encompassing proximal lower limb muscles from the beginning. One of our patients had moderately decreased pulmonary function but no respiratory symptoms; others had no respiratory findings which is consistent with majority of the earlier described DNAJB6 patients [6]. Dysphagia has been reported in several DNAJB6-mutated patients and was present in one of the patients reported here. Cardiomyopathy has so far not been reported in any DNAJB6 patient, although DNAJB6 has been identified as a cardiomyopathy susceptibility gene [32].
On muscle biopsy, the findings from individuals with the novel J-domain mutations appear similar to those observed in G/F-domain mutation patients [1,5,14,28], with myofibrillar protein accumulations and at later stages rimmed vacuoles.
The canonical J domain is the defining feature of J-protein co-chaperones and mediates their interaction with the HSPA chaperones. The mutated residues A50 and E54 are within the J domain but not in close proximity to the HPD motif required for HSPA binding [30]. The residues are, according to our structural homology model, located at the interface between the J and G/F domains, suggesting that the mutations interfere with interdomain interactions rather than protein-protein interactions. Interestingly, they both appear to interact with F91, a residue mutated in a severe form of LGMD1D [5,7]. This interface area has been suggested [6] to be important for proper function of DNAJB6. Our results are in line with this idea and may imply that the disease-causing mutations in the J and G/F domains act through a common downstream pathomechanism. Furthermore, the additive effect of p.H31Q and p.A50V on DNAJB6b polyQ anti-aggregation activity, and the lack of effect of p.H31Q compared to the other mutant constructs in the TDP-43 assay suggest that the myopathy-causing mutations do not simply inactivate the HSPA-dependent function of DNAJB6, but affect the HSPA-independent function.
The c.149C>T (p.A50V) variant is reported once in gnomAD (rs575604496) [33]. As indicated above, due to late onset and mild symptoms this mutation can occur in younger individuals without subjective problems. Here we identified the p.A50V variant in four separate families using targeted sequencing and verified this finding as well as the segregation in all available family members with Sanger sequencing. Together with the functional effects observed with two independent methods, this indicates that p.A50V is a pathogenic variant and the cause of distal myopathy in these families.
The c.161A>C (p.E54A) mutation is to date not found in any database. Pathogenicity of this variant is supported by our functional analyses where this variant behaved comparably to p.A50V. However, with segregation in just one small family it currently only reaches the criteria of likely pathogenic according to the ACMG guidelines [34].
The c.170C>T (p.S57L) variant was originally identified in a single LGMD patient [29], but later genetic analysis of family members revealed that this variant did not segregate with the disease (Savarese, personal communication). The variant is found in 3 of 243,726 alleles in gnomAD. The residue S57 is not located in the domain interface and mutation at this location will therefore probably not affect the interaction between the J and G/F domains. So far all experimentally tested G/F-domain mutations have shown loss of chaperone function in filter trap assay using 120Q-HTT, but p.S57L performed as efficiently as wild-type in this assay. In the TDP-43 assay, however, p.S57L behaved similarly to the other DNAJB6 variants analyzed. We hence cannot rule out that p.S57L can affect DNAJB6 function under some circumstances, and it should be considered as a variant of unknown significance. Nevertheless, our results indicate that p.S57L acts differently from the other J-domain mutations described here, and the lack of segregation strongly suggests that this variant is not the cause of disease in the original report [29].
Our findings expand the mutational spectrum from being restricted to the G/F domain and into the J domain and underline the importance of DNAJB6 mutations in distal myopathies. Mutations in DNAJB6 should therefore be considered not only in LGMD but also in other myopathies especially with aggregates and rimmed vacuolar pathology.
Supplementary Material
Acknowledgments
We are grateful to Helena Luque, Merja Soininen and Sabita Kawan for technical assistance and Professor Bruno Eymard for assistance with Family 4. CSC – IT Center for Science Ltd. provided computational resources. JP received research funding from the Finnish Medical Foundation. MI, MN, SN and IN received funding from an Intramural Research Grant (29-4) for Neurological and Psychiatric Disorders of NCNP, and AMED [grant Numbers JP17ek0109285h0001 and JP17kk0205001s0202]. MO received funding from ISCIII PI14/00738, ERDF funds “a way to achieve Europe” and PH from Jane and Aatos Erkko Foundation. CCW received funding from NIH [grant numbers R01AR068797 and K24AR073317] and BU from the Academy of Finland, the Sigrid Jusélius Foundation, and the Folkhälsan Research Foundation.
Abbreviations
- G/F
glycine/phenylalanine
- GFP
green fluorescent protein
Footnotes
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.nmd.2019.11.005.
References
- [1].Sarparanta J, Jonson PH, Golzio C, Sandell S, Luque H, Screen M, et al. Mutations affecting the cytoplasmic functions of the co-chaperone DNAJB6 cause limb-girdle muscular dystrophy. Nat Genet 2012;44:450–5. doi: 10.1038/ng.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Harms MB, Sommerville RB, Allred P, Bell S, Ma D, Cooper P, et al. Exome sequencing reveals DNAJB6 mutations in dominantly-inherited myopathy. Ann Neurol 2012;71:407–16. doi: 10.1002/ana.22683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Sato T, Hayashi YK, Oya Y, Kondo T, Sugie K, Kaneda D, et al. DNAJB6 myopathy in an Asian cohort and cytoplasmic/nuclear inclusions. Neuromuscul Disord 2013;23:269–76. doi: 10.1016/j.nmd.2012.12.010. [DOI] [PubMed] [Google Scholar]
- [4].Suarez-Cedeno G, Winder T, Milone M. DNAJB6 myopathy: a vacuolar myopathy with childhood onset. Muscle Nerve 2014;49:607–10. doi: 10.1002/mus.24106. [DOI] [PubMed] [Google Scholar]
- [5].Palmio J, Jonson PH, Evilä A, Auranen M, Straub V, Bushby K, et al. Novel mutations in DNAJB6 gene cause a very severe early-onset limb-girdle muscular dystrophy 1D disease. Neuromuscul Disord 2015;25:835–42. doi: 10.1016/j.nmd.2015.07.014. [DOI] [PubMed] [Google Scholar]
- [6].Ruggieri A, Brancati F, Zanotti S, Maggi L, Pasanisi MB, Saredi S, et al. Complete loss of the DNAJB6 G/F domain and novel missense mutations cause distal-onset DNAJB6 myopathy. Acta Neuropathol Commun 2015;3:44. doi: 10.1186/s40478-015-0224-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nam TS, Li W, Heo SH, Lee KH, Cho A, Shin JH, et al. A novel mutation in DNAJB6, p.(Phe91Leu), in childhood-onset LGMD1D with a severe phenotype. Neuromuscul Disord 2015;25:843–51. doi: 10.1016/j.nmd.2015.08.002. [DOI] [PubMed] [Google Scholar]
- [8].Monies D, Alhindi HN, Almuhaizea MA, Abouelhoda M, Alazami AM, Goljan E, et al. A first-line diagnostic assay for limb-girdle muscular dystrophy and other myopathies. Hum Genom 2016;10:32. doi: 10.1186/s40246-016-0089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jonson PH, Palmio J, Johari M, Penttilä S, Evilä A, Nelson I, et al. Novel mutations in DNAJB6 cause LGMD1D and distal myopathy in French families. Eur J Neurol 2018;25:790–4. doi: 10.1111/ene.13598. [DOI] [PubMed] [Google Scholar]
- [10].Nallamilli BRR, Chakravorty S, Kesari A, Tanner A, Ankala A, Schneider T, et al. Genetic landscape and novel disease mechanisms from a large LGMD cohort of 4656 patients. Ann Clin Transl Neurol 2018;5:1574–87. doi: 10.1002/acn3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bohlega SA, Alfawaz S, Abou-AlShaar H, Al-Hindi HN, Murad HN, Bohlega MS, et al. LGMD1D myopathy with cytoplasmic and nuclear inclusions in a Saudi family due to DNAJB6 mutation. Acta Myol 2018;37:221–6. [PMC free article] [PubMed] [Google Scholar]
- [12].Tsai PC, Tsai YS, Soong BW, Huang YH, Wu HT, Chen YH, et al. A novel DNAJB6 mutation causes dominantly inherited distal-onset myopathy and compromises DNAJB6 function. Clin Genet 2017;92:150–7. doi: 10.1111/cge.13001. [DOI] [PubMed] [Google Scholar]
- [13].Kim K, Park HJ, Lee JH, Hong J, Ahn SW, Choi YC. Two Korean families with limb-girdle muscular dystrophy type 1D associated with DNAJB6 mutations. Yonsei Med J 2018;59:698–701. doi: 10.3349/ymj.2018.59.5.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sandell S, Huovinen S, Palmio J, Raheem O, Lindfors M, Zhao F, et al. Diagnostically important muscle pathology in DNAJB6 mutated LGMD1D. Acta Neuropathol Commun 2016;4:9. doi: 10.1186/s40478-016-0276-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol 2010;11:579–92. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Izawa I, Nishizawa M, Ohtakara K, Ohtsuka K, Inada H, Inagaki M. Identification of MRJ, a DNAJ/HSP40 family protein, as a keratin 8/18 filament regulatory protein. J Biol Chem 2000;275:34521–7. doi: 10.1074/jbc.M003492200. [DOI] [PubMed] [Google Scholar]
- [17].Chuang JZ, Zhou H, Zhu M, Li SH, Li XJ, Sung CH. Characterization of a brain-enriched chaperone, MRJ, that inhibits huntingtin aggregation and toxicity independently. J Biol Chem 2002;277:19831–8. doi: 10.1074/jbc.M109613200. [DOI] [PubMed] [Google Scholar]
- [18].Hageman J, Rujano MA, van Waarde MA, Kakkar V, Dirks RP, Govorukhina N, et al. A DNAJB chaperone subfamily with HDAC-dependent activities suppresses toxic protein aggregation. Mol Cell 2010;37:355–69. doi: 10.1016/j.molcel.2010.01.001. [DOI] [PubMed] [Google Scholar]
- [19].Stein KC, Bengoechea R, Harms MB, Weihl CC, True HL. Myopathy-causing mutations in an HSP40 chaperone disrupt processing of specific client conformers. J Biol Chem 2014;289:21120–30. doi: 10.1074/jbc.M114.572461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Evilä A, Arumilli M, Udd B, Hackman P. Targeted next-generation sequencing assay for detection of mutations in primary myopathies. Neuromuscul Disord 2016;26:7–15. doi: 10.1016/j.nmd.2015.10.003. [DOI] [PubMed] [Google Scholar]
- [21].Nishikawa A, Mitsuhashi S, Miyata N, Nishino I. Targeted massively parallel sequencing and histological assessment of skeletal muscles for the molecular diagnosis of inherited muscle disorders. J Med Genet 2017;54:104–10. doi: 10.1136/jmedgenet-2016-104073. [DOI] [PubMed] [Google Scholar]
- [22].Zhang Y I-TASSER server for protein 3D structure prediction. BMC Bioinform 2008;9:40 2105-9-40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 2010;5:725–38. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER suite: protein structure and function prediction. Nat Methods 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 1997;18:2714–23. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- [26].Hasholt L, Abell K, Nørremølle A, Nellemann C, Fenger K, Sørensen SA. Antisense downregulation of mutant huntingtin in a cell model. J Gene Med 2003;5:528–38. doi: 10.1002/jgm.378. [DOI] [PubMed] [Google Scholar]
- [27].Udan-Johns M, Bengoechea R, Bell S, Shao J, Diamond MI, True HL, et al. Prion-like nuclear aggregation of TDP-43 during heat shock is regulated by HSP40/70 chaperones. Hum Mol Genet 2014;23:157–70. doi: 10.1093/hmg/ddt408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bengoechea R, Pittman SK, Tuck EP, True HL, Weihl CC. Myofibrillar disruption and RNA-binding protein aggregation in a mouse model of limb-girdle muscular dystrophy 1D. Hum Mol Genet 2015;24:6588–602. doi: 10.1093/hmg/ddv363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Savarese M, Di Fruscio G, Mutarelli M, Torella A, Magri F, Santorelli FM, et al. MotorPlex provides accurate variant detection across large muscle genes both in single myopathic patients and in pools of DNA samples. Acta Neuropathol Commun 2014;2:100 014-0100-3. doi: 10.1186/s40478-014-0100-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Mayer MP, Laufen T, Paal K, McCarty JS, Bukau B. Investigation of the interaction between DNAK and DNAJ by surface plasmon resonance spectroscopy. J Mol Biol 1999;289:1131–44. doi: 10.1006/jmbi.1999.2844. [DOI] [PubMed] [Google Scholar]
- [31].Kakkar V, Kuiper EF, Pandey A, Braakman I, Kampinga HH. Versatile members of the DNAJ family show Hsp70 dependent anti-aggregation activity on RING1 mutant Parkin C289G. Sci Rep 2016;6:34830. doi: 10.1038/srep34830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ding Y, Long PA, Bos JM, Shih YH, Ma X, Sundsbak RS, et al. A modifier screen identifies DNAJB6 as a cardiomyopathy susceptibility gene. JCI Insight 2016;1:e88797. doi: 10.1172/jci.insight.88797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.