Abstract
Tissue stem cells (SCs) divide infrequently as a protective mechanism against internal and external stresses associated with aging. Here, we demonstrate that slow‐ and fast‐cycling SCs in the mouse skin epidermis undergo distinct aging processes. Two years of lineage tracing reveals that Dlx1+ slow‐cycling clones expand into the fast‐cycling SC territory, while the number of Slc1a3+ fast‐cycling clones gradually declines. Transcriptome analysis further indicate that the molecular properties of each SC population are altered with age. Mice lacking fibulin 7, an extracellular matrix (ECM) protein, show early impairments resembling epidermal SC aging, such as the loss of fast‐cycling clones, delayed wound healing, and increased expression of inflammation‐ and differentiation‐related genes. Fibulin 7 interacts with structural ECM and matricellular proteins, and the overexpression of fibulin 7 in primary keratinocytes results in slower proliferation and suppresses differentiation. These results suggest that fibulin 7 plays a crucial role in maintaining tissue resilience and epidermal SC heterogeneity during skin aging.
Keywords: epidermal stem cells, extracellular matrix, skin aging, slow‐cycling cell, stem cell aging
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Signal Transduction; Stem Cells & Regenerative Medicine
The matricellular protein fibulin 7 is required for the long‐term maintenance of epidermal stem cell heterogeneity and skin regenerative capacity by supporting ECM organization.
Introduction
Stem cells (SCs) in the skin are activated in response to external and internal factors, such as physical injury, inflammation, and mitotic stress, for the promotion of tissue remodeling. Skin becomes thinner as we age, and its ability to recover from damage or stress declines. The skin epithelium consists of the interfollicular epidermis (IFE) and its appendages (i.e., hair follicles [HFs], sebaceous glands), each of which is maintained by its own SC population (Gonzales & Fuchs, 2017). In homeostatic IFE, epidermal SCs divide and differentiate upward to provide barrier functions and repair damage. Hair follicle stem cells (HFSCs) behave independently of epidermal SCs during homeostasis but have the plasticity to switch their fate to the epidermal lineage during skin injury or tumorigenesis (Ito et al, 2005; Ge et al, 2017; Gonzales et al, 2021). In chronologically aged skin, ectopic differentiation, migration, and loss of HFSCs, melanocytes, or epidermal SCs have been observed (Inomata et al, 2009; Matsumura et al, 2016; Liu et al, 2019; Zhang et al, 2021). However, whether skin aging occurs primarily due to SC‐intrinsic factors or changes in the microenvironment is still debated (Ge et al, 2020).
SC adhesion with the ECM is crucial in self‐renewal and fate regulation, and alterations in the quality and quantity of ECM can induce aging‐associated skin dysfunction (Watt & Fujiwara, 2011; Egbert et al, 2014; Watanabe et al, 2017; Liu et al, 2019; Ge et al, 2020; Koester et al, 2021; Ichijo et al, 2022). Skin aging leads to DNA damage and proteolysis of collagen XVII, a hemidesmosome component important for HFSC and epidermal SC maintenance, proliferation, polarity, and epidermal patterning (Matsumura et al, 2016; Watanabe et al, 2017, 2021; Liu et al, 2019; Wang et al, 2022). This is associated with reduced wound healing ability (Keyes et al, 2016; Liu et al, 2019). However, little is known about the factors that regulate ECM changes in epidermal SCs during aging.
Tissue SCs are thought to be protected against aging by their less frequent division. SC aging may be caused by repeated replication stress and accumulation of DNA damage (Behrens et al, 2014), which culminates in metabolic and epigenetic alterations, aberrant proliferation and differentiation, and depletion of SC pools (Ermolaeva et al, 2018). The slow‐cycling populations of HF (Keyes et al, 2013; Lay et al, 2016; Wang et al, 2016) and hematopoietic (Sacma et al, 2019) SCs have protective mechanisms against aging. Although the slow‐cycling nature of SCs can minimize DNA damage, mutations acquired in slow‐cycling SCs are more likely to accumulate over time due to the error‐prone DNA repair pathway non‐homologous end joining, in contrast to the homologous recombination that predominantly occurs in faster‐cycling SCs (Tumpel & Rudolph, 2019). Whether this slow‐cycling speed serves as a mechanism to maintain long‐term SC potential and delay SC aging in the skin remains unclear.
The slow‐ and fast‐cycling SCs in the IFE of mouse skin enable us to study the relationship between cell division frequency and aging by directly comparing aging processes within the same tissue environment. Using the H2B‐GFP system, we previously reported that slow‐cycling (label‐retaining cells; LRCs) and fast‐cycling (non‐label‐retaining cell; nLRC) populations were molecularly distinct and highly compartmentalized in the IFE of mouse skin (Sada et al, 2016; Figs 1A and EV1A). Slow‐cycling SCs express Dlx1, while fast‐cycling SCs express Slc1a3. Similar SCs heterogeneity and compartmentalization have also been described in the human skin (Ghuwalewala et al, 2022) and the eye epithelium (Ishii et al, 2020; Altshuler et al, 2021; Farrelly et al, 2021). Slow‐ and fast‐cycling SCs undergo distinct differentiation programs to give rise to the keratin (K)10+ interscale and K31+ scale structures of the tail skin, respectively (Gomez et al, 2013; Sada et al, 2016). Although the fast‐cycling SCs proliferate faster, they are not committed progenitors, and both slow‐ and fast‐cycling populations express undifferentiated SC markers (Sada et al, 2016; Ghuwalewala et al, 2022). They stay within their territorial boundaries in homeostasis but retain plasticity to contribute to each other's territory during injury repair (Sada et al, 2016). However, it remains unknown how these two SC populations age and whether they are controlled by different molecular mechanisms. Here, we found that aging led to disruption of the SC compartment in the IFE, along with the reduced number of fast‐cycling SC clones and diminished skin resilience to external tissue damage and inflammation. We further identified fibulin 7 as a regulator of the SC aging process that controls long‐term SC potential and ECM maintenance.
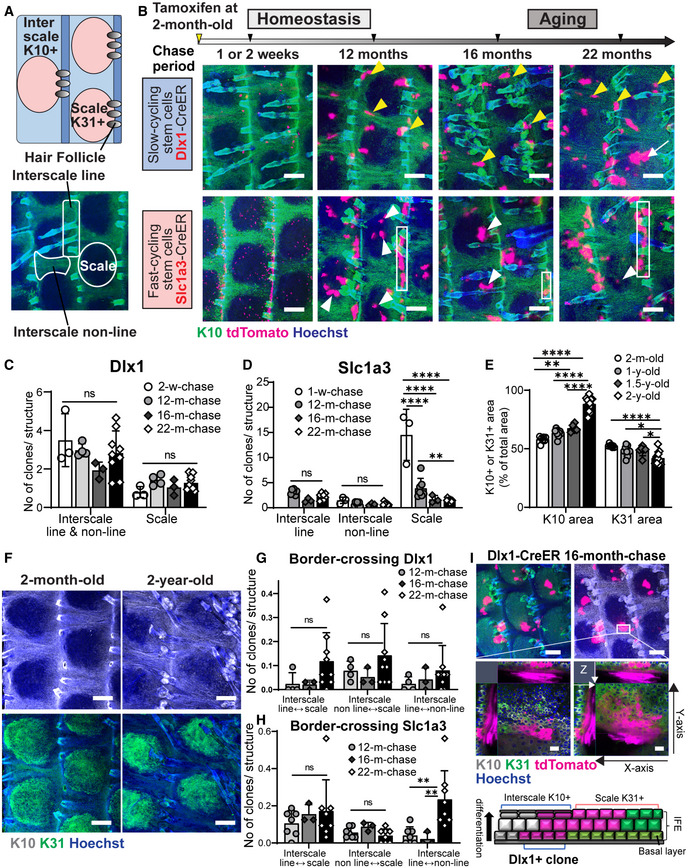
Figure 1. Long‐term lineage tracing depicting depletion of fast‐cycling stem cell clones and dysregulation of the stem cell compartmentalization in chronological aging.
-
AUpper diagram describes the interscale and scale localizations in the interfollicular epidermis (IFE), marked by keratin (K) 10 and K31, respectively. The lower wholemount (WM) tail epidermis image shows delineations of areas defined as scale, interscale line, and interscale non‐line.
-
BLow‐dose tamoxifen was administered once at 2 months of age for single‐cell labeling and samples were collected after 1–2‐week‐, 12‐, 16‐, and 22‐month‐chases. WM staining of the tail epidermis with tdTomato, K10, and Hoechst. Scale: bar: 200 μm. Yellow and white arrowheads are examples of Dlx1CreER+ interscale and Slc1a3CreER+ scale clones, respectively. White boxes indicate examples of Slc1a3CreER+ interscale line clones. A white arrow indicates a clone that is crossing the boundary.
-
CThe number of Dlx1CreER+ clones per interscale or scale structure. ns., not significant. The number of mice and chase time: N = 3 (2 weeks), N = 4 (12 months), N = 3 (16 months), and N = 9 (22 months). w, week; m, month. ns, not significant.
-
DThe number of Slc1a3CreER+ clones per interscale or scale structure. ****P < 0.0001. **P < 0.01. The number of mice and chase time: N = 3 (1 week), N = 7 (12 months), N = 3 (16 months), and N = 8 (22 months).
-
EQuantitation of K10+ or K31+ areas normalized to total IFE areas. The number of mice at age 2 months N = 7, 1 year N = 9, 1.5 years N = 6, 2 years N = 12. *P < 0.05. y, year.
-
FWM staining of K10 and K31 in 2‐month‐old compared with 2‐year‐old mice. Scale bar: 200 μm.
-
G, HThe number of Dlx1CreER+ and Slc1a3CreER+ clones crossing the interscale–scale boundaries. The number of mice used is the same as in (C–D). **P < 0.01.
-
IConfocal imaging of representative clones at 1.5‐year‐chase, stained with K10 and K31. White box indicates area that is enlarged in the lower panel. Z‐stack images show the clone originating from the basal layer and expanding into the upper differentiated layers. Cartoon summarizes the sagittal view of the clones. Scale bars: 200 μm (upper panel), 20 μm (lower panel).
Data information: Tests performed in (CE, G, H): two‐way ANOVA, Tukey's multiple comparisons. Graphs show mean SD (N reflects the number of biological replicates summarized from at least two independent experiments). K10 and K31 intensities in (B, F, I) were adjusted to similar levels between samples.
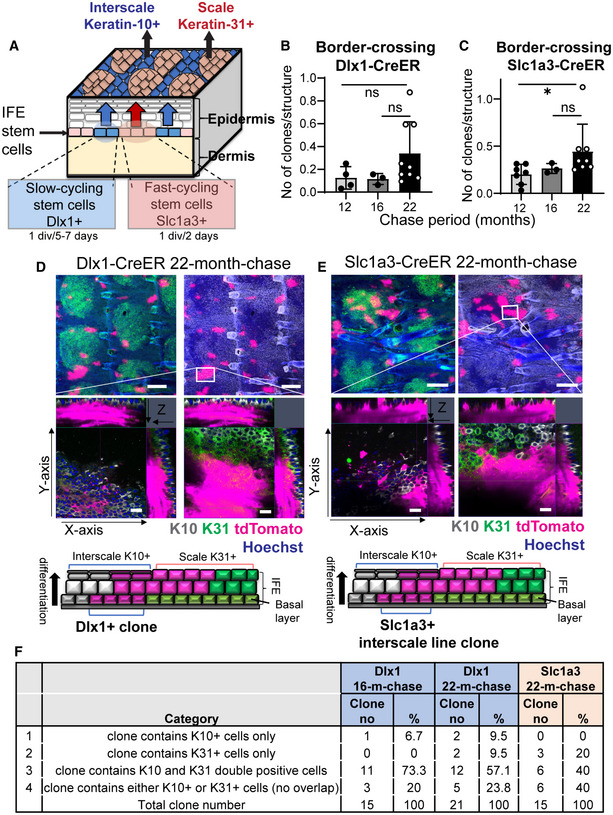
Figure EV1. Disruptions in the epidermal stem cell compartments in aging skin.
-
AIllustration of a mouse tail skin showing the slow‐ and fast‐cycling stem cells in the interfollicular epidermis (IFE) and their distinct differentiation program, giving rise to the interscale and scale regions, respectively.
-
B, CThe total number of boundary‐crossing clones in Dlx1CreER and Slc1a3CreER lineage‐tracing mice. Dlx1CreER mice and chase time: N = 4 (12 months), N = 3 (16 months), and N = 9 (22 months). Slc1a3CreER mice and chase time: N = 7 (12 months), N = 3 (16 months), and N = 8 (22 months). One‐way ANOVA, Dunn's multiple comparisons test. ns, not significant; *P < 0.05. Data show mean ± SD. N reflects biological replicates, which are summarized from at least two independent experiments.
-
D, EConfocal imaging of representative clones at 2‐year‐chase, stained with K10 and K31. White boxes indicate areas that are enlarged in the lower panels. Z‐stack images show that the clones are originating from the basal layer and expanding into the upper differentiated layers. Cartoons summarize the sagittal view of the clones. Scale bar: 200 μm (upper panels), 20 μm (lower panels). K10 and K31 intensities were adjusted to similar levels between samples.
-
FClassification of the border‐crossing clones from Dlx1CreER and Slc1a3CreER lineage‐tracing mice at 16‐month‐ and 22‐month‐chases. Images of these clones were represented in Fig 1I and EV1D and E.
Results
Fast‐cycling stem cells are gradually lost during aging
To address long‐term SC potential and its behavioral changes in aging, lineage tracing was performed using Dlx1CreER and Slc1a3CreER to mark slow‐ and fast‐cycling SCs, respectively. As previously observed (Sada et al, 2016), from 1‐week (1‐w) to 1‐year (1‐y) post‐labeling in homeostasis, Dlx1 clones were enriched in the interscale (yellow arrowheads, Fig 1B), whereas Slc1a3 clones were preferentially localized in the scale and interscale line regions (white arrowheads and white boxes, Fig 1B). We found that the number and localization of Dlx1 clones were less affected during aging (Fig 1B and C), but the number of Slc1a3 clones in the scale was significantly decreased, starting at a 1‐y chase (Fig 1B and D). It is noteworthy that while Slc1a3‐labeled fast‐cycling SC clones in the scale were gradually lost during aging, the small number of Slc1a3 clones in the interscale continued to thrive and expand within the interscale line and non‐line regions (Fig 1B and D). Hence, fast‐cycling SC clones were gradually depleted during aging, but slow‐cycling SC lineages were maintained, suggesting that an imbalance in epidermal SC populations may occur in aged skin.
Along with changes at the SC level, wholemount images of K10 and K31 staining (the regional differentiation markers of interscale and scale, respectively) showed a significant age‐dependent increase in the K10+ interscale area in 2‐y‐old mice compared with 2‐month (2‐m)‐old mice (Fig 1B, E, and F). Although there was a significant decrease in the number of fast‐cycling SC clones in the scale, effects on the size of the K31+ scale region were minimal (Fig 1E and F), suggesting a possible change of the slow‐cycling SC fate to the scale lineage occurring at the interscale‐scale boundary. Indeed, a small number of clones were found to cross the borders between the interscale and scale regions over time, and the frequency of these border‐crossing clones tended to increase in 2‐y‐old mice (arrow in Figs 1B, G, and H and EV1B and C). Using higher magnification Z‐stack images from Dlx1 aging clones at 16‐ and 22‐m‐chases, it became apparent that the cells crossing the interscale‐scale borders mainly consisted of upper differentiated cells (expressing both K10 and K31), whereas basal cells were located in the interscale (Figs 1I and EV1D and F). Thus the conversion from slow‐ to fast‐cycling lineage may not be occurring at the SC level. At a 22‐m‐chase, we also found Slc1a3 clones that expanded beyond the boundary from the interscale line region to scale, which may have compensated for the decrease of scale clones (Fig EV1E and F). This result suggests that aging alters the unique lineage fate of epidermal SCs and disrupts their compartmentalization within the tissue.
Aging induces alterations in molecular properties of slow‐ and fast‐cycling stem cells
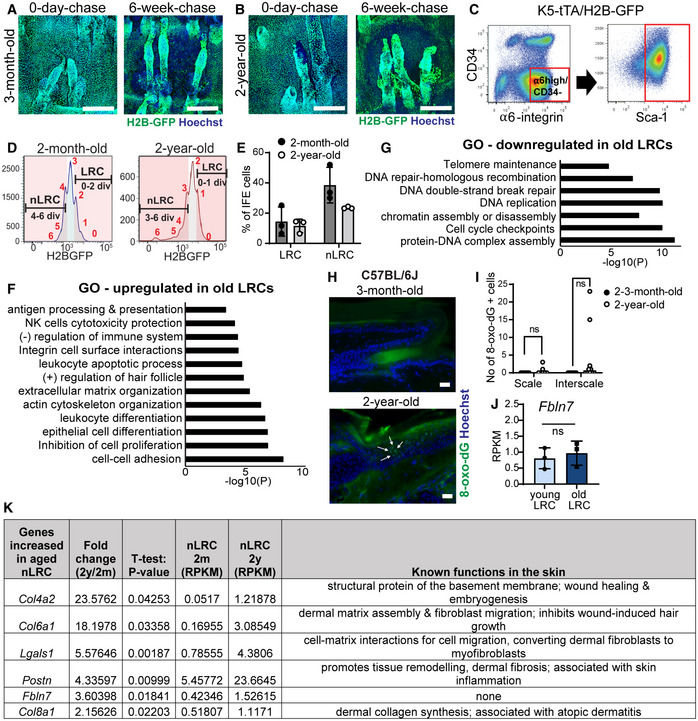
To investigate age‐dependent molecular changes, bulk RNA‐sequencing (RNA‐seq) was performed using slow‐ and fast‐cycling IFE populations isolated from the tail skin of 2‐m‐ and 2‐y‐old mice (Tumbar et al, 2004; Sada et al, 2016). Since most fast‐cycling clones labeled with Slc1a3CreER in the scale region were depleted with age, making cell isolation difficult, we used the K5‐tTA/pTRE‐H2B‐GFP mouse model to isolate a fast‐cycling cell population (nLRCs) at 2 years of age. Wholemount images showed differences in GFP dilution rates after doxycycline administration in interscale LRCs and scale nLRCs in both 3‐m‐old and 2‐y‐old mice (Fig EV2A and B). Epidermal SCs (α6‐integrinhigh/CD34−/Sca1+) from 2‐m‐old mice that underwent 0–2 cell divisions were defined as LRCs, whereas nLRCs were those that experienced 4 divisions (Fig EV2A, C, and D‐left panel). In 2‐y‐old mice, due to slower basal cell proliferation, LRCs and nLRCs were defined as cells with 0–1 divisions and 3 divisions, respectively (Fig EV2B–D‐right panel). The slowest dividing 10–15% and fastest dividing 20–30% of the total basal IFE population were isolated in both age groups (Fig EV2E).
Figure EV2. Isolation of slow‐ and fast‐cycling epidermal stem cells from H2B‐GFP mice and changes in their signature gene expression during aging.
-
A, BConfocal imaging from wholemount tail epidermis showing GFP expression level in the basal cells before and after 6 weeks of doxycycline chase in the 3‐month‐old (A) and 2‐year‐old mice (B). Scale bar: 200 μm.
-
CFACS plots show isolation strategy of epidermal basal cells (α6‐integrinhigh/CD34−/Sca‐1+).
-
DFACS histograms illustrate isolation of label‐retaining cells (LRCs) and non‐label‐retaining cells (nLRCs) based on their GFP signal peaks representative of the number of cell divisions from young (2‐month‐old) and old (2‐year‐old) H2B‐GFP mice at 2 weeks doxycycline chase.
-
EGraph describes LRC or nLRC defined from their GFP dilution in (D) as percentages of basal interfollicular epidermal (IFE) cells in young and old H2B‐GFP mice. N = 3 mice per group.
-
F, GGene ontology (GO) analysis obtained from ≥twofold differentially regulated genes (P < 0.05) in 2‐year‐old LRCs compared to 2‐month‐old mice.
-
HImmunostaining of DNA oxidation marker 8‐oxo‐dG in tail epidermis of 3‐month‐old versus 2‐year‐old C57BL6J mice. White arrows indicate positively stained cells in the interscale region. Scale bar: 20 μm.
-
IGraph summarizes the number of 8‐oxo‐dG positively stained cells in the scale or interscale regions of 2–3‐month‐old (N = 7) or 2‐year‐old mice (N = 10). Mann–Whitney test. ns, not significant. N reflects number of biological replicates summarized from two independent experiments.
-
JFbln7 gene expression in 2‐month‐ versus 2‐year‐old LRCs. N = 3 mice per group (t‐test). Data show mean ± SD.
-
KTable shows 6 ECM genes significantly upregulated in the 2‐year‐old (aged) nLRCs and their known function in the skin (Abreu‐Velez & Howard, 2012; Theocharidis et al, 2016; Kuwatsuka & Murota, 2019; Pasmatzi et al, 2019; Strafella et al, 2019). These genes were shortlisted from the 466 genes increased in the aged nLRC (Dataset EV1) according to ECM structural protein and matricellular protein category and RPKM values of ≥1 in 2‐year‐old nLRC. 2 y; 2‐year‐old. 2 m; 2‐month‐old.
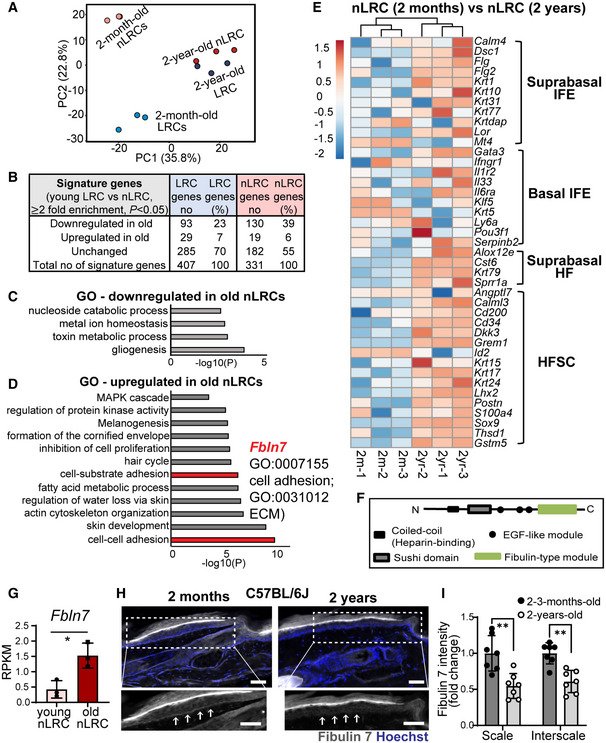
In principal component analysis (PCA), LRCs and nLRCs showed clustering separation, indicating their unique transcriptome identities and age‐dependent changes (Fig 2A; Dataset EV1 and EV2). We analyzed aging‐induced changes in signature gene expression for young, 2‐m‐old LRCs (407 genes) and nLRCs (331 genes) (Fig 2B). The following percentages of these signature genes were altered with age: 23% of young LRC genes and 39% of young nLRC genes were downregulated, whereas 7% of young LRC genes and 6% of young nLRC genes were upregulated in the 2‐y‐old counterparts (Fig 2B; Appendix Fig S1A–D; Dataset EV2). Nonetheless, some of the 2‐m‐old signature gene expression patterns were maintained during aging (Appendix Fig S1E and F; Dataset EV2), suggesting that some characteristics of young LRCs or nLRCs are retained.
Figure 2. Global gene expression analysis reveals molecular changes in slow‐ and fast‐cycling epidermal stem cells during aging.
-
APrincipal component analysis map describes the transcriptomic clustering of label‐retaining cells (LRCs) and non‐label retaining cells (nLRCs) constructed from threefold differentially expressed genes among 2‐month‐ (N = 3) and 2‐year‐old (N = 3) mice.
-
BTable summarizes the number of signature genes in young LRCs versus nLRCs and their respective changes in 2‐year‐old tail skin.
-
C, DGene ontology (GO) analysis obtained from ≥twofold differentially regulated genes (P < 0.05) in 2‐year‐old nLRCs compared with 2‐month‐old mice.
-
EHeatmap shows basal and suprabasal signature genes of epidermal stem cells and HFSCs (Ge et al, 2020) in 2‐month‐old versus 2‐year‐old nLRCs. Scale bar reflects Z‐score. IFE, interfollicular epidermis; HF, hair follicle; HFSCs, hair follicle stem cells.
-
FSchematic of fibulin 7 protein structure.
-
GFbln7 gene expression in 2‐month‐ versus 2‐year‐old nLRCs. N = 3 mice per group (t‐test). *P < 0.05. Data show mean ± SD.
-
H, IFibulin 7 immunostaining in 2‐month‐ versus 2‐year‐old tail section (H) and its intensity quantification per basal epidermal stem cell/basement membrane (normalized to 2‐month‐old) (I). Dotted box areas were enlarged in the lower panels. White arrows indicate fibulin 7 basement membrane staining. Scale bar: 50 μm. Data show mean ± SD. **P < 0.01 (Mann–Whitney test). N = 7 mice per age group (biological replicates).
To determine molecular factors that contribute to the loss of epidermal SC heterogeneity during aging, particularly the depletion of fast‐cycling SCs, we further interrogated gene expression changes in 2‐y‐old LRCs and nLRCs. Gene ontology (GO) analysis showed the 2‐y‐old LRCs upregulated genes related to immune response and extracellular organization (Fig EV2F; Dataset EV1). Moreover, a group of genes involved in DNA damage repair, telomere maintenance, DNA replication, and chromatin regulation were markedly reduced in old LRCs, suggesting that aged slow‐cycling epidermal SCs may be more prone to accumulating DNA damage (Fig EV2G, Dataset EV1). To clarify if aged LRCs have more DNA damage, immunostaining was performed with DNA oxidation marker 8‐oxo‐dG, which suggests that cells detected with this marker were rare, although it is absent in the young skin (Fig EV2H and I). DNA‐damaged SCs could be eliminated from tissues via differentiation (Kato et al, 2021), which may also account for the relatively low incidence of skin cancer compared to the rate of mutations occurring (Martincorena et al, 2015). Further study is needed to determine whether there are differences in DNA damage repair mechanisms or the behavior of DNA‐damaged cells between slow‐ and fast‐cycling SC populations.
The nLRCs, on the other hand, were characterized by changes in genes related to cell metabolism (Fig 2C and D; Dataset EV1), which is implicated in controlling SC proliferative heterogeneity and aging (Nakamura‐Ishizu et al, 2020). Over‐representation of processes that were common to the 2‐y‐old LRCs or nLRCs versus the young LRCs or nLRCs was also observed, including cell adhesion, ECM, actin cytoskeleton dynamics, inhibition of proliferation, induction of differentiation, and HF development (Figs 2D and EV2F; Dataset EV1). To evaluate whether an imbalance between SC self‐renewal and differentiation contributes to the age‐related clonal decline in fast‐cycling epidermal SCs, we examined changes in the expression patterns of established epidermal lineage marker genes (Ge et al, 2020). We found that the expression of IFE differentiation genes and HF‐lineage markers, which were repressed in young nLRCs, were enhanced in older ones, suggesting that the maintenance of undifferentiated status and lineage identity may be compromised in aged fast‐cycling epidermal SCs (Fig 2E). Hence, the RNA‐seq results and lineage tracing analysis suggest that epidermal SCs with different division frequencies undergo distinct cellular and molecular processes and show functional decline with aging.
Loss of Fbln7 accelerates age‐dependent depletion of fast‐cycling stem cell clones and delays wound healing
Because both microenvironment and SC‐intrinsic mechanisms govern the SC aging process, we focused on the secreted ECM genes of 2‐y‐old fast‐cycling SCs, whose SC potential/lineage is compromised in aged skin. Among the genes upregulated in the 2‐y‐old nLRCs with unknown function in the skin was fibulin 7 (gene symbol Fbln7), a secreted glycoprotein belonging to the short fibulin family of ECM proteins (Figs 2D, F, and G and EV2J and K). Fibulin 7 regulates cell differentiation and migration through interaction with other ECM proteins, heparin, and cell surface receptors such as β1 integrins to mediate binding to odontoblasts, monocytes, and endothelial cells (de Vega et al, 2007; Ikeuchi et al, 2018; Sarangi et al, 2018; Tsunezumi et al, 2018). However, the role of fibulin 7 in the skin remains largely unknown (Fig EV2K).
We observed modest staining but distinct localization of fibulin 7 protein to the basement membrane (BM) in both the scale and interscale regions (Fig EV3A and B, white arrows), and the expression was significantly lower in the Fbln7 KO compared to wild‐type (WT) controls (Fig EV3A–E), although non‐specific staining was noted in the stratum corneum (asterisks). Surprisingly, fibulin 7 protein level was significantly decreased in the 2‐year‐old tail skin of C57BL/6J (B6) WT mice compared to the 2–3‐m‐old counterparts (Fig 2H and I). This suggests that although Fbln7 mRNA was induced (Fig 2G), fibulin 7 function may actually decrease in aged skin due to its lower protein abundance, in line with the observed loss of fast‐cycling SCs in 2‐y‐old skin (Fig 1B and D).
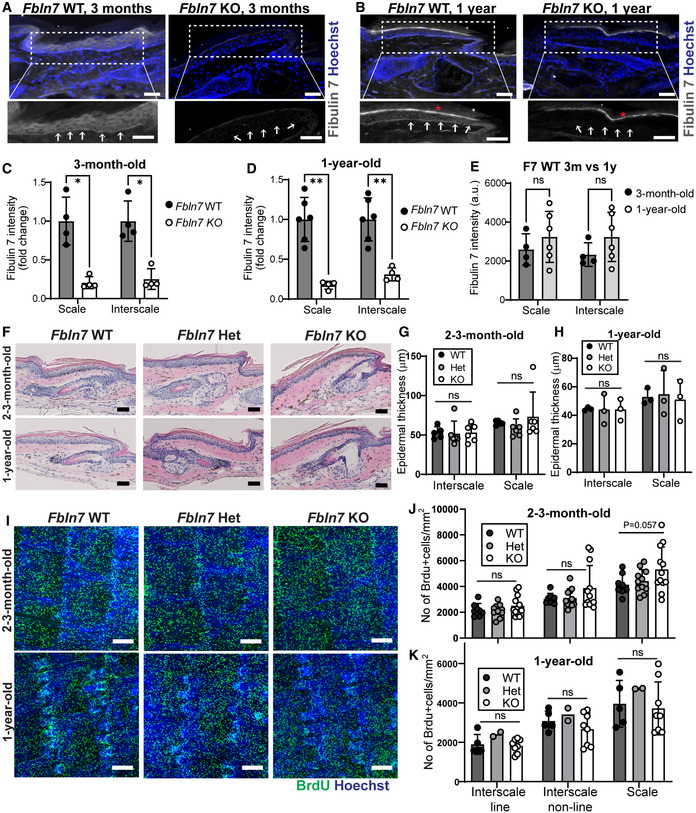
Figure EV3. Skin histology and cell proliferation assessment in Fbln7 knockout mice.
-
A, BFibulin 7 immunostaining in 3‐month‐ (A) and 1‐year‐old tail section (B) in Fbln7 WT versus KO mice. Dotted box areas were enlarged in the lower panels. White arrows indicate fibulin 7 basement membrane staining. Signal in the uppermost stratum corneum is background (asterisk). Scale bar: 50 μm.
-
C–EFibulin 7 intensity quantification per basal epidermal stem cell/basement membrane, normalized to WT (C, D) or in plain intensity measurement (E). a.u., arbitrary unit. Data show mean ± SD. **P < 0.01; *P < 0.05; ns, not significant (Mann–Whitney test). N = 4 mice per group in 3‐month‐old, N = 6 WT and 4 KO in 1‐year‐old mice (C, D). N = 4 (3‐month‐old) and 6 WT mice (1‐year‐old) (E).
-
FHematoxylin and eosin staining from tail sections of 2‐month‐ and 2‐year‐old mice. Scale bar: 50 μm. Het, heterozygous.
-
G, HEpidermal thickness measurements from the scale and interscale areas of 2‐ to 3‐month‐old (G) and 1‐year‐old (H) mice. No significant changes were observed among the Fbln7 WT, het, and KO mice (N = 5 WT, N = 6 het, N = 6 KO in 2‐ to 3‐month‐old mice; and N = 3 in 1‐year‐old WT/het/KO). Data show mean ± SD. Mann–Whitney test.
-
IWholemount immunostaining from tail epidermis labeled with BrdU and Hoechst nuclear staining. Scale bar: 200 μm.
-
J, KQuantitation of BrdU+ cells per mm2 structure area in 2‐ to 3‐month‐old mice (J) and 1‐year‐old mice (K). For 2‐ to 3‐month‐old mice, N = 8 (WT), N = 11 (het), N = 12 (KO). For 1‐year‐old mice, N = 5 (WT), N = 2 (het), and N = 8 (KO). P = 0.057 in 2‐ to 3‐month‐old Fbln7 WT versus KO mice from Welch's t‐test.
Data information: All graphs indicate mean SD. N reflects the number of biological replicates summarized from at least two independent experiments.
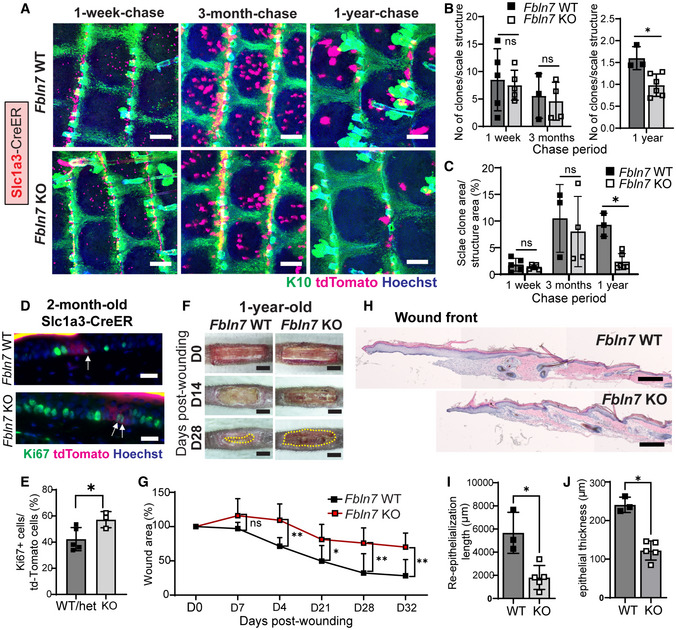
Loss of Fbln7 did not exhibit apparent changes in skin histology or epidermal thickness (Fig EV3F–H) but a modest trend toward increased proliferation in the 2‐ to 3‐m‐old Fbln7 KO scale IFE was observed, as indicated by BrdU labeling (Fig EV3I–K). To test whether fibulin 7 modulates epidermal SC heterogeneity and behavior during aging, we labeled fast‐cycling SCs with Slc1a3CreER in the Fbln7 KO background. Fast‐cycling SC clones were localized in the scale region at 1‐w or 3‐m post‐labeling, and some clones persisted for up to 1‐y in Fbln7 WT mice (Fig 3A‐upper panel, B, and C). Notably, at 1‐w post‐labeling (in 2‐m‐old mice), there was more proliferation observed in the Fbln7 KO fast‐cycling SCs as examined by Ki67 marker (Fig 3D and E), in line with the data from BrdU labeling (Fig EV3I and J). In contrast, after 1 year, the number and size of fast‐cycling SC clones in the scale region of Fbln7 KO mice were significantly decreased compared with Fbln7 WT mice (Fig 3A‐lower panel, B, and C). Hence, Fbln7 exhibited an age‐dependent function in the long‐term maintenance of the fast‐cycling SC population. It is probable that negative impacts on fast‐cycling SCs at 1‐y‐old were linked to the earlier replication stress due to loss of fibulin 7 that induced more cell divisions at 2–3‐m‐old.
Figure 3. Fbln7 knock‐out leads to aggravated fast‐cycling stem cell loss and impaired injury repair.
-
ASlc1a3CreER lineage tracing in the Fbln7 WT and KO mice. Low‐dose tamoxifen was administered once at 2 months of age and samples were collected after 1‐week, 3‐month, and 1‐year chases. Wholemount staining of tail epidermis with tdTomato, K10, and Hoechst. Scale bar: 200 μm. WT, wild‐type; KO, knock‐out.
-
BThe number of Slc1a3CreER+ clones per scale structure in 1‐week and 3‐month chases (left panel) and 1‐year chase (right panel).
-
CQuantitation of the area of Slc1a3CreER+ clones per total area of scale structure. Fbln7 WT mice for 1‐week (N = 5), 3‐month (N = 3), and 1‐year chases (N = 3). Fbln7 KO mice for 1‐week (N = 5), 3‐month (N = 4), and 1‐year chases (N = 6).
-
D, EKi67 immunostaining in 1‐week‐chase mice (D) and its quantitation (E). White arrows show Ki67 and tdTomato double positive cells, which were counted and summarized in (E). Scale bar: 20 μm. N = 5 WT/het and 3 KO mice. Welch's t‐test.
-
FRepresentative pictures from tail wounds of 1‐year‐old Fbln7 WT and KO mice. Scale Bar: 4 mm.
-
GMeasurements of wound area over time in Fbln7 WT (N = 5) and KO (N = 7) mice. Two‐way ANOVA, Tukey's multiple comparisons test.
-
H–JHematoxylin and eosin staining from tail sections of wounded Fbln7 WT versus KO mice (H) and quantitation of the re‐epithelialization length (I) and thickness at the healing front (J). Scale bar: 200 μm. N = 3 WT and 5 KO mice.
Data information: All graphs show mean SD. N reflects the number of biological replicates, which are summarized from at least two independent experiments. *P < 0.05. **P < 0.01. (B, C, I, J) Mann–Whitney test. One sample from 1‐year KO group was removed after an outlier test was performed in Graphpad Prism.
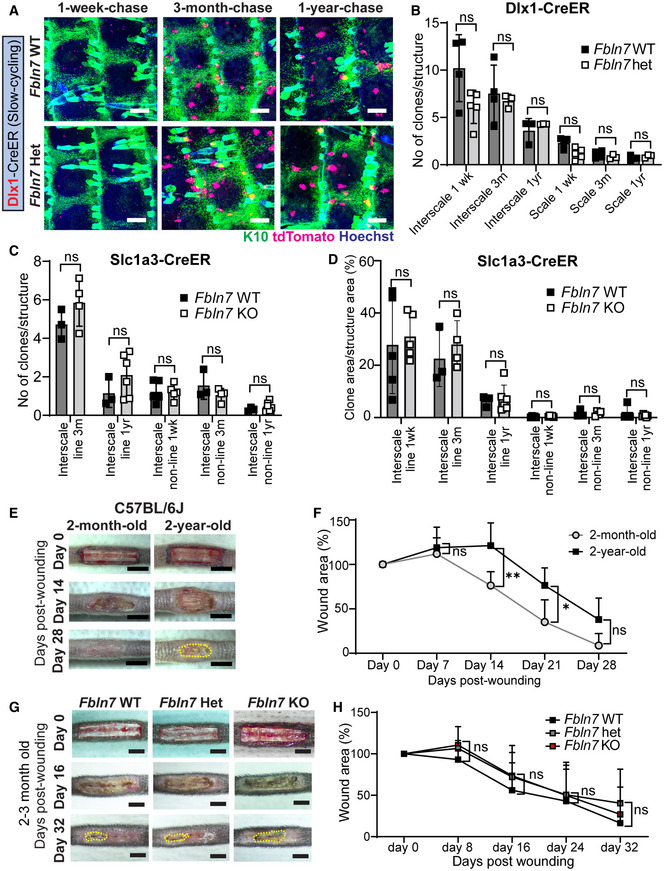
Next, we attempted to compare the slow‐cycling SCs labeled with Dlx1CreER in WT and Fbln7 KO mice; however, due to the proximity of the Dlx1 promoter to the Fbln7 gene locus on chromosome 2, we could not obtain Fbln7 KO mice harboring Dlx1CreER. Comparisons between Fbln7 WT and Fbln7 heterozygous (Het) mice from 1‐w, 3‐m, and 1‐y chases revealed that Dlx1 clones were enriched in the interscale region but their quantity and distribution showed no differences between WT and Het mice (Fig EV4A and B). Similarly, a smaller number of Slc1a3 clones in the interscale and their respective clonal areas were not affected by the partial loss of Fbln7 (Fig EV4C and D).
Figure EV4. Fbln7 knockout does not affect the maintenance of slow‐cycling stem cells or wound healing in young mice.
-
ADlx1CreER lineage tracing in the Fbln7 WT and Het backgrounds. Low‐dose tamoxifen was administered once at 2 months of age and samples were collected after 1‐week, 3‐month, and 1‐year chases. Wholemount staining of tail epidermis with tdTomato, K10, and Hoechst. Scale bar: 200 μm.
-
BThe number of Dlx1CreER clones in the scale or interscale (both line and non‐line) of Fbln7 WT mice for 1‐week (N = 4), 3‐month (N = 4), 1‐year chase (N = 3) and Fbln7 het mice for 1‐week (N = 5), 3‐month (N = 3), and 1‐year (N = 3) chase. ns, not significant. Mann–Whitney test.
-
CThe number of Slc1a3CreER clones in the interscale non‐line or interscale line regions for 1‐week, 3‐month or 1‐year chase. Fbln7 WT mice for 1‐week (N = 5), 3‐month (N = 3), and 1‐year chase (N = 3). Fbln7 KO mice for 1‐week (N = 5), 3‐month (N = 4), and 1‐year (N = 6) chase.
-
DQuantitation of the area of Slc1a3CreER clones per structural area from the same experiment as in (C).
-
E, FRepresentative pictures from tail wounds of 2‐month versus 2‐year‐old C57BL/6J wild‐type mice (E) and measurements of wound area over time (F). N = 6 (2‐month‐old) and N = 5 (2‐year‐old). Scale bar: 4 mm. **P < 0.01; *P < 0.05.
-
G, HRepresentative pictures from tail wound healing experiment in 2‐ to 3‐month‐old Fbln7 mice (G) and the wound area quantitation over time (H). N = 4 (WT), N = 8 (Het), and N = 7 (KO). Scale bar: 4 mm.
Data information: All graphs show mean SD. N reflects the number of biological replicates summarized from at least two independent experiments. Mann–Whitney test was performed for (B–D). two‐way ANOVA (Tukey's multiple comparison test) was performed for (F, H).
Slow‐ and fast‐cycling SCs cooperate in response to skin damage (Sada et al, 2016), and wound healing is delayed in aging skin (Keyes et al, 2016; Liu et al, 2019). We also confirmed that wound healing speed was significantly delayed in the tail skin of 2‐y‐old mice compared to the 2‐m‐old B6 WT mice (Fig EV4E and F). We hypothesized that disturbed epidermal SC heterogeneity in the absence of Fbln7 might reduce skin regenerative capacity upon injury. To test this, a wound‐healing assay was performed in the tail skin of 2‐ to 3‐m‐old and 1‐y‐old mice in the presence or absence of Fbln7 (Fig 3F and G and EV4G and H). While no apparent difference was observed in the wound healing ability of Fbln7 WT or KO mice at 2–3‐m of age (Fig EV4G and H), 1‐y‐old Fbln7 KO mice showed impaired wound closure compared with Fbln7 WT mice (Fig 3F–J). Further examinations of the skin histology at the wound front indicate that the re‐epithelialization process was hampered in 1‐y‐old skin lacking Fbln7, as shown by the newly formed epidermis that was shorter and thinner in the Fbln7 KO mice (Fig 3H–J). Altogether, the expression of Fbln7 in aging skin may minimize the loss of epidermal SC heterogeneity, supporting resilience against skin tissue damage.
Fibulin 7 modulates stem cell inflammatory stress response and fate specification
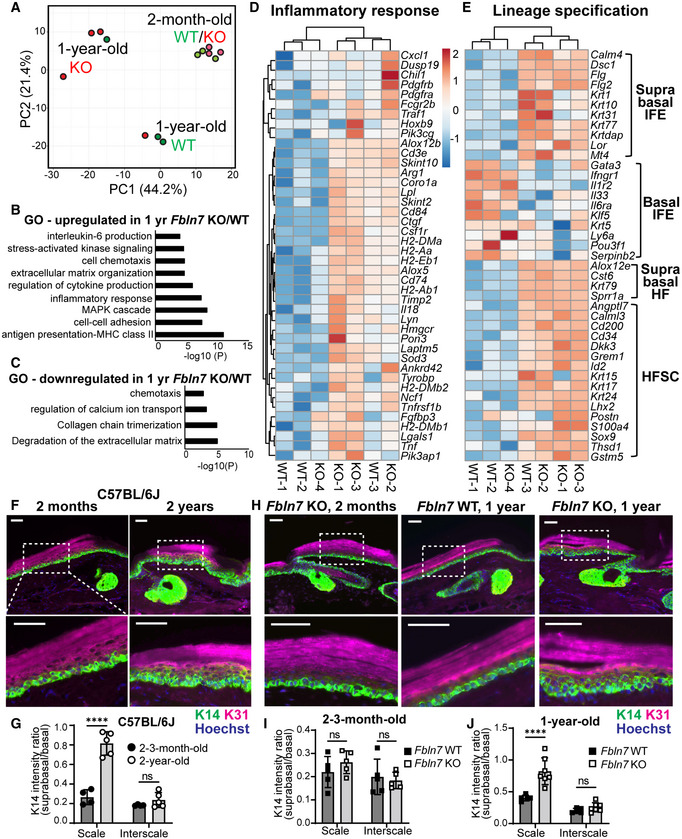
To further understand how fibulin 7 supports long‐term SC maintenance, RNA‐seq was performed using basal epidermal SCs isolated from the dorsal skin of Fbln7 WT and KO mice at 2‐m or 1‐y of age. The dorsal skin was used as it is the largest skin area containing the slow‐ and fast‐cycling SC populations in the IFE (Sada et al, 2016). PCA analysis showed that the transcriptomic difference between Fbln7 WT and KO mice occurred at 1 year but not at 2 month of age, supporting our previous findings from lineage tracing and wound healing assays (Fig 4A, Dataset EV3). Intriguingly, in the 1‐y‐old Fbln7 KO mice, inflammatory response genes were upregulated, including antigen presentation, the MAPK cascade, and cytokine production while chemotaxis was both up‐ and downregulated (Fig 4B–D, Dataset EV3). Lineage fate changes were reported as part of the inflammatory response (Ge et al, 2017), and an analysis of these gene lists (Ge et al, 2020) further underscored their alteration in 1‐y‐old Fbln7 KO mice (Fig 4E). The changes resemble some features of the 2‐y‐old nLRCs in the tail (Fig 2E), such as enhanced hair lineage and IFE differentiation markers, albeit with some mouse‐to‐mouse variations (Fig 4E).
Figure 4. Loss of Fbln7 is associated with increased inflammatory response in aging epidermal stem cells and lineage misregulation.
-
APrincipal component analysis map constructed from threefold differentially expressed genes in epidermal stem cells from Fbln7 WT versus KO dorsal skin in 2‐month‐ and 1‐year‐old mice. Each dot represents one mouse (N = 3 per group, except 1‐year‐old Fbln7 KO N = 4).
-
B, CGene ontology (GO) analysis from ≥twofold upregulated (B) or downregulated (C) genes in 1‐year‐old Fbln7 KO mice compared with WT.
-
D, EHeatmap illustrating changes in inflammatory response genes (D) and basal or suprabasal signature genes of epidermal stem cells (IFEs) and HFSCs (Ge et al, 2020) (E) in 1‐year‐old Fbln7 WT versus KO mice, constructed from ≥twofold differentially expressed genes obtained from GO analysis in (B). Scale: Z‐score.
-
F, GK31 and K14 immunostaining in tail sections of 2‐month‐ versus 2‐year‐old C57BL/6J WT mice (F) and the quantification (G). Dotted box areas are enlarged in the lower panels. Scale bar: 50 μm. Quantification of K14 intensity in the suprabasal area was normalized to basal area per cell. N = 4 (2–3‐month‐old) and N = 5 (2‐year‐old) C57BL/6J mice (G).
-
H–JK31 and K14 immunostaining in tail sections of 1‐year‐old Fbln7 WT versus KO compared with 2‐month‐old Fbln7 KO mice (H) and the quantitation (I, J). Scale bar: 50 μm. N = 5 for both WT and KO of 2–3‐month‐old mice (I). N = 5 and N = 7 for 1‐year‐old WT versus KO, respectively (J).
Data information: (F–J) Two‐way ANOVA, Tukey's multiple comparisons test. All graphs show mean SD. N reflects the number of biological replicates which are summarized from at least two independent experiments. ****P < 0.0001, ns; not significant.
Increased inflammation due to DNA damage has been linked to SC fate misspecification, which is manifested in the suprabasal expression of basal marker K14 (Seldin & Macara, 2020). We tested this in our model and found that unlike the K14 restriction to the basal layer in the 2‐m‐old B6 WT tail epidermis, K14 distribution significantly expanded from basal‐to‐suprabasal layers primarily in the scale region of the 2‐y‐old counterparts (Fig 4F and G). Likewise, there was a significant increase in basal‐to‐suprabasal expansion of K14 in the scale region of the 1‐y‐old Fbln7 KO mice but not in the 2‐m‐old Fbln7 KO mice (Fig 4H–J). Although suprabasal K14 was not observed in aged dorsal skin (Keyes et al, 2016), it has been reported that aging promotes an inflammatory environment in skin (Doles et al, 2012; Hu et al, 2017); fast‐cycling SCs in the tail scale may thus be more susceptible to this change. Therefore, our results indicate that Fbln7 loss increased the expression of inflammatory response genes in epidermal SCs, which may, in turn, influence the specification of SC fate.
Fibulin 7 maintains the extracellular environment of epidermal stem cells by regulating basement membrane proteins
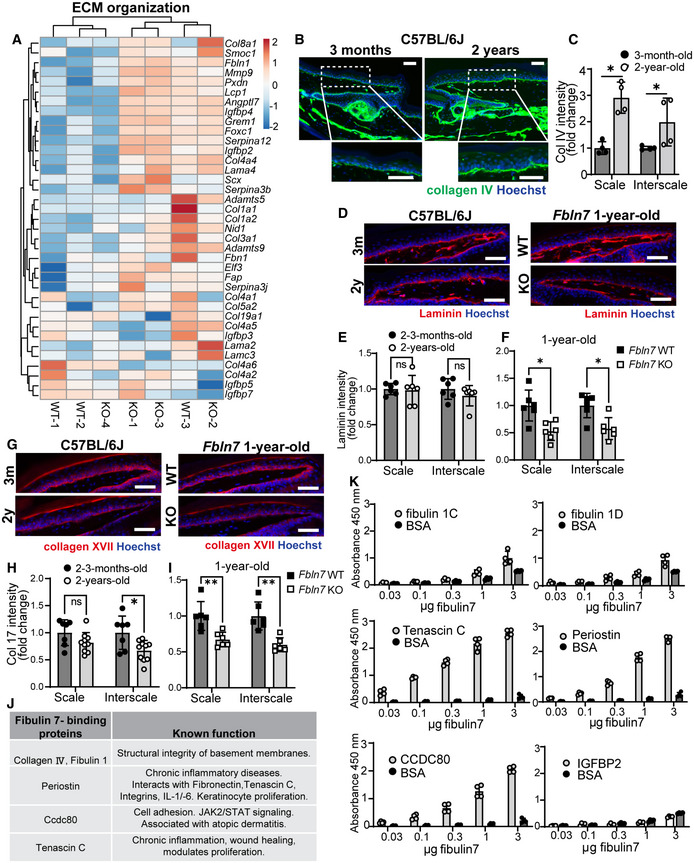
The structural proteins of the BM are essential in maintaining epidermal SCs in physiological and pathological conditions (Abreu‐Velez & Howard, 2012; Watanabe et al, 2017; Chermnykh et al, 2018; Gatseva et al, 2019). In the 1‐y‐old Fbln7 KO mice, in addition to inflammatory response genes, ECM‐related genes were also affected, with changes impacting BM components (collagen IV [col IV], nidogen 1, laminins, fibulin 1) and ECM remodeling genes (Fig EV5A). To confirm this, immunostainings of col IV and laminins as major components of the BM were performed. Col IV staining was increased along the BM of 2‐y‐old versus 3‐m‐old B6 WT mice (Fig EV5B). A similar pattern was observed in the epidermis of 1‐y‐old Fbln7 KO mice compared to WT (Fig 5A). This aligns with a previous report describing the upregulation of col IV in thickened BM, which led to niche stiffness in aged skin (Koester et al, 2021). Quantifications of BM col IV intensity demonstrated a significant increase in the scale and interscale of 2‐y‐old B6 WT; however, the increase was only significant in the scale of 1‐y‐old Fbln7 KO epidermis and not in the interscale (Fig 5A and B and EV5C). On the contrary, laminin BM staining was unchanged in the 2‐y‐old B6 WT mice versus 2–3‐m‐old mice, but its protein abundance was significantly lower in the Fbln7 KO BM in both scale and interscale regions (Fig EV5D–F). Moreover, collagen XVII proteolysis promotes skin aging (Liu et al, 2019), and we confirmed that collagen XVII staining was significantly decreased in the interscale region of 2‐y‐old B6 WT mice as well as in the scale and interscale regions of 1‐y‐old Fbln7 KO mice (Fig EV5G–I). These results suggest that the loss of fibulin 7 affects BM composition, favoring the aging‐like conditions.
Figure EV5. Transcriptome of Fbln7 KO mice and fibulin 7‐binding assays indicate fibulin 7 mechanism of action through ECM regulation.
-
AHeatmap of ≥twofold upregulated or downregulated genes related to ECM from gene ontology analysis in Fig 4B and C. Scale reflects Z‐score.
-
BCollagen IV immunostaining in tail sections of 3‐month‐old versus 2‐year‐old C57BL/6J mice. Dotted box regions are enlarged.
-
CQuantification of Collagen IV basement membrane intensity per cell in 2‐year‐old mice normalized to 3‐month‐old mice in C57BL/6J mice (B). *P < 0.05. N = 4 mice per age group (biological replicates).
-
DLaminin immunostaining in tail sections of 3‐month‐old versus 2‐year‐old C57BL/6J mice (left panels) and 1‐year‐old Fbln7 WT versus KO (right panels).
-
E, FQuantification of laminin basement membrane intensity per cell in 2‐year‐old mice normalized to 3‐month‐old mice in C57BL/6J mice (E) or in Fbln7 KO mice normalized to WT mice (F). N = 6 per group for all mice. ns; not significant.
-
GCollagen XVII immunostaining in tail sections of 3‐month‐old versus 2‐year‐old C57BL/6J mice (left panels) and 1‐year‐old Fbln7 WT versus KO (right panels).
-
H, IQuantification of Collagen XVII intensity per cell in 2‐year‐old mice normalized to 3‐month‐old mice in C57BL/6J mice (H) or in Fbln7 KO mice normalized to WT mice (I). N = 7 (2–3 month‐old) and N = 10 (2‐year‐old) C57BL/6J mice; N = 6 per group in Fbln7 mice. **P < 0.01.
-
JShortlisted fibulin 7‐binding protein candidates (from Fig 5D and E) and their reported functions.
-
KSolid‐phase binding assays using recombinant fibulin 7 as liquid phase and purified or recombinant ECM proteins as solid phase. X‐axis shows increasing doses of fibulin 7 (μg/well). Bovine serum albumin (BSA) was used as the control liquid phase and added in the same amounts as fibulin 7. Data are from four technical repeats in two independent experiments.
Data information: All graphs show mean SD with Mann–Whitney test. N reflects the number of biological replicates summarized from at least two independent experiments. All scale bars: 50 μm.
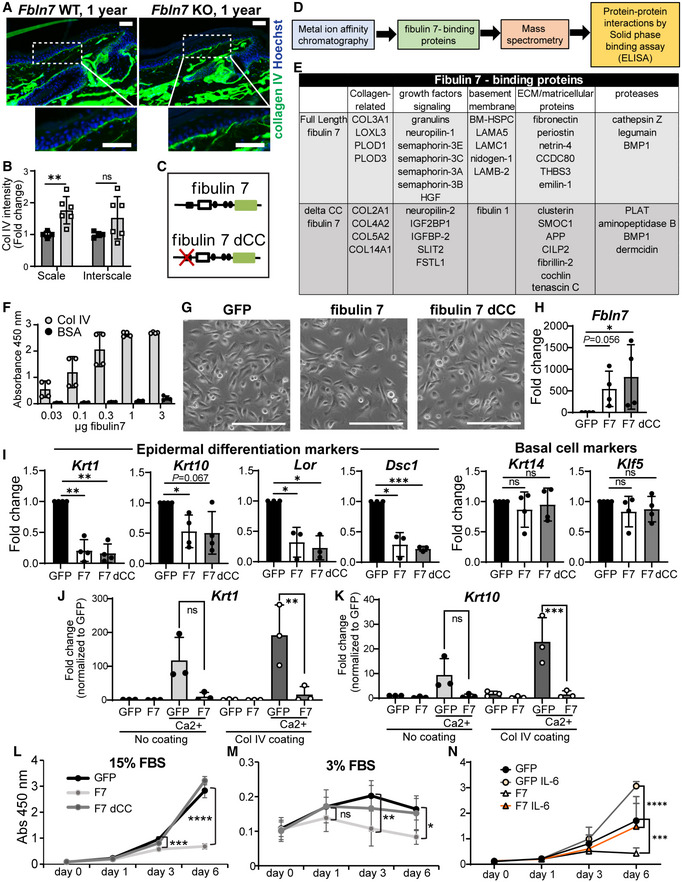
Figure 5. Fibulin 7 modulates Collagen IV and Fbln7 overexpression maintains the undifferentiated, slower proliferation state in primary keratinocytes.
-
ACollagen IV immunostaining in tail sections of 1‐year‐old Fbln7 WT versus KO. Dotted box regions are enlarged. Scale bar: 50 μm.
-
BQuantification of Collagen IV basement membrane intensity per cell in Fbln7 KO mice normalized to WT mice. N = 4 WT and N = 6 KO mice. Graphs show mean SD. Mann–Whitney test. **P < 0.01, ns, not significant.
-
C–EDiagram of proteins binding to full‐length fibulin 7 or fibulin 7 with coiled‐coil deletion mutant (delta CC) (C) and workflow chart of screening experiments to investigate secreted fibulin 7‐interacting proteins (D). List of the fibulin‐7 binding proteins obtained from mass‐spectrometry (E).
-
FSolid‐phase binding assay using recombinant fibulin 7 as liquid phase and purified collagen IV protein as solid phase. X‐axis shows increasing doses of fibulin 7 (μg/well). Bovine serum albumin (BSA) was used at the same amounts as control liquid phase. Graph shows mean SD from four technical repeats in two independent experiments.
-
GBrightfield images of mouse primary keratinocytes overexpressing GFP, full‐length fibulin 7 (F7), or coiled‐coil deletion mutant (F7 dCC) as shown in (C). Scale bar: 200 μm.
-
H–KQuantitative RT–PCR shows fold changes in gene expression of F7 or F7 dCC compared with GFP control. Keratinocytes were induced with CaCl2 for 1 day with or without Collagen IV coating prior to quantitative RT–PCR (J, K). N = at least three biological repeats. Statistical test: One‐way ANOVA with Dunn's multiple comparison test (H); Welch's t‐test. ***P < 0.001. **P < 0.01. *P < 0.05; ns, not significant (I). One‐way ANOVA (Tukey's multiple comparisons test) was used for (J, K).
-
L, MCell proliferation assays comparing growth rates of GFP overexpression control with F7 and F7 dCC over 6 days in 15% FBS growth medium (L) or 3% reduced growth medium (M). FBS, fetal bovine serum. Abs, absorbance.
-
NCell proliferation assay comparing GFP or F7 overexpressing keratinocytes treated with 5 ng/ml IL‐6 over 6 days. Statistical test two‐way ANOVA (Tukey's multiple comparisons test). ****P < 0.0001; **P < 0.01. *P < 0.05; ns, not significant (L–N).
Data information: All graphs describe mean values SD from at least three biological repeats.
To further characterize the biochemical functions of fibulin 7, we screened for secreted fibulin 7‐binding proteins using conditioned media prepared from heparin‐treated cells overexpressing full‐length (FL) fibulin 7 or cells overexpressing fibulin 7 but lacking the N‐terminal heparin‐binding coiled‐coil domain (dCC) (Tsunezumi et al, 2018; Fig 5C–E). The CC domain mediates binding to heparin and is important for the pericellular tethering of fibulin 7 (Fig 5C; Tsunezumi et al, 2018). The putative fibulin 7‐binding proteins were co‐eluted with fibulin 7 from a metal ion affinity column and identified via mass spectrometry (Fig 5D). The following classes of candidate proteins were selected: those with functions in the structural integrity of the BM, proliferation‐modulating proteins (growth factors); matricellular proteins associated with wound healing; and extracellular proteases involved in ECM remodeling (Fig 5E, shortlisted in EV5J). We confirmed via solid‐phase binding assay the dose‐dependent interactions of fibulin 7 with BM components such as col IV (Fig 5F) and, to a lesser extent, the fibulin 1C and 1D isoforms (Fig EV5K). Fibulin 7 also exhibited direct binding to matricellular proteins that are upregulated upon wound healing or inflammation, such as tenascin C, periostin, and Ccdc80 (Fig EV5K; Tremblay et al, 2009; Hirota et al, 2012; Midwood et al, 2016; Nikoloudaki et al, 2020), but not to insulin growth factor binding protein‐2 (IGFBP‐2, Fig EV5K). These results suggest that fibulin 7 physically interacts with ECM proteins that regulate BM composition and integrity, supporting the extracellular environment of epidermal SCs.
Fibulin 7 regulates the undifferentiated state and proliferative response to growth factors in primary keratinocytes
In vivo phenotype analysis has so far suggested that fibulin 7 may function as a protective factor for epidermal SCs during physiological aging, maintaining their long‐term SC potential. To investigate the function of fibulin 7 in primary mouse keratinocytes, we overexpressed the FL or dCC mutant (Fig 5G and H). Overexpression of fibulin 7, which interacts with col IV (Fig 5E and F), suppressed differentiation markers (Krt1, Krt10, Lor, Dsc1), independently of its CC domain. The SC markers (Krt14, Klf5) and cobblestone‐like morphology of keratinocytes were unchanged, suggesting that fibulin 7 maintains primary keratinocytes in an undifferentiated state (Fig 5G and I). Fibulin 7 overexpression was able to inhibit differentiation markers (Krt1 and Krt10) not only in the basal state (Fig 5I) but also during calcium induction in the presence or absence of col IV coating, albeit a more significant suppression was observed with col IV coating (Fig 5J and K). Binding with col IV may enhance fibulin 7 function through their protein–protein interactions.
We further investigated the role of fibulin 7 in keratinocytes exposed to a high or low amount of serum growth factors, mimicking the aging skin microenvironment. The proliferation assays in high (15%) or low (3%) serum conditions demonstrated that fibulin 7 maintains a lower cell division frequency of keratinocytes and that this effect required the CC domain (Fig 5L and M). GO analysis from 1‐y‐old Fbln7 KO RNA‐seq data indicated an upregulation of genes associated with interleukin‐6 (IL‐6) production (Fig 4B). IL‐6 induces hyperproliferation of keratinocytes in vitro and is associated with skin aging, wounding, and psoriasis (Grossman et al, 1989; Barrientos et al, 2008; Doles et al, 2012; Taniguchi et al, 2014; Hu et al, 2017). We found that responses to growth cues by this cytokine were blunted in fibulin 7‐overexpressing keratinocytes (Fig 5N). Thus, fibulin 7 gain‐of‐function indicates that fibulin 7 may be beneficial for suppressing differentiation and maintaining slower proliferation status in epidermal SCs under high‐ or low‐growth‐inducing conditions.
Discussion
Young skin is supported by heterogeneous SC populations having the high capacity for injury recovery and resilience to stress (Sada et al, 2016; Ghuwalewala et al, 2022). The current study highlights the importance of epidermal SC heterogeneity and its dysregulation during physiological aging. We demonstrated that the Slc1a3+ fast‐cycling epidermal SC clones were decreased and the cellular and molecular properties of epidermal SCs were impaired in aged skin. The deletion of Fbln7, an ECM gene, accelerated the age‐dependent loss of fast‐cycling SC clones, accompanied by defective skin regeneration after tissue damage. Fibulin 7 maintains epidermal SCs in part by regulating inflammatory stress responses and the fate balance of SCs through direct binding to ECM proteins. Our results suggest that regulation of the extracellular environment by fibulin 7 maintains heterogeneous epidermal SC populations over the long term, which may be an essential mechanism in controlling skin aging.
ECM composition is modified tremendously during skin aging, an alteration that is partially caused by ECM gene expression changes (Dataset EV1; Marsh et al, 2018; Liu et al, 2019; Ge et al, 2020; McCabe et al, 2020; Koester et al, 2021). Interestingly, the loss of Fbln7 showed an age‐dependent effect at 1‐y‐old but not at 2–3‐m‐old and while its mRNA expression was increased in the 2‐y‐old fast‐cycling SCs, its protein amount was decreased in the aged basal SCs. This may be due to the increased inflammatory environment during chronological aging and fibulin 7 may protect epidermal SCs from the adverse consequences of chronic inflammation, such as inhibition of self‐renewal and skewed SC lineages (Tumpel & Rudolph, 2019). Future studies should evaluate whether the binding of fibulin 7 to BM proteins supports BM structural integrity and remodeling under physiological and pathological conditions of the skin. The binding of fibulin 7 to β1 integrin (de Vega et al, 2007; Ikeuchi et al, 2018; Sarangi et al, 2018) could also be relevant in maintaining epidermal SCs. β1 integrin plays an immuno‐protective role by inhibiting inflammation‐induced ECM degradation in the BM (Kurbet et al, 2016), and its loss results in IFE proliferation and differentiation aberrations, BM abnormalities, and defective injury repair (Watt, 2002; Lopez‐Rovira et al, 2005; Margadant et al, 2010). It is worth noting that the reported IFE changes due to β1 integrin loss (Lopez‐Rovira et al, 2005) bear some similarities to our description of aging tail skin, such as K10‐marked interscale expansion (Fig 1E and F) and increased col IV deposition in the BM (Fig 5A and B).
Overexpression of fibulin 7 slowed proliferation and enhanced the undifferentiated state of primary keratinocytes (Fig 5G–N), in contrast to the differentiation‐biased state caused by loss of fibulin 7 (Fig 4E). In line, Fbln7 loss in the young 2–3‐m‐old mice was also shown to increase fast‐cycling SC proliferation (Fig 3D and E and EV3I–J). Of note, the slower cycling rate induced by fibulin 7 overexpression was only partially enhanced by IL‐6, a positive regulator of keratinocyte proliferation (Taniguchi et al, 2014). This indicates that fibulin 7 may dampen keratinocyte response to excess inflammatory cytokine signaling. Fibulin 7 could represent a potential molecule for protecting epidermal SCs against the detrimental effects of aging‐associated inflammation, lineage misspecifications, and a premature differentiation‐like state. Pending future research, fibulin 7 could be a valuable candidate for the intervention of chronic wounds observed more frequently in older patients.
Limitations of the study
Since basal cells in the scale proliferate less than those in the interscale in aged mice (Giangreco et al, 2008; Changarathil et al, 2019), separation by cell division frequency in the H2B‐GFP system became more ambiguous and may not exactly represent the same SC populations in the young mice. Therefore, although our interpretation of RNA‐seq data largely reflects molecular changes that have occurred in slow‐ and fast‐cycling SCs, in‐depth analysis by single‐cell analysis or isolation using specific cell surface markers will advance the understanding of epidermal SC aging.
In our RNA‐seq analysis, we used tail and back skin which had distinct gene expression profiles and responses to inflammation (Quigley et al, 2016) due to different originating skin locations. Although the effects of aging may differ between dorsal skin (Keyes et al, 2016; Ge et al, 2020) and tail skin, it is worth noting that Fbln7 KO back skin showed molecular changes (Fig 4E) comparable to those of aging tail skin (Fig 2E). In the future, RNA‐seq using tail skin from Fbln7 KO will allow for direct comparison with physiological aging.
Finally, since we use a whole‐body knockout of Fbln7, we cannot definitively determine if Fbln7 functions primarily in the dermis or immune cells rather than epidermal SCs, although we have detected fibulin 7 protein in the BM region and experiments with keratinocytes support the intrinsic effects of Fbln7. Future phenotype analysis using conditional knockout mice is needed to investigate the cell type‐specific functions of Fbln7.
Materials and Methods
Mice
All animal experiments were performed according to the Institutional Animal Experiment Committee guidelines at the University of Tsukuba, Kumamoto University, and Kyushu University of Health and Welfare. The generation of Fbln7 KO mice (129SvEv;C57BL/6J) was previously reported (Tsunezumi et al, 2018). K5‐tTA (C57BL/6J) (Diamond et al, 2000)/pTRE‐H2B‐GFP (CD1;C57BL/6J) (Tumbar et al, 2004) double transgenic mice were used for the isolation of LRCs and non‐LRCs from young and old mice (Sada et al, 2016). WT C57BL/6J mice were purchased from Charles River Laboratories or Japan SLC, Inc. For the lineage‐tracing experiment, Dlx1CreER (C57BL/6J) (The Jackson Laboratory, no. 014551) or Slc1a3CreER (C57BL/6J) (The Jackson Laboratory, no. 012586) mice were crossed with Rosa‐tdTomato reporter mice (C57BL/6J) (The Jackson Laboratory, no. 007905). CreER/Rosa‐tdTomato was introduced in the Fbln7 knockout background. Mice of both sexes were used for experiments and the allocation of groups was done randomly. Mice were maintained in animal housing facilities with 12 h of dark and light cycles with regular caging/food/water maintenance every week.
Tamoxifen, BrdU, and doxycycline administration
Tamoxifen (Sigma) was injected intraperitoneally at a single dose of 25 μg or 50 μg/g body weight (BW) of Slc1a3CreER and Dlx1CreER mice, respectively, at 2 months of age for lineage tracing experiments. BrdU (5‐Bromo‐2′‐deoxyuridine; Sigma‐Aldrich, B5002) was administered in drinking water (0.8 mg/ml) 2 days before sacrifice to label proliferative cells. For H2B‐GFP pulse‐chase, 2‐month‐old or 22‐month‐old mice were fed with doxycycline chow (1 g/kg, Oriental Kobo Inc.) for 2 weeks before sacrifice.
FACS isolation
The subcutaneous and fat tissues were removed from the skin and incubated overnight at 4°C in 0.25% trypsin/EDTA and the next day at 37°C for 30 min. Single‐cell suspensions were prepared by gentle scraping of the epidermis and subsequent filtering with 70 and 40 μm filters. Cells were stained with the following antibodies for 30 min on ice: CD34‐biotin (1:50, eBioscience, 13‐0341), Streptavidin‐APC (1:100, BD Biosciences, 554067), α6‐integrin‐BUV395 (1:100, BD Biosciences, custom order), and Sca1‐BV421 (1:100, BD Biosciences, 562729). Dead cells were excluded by propidium iodide (P4864, Sigma) staining. Cells from mouse tail skin (aging RNA‐seq) and dorsal/ventral skin (fibulin 7 RNA‐seq) were isolated using FACS Aria flow cytometer (BD Biosciences). Data were analyzed using FlowJo software (BD Biosciences).
RNA‐sequencing
FACS‐isolated cells from K5‐tTA/pTRE‐H2B‐GFP mice and Fbln7 WT and KO mice were directly sorted into Trizol (Ambion, 10296028) and submitted to Tsukuba i‐Laboratory LLP, the University of Tsukuba, for further analysis. RNA integrity was analyzed with Agilent 2100 bioanalyzer. The RNA‐seq library was prepared with SMARTer® Stranded Total RNA‐Seq Kit v2 – Pico Input Mammalian (Takara), and sequencing was performed in the Next Generation Sequencing platform using NextSeq500 (Illumina). The data were analyzed with CLC genomics 11 software (Qiagen). After normalization, genes that showed ≥twofold changes were selected, and hierarchical clustering and GO analysis were performed using the online tools VENNY 2.1 (Oliveros, 2007–2015; https://bioinfogp.cnb.csic.es/tools/venny/index.html), Morpheus (https://software.broadinstitute.org/morpheus), Clustvis, Panther (www.geneontology.org; Ashburner et al, 2000; Mi et al, 2019a; Gene Ontology, 2021), and Metascape (Metsalu & Vilo, 2015; Zhou et al, 2019; Mi et al, 2019b).
Wholemount and section immunostaining of mouse tail epidermis
Tail skin wholemount and frozen section staining procedures were performed as previously described (Sada et al, 2016). The primary antibodies and dilutions used included the following: rat anti‐BrdU (1:300, Abcam, ab6326), rabbit anti‐K14 (1:1,000, BioLegend, 905304), mouse anti‐K10 (1:100, BioLegend, 904301, or 1:100, Abcam, ab9026), guinea pig anti‐K31 (1:100, PROGEN Biotechnik, GP‐hHa1), rabbit anti‐collagen IV (1:200, Millipore, AB756P), chicken anti‐GFP (1:500, AB13970), monoclonal mouse anti‐fibulin 7 (1:800), collagen XVII (1:500, Abcam, ab184996), rabbit anti‐laminin (1:100, abcam, ab11575), mouse anti‐DNA/RNA damage antibody (8‐oxo‐dG, 1:700, Abcam, ab62623), and rat anti‐Ki67 (1:100, eBioscience, 14‐5698‐82). Mouse on Mouse (M.O.M) kit reagents (Vector Laboratories) were added for blocking when a primary mouse antibody was used. For 8‐oxo‐dG staining, primary antibody incubation was performed at room temperature (RT) for 30 min. For fibulin 7 staining, skin sections were fixed in ice‐cold methanol for 10 min prior to blocking supplemented with goat F(ab) anti‐mouse IgG HL (1:10, Abcam, ab6668) overnight at 4°C. Incubation with fibulin 7 antibody conjugated to hilyte555 was done for 30 min at RT.
Secondary antibodies (Alexa 488, 546, 647, Invitrogen) were diluted at 1:200. All samples were counterstained with Hoechst nuclear stain (Sigma, B2261) before mounting. Before anti‐BrdU antibody incubation, samples were treated with 2 N HCl at 37°C for 1 h. The stained wholemount epidermis was observed under a confocal microscope (Zeiss LSM 700) and images were captured and analyzed using ZEN 2010 software. All wholemount pictures are shown as projected Z‐stack images, viewed from the basal side. A Zeiss Axio Imager Z2 fluorescence microscope was used for stained sections and images were acquired with Zen 2.3 Pro software with final adjustments in Adobe Photoshop.
Fibulin 7 antigen purification and hybridoma production
The human fibulin‐7 (FBLN7) coding sequence with N‐terminal 6xHN, streptavidin‐binding protein (SBP) and human rhinovirus 3C (3C) protease recognition sequences were inserted between the SBP tag and FBLN7, allowing removal of the N‐terminal tags from the purified protein, was cloned by PCR and ligated into the pEF6/V5‐His TOPO plasmid vector (Invitrogen). The 6HN‐SBP‐3C‐FBLN7 vector was transfected into CHO‐K1 cells (ATCC; CCL‐61) to express human fibulin 7 as an amino ‐terminally 6HN‐SBP tagged recombinant protein (6HN‐SBP‐3C‐FBLN7‐CHO‐K1).
6HN‐SBP‐FBLN7‐CHO‐K1 cells were maintained in a mixture of Dulbecco's‐modified Eagle's medium and Ham's F‐12 medium (DMEM/F12) (Invitrogen; Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS). Cells were grown to confluence and incubated in the serum‐free medium, and the resulting serum‐free conditioned medium (CM) was harvested every 2 days. The collected CM was concentrated by ammonium sulfate (Sigma). To perform 6HN‐SBP‐FBLN7 protein purification, the concentrated CM was subjected to Nickel, heparin and streptavidin sepharose columns. The final eluted FBLN7 was cleaved by 3C protease to remove N‐terminal tags.
Female Balb/c mice (6 weeks old) were inoculated subcutaneously with FBLN7 in Freund's complete adjuvant and then boosted three times at intervals of 2 weeks. Mice were euthanized 3 days after the final immunization and splenectomy was performed. To prepare splenocytes, a small hole was made in the spleen using 27G needle and the medium was injected into the spleen and cells were harvested. Myeloma cells (P3U1; P3X63Ag8U.1, ATCC [CRL‐1597]) maintained in RPMI1640‐10% FBS and the splenocyte were mixed and fused by polyethylene glycol (Sigma). Fused cells were selected by HAT (hypoxanthine–aminopterin–thymidine) containing medium. Hybridomas were monocloned and their supernatants were assayed against human and mouse fibulin 7 by ELISA. Hybridomas producing antibodies against fibulin 7 were maintained in RPMI1640‐10% FBS containing Briclone (NICB).
Hematoxylin and eosin staining
Tail skin was directly embedded in optimal cutting temperature compound (Tissue‐Tek, Sakura). Ten micrometre sections were fixed in 4% paraformaldehyde at RT for 10 min. Sections were stained with hematoxylin (Wako, 131‐09665) for 20 min and eosin Y (Wako, 058‐00062) for 15 s before dehydration and mounting in Entellan solution (Merck Millipore, HX73846161). Images were captured using a Zeiss Axio Imager Z2 microscope and Zen 2.3 Pro software.
Quantification and statistical analysis
All quantifications were independently performed on at least three mice. Blinded analysis was not performed due to the limited number of investigators. Data are shown as mean ± standard deviation (SD). In H&E‐stained tail sections, an epidermal unit was defined as the IFE region comprising one scale and one interscale structure, often between two HFs. The epidermal thickness was quantified from six epidermal units per mouse (Changarathil et al, 2019). The scale and interscale IFE thicknesses were measured at the center of an epidermal unit and the IFE region adjacent to the HF, respectively. K10/K31‐positive areas, the number of tdTomato positive cell clones, clonal areas, and BrdU+ cells were manually scored from 4 to 8 images consisting of 16–32 interscale or scale IFE structures per mouse in projected Z‐stack images using ImageJ software (NIH). Using the same software, K14 intensity ratio between suprabasal/basal cells; Collagen IV, laminin BM intensity, collagen XVII and fibulin 7 cell, and BM intensity were measured from five cells/structure/picture, and four pictures were used to obtain an average score per mouse. Ki67‐positive cells were scored from about 100 tdTomato‐positive cells in the scale regions of nine tail sections per mouse, whereas 8‐oxo‐dG‐positive cells were scored from analyzing about 50–90 scale/interscale structures per mouse. In wound healing quantification, re‐epithelialization length was measured from the end of the last intact epidermal unit until the end of the newly formed “epithelial tongue.” The thickness of each epithelial tongue was measured at three random sites along its length. At least two epithelial tongues were measured per mouse. Statistical tests are described in the respective figure legends. Each dot in all graphs represents mean value from each mouse.
Wound healing assay
Full‐thickness tail wounding was performed as described previously at 10 mm length × 3 mm width on the dorsal side of the tail (Liu et al, 2019). Ketoprofen (2 μg/g BW, Sigma) and amoxicillin (100 μg/g BW, Sigma) were intraperitoneally injected prior to tail cutting. After wound pressurization to stop bleeding, film spray dressing (Cavilon, 3M) was applied to the wound surface, and mice were monitored until recovery. To follow up on wound size over time, images were taken with a Zeiss Stemi305 microscope with an Axiocam208 camera every 7 or 8 days until day 32 post‐wounding, and the wound area was measured by ImageJ.
Lentivirus induction of Fbln7 overexpression in primary mouse keratinocytes
Isolation of 2‐day‐old WT C57BL/6J mouse primary keratinocytes was performed as previously reported (Lichti et al, 2008). Keratinocytes were cultured in low‐calcium (0.05 mM) E‐medium containing 15% chelex‐treated FBS and used for experiments from passage 7 to 15. Cell images were acquired using an Evos FL cell imaging system (Thermo Fischer Scientific).
Mouse FL or dCC mutant Fbln7 with V5‐His tag (Tsunezumi et al, 2018) was subcloned into a CSII‐CMV‐MCS‐IRES2‐Bsd vector (RIKEN, Tsukuba, Japan). The CSII‐CMV‐MCS‐IRES2‐Bsd vector (Fbln7 FL, dCC, or control eGFP) was transfected into 293T cells (ATCC; CRL‐3216) using polyethylenimine reagent (PEI, Polysciences Inc.) together with lentivirus packaging vectors (pRSV‐Rev, pMD2.G, and pMDLg/pRRE) (Addgene). Lentivirus‐containing medium was concentrated using Lenti‐X‐concentrator (Takara Bio). Six‐well plates were seeded with 100,000 primary keratinocytes per well and transduced with 150 μl each of lenti‐eGFP, lenti‐Fbln7, or lenti‐Fbln7 dCC and 4 μg/ml polybrene (Sigma). The medium was changed the next day, and cells were cultured for 2–3 days before selection with blasticidin at a concentration of 1 μg/ml.
Cell proliferation and differentiation assay
Primary keratinocytes were seeded at 2,000 cells/well in 96‐well plates with technical duplicates or triplicates. Experiments were repeated three times unless indicated otherwise in the figure legend. Cell proliferation was measured using Cell Counting Kit‐8 (CCK‐8, Dojindo) as previously described (Oinam et al, 2020). Recombinant mouse interleukin‐6 (R&D Systems, 406‐ML) was added to the culture medium at the indicated concentration. For differentiation assay, cells were seeded on plates pre‐coated with 50 μg/ml col IV (Sigma) and treated with 1.2 mM CaCl2 (Wako) for 24 h prior to cell lysis and RNA extraction.
Quantitative RT–PCR
RNA was isolated using RNeasy mini kit (Qiagen). cDNA was synthesized using iScript cDNA synthesis kit. RT–PCR was performed using iTaq Universal SYBR green supermix (Bio‐Rad). Mouse primer sequences are as follows: Klf5 forward (F) 5′‐GGCTCTCCCCGAGTTCACTA‐3′ and reverse (R) 5′‐ATTACTGCCGTCTGGTTTGTC‐3′; Krt14 F 5′‐AAGGTCATGGATGTGCACGAT‐3′ and R 5′‐CAGCATGTAGCAGCTTTAGTTCTTG‐3′; Krt1 F 5′‐AACCCGGACCCAAAACTTAG‐3′ and R 5′‐CCGTGACTGGTCACTCTTCA‐3′; Krt10 F 5′‐GGAGGGTAAAATCAAGGAGTGGTA‐3′ and R 5′‐TCAATCTGCAGCAGCACGTT‐3′; Lor F 5′‐TCACTCATCTTCCCTGGTGCTT‐3′ and R 5′‐GTCTTTCCACAACCCACAGGA‐3′; Dsc1 F 5′‐GGGAGCACCTTCTCTAAGCA‐3′ and R 5′‐CACTCTTCCAGATTCACTTTGCC‐3′; Fbln7 F 5′‐GAGGAGGCTTCCAGTGTGTC‐3′ and R 5′‐AATGGAAGGAGATGGTCTTGG‐3′; Gapdh F 5′‐TGCCCAGAACATCATCCCT‐3′ and R 5′‐GGTCCTCAGTGTAGCCCAAG‐3′.
Metal ion affinity chromatography and mass spectrometry
Mouse fibulin 7 (Fbln7) coding sequence (Tsunezumi et al, 2018) or human FBLN7 lacking the coiled‐coil domain (ΔCC) (Primers: F 5′‐GAACTGTCCAGATGCCCTTCCAGTT‐3′ and R 5′‐GCATCTGGACAGTTCTGGGAAGCCCG‐3′) was cloned by PCR and ligated into the pEF6/V5‐His TOPO plasmid vector (Invitrogen). FBLN7‐V5‐His vector was transfected into CHO‐K1 cells (FBLN7‐CHO‐K1) and ΔCC‐V5‐His vector was transfected into HEK293T cells (ΔCC‐293T) to express fibulin 7 as a COOH‐terminally V5‐His‐tagged recombinant protein. Both cell types were maintained in a mixture of (DMEM/F12) (Invitrogen) supplemented with 10% FBS.
FBLN7‐CHO‐K1 cells were grown to confluence and the resulting serum‐free CM was harvested every 2 days. The collected CM was concentrated by protein precipitation with 80% saturated ammonium sulfate (Sigma) and dissolved in PBS without calcium and magnesium [PBS (−)] supplemented with 0.1% Triton X‐100 and a proteinase inhibitor mixture [0.2 mM 4‐(2‐aminoethyl) benzenesulfonyl fluoride (AEBSF), 0.16 μM aprotinin, 0.025 mM bestatin, 7.5 μM E‐64, 0.01 mM leupeptin, and 5 μM pepstatin] (Calbiochem). The precipitants were dialyzed against a 400 times volume of PBS (−) plus Triton X‐100 and then applied to a Nickel (Ni)‐Sepharose Histidine‐affinity column (His GraviTrap; GE Healthcare). Bound proteins were eluted with PBS (−) supplemented with 500 mM NaCl, 500 mM imidazole, and 0.01% Triton X‐100. To exclude non‐specific bound proteins, the fractions containing FBLN7 were applied to a heparin‐Sepharose‐6B column (GE Healthcare). FBLN7 bound to the column and mainly eluted at 0.6 M NaCl. This fraction was further purified by Ni‐Sepharose column chromatography and the final elute fraction containing FBLN7 and its putative high‐affinity binding proteins was subjected to SDS–PAGE. Separated proteins were detected by staining with Coomassie Brilliant Blue R250.
293T and ΔCC‐293T cells were grown to confluence and the resulting serum‐free CM was harvested every 2 days. The collected CM was concentrated by protein precipitation with 80% saturated ammonium sulfate (Sigma) and dissolved in 50 mM Tris–HCl (pH 7.4) containing 150 mm NaCl supplemented with 5 mM CaCl2, 0.1% Triton X‐100, and a proteinase inhibitor mixture (as previously described). The precipitants were dialyzed against a 400 times volume of diluent buffer. The dialyzed 293T CM was passed through a Cobalt‐affinity column (ThermoFisher Scientific) once to eliminate non‐specific bound proteins, and the flow‐through fraction was collected and mixed with ΔCC, and then applied to the column chromatography. ΔCC and its affinity‐binding proteins bound to the column, and 1,200 mM NaCl was used to elute the proteins that were associated with ΔCC. The eluted samples were subjected to SDS–PAGE and separated proteins were visualized by Coomassie staining as previously described. The excised proteins from both preparations were submitted to the Proteomics Core at the University of Texas Southwestern Medical Center for analysis.
Solid‐phase binding assay
Ninety‐six‐well flat‐bottom MaxiSorp Nunc plates (Invitrogen) were coated with 1 μg/well of recombinant mouse periostin, Ccdc80 (R&D Systems), mouse fibulin 1C, fibulin 1D, human collagen IV (Sasaki et al, 1995), human tenascin C (Matsui et al, 2018), or BSA controls (Sigma) in bicarbonate buffer (15 mM Na2CO3, 35 mM NaHCO3, pH 9.2) at 4°C for 16 h. All incubations thereafter were performed at room temperature. After three washes with Ca2+‐Mg2+ free Dulbecco phosphate ‐buffered saline (DPBS, Gibco), wells were blocked with 5% non‐fat dry milk in Tris‐buffered saline (TBS) for 1 h. As a liquid phase, 0.03–3 μg/well of the soluble mouse or human HA‐tagged recombinant fibulin 7 (R&D Systems) in 2% dry milk/TBS and 2 mM CaCl2 were added to the wells and incubated for 3 h. Wells were then washed five times with wash buffer (TBS, 0.025% Tween‐20, 2 mM CaCl2). Anti‐HA antibody (3724S, Cell Signaling Technology) was diluted 1/1,500 in 2% dry milk TBS buffer and added to the wells for 1.5 h. After five washes, secondary anti‐rabbit antibody (170‐6515, Bio‐Rad) was applied at 1/1,000 dilution for 1 h, followed by another five washes. Colorimetric reaction was performed using Substrate Reagent Pack (DY999, R&D Systems) according to the manufacturer's protocol and absorbance was read at 450 nm using an xMark microplate reader (Bio‐Rad).
Author contributions
Erna Raja: Conceptualization; formal analysis; funding acquisition; validation; investigation; visualization; writing—original draft. Gopakumar Changarathil: Formal analysis; investigation. Lalhaba Oinam: Formal analysis; investigation. Jun Tsunezumi: Resources; formal analysis; funding acquisition; investigation. Yen Xuan Ngo: Resources; funding acquisition; investigation. Ryutaro Ishii: Investigation. Takako Sasaki: Resources. Kyoko Imanaka‐Yoshida: Resources. Hiromi Yanagisawa: Conceptualization; supervision; funding acquisition; project administration; writing—review and editing. Aiko Sada: Conceptualization; supervision; funding acquisition; investigation; visualization; project administration; writing—review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
PDF+
Acknowledgments
We thank the Animal Resource Center at the University of Tsukuba and the Center for Animal Resources and Development at Kumamoto University for their excellent mouse care, M. Higashi and T. Keida for technical help and M. Goodarzi and A. Lemoff and the Proteomics Core Laboratory at the University of Texas Southwestern Medical Center for proteomics data analysis. We would like to thank Dr. M. Kato, Dr. H. Suzuki, and Dr. Y. Watanabe (University of Tsukuba) for their help in lentivirus production. We also thank Dr. T. Suda (Kumamoto University) for critical reading of this manuscript and Dr. M. Muratani (University of Tsukuba) for his advice on RNA‐seq analysis. This work was supported by AMED‐PRIME, AMED (JP21gm6110016) (to AS), AMED under Grant Number 21bm0704067 (to AS), Grant‐in‐Aid for Scientific Research (B) (20H03266) (to AS), Grant‐in‐Aid for Scientific Research on Innovative Areas “Stem Cell Aging and Disease” (17H05631) (to AS), Grant‐in‐Aid for Research Activity Start‐up (16H06660) (to AS) and (20K22659) (to ER), and Grant‐in‐Aid for Early‐Career Scientists (18K14709) (to AS). This work was supported by the following research grants: The American Heart Association (12EIA8190000), The Mizutani Foundation for Glycoscience, The Uehara Memorial Foundation, and The Tokyo Biochemical Research Foundation to HY, and The Naito Foundation, The Nakajima Foundation, The Astellas Foundation for Research on Metabolic Disorders, The Uehara Memorial Foundation, The Inamori Foundation, The Takeda Science Foundation, The Koyanagi Foundation, The Nakatomi Foundation, The NOVARTIS Foundation (Japan) for the Promotion of Science, Leave a Nest Co., Ltd. for IKEDARIKA award, and Basic Research Support Program Type A, University of Tsukuba (all to AS), and the Cooperative Research Project Program of Life Science Center for Survival Dynamics, Tsukuba Advanced Research Alliance (TARA Center) (to AS and JT). This work was also supported by JST SPRING, Grant Number JPMJSP2124 (to YXN).
EMBO reports (2022) 23: e55478
Contributor Information
Hiromi Yanagisawa, Email: hkyanagisawa@tara.tsukuba.ac.jp.
Aiko Sada, Email: aisada@kumamoto-u.ac.jp.
Data availability
RNA‐sequencing data are deposited in the Gene expression omnibus (GEO) under accession numbers, GSE185086 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE185086) and GSE185087 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE185087). All raw mass spectrometry data files have been deposited to the Mass spectrometry Interactive Virtual Environment (MassIVE; Center for Computational Mass Spectrometry at the University of California, San Diego) and can be accessed using the MassIVE ID MSV000088142 (https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=a25ecf946b83414689b0bc9611d892e5).
References
- Abreu‐Velez AM, Howard MS (2012) Collagen IV in Normal skin and in pathological processes. N Am J Med Sci 4: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler A, Amitai‐Lange A, Tarazi N, Dey S, Strinkovsky L, Hadad‐Porat S, Bhattacharya S, Nasser W, Imeri J, Ben‐David G et al (2021) Discrete limbal epithelial stem cell populations mediate corneal homeostasis and wound healing. Cell Stem Cell 28: 1248–1261.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT et al (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos S, Stojadinovic O, Golinko MS, Brem H, Tomic‐Canic M (2008) Growth factors and cytokines in wound healing. Wound Repair Regen 16: 585–601 [DOI] [PubMed] [Google Scholar]
- Behrens A, van Deursen JM, Rudolph KL, Schumacher B (2014) Impact of genomic damage and ageing on stem cell function. Nat Cell Biol 16: 201–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changarathil G, Ramirez K, Isoda H, Sada A, Yanagisawa H (2019) Wild‐type and SAMP8 mice show age‐dependent changes in distinct stem cell compartments of the interfollicular epidermis. PLoS One 14: e0215908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chermnykh E, Kalabusheva E, Vorotelyak E (2018) Extracellular matrix as a regulator of epidermal stem cell fate. Int J Mol Sci 19: 1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vega S, Iwamoto T, Nakamura T, Hozumi K, McKnight DA, Fisher LW, Fukumoto S, Yamada Y (2007) TM14 is a new member of the fibulin family (fibulin‐7) that interacts with extracellular matrix molecules and is active for cell binding. J Biol Chem 282: 30878–30888 [DOI] [PubMed] [Google Scholar]
- Diamond I, Owolabi T, Marco M, Lam C, Glick A (2000) Conditional gene expression in the epidermis of transgenic mice using the tetracycline‐regulated transactivators tTA and rTA linked to the keratin 5 promoter. J Invest Dermatol 115: 788–794 [DOI] [PubMed] [Google Scholar]
- Doles J, Storer M, Cozzuto L, Roma G, Keyes WM (2012) Age‐associated inflammation inhibits epidermal stem cell function. Genes Dev 26: 2144–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egbert M, Ruetze M, Sattler M, Wenck H, Gallinat S, Lucius R, Weise JM (2014) The matricellular protein periostin contributes to proper collagen function and is downregulated during skin aging. J Dermatol Sci 73: 40–48 [DOI] [PubMed] [Google Scholar]
- Ermolaeva M, Neri F, Ori A, Rudolph KL (2018) Cellular and epigenetic drivers of stem cell ageing. Nat Rev Mol Cell Biol 19: 594–610 [DOI] [PubMed] [Google Scholar]
- Farrelly O, Suzuki-Horiuchi Y, Brewster M, Kuri P, Huang S, Rice G, Bae H, Xu J, Dentchev T, Lee V et al (2021) Two-photon live imaging of single corneal stem cells reveals compartmentalized organization of the limbal niche. Cell Stem Cell 28: 1233–1247.e1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatseva A, Sin YY, Brezzo G, Van Agtmael T (2019) Basement membrane collagens and disease mechanisms. Essays Biochem 63: 297–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Gomez NC, Adam RC, Nikolova M, Yang H, Verma A, Lu CP, Polak L, Yuan S, Elemento O et al (2017) Stem cell lineage infidelity drives wound repair and cancer. Cell 169: 636–650.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Miao Y, Gur‐Cohen S, Gomez N, Yang H, Nikolova M, Polak L, Hu Y, Verma A, Elemento O et al (2020) The aging skin microenvironment dictates stem cell behavior. Proc Natl Acad Sci USA 117: 5339–5350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology C (2021) The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res 49: D325–D334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghuwalewala S, Lee SA, Jiang K, Baidya J, Chovatiya G, Kaur P, Shalloway D, Tumbar T (2022) Binary organization of epidermal basal domains highlights robustness to environmental exposure. EMBO J 41: e110488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangreco A, Qin M, Pintar JE, Watt FM (2008) Epidermal stem cells are retained in vivo throughout skin aging. Aging Cell 7: 250–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez C, Chua W, Miremadi A, Quist S, Headon DJ, Watt FM (2013) The interfollicular epidermis of adult mouse tail comprises two distinct cell lineages that are differentially regulated by Wnt, Edaradd, and Lrig1. Stem Cell Reports 1: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales KAU, Fuchs E (2017) Skin and its regenerative powers: an Alliance between stem cells and their niche. Dev Cell 43: 387–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales KAU, Polak L, Matos I, Tierney MT, Gola A, Wong E, Infarinato NR, Nikolova M, Luo S, Liu S et al (2021) Stem cells expand potency and alter tissue fitness by accumulating diverse epigenetic memories. Science 374: eabh2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman RM, Krueger J, Yourish D, Granelli‐Piperno A, Murphy DP, May LT, Kupper TS, Sehgal PB, Gottlieb AB (1989) Interleukin 6 is expressed in high levels in psoriatic skin and stimulates proliferation of cultured human keratinocytes. Proc Natl Acad Sci USA 86: 6367–6371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Sakashita M, Yamada T, Fujieda S, Tanaka S, Doi S et al (2012) Genome‐wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nat Genet 44: 1222–1226 [DOI] [PubMed] [Google Scholar]
- Hu L, Mauro TM, Dang E, Man G, Zhang J, Lee D, Wang G, Feingold KR, Elias PM, Man MQ (2017) Epidermal dysfunction leads to an age‐associated increase in levels of serum inflammatory cytokines. J Invest Dermatol 137: 1277–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichijo R, Maki K, Kabata M, Murata T, Nagasaka A, Ishihara S, Haga H, Honda T, Adachi T, Yamamoto T et al (2022) Vasculature atrophy causes a stiffened microenvironment that augments epidermal stem cell differentiation in aged skin. Nature Aging 2: 592–600 [DOI] [PubMed] [Google Scholar]
- Ikeuchi T, de Vega S, Forcinito P, Doyle AD, Amaral J, Rodriguez IR, Arikawa‐Hirasawa E, Yamada Y (2018) Extracellular protein Fibulin‐7 and its C‐terminal fragment have In vivo antiangiogenic activity. Sci Rep 8: 17654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inomata K, Aoto T, Binh NT, Okamoto N, Tanimura S, Wakayama T, Iseki S, Hara E, Masunaga T, Shimizu H et al (2009) Genotoxic stress abrogates renewal of melanocyte stem cells by triggering their differentiation. Cell 137: 1088–1099 [DOI] [PubMed] [Google Scholar]
- Ishii R, Yanagisawa H, Sada A (2020) Defining compartmentalized stem cell populations with distinct cell division dynamics in the ocular surface epithelium. Development 147: dev197590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Liu Y, Yang Z, Nguyen J, Liang F, Morris RJ, Cotsarelis G (2005) Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med 11: 1351–1354 [DOI] [PubMed] [Google Scholar]
- Kato T, Liu N, Morinaga H, Asakawa K, Muraguchi T, Muroyama Y, Shimokawa M, Matsumura H, Nishimori Y, Tan LJ et al (2021) Dynamic stem cell selection safeguards the genomic integrity of the epidermis. Dev Cell 56: 3309–3320.e5 [DOI] [PubMed] [Google Scholar]
- Keyes BE, Liu S, Asare A, Naik S, Levorse J, Polak L, Lu CP, Nikolova M, Pasolli HA, Fuchs E (2016) Impaired epidermal to dendritic T cell signaling slows wound repair in aged skin. Cell 167: 1323–1338.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes BE, Segal JP, Heller E, Lien WH, Chang CY, Guo X, Oristian DS, Zheng D, Fuchs E (2013) Nfatc1 orchestrates aging in hair follicle stem cells. Proc Natl Acad Sci USA 110: E4950–E4959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koester J, Miroshnikova YA, Ghatak S, Chacon‐Martinez CA, Morgner J, Li X, Atanassov I, Altmuller J, Birk DE, Koch M et al (2021) Niche stiffening compromises hair follicle stem cell potential during ageing by reducing bivalent promoter accessibility. Nat Cell Biol 23: 771–781 [DOI] [PubMed] [Google Scholar]
- Kurbet AS, Hegde S, Bhattacharjee O, Marepally S, Vemula PK, Raghavan S (2016) Sterile inflammation enhances ECM degradation in integrin beta1 KO embryonic skin. Cell Rep 16: 3334–3347 [DOI] [PubMed] [Google Scholar]
- Kuwatsuka Y, Murota H (2019) Involvement of Periostin in skin function and the pathogenesis of skin diseases. Adv Exp Med Biol 1132: 89–98 [DOI] [PubMed] [Google Scholar]
- Lay K, Kume T, Fuchs E (2016) FOXC1 maintains the hair follicle stem cell niche and governs stem cell quiescence to preserve long‐term tissue‐regenerating potential. Proc Natl Acad Sci USA 113: E1506–E1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichti U, Anders J, Yuspa SH (2008) Isolation and short‐term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat Protoc 3: 799–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Matsumura H, Kato T, Ichinose S, Takada A, Namiki T, Asakawa K, Morinaga H, Mohri Y, De Arcangelis A et al (2019) Stem cell competition orchestrates skin homeostasis and ageing. Nature 568: 344–350 [DOI] [PubMed] [Google Scholar]
- Lopez‐Rovira T, Silva‐Vargas V, Watt FM (2005) Different consequences of beta1 integrin deletion in neonatal and adult mouse epidermis reveal a context‐dependent role of integrins in regulating proliferation, differentiation, and intercellular communication. J Invest Dermatol 125: 1215–1227 [DOI] [PubMed] [Google Scholar]
- Margadant C, Charafeddine RA, Sonnenberg A (2010) Unique and redundant functions of integrins in the epidermis. FASEB J 24: 4133–4152 [DOI] [PubMed] [Google Scholar]
- Marsh E, Gonzalez DG, Lathrop EA, Boucher J, Greco V (2018) Positional stability and membrane occupancy define skin fibroblast homeostasis in vivo. Cell 175: 1620–1633.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, Wedge DC, Fullam A, Alexandrov LB, Tubio JM et al (2015) Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348: 880–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y, Hasegawa M, Iino T, Imanaka‐Yoshida K, Yoshida T, Sudo A (2018) Tenascin‐C prevents articular cartilage degeneration in murine osteoarthritis models. Cartilage 9: 80–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura H, Mohri Y, Binh NT, Morinaga H, Fukuda M, Ito M, Kurata S, Hoeijmakers J, Nishimura EK (2016) Hair follicle aging is driven by transepidermal elimination of stem cells via COL17A1 proteolysis. Science 351: aad4395 [DOI] [PubMed] [Google Scholar]
- McCabe MC, Hill RC, Calderone K, Cui Y, Yan Y, Quan T, Fisher GJ, Hansen KC (2020) Alterations in extracellular matrix composition during aging and photoaging of the skin. Matrix Biol Plus 8: 100041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metsalu T, Vilo J (2015) ClustVis: a web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res 43: W566–W570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD (2019a) PANTHER version 14: more genomes, a new PANTHER GO‐slim and improvements in enrichment analysis tools. Nucleic Acids Res 47: D419–D426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Huang X, Ebert D, Mills C, Guo X, Thomas PD (2019b) Protocol update for large‐scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat Protoc 14: 703–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midwood KS, Chiquet M, Tucker RP, Orend G (2016) Tenascin‐C at a glance. J Cell Sci 129: 4321–4327 [DOI] [PubMed] [Google Scholar]
- Nakamura‐Ishizu A, Ito K, Suda T (2020) Hematopoietic stem cell metabolism during development and aging. Dev Cell 54: 239–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoloudaki G, Creber K, Hamilton DW (2020) Wound healing and fibrosis: a contrasting role for periostin in skin and the oral mucosa. Am J Physiol Cell Physiol 318: C1065–C1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oinam L, Changarathil G, Raja E, Ngo YX, Tateno H, Sada A, Yanagisawa H (2020) Glycome profiling by lectin microarray reveals dynamic glycan alterations during epidermal stem cell aging. Aging Cell 19: e13190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasmatzi E, Papadionysiou C, Monastirli A, Badavanis G, Tsambaos D (2019) Galectin 1 in dermatology: current knowledge and perspectives. Acta Dermatovenerol Alp Pannonica Adriat 28: 27–31 [PubMed] [Google Scholar]
- Quigley DA, Kandyba E, Huang P, Halliwill KD, Sjolund J, Pelorosso F, Wong CE, Hirst GL, Wu D, Delrosario R et al (2016) Gene expression architecture of mouse dorsal and tail skin reveals functional differences in inflammation and cancer. Cell Rep 16: 1153–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacma M, Pospiech J, Bogeska R, de Back W, Mallm JP, Sakk V, Soller K, Marka G, Vollmer A, Karns R et al (2019) Haematopoietic stem cells in perisinusoidal niches are protected from ageing. Nat Cell Biol 21: 1309–1320 [DOI] [PubMed] [Google Scholar]
- Sada A, Jacob F, Leung E, Wang S, White BS, Shalloway D, Tumbar T (2016) Defining the cellular lineage hierarchy in the interfollicular epidermis of adult skin. Nat Cell Biol 18: 619–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarangi PP, Chakraborty P, Dash SP, Ikeuchi T, de Vega S, Ambatipudi K, Wahl L, Yamada Y (2018) Cell adhesion protein fibulin‐7 and its C‐terminal fragment negatively regulate monocyte and macrophage migration and functions in vitro and in vivo. FASEB J 32: 4889–4898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Kostka G, Gohring W, Wiedemann H, Mann K, Chu ML, Timpl R (1995) Structural characterization of two variants of fibulin‐1 that differ in nidogen affinity. J Mol Biol 245: 241–250 [DOI] [PubMed] [Google Scholar]
- Seldin L, Macara IG (2020) DNA damage promotes epithelial hyperplasia and fate mis‐specification via fibroblast inflammasome activation. Dev Cell 55: 558–573.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strafella C, Caputo V, Minozzi G, Milano F, Arcangeli M, Sobhy N, Abdelmaksood R, Hashad D, Vakirlis E, Novelli G et al (2019) Atopic eczema: genetic analysis of COL6A5, COL8A1, and COL10A1 in Mediterranean populations. Biomed Res Int 2019: 3457898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K, Arima K, Masuoka M, Ohta S, Shiraishi H, Ontsuka K, Suzuki S, Inamitsu M, Yamamoto KI, Simmons O et al (2014) Periostin controls keratinocyte proliferation and differentiation by interacting with the paracrine IL‐1alpha/IL‐6 loop. J Invest Dermatol 134: 1295–1304 [DOI] [PubMed] [Google Scholar]
- Theocharidis G, Drymoussi Z, Kao AP, Barber AH, Lee DA, Braun KM, Connelly JT (2016) Type VI collagen regulates dermal matrix assembly and fibroblast motility. J Invest Dermatol 136: 74–83 [DOI] [PubMed] [Google Scholar]
- Tremblay F, Revett T, Huard C, Zhang Y, Tobin JF, Martinez RV, Gimeno RE (2009) Bidirectional modulation of adipogenesis by the secreted protein Ccdc80/DRO1/URB. J Biol Chem 284: 8136–8147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunezumi J, Sugiura H, Oinam L, Ali A, Thang BQ, Sada A, Yamashiro Y, Kuro OM, Yanagisawa H (2018) Fibulin‐7, a heparin binding matricellular protein, promotes renal tubular calcification in mice. Matrix Biol 74: 5–20 [DOI] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E (2004) Defining the epithelial stem cell niche in skin. Science 303: 359–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumpel S, Rudolph KL (2019) Quiescence: good and bad of stem cell aging. Trends Cell Biol 29: 672–685 [DOI] [PubMed] [Google Scholar]
- Wang L, Siegenthaler JA, Dowell RD, Yi R (2016) Foxc1 reinforces quiescence in self‐renewing hair follicle stem cells. Science 351: 613–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kitahata H, Kosumi H, Watanabe M, Fujimura Y, Takashima S, Osada SI, Hirose T, Nishie W, Nagayama M et al (2022) Collagen XVII deficiency alters epidermal patterning. Lab Invest 102: 581–588 [DOI] [PubMed] [Google Scholar]
- Watanabe M, Kosumi H, Osada SI, Takashima S, Wang Y, Nishie W, Oikawa T, Hirose T, Shimizu H, Natsuga K (2021) Type XVII collagen interacts with the aPKC‐PAR complex and maintains epidermal cell polarity. Exp Dermatol 30: 62–67 [DOI] [PubMed] [Google Scholar]
- Watanabe M, Natsuga K, Nishie W, Kobayashi Y, Donati G, Suzuki S, Fujimura Y, Tsukiyama T, Ujiie H, Shinkuma S et al (2017) Type XVII collagen coordinates proliferation in the interfollicular epidermis. Elife 6: e26635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt FM (2002) Role of integrins in regulating epidermal adhesion, growth and differentiation. EMBO J 21: 3919–3926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt FM, Fujiwara H (2011) Cell‐extracellular matrix interactions in normal and diseased skin. Cold Spring Harb Perspect Biol 3: a005124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Wang D, Wang J, Wang L, Qiu W, Kume T, Dowell R, Yi R (2021) Escape of hair follicle stem cells causes stem cell exhaustion during aging. Nat Aging 1: 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK (2019) Metascape provides a biologist‐oriented resource for the analysis of systems‐level datasets. Nat Commun 10: 1523 [DOI] [PMC free article] [PubMed] [Google Scholar]


