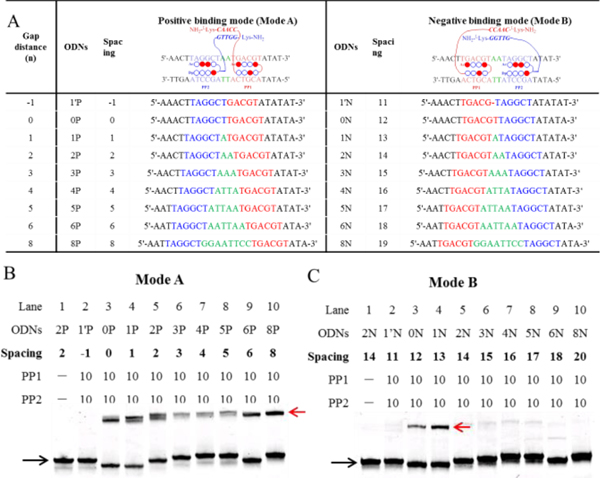
Figure 4.
Spacing-dependent manner of cooperative binding of Pip–NaCo. (A) The DNA oligomers (ODNs) used in the Tm assay, including positive (Mode A, ODN1′P–ODN8P) and negative (Mode B, ODN1′N–ODN8N) binding sequences. The gap distance (green) is the number of base pairs between the binding sites of PP1 (blue) and PP2 (red). Spacing is the distance between two PNA conjugation sites: i.e., spacing equals the gap distance in Mode A, but in Mode B, it equals the gap distance plus two PIP-binding sites. The upper chart shows only the forward DNA strand and omits the complementary DNA strand. (B, C) The gel-shift behavior of all the positive-binding sequences in Mode A (B) and negative-binding sequences in Mode B (C) with PP1–PP2. ODN concentration: 1.0 μM. Compound concentration: 10.0 μM. Black arrow: ODN2P; red: ODN2P/PP1–PP2. Except special illustration, the gel bands were stained with SYBR gold and quantified with a FujiFilm FLA-3000G fluorescent imaging analyzer. Unless otherwise stated, all samples used in the electrophoretic mobility shift assay measurements were prepared in sodium phosphate buffer (10 mM sodium phosphate, 0.1 mM EDTA, 100 mM NaCl, pH 7.2).