Abstract
The central melanocortin system is fundamentally important for controlling food intake and energy homeostasis. Melanocortin-3 receptor (MC3R) is one of two major receptors of the melanocortin system found in the brain. In contrast to the well-characterized melanocortin-4 receptor (MC4R), little is known regarding the organization of MC3R-expressing neural circuits. To increase our understanding of the intrinsic organization of MC3R neural circuits, identify specific differences between males and females, and gain a neural systems level perspective of this circuitry, we conducted a brain-wide mapping of neurons labeled for MC3R and characterized the distribution of their projections. Analysis revealed MC3R neuronal and terminal labeling in multiple brain regions that control a diverse range of physiological functions and behavioral processes. Notably, dense labeling was observed in the hypothalamus, as well as areas that share considerable connections with the hypothalamus, including the cortex, amygdala, thalamus, and brainstem. Additionally, MC3R neuronal labeling was sexually dimorphic in several areas, including the anteroventral periventricular area, arcuate nucleus, principal nucleus of the bed nucleus of the stria terminalis, and ventral premammillary region. Altogether, anatomical evidence reported here suggests that MC3R has the potential to influence several different classes of motivated behavior that are essential for survival, including ingestive, reproductive, defensive, and arousal behaviors, and is likely to modulate these behaviors differently in males and females.
Keywords: clearing, lightsheet, M3R, melanocortin, RNAscope, sex differences
1 |. INTRODUCTION
The central melanocortin system is fundamentally important for controlling energy balance and maintaining homeostasis. This system consists of four endogenous agonists, α-melanocyte-stimulating hormone (MSH), β-MSH, γ-MSH, and adrenocorticotropin (ACTH), which are derived from the preprohormone, proopiomelanocortin (POMC). Agouti-related protein (AgRP) is uniquely expressed in neurons in the arcuate hypothalamic nucleus (ARH) that also contain neuropeptide Y and functions as an endogenous antagonist for melanocortin-3 receptor (MC3R) and melanocortin-4 receptors (MC4R), which are the predominant melanocortin receptors expressed in the brain (Cone, 2006; Garfield et al., 2009; Yang & Tao, 2016).
While there is abundant evidence that MC4R plays a determinant role in the central regulation of food intake and energy homeostasis (Butler et al., 2001; Cone, 2006; Fan et al., 2000, 2004; Huszar et al., 1997; Morens et al., 2005), the homeostatic functions of MC3R remain to be thoroughly characterized. MC3R KO mice exhibit late onset obesity (Butler et al., 2000), reduced lean mass (Butler et al., 2000; A. S. Chen et al., 2000; Renquist et al., 2011), heightened diet-induced obesity linked to dysfunction in physical activity and nutrient partitioning (A. S. Chen et al., 2000; Ellacott et al., 2007; G. M. Sutton et al., 2006;), disrupted daily feeding patterns (Butler, 2006), and decreased locomotor activity (Begriche et al., 2011; Butler et al., 2000; A. S. Chen et al., 2000; Ellacott et al., 2007; Pei et al., 2019; G. M. Sutton et al., 2006, 2008). Increased adipose mass and reduced lean mass have also been observed in humans with reduced or absent MC3R expression (Lam et al., 2021). Recently, MC3R has also been implicated in the bidirectional control of responses to homeostatic challenges, providing rheostatic control on energy storage (Ghamari-Langroudi et al., 2018) and is a potential pharmacological target for the treatment of anorexia (Sweeney et al., 2021). Additionally, MC3R has been implicated in the regulatory control of the hypothalamic-pituitary-thyroid axis (Kim et al., 2002), inflammation (Catania et al., 2004; Getting et al., 2008), cardiovascular function (Mioni et al., 2003; Versteeg et al., 1998), glucose disposal (A. K. Sutton et al., 2021), natriuresis (Chandramohan et al., 2009), and reward (Lippert et al., 2014; Mavrikaki et al., 2016; Pandit et al., 2016; Shanmugarajah et al., 2017; West et al., 2019).
While it is clearly evident that MC3R modulates critical physiological and behavioral processes, little is known regarding the organization of MC3R-expressing neural circuits regulating these processes. The overall distribution of Mc3r mRNA in the rat brain has been reported (Roselli-Rehfuss et al., 1993), but a detailed mapping of MC3R-expressing neurons or a detailed examination of their neural projections is lacking.
To increase our understanding of the intrinsic organization of MC3R neural circuits, characterize differences that may exist between males and females, and gain a neural systems level perspective of this circuitry, we have undertaken a brain-wide mapping of neurons labeled for MC3R and their projections through the application of immunohistochemistry, genetically targeted axonal labeling, tissue clearing, light-sheet microscopy, and registration of labeled neurons to the Allen Brain Atlas Common Coordinate Framework.
2 |. MATERIALS AND METHODS
2.1 |. Animals
Transgenic BAC mice expressing Cre recombinase under the control of the Mc3r promoter, MC3R-Cre mice, are maintained in our colony at Vanderbilt University (Ghamari-Langroudi et al., 2018). Mice expressing the Cre-dependent fluorescent reporters tdTomato (tdTom mice; Ai14D-Gt(Rosa)26Sor; stock number: 007914) and synaptophysin-tdTomato (SynTom mice; Ai34D-Rosa-CAG-LSL-Synaptophysin-tdTomato-WPRE; stock number: 012570; MGI:J:170755) were obtained from The Jackson Laboratory (Bar Harbor, ME). Wild-type C57BL/6J mice (stock number: 000664) were also obtained from The Jackson Laboratory. To visualize neurons that express MC3R, MC3R-Cre mice were crossed with tdTom mice to generate MC3R-Cre::tdTom mice. To visualize MC3R inputs, MC3R-Cre mice were crossed with SynTom mice to generate MC3R-Cre::SynTom mice, as described previously (Biddinger et al., 2020).
All animal care and experimental procedures were performed in accordance with the guidelines of the National Institutes of Health and the Institutional Care and Use Committee of Vanderbilt University. Mice were housed at 22°C on a 12:12 h light:dark cycle (lights on at 6:00 a.m.: lights off at 6:00 p.m.). Mice were provided ad libitum access to a standard chow diet (PicoLab Rodent Diet 20 #5053). Mice were weaned at P22 and maintained with mixed genotype littermates until used for experiments.
2.2 |. Immunohistochemistry, image acquisition, and analysis
2.2.1 |. MC3R neuronal distribution and postsynaptic target visualization
To visualize MC3R neuronal labeling as well as MC3R neural inputs, brains from adult MC3R-Cre::tdTomato and MC3R-Cre::SynTom mice were collected and processed for immunofluorescence. Mice were anesthetized with tribromoethanol and transcardially perfused with saline followed by fixative (4% paraformaldehyde in borate buffer, pH 9.5). Brains were postfixed in a solution of 20% sucrose in fixative and cryoprotected in 20% sucrose in 0.2 M potassium phosphate buffered saline (KPBS). Four series of 30 μm-thick frozen sections were collected using a sliding microtome. One full series of sections from each animal was rinsed in KPBS and blocked in 2% normal goat serum containing 0.3% Triton X-100 overnight at 4°C. Sections were then incubated in primary antibody (rabbit anti-RFP, 1:10,000; Rockland Immunochemicals Inc., Limerick, PA, USA) for 48 h at 4°C. Following primary antibody incubation, sections were rinsed several times in KPBS and incubated in Alexa 568 goat anti-rabbit secondary antibody (1:500; Life Technologies, Carlsbad, CA, USA) for 1 h at room temperature. Sections were rinsed several times in KPBS and incubated in NeuroTrace 500/524 Green fluorescent Nissl stain (1:500; ThermoFisher Scientific, Waltham, MA, USA) for 20 min at room temperature to aid in visualization of brain cytoarchitecture. Sections were rinsed several times in KPBS, mounted on gelatin-subbed slides and coverslipped using ProLong mounting medium (Life Technologies).
Images from each animal were obtained using a laser scanning confocal microscope (Zeiss LSM 800; Zeiss, Oberkochen, Germany). For digital maps, confocal image stacks were collected through the z-axis at a frequency of 10.00 μm using a 10× objective (NA 0.45). A panoramic tile of a coronal hemisection was captured using the image stitching module in Zen Blue. A tile of the Nissl stain for each section was flipped horizontally in Photoshop (version 20.0.9; Adobe, San Jose, CA, USA) to mirror MC3R labeling in the same section so that cytoarchitectonic features and MC3R neuronal and fiber labeling could be observed simultaneously. Image stacks captured to compare neuronal/fiber labeling, density, and distribution were collected through the z-axis at a frequency of 3.41 μm using a 10× objective (NA 0.45) or 0.8 μm using a 20× objective (NA 0.8).
2.2.2 |. Sexually dimorphic distributions of MC3R neurons
To determine whether MC3R labeling is sexually dimorphic in specific anatomical areas, brains from male and female adult MC3R-Cre::tdTomato mice were collected and processed for immunofluorescence. Tissue was collected and sectioned as described above. Sections containing the anteroventral periventricular area (AVPV), principal nucleus of the bed nucleus of the stria terminalis (BSTPr), ventromedial hypothalamus (VMH), ventrolateral-ventromedial hypothalamus (VL-VMH), arcuate nucleus (ARH), posterodorsal nucleus of the medial amygdala (MEApd), and ventral premammillary nucleus (PMv) were selected and processed as described above.
For a quantitative comparison of cell densities in male and female mice, confocal image stacks were collected through the z-axis at a frequency of 0.8 μm using a 20× objective (NA 0.8). Cytoarchitectonic features of each specific anatomical area analyzed, visualized with the NeuroTrace fluorescent Nissl stain, were used to define matching regions of interest (ROI), and three-dimensional representations of labeled cells from matched sections were digitally rendered using Imaris software (version 9.5.1; Bitplane, Zurich, Switzerland). The total number of MC3R neurons in each area was quantified using the spots function in Imaris.
2.3 |. Tissue clearing and light-sheet microscopy
2.3.1 |. Tissue collection, preparation, and clearing
To spatially map MC3R labeling in intact tissue, brains derived from MC3R-Cre::tdTomato and MC3R-Cre::SynTom mice were stabilized and cleared using stabilization under harsh conditions via intramolecular epoxide linkages to prevent degradation (SHIELD) protocol (Park et al., 2019). Mice were anesthetized with tribromoethanol and transcardially perfused with saline followed by fixative (4% paraformaldehyde in borate buffer, pH 9.5). Brains were incubated in fixative overnight and then transferred to SHIELD-OFF solution (LifeCanvas Technologies, Cambridge, MA, USA) and incubated at 4°C for 4 days. Brains were next incubated in SHIELD-ON solution (LifeCanvas Technologies) for 24 h at 37°C. SHIELD-fixed brains were rapidly cleared using stochastic electrotransport to remove electromobile molecules (SmartClear II Pro; LifeCanvas Technologies; limit = 90 V, current = 1500 mA, buffer A temp. = 42°C, buffer B temp. = 30–40°C). Once the samples were delipidated, the tissue was incubated in EasyIndex (LifeCanvas Technologies) to render the sample optically transparent for imaging.
2.3.2 |. Imaging and data processing
SHIELD brains were imaged using an axially swept light-sheet microscope (SmartSPIM; LifeCanvas Technologies) equipped with a 3.6×objective (NA 0.2, uniform axial resolution ~4 μm) and sCMOS camera with a rolling shutter (Dean et al., 2015; Hedde & Gratton, 2018). Following acquisition, images were stitched to generate composite TIFF images using a modified version of Terastitcher (Bria et al., 2019). Stitched TIFF images were converted to Imaris files using Imaris File Converter 9.2.1, and 3D renderings of brains were visualized using Imaris software.
2.4 |. Brain registration and volumetric quantification of MC3R neurons and fibers
The distribution of neurons in SHIELD-cleared MC3R-Cre::tdTomato brains was registered to the Allen Common Coordinate Reference Framework (ACCF, V3), and regional densities of labeled cells were quantified through the application of machine learning filters developed from a user-classified training set using NeUroGlancer Ground Truth (NUGGT), with custom modifications provided by LifeCanvas Technologies (Swaney et al., 2019). A minimum of 200 cells per brain were trained using the NUGGT pipeline. Registration to the ACCF was also used to define the regional mean fluorescence intensity of labeled fibers in brains collected from MC3R-Cre::SynTom mice.
2.5 |. RNAscope fluorescence in situ hybridization
To visualize the overall Mc3r mRNA abundance and compare it to MC3R-Cre::tdTomato expression, an RNAscope fluorescence multiplex assay (V2, Advanced Cell Diagnostics, Newark, CA, USA) was used. MC3R-Cre::tdTomato mice were anesthetized with tribromoethanol and transcardially perfused with saline followed by fixative (4% paraformaldehyde in borate buffer, pH 9.5). Brains were postfixed in a solution of 20% sucrose in fixative and cryoprotected in 20% sucrose in KPBS. Four series of 20 μm-thick frozen sections were collected using a sliding microtome. One series of sections from each animal were mounted onto SuperFrost Plus slides (Fisher Scientific), and in situ hybridization was performed according to the RNAscope fluorescent multiplex kit V2 user manual for fixed frozen tissue (Advanced Cell Diagnostics,) using RNAscope Probe-Mm-Mc3r-C1 (cat# 412541). Following in situ hybridization, slides were run through the same immunohistochemistry protocol described above. Slides were coverslipped using ProLong mounting medium (Life Technologies). Images from each animal were obtained using a laser scanning confocal microscope (Zeiss LSM 800). Confocal image stacks were collected through the z-axis at a frequency of 0.41 μm using a 40× objective (NA 1.40).
2.6 |. Experimental design and statistical analyses
Group data are presented as the mean values ± SEM. Statistical significance was determined using GraphPad Prism software (GraphPad Software, San Diego, CA). Student’s unpaired t-test was used to compare data within two groups. p-Values < .05 were considered statistically significant.
3 |. RESULTS
To validate the accuracy of the genetically targeted label, we used fluorescence in situ hybridization (FISH) combined with immunohistochemistry to compare the distribution of neurons that express Mc3r mRNA with that of labeled neurons in MC3R-Cre::tdTomato mice (Figure 1a–i). Particular attention was given to regions that contained neurons with tdTomato labeling but in which MC3R expression had not been reported previously (Figure 1a,b, e–g). The distribution of neurons labeled with tdTomato fluorescence matched that of Mc3r mRNA expression (Figure 1), although we cannot rule out the possibility that in some cells, MC3R-Cre expression is transient.
FIGURE 1.
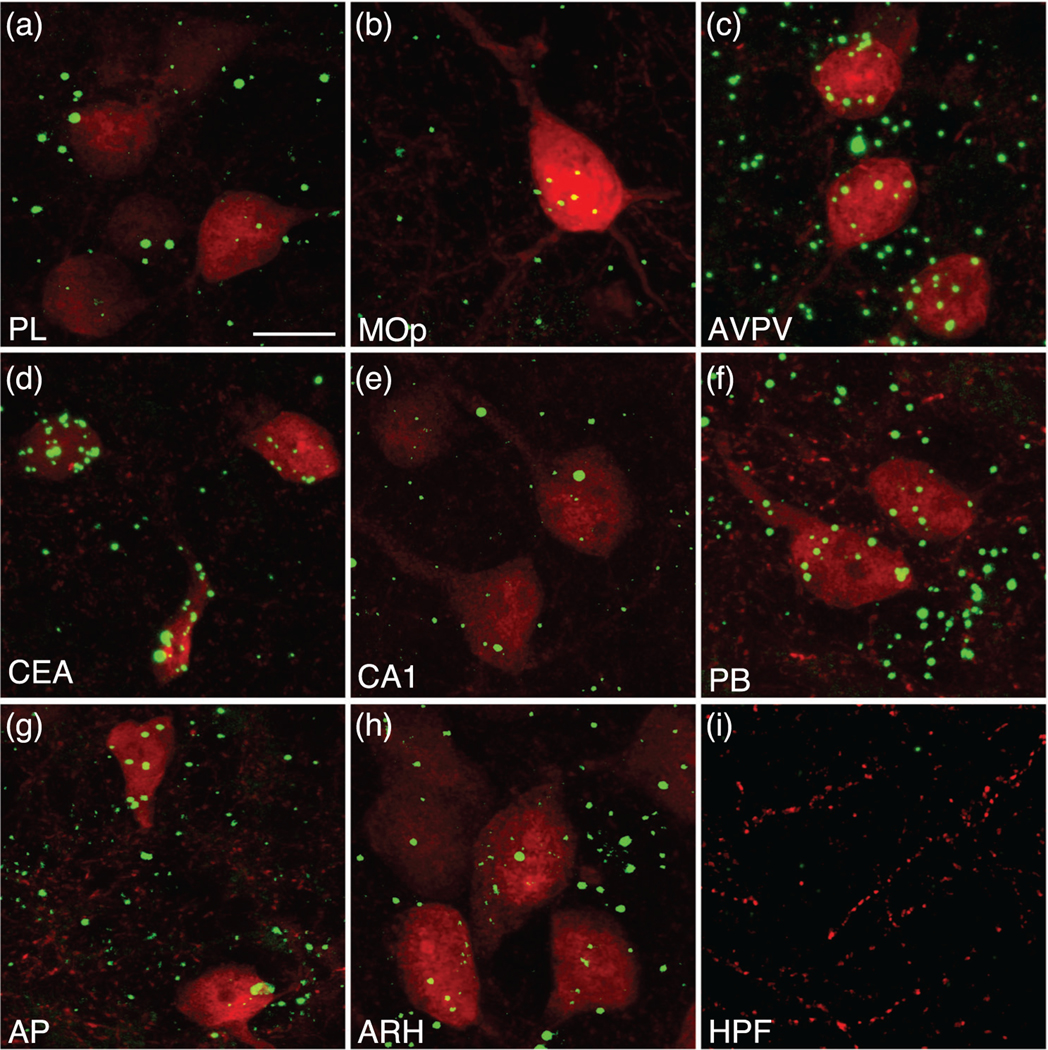
Analysis of Mc3r mRNA colocalization in melanocortin-3 receptor (MC3R) cells labeled with tdTomato in the mouse brain. High magnification images of Mc3r mRNA (green) and tdTomato labeling (red) in an MC3R-Cre::tdTomato mouse in major brain regions, including the cortex (a and b), hypothalamus (c), amygdala (d), hippocampus (e), midbrain (f), and brainstem (g). Mc3r abundance in the arcuate (h) and hippocampal formation (i) serve as anatomical areas exhibiting high (arcuate) and little to no (hippocampal formation) Mc3r expression. Estimates of coexpression of Mc3r mRNA in tdTomato-labeled cells measured in representative sections of male mice was 90% in the prelimbic area (PL) (a), 100% in the primary motor area (Mop) (b), anteroventral periventricular nucleus hypothalamus (AVPV) (c), central nucleus amygdala (CEA) (d), CA1 (e), parabrachial nucleus (PB) (f), and area postrema (AP) (g), and 94% in the arcuate nucleus hypothalamus (ARH) (h). Scale bar, 20 μm
3.1 |. Distribution of cell bodies labeled for MC3R
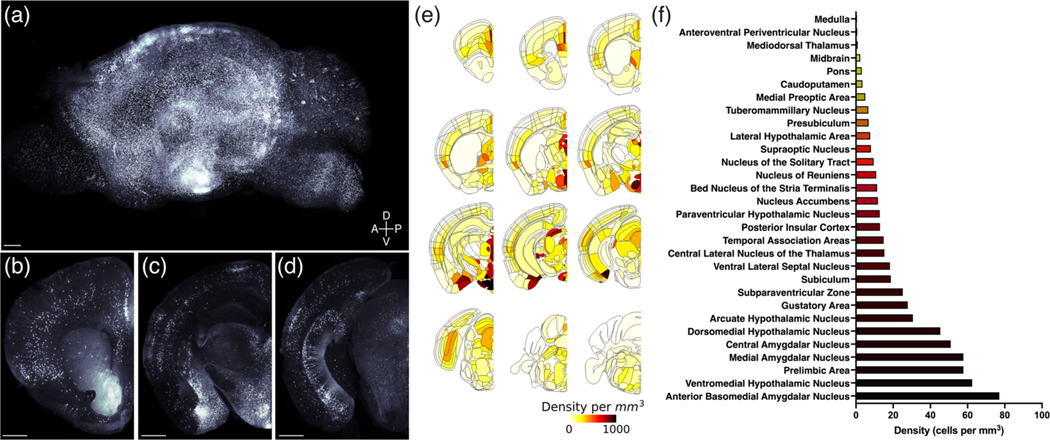
The organization of MC3R neurons was visualized in both intact brains through application of optical clearing and light-sheet microscopy (Figure 2, Videos S1–S3) as well as in tissue sections derived from MC3R-Cre::tdTom mice (Kamitakahara et al., 2016; Biddinger et al., 2020). Visualization, (Figure 2a–d, Videos S1–S3) brain registration, and quantification of MC3R neurons in whole brain tissue (Figure 2e,f) facilitated appreciation of the spatial context of systems-level molecular and anatomical features and underscored the complexity of anatomical relationships between MC3R neurons in distinct brain areas. The distribution of tdTomato-labeled neuronal cell bodies in the brains of MC3R::tdTom mice extends from the rostral pole of the cerebral cortex to the caudal brainstem, and unambiguously labeled neurons were located in each major subdivision of the central nervous system (Figures 1 and 2a–i, Videos S1–S3). A striking feature of MC3R neuronal labeling is that high densities of neurons and equally profuse labeling of their targets involve brain regions that not only impact energy balance but also contribute to a diverse range of well-characterized physiological functions (Figures 2 and 3). However, the highest densities of labeled neurons were located in the hypothalamus and in regions that shared strong connections with the hypothalamus.
FIGURE 2.
Brain-wide mapping of melanocortin-3 receptor (MC3R) neurons. Three-dimensional rendering of an MC3R-Cre::tdTomato brain viewed in the sagittal plane (a). Scale bar, 500 μm. Coronal view of MC3R labeling in the same brain at the level of the prelimbic and insular cortex (b), arcuate nucleus, ventromedial hypothalamus and basomedial, central and medial amygdala (c), and subiculum and periaqueductal gray (d). Scale bars, 500 μm. MC3R cell density in the whole brain was quantified by registering cell locations to the Allen Common Coordinate Reference Framework (e and f)
FIGURE 3.
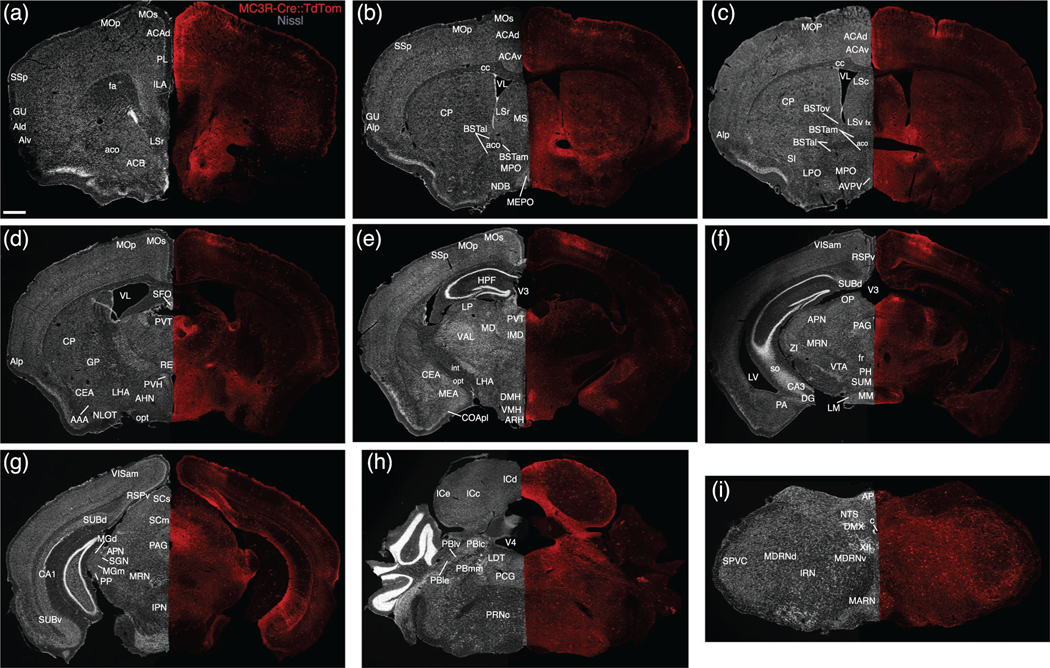
Two-dimensional atlas of MC3R neurons. Low magnification stitched tile images of melanocortin-3 receptor (MC3R) labeling in an MC3R-Cre::tdTomato brain (a–i). Scale bar, 1 mm
3.1.1 |. Hypothalamus
Most hypothalamic nuclei contained at least a few labelled neurons with the notable exception of the suprachiasmatic nucleus, and a diffuse distribution of labeled cells was observed throughout the hypothalamus, with clusters of neurons in discrete nuclei (Figures 2a,c,e,f, and 3b–f, Videos S1–S3). The greatest density of labeled neurons was present within nuclei located in the periventricular and medial zones of the hypothalamus (Simerly, 2005). The medial preoptic nucleus contained a high density of labeled neurons (Figures 2f and 3b–c, Video S2), especially the medial component, but the density of labeled neurons was somewhat lower in the anterior hypothalamic nucleus (Figure 3d, Video S2). Caudal to the anterior hypothalamic region, the overall density of labeled neurons decreased, except in discrete nuclear cell groups (Video S2). The arcuate nucleus, ventromedial hypothalamus and dorsomedial hypothalamus contained the highest densities of labeled neurons (Figures 2c,e,f and 3e, Video S2), whereas fewer labeled cells were located in the paraventricular nucleus of the hypothalamus (Figures 2e,f and 3d, Video S2). Intermediate densities of labeled neurons were observed in the lateral preoptic (Figures 2e and 3c, Video S2) and hypothalamic areas (Figures 2c,e,f and 3d,e, Video S2), and in the ventral premammillary nucleus (Figure 2e, Video S2). Fewer cells were detected in the dorsal premammillary nucleus (Figure 2e, Video S2).
3.1.2 |. Amygdala
Labeling in the amygdala was extensive. At rostral levels through the amygdala, scattered labeled neurons were distributed throughout the anterior area of the amygdala (Figures 2e and 3d, Video S2), excluding the nucleus of the lateral olfactory tract (Figures 2e and 3d, Video S2). The highest densities of labeled neurons in the amygdala were found in the medial (especially its posterodorsal part) and basomedial nuclei (Figures 2c,e,f and 3e, Video S2). Abundant labeling was also present in the central amygdalar nucleus, where a majority of cells were localized to the lateral subdivision (Figures 2c,e,f and 3d,e, Video S2). Scattered neurons with light to moderate labeling were observed in select cortical amygdalar nuclei, including the lateral zone of the posterior part of the cortical amygdalar nucleus (Figures 2e and 3e, Video S2). Only low densities of labeled neurons were detected in the lateral, basolateral, or posterior amygdalar nuclei (Figures 2e, 3d–f, and 4a, Video S2).
FIGURE 4.
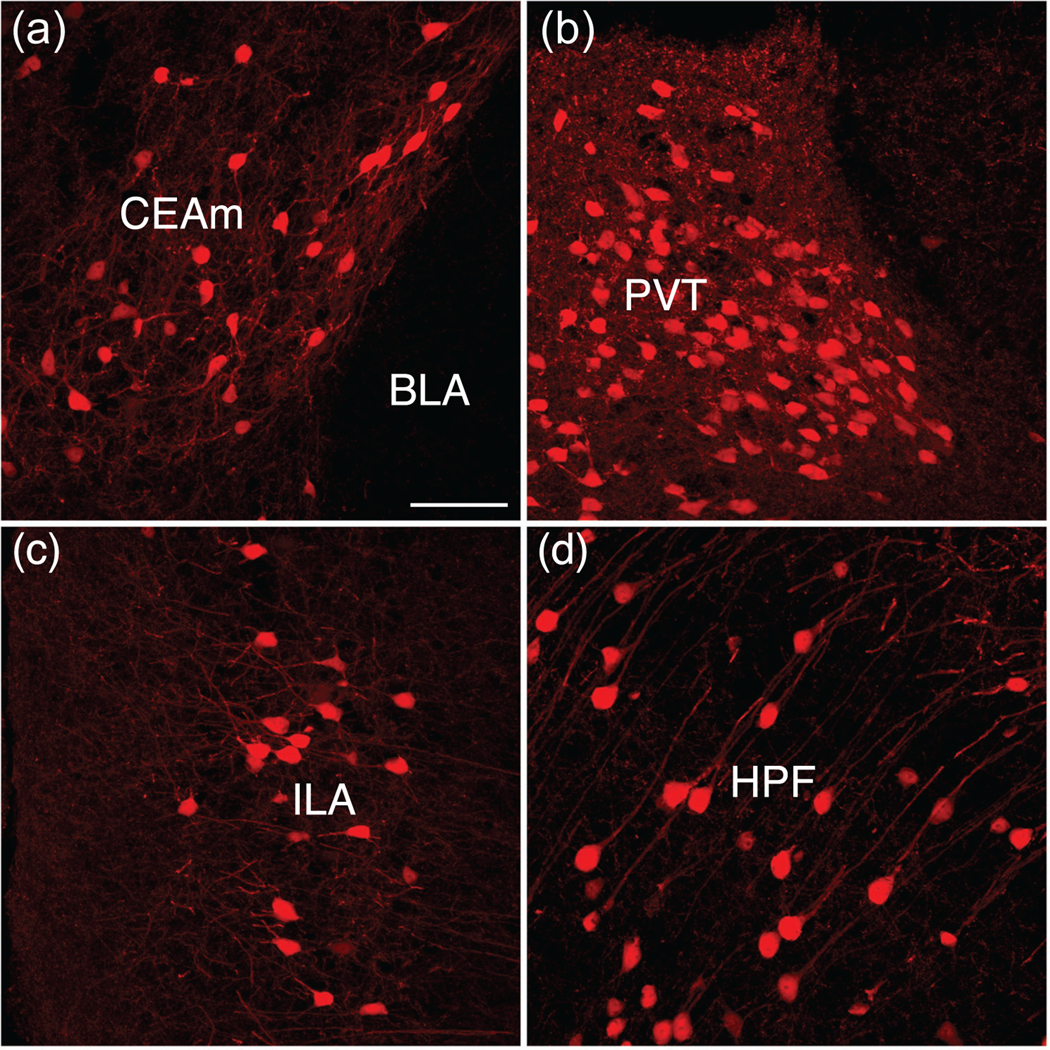
Comparison of melanocortin-3 receptor (MC3R) cellular architecture, morphology, and distribution. Prominent MC3R labeling in the central amygdala juxtaposed with the lack of labeling in the adjacent basolateral amygdala (a). Dense cellular and fiber labeling in the paraventricular thalamus (b). MC3R expression in layer 2/3 of the infralimbic area (c). Pyramidal cells in the pyramidal layer of CA1 in the hippocampus (d). Scale bar, 100 μm
3.1.3 |. Septal nuclei, bed nuclei of the stria terminalis, and pallidum
The septal nuclei contained clusters of labeled neurons with heavily labeled dendrites extending from the cell bodies (Figure 3b,c). In contrast to the low density of neurons in the medial septal nucleus (Figures 2e and 3b,c, Video S2), the lateral nucleus of the septum contained dense clusters of labeled cells, with the highest density in its ventral component, although the caudal part of the lateral septal nucleus also contained a high density of labeled neurons (Figures 2e and 3b,c, Video S2). In contrast to the septal nuclei, most parts of the bed nuclei of the stria terminalis (BST) contained only low to moderate densities of labeled neurons (Figures 2e,f and 3b,c, Video S2). A notable exception is the principal nucleus of the BST (Video S2); its high density of labeled neurons contrasted with the significantly lower levels of labeling observed in the anterior division of the BST. Only a few scattered cells were detected in the globus pallidus or diagonal band nucleus (Figures 2e and 3b,d, Video S2).
3.1.4 |. Thalamus and striatum
The highest densities of labeled neurons in the thalamus were primarily in midline nuclei (Figures 2c,e and 3d,e, Video S2). Heavily labeled and densely packed cells were found throughout the rostrocaudal extent of the paraventricular nucleus of the thalamus (Figures 2c,e, 3d,e, and 4b, Video S2). Additionally, the intermediodorsal nucleus, mediodorsal nucleus, and lateral dorsal nucleus exhibited moderate to dense concentrations of labeled neurons (Figures 2e and 3d,e, Video S2), and a high to moderate density of cells was found in the nucleus reuniens (Figures 2e and 3d, Video S2). Distinct clusters of labeled neurons were also found in the more posterior areas of the thalamus, including the medial geniculate complex and suprageniculate nucleus (Figures 2e and 3g, Video S2). Overall, cellular labeling in the striatum was relatively sparse when compared to other brain regions (Figures 2b,e,f and 3a, Video S2). The caudoputamen exhibits the densest labeling in the striatum, with widely scattered neurons with intensely labeled dendrites (Figures 2b,e,f and 3b–d, Video S2). The nucleus accumbens contained few labeled neurons, which stood in stark contrast to the apparent intensity of its innervation (see below).
3.1.5 |. Cortical regions
Clearly labeled MC3R neurons were present in discrete cortical regions, with the most extensive labeling observed in the prefrontal region of the cerebral cortex and in the ventral subiculum (Figures 2b,d–f and 3a,g, Video S2). In the prefrontal region, a high density of labeled neurons was largely confined to layers II and III of the prelimbic cortex, with significant labeling also in the infralimbic cortex (Figures 2b,e,f, 3a and 4c, Video S2). A similar distribution was apparent in parts of the motor cortex, with scattered neurons present in deeper cortical layers as well (Figures 2e and 3a–e, Video S2). A moderate density of labeled neurons was present in cortical regions associated with decision-making, spatial orientation, and sensory processes, including the anterior cingulate area (Figures 2e and 3a–c, Video S2), insula (Figures 2b,e,f and 3a–d, Video S2), gustatory area (Figures 2b,e,f and 3a,b, Video S2), primary somatosensory area (Figures 2b,e and 3a,b,e, Video S2), and ventral retrosplenial area (Figures 2d,e and 3f,g, Video S2). In the hippocampal formation, the highest density of labeled neurons was in the pyramidal layer of the ventral subiculum (Figures 2d–f and 3g, Video S2). Lower densities of labeled neurons were found in the dorsal subiculum, and a few scattered neurons were distributed in the hippocampus proper (Ammon’s horn; Figures 2d,e, 3f,g, and 4d, Video S2). We did not detect labeling in dentate granule cells.
3.1.6 |. Midbrain and medulla
Labeled MC3R neurons were found in the brainstem from the midbrain through the caudal medulla with particular clusters in regions associated with sensory, motor, and autonomic functions (Figures 2d–f and 3f,i, Video S2). In the midbrain, the periaqueductal gray contained clearly labeled cells with the highest density located in the dorsal half of the PAG, a region with strong projections to the hypothalamus (Figures 2d,e and 3f,g, Video S2). The parabrachial nucleus, another region that sends projections to the hypothalamus, exhibited a moderate density of labeled neurons, with the highest density in the lateral and medial components (Figure 2e and 3h, Video S2). Low levels of labeling were observed in the locus coeruleus and raphe nuclei (Figures 2e and 3h, Video S2). Regions known to relay visceral sensory information rostrally, including the nucleus of the solitary tract (Figures 2e,f and 3i, Video S2), exhibited moderate to light labeling, whereas labeling was almost completely absent in the dorsal motor nucleus of the vagus nerve (Figures 2e and 3i, Video S2). In the area postrema, a circumventricular organ, moderate to heavy labeling was apparent (Figures 2e and 3i, Video S2). Regions associated with visual, auditory, or somatosensory processes also contained high to moderate amounts of labeled neurons, including the anterior pretectal nucleus (Figures 2e and 3g, Video S2), inferior and superior colliculus (Figures 2e and 3g,h, Video S2), and spinal nucleus of the trigeminal nerve (Figures 2e and 3i, Video S2), with a very dense cluster of labeled neurons in the olivary pretectal nucleus (Figures 2e and 3f, Video S2). A moderate density of diffusely distributed labeled cells was found in nuclei of the reticular formation, including the intermediate, medullary, and pontine reticular nuclei (Figures 2e and 3f,i, Video S2).
3.2 |. Sexually dimorphic distribution of MC3R neurons
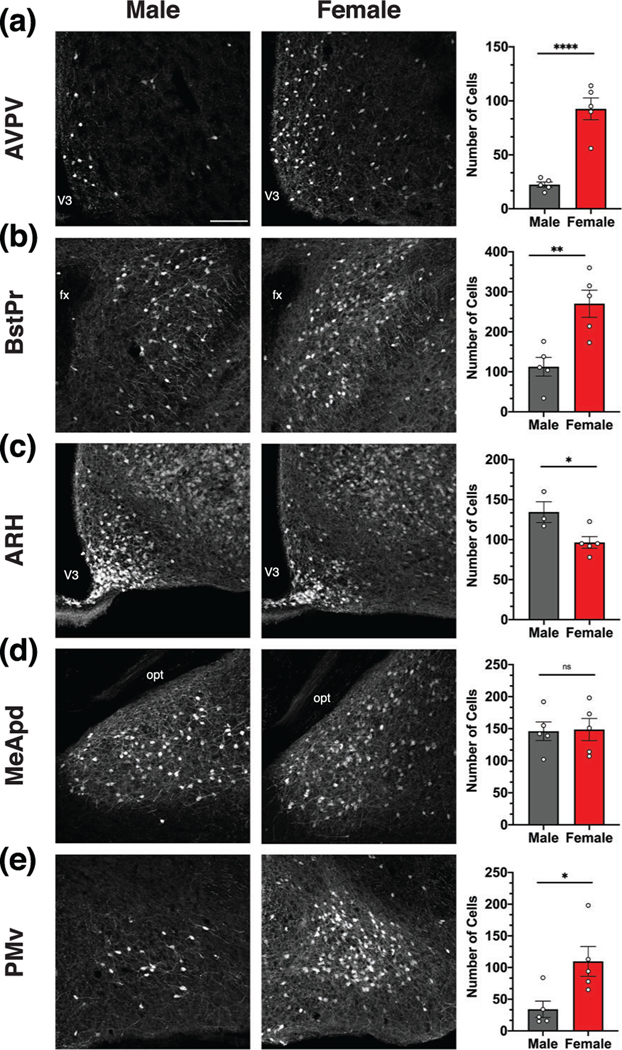
To determine if MC3R neuronal labeling is significantly different between males and females, the number of MC3R neurons was compared between male and female MC3R-Cre::tdTomato mice in anatomical areas where sexually dimorphic expression patterns have been previously reported (Figure 5a–e) (Gu et al., 2003; Hutton et al., 1998; Polston & Simerly, 2003; Polston et al., 2004; Simerly, 1998; Sweeney et al., 2021). In the AVPV, BSTPr, and PMv, females exhibited significantly more MC3R neurons than males (Figure 5a,b,e). Alternatively, in the ARH, males exhibited significantly more MC3R neurons than females (Figure 5c). No differences in the number of labeled neurons were found between males and females in the MeApd (Figure 5d), VMH, or VL-VMH.
FIGURE 5.
Melanocortin-3 receptor (MC3R) labeling is sexually dimorphic. Low magnification images (left, middle column) and comparison of the number of MC3R neurons (right column) in male and female MC3R-Cre::tdTomato mice (a–e). The number of MC3R neurons was significantly higher in the anteroventral periventricular nucleus hypothalamus (AVPV) (a), principal nucleus of the bed nucleus of the stria terminalis (BSTPr) (b), and ventral premammillary nucleus (PMv) (e) in females. In contrast, the number of MC3R neurons was significantly higher in males in the arcuate nucleus hypothalamus (ARH) (c), and no differences were found in the MeApd (d). Scale bar, 200 μm. Data were analyzed with Student’s unpaired t-test. *p < .05, **p < .01, ****p < .0001
3.3 |. Distribution of MC3R-labeled fibers and terminals
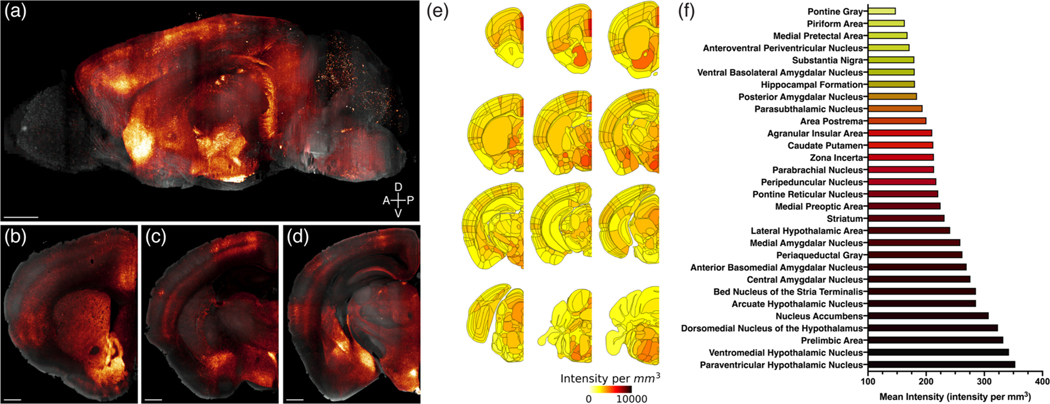
In addition to mapping labeled MC3R neuronal cell bodies, MC3R axonal projections were effectively visualized in whole brains and tissue sections derived from MC3R-Cre::SynTom mice. Based on the distribution of MC3R neuronal labeling described above, MC3R terminals were apparent throughout the brain, forming dense terminal fields in regions that represent a diversity of functional neural systems (Figures 6 and 7, Videos S4–S5). Several regions of the hypothalamus exhibited profuse axonal labeling, as did a number of forebrain and brainstem regions that shared strong connections with the hypothalamus.
FIGURE 6.
Brain-wide mapping of melanocortin-3 receptor (MC3R) projections. Three-dimensional rendering of an MC3R-Cre::SynTom brain viewed in the sagittal plane. Scale bar, 500 μm. Coronal view of MC3R projections in the same brain at the level of the nucleus accumbens (b), arcuate nucleus, ventromedial hypothalamus and basomedial, central and medial amygdala (c), and brainstem (d). Scale bars, 500 μm. MC3R fiber density in the whole brain was quantified by registering the mean fluorescence intensity in each brain region to the Allen Common Coordinate Reference Framework (e and f)
FIGURE 7.
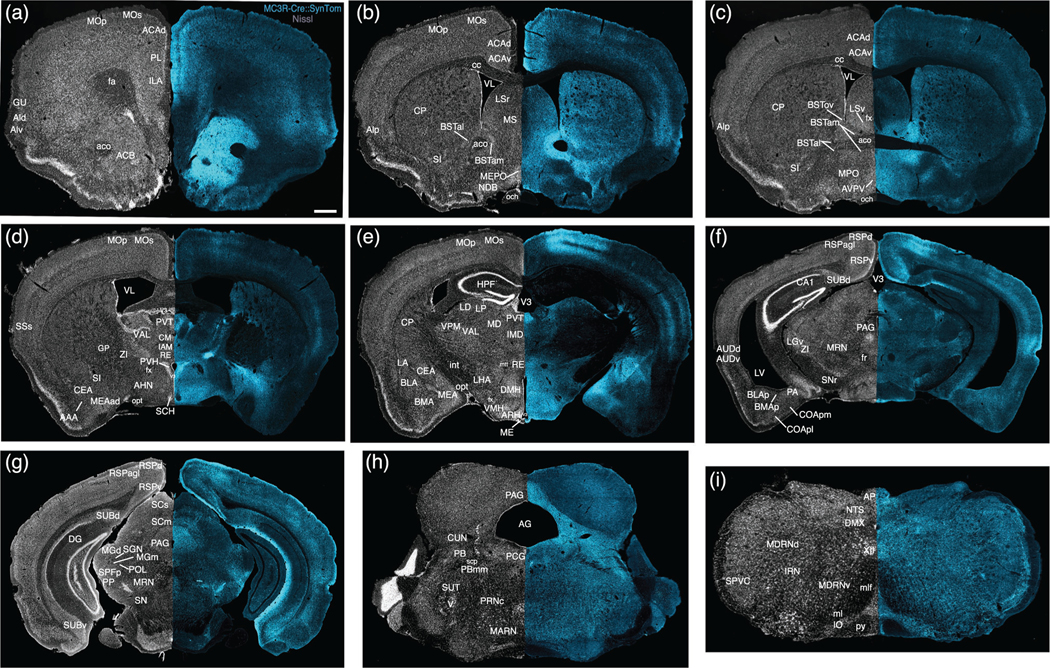
Two-dimensional atlas of melanocortin-3 receptor (MC3R) projections. Low magnification stitched tile images of MC3R projections in an MC3R-Cre::SynTom brain (a–i). Scale bar, 1mm
3.3.1 |. Hypothalamus
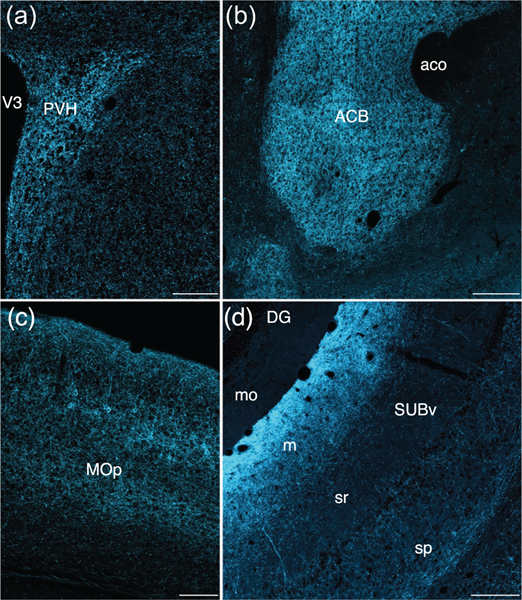
Most hypothalamic nuclei contained at least a moderate density of labeled fibers, with the notable exception of the suprachiasmatic nucleus, which was nearly devoid of labeling (Figures 6c,e and 7d, Video S5). Given the high density of labeled neurons in regions that provide strong hypothalamic projections, the highest densities of labeled fibers extended throughout most of the periventricular and medial zones of the hypothalamus. Several nuclei contained dense plexuses of labeled fibers, including the paraventricular nucleus of the hypothalamus (Figures 6c,e,f, 7d, and 8a, Video S5), the arcuate nucleus (Figures 6d–f and 7e, Video S5), the anteroventral periventricular nucleus (Figures 6e,f and 7c, Video S5), the medial preoptic nucleus (Figures 6e and 7c, Video S5), and the dorsomedial and ventromedial hypothalamic nuclei (Figures 6d–f and 7e, Video S5). The lateral hypothalamus contained a lower density of labeled fibers (Figures 6c–f and 7d,e, Video S5), but at least a moderate density was present in each major subdivision (Hahn & Swanson, 2012).
FIGURE 8.
Comparison of melanocortin-3 receptor (MC3R) fiber projection pattern, density, and distribution. Abundant MC3R fiber labeling is apparent in the paraventricular hypothalamus (a) and nucleus accumbens (b). Scale bar in (a) is 200 μm. Scale bar in (b) is 500 μm. In the primary motor cortex, fiber density is particularly prominent in layer 2/3 (c). Scale bar, 200 μm. MC3R fiber labeling is incredibly dense in the molecular layer of the ventral subiculum when compared to the stratum radiatum or pyramidal layer (d). Scale bar, 500 μm
3.3.2 |. Amygdala
Amygdalar nuclei, particularly the medial and central zones, contained intense terminal labeling (Figures 6d–f and 7d,e, Video S5). High densities of labeled fibers extended throughout the rostrocaudal extent of the medial amygdala, with the posterodorsal part of the nucleus exhibiting the most profuse labeling (Figures 6d–f and 7e, Video S5). Heavy labeling was also observed in the central amygdala, especially in the medial and capsular subdivisions (Figures 6d–f and 7d,e, Video S5). A moderate density of labeled axons was found in the basolateral nucleus, with the highest levels of labeling in its anterior part (Figures 6e,f and 7e,f, Video S5). The anterior (Figures 6c,e and 7d, Video S5) and lateral amygdalar nuclei (Figures 6d,e and 7e, Video S5) also exhibited moderate levels of axonal labeling. Labeling in cortical amygdalar areas was generally sparse (Figure 7f, Video S5).
3.3.3 |. Septal nuclei, bed nuclei of the stria terminalis, and pallidum
Moderate to high densities of labeled axonal projections were present in every part of the bed nuclei of the stria terminalis, with the greatest density of labeling observed in the anteromedial and principal nuclei (Figures 6e,f and 7b,c, Video S5). In the septum, the highest density of labeling was found in the ventral part of the lateral septum, with sparser (but notable) labeled inputs in the dorsal part of the lateral septal nucleus (Figures 6e and 7b,c, Video S5). These labeling patterns contrasted with the low levels of labeled axons in the medial septum (Figures 6e and 7b,c, Video S5). The substantia innominate contains a high density of labeled axons, especially in its anterior and medial portions (Figures 6e and 7b–d, Video S5). Only meagre labeling was present in the diagonal band (Figures 6e and 7b, Video S5), and few to no labeled projections were found in the globus pallidus (Figures 6e and 7d, Video S5).
3.3.4 |. Thalamus and striatum
Rich terminal fields were mainly present in the mediodorsal and caudal nuclei of the thalamus (Figures 6e and 7d,e,g, Video S5). Profuse labeling was apparent throughout the rostrocaudal extent of the paraventricular thalamus (Figures 6e and 7d,e, Video S5), and relatively dense labeling was also present in the nucleus reuniens (Figures 6e and 7d,e, Video S5). Moderate densities of labeled fibers were found in the centromedial, intermediodorsal, mediodorsal, lateral dorsal, and lateral posterior nuclei (Figures 6e and 7d,e, Video S5). Multiple caudal nuclei also exhibited dense to moderate labeling, including the medial geniculate complex, suprageniculate nucleus, peripeduncular nucleus, parvicellular part of the subparafascicular nucleus, and posterior limiting nucleus (Figures 6e and 7g, Video S5). Most lateral and ventral nuclei of the thalamus exhibited little to no fiber labeling (Figures 6e and 7d,e, Video S5).
In the striatum, both the caudoputamen and nucleus accumbens appear to be heavily innervated by MC3R neurons (Figures 6b,e,f and 7a, Video S5). Very dense fiber labeling was found in both the core and shell of the nucleus accumbens (Figures 6b, 7a, and 8b, Video S5), abundant labeled axons were found throughout the rostral caudoputamen, and the density was especially high in dorsal and medial regions of the caudoputamen (Figures 6e,f and 7b,c, Video S5).
3.3.5 |. Cortical regions
MC3R axonal projections appear to innervate select cortical regions, with the most extensive labeling observed in the prefrontal cortex (Figures 6b,e,f and 7a–c, Video S5) and in the hippocampal formation (Figures 6e and 7g, Video S5). In the prefrontal region, layers I-III of the infralimbic, prelimbic, and anterior cingulate areas contained high densities of labeled fibers (Figures 6b,e,f and 7a–c, Video S5). A similar distribution in superficial layers was apparent in the motor cortex (Figures 6c,e 7a,b,d,e, and 8c, Video S5), although abundant labeling was also present in layer V, which contrasts with the sparse labeling characteristic of layer IV. Moderate densities of labeled terminals were present in cortical regions associated with decision-making, spatial orientation, and interosensory processes, including the insula (Figures 6b,e,f and 7a–c, Video S5), gustatory area (Figures 6b,e and 7a, Video S5), and secondary somatosensory area (Figures 6c,e and 7d, Video S5). In the hippocampal formation, a dense cluster of labeled fibers was largely localized to the stratum lacunosum moleculare in CA1 of Ammon’s Horn (Figure 7g and 8d, Video S5). A lower density of fibers was found in the stratum radiatum (Figures 7g and 8d, Video S5). Labeled axons were scarce in CA3 of Ammon’s horn and the dentate gyrus (Figures 7g and 8d, Video S5). The subiculum contained the most intense fiber labeling, with the highest densities located in the molecular and pyramidal layers of the ventral subiculum (Figures 6e, 7g and 8d, Video S5).
3.3.6 |. Midbrain and medulla
Clearly labeled axons were distributed throughout most brainstem structures, although distinct clusters of high-density labeling were apparent in various regions (Figures 6e,f and 7f–i, Video S5). In the midbrain, the periaqueductal gray contained heavy terminal labeling, with the highest density of labeling in the dorsal and posterior portions of the periaqueductal gray (Figures 6e,f and 7f,g, Video S5). Rich axonal projections were also found in the parabrachial nucleus, especially the lateral subdivision (Figures 6e,f and 7h, Video S5). In the medulla, moderate to heavy labeling was apparent in areas that modulate visceral sensory and motor information, including the area postrema, nucleus of the solitary tract, dorsal motor nucleus of the vagus nerve, and hypoglossal nucleus (Figures 6e and 7i, Video S5). The medullary and intermediate reticular nuclei also exhibited considerable fiber labeling (Figures 6e,f and 7f–i, Video S5), whereas scarce and scattered labeling was apparent in the spinal nucleus of the trigeminal nerve (Figures 6e and 7i, Video S5).
4 |. DISCUSSION
While MC3R deletion is associated with obesity in humans and mice, MC3RKO mice exhibit increased anorexia and weight loss in response to multiple challenges, including fasting (Ghamari-Langroudi et al., 2018), behavioral stressors (Sweeney et al., 2021), cachexigenic agents (Marks & Cone, 2003; Marks et al., 2003), and GLP1R agonists (Sweeney et al., 2021). Due to its expression in key populations of neurons located in the arcuate and ventromedial nuclei of the hypothalamus, most functional studies have focused on the role of MC3R in the regulation of food intake. Here, we present a detailed mapping of the distribution of neurons that express MC3R, and the results suggest that expansion of their functional role may be in order, as we discovered that these neurons are significant components of each major subdivision of the central nervous system, including the cortex, hippocampal formation, amygdala, basal ganglia, thalamus, hypothalamus, and brainstem. Although the present findings correspond well with previous reports that relied on in situ hybridization for localization of MC3R expression (Gantz et al., 1993; Lein et al., 2007; Roselli-Rehfuss et al., 1993), detection of MC3R neurons in MC3R-Cre::tdTomato mice allows a more comprehensive examination of their distribution and enabled discovery of new sites, which were confirmed with RNAscope FISH. In addition, new information about possible projection pathways emanating from MC3R neurons was derived from independent conditional visualization of labeled axons. It is also clear that the distribution of neurons expressing MC3R is quite distinct from that of neurons that express MC4R (Mountjoy et al., 1994).
MC3R deletion results in elevated adiposity, but MC3R null mice appear to be relatively protected from the development of the most severe aspects of metabolic syndrome (Butler et al., 2000; Butler & Cone, 2002; G. M. Sutton et al., 2008). However, these mice do appear to be hypersensitive to social isolation, restraint stress, and anxiolytic factors (Sweeney et al., 2021), suggesting an important role for MC3R in modulating the transmission of exterosensory information to hypothalamic neural systems more directly involved in the regulation of food intake. The abundant expression of MC3R in septohippocampal and strial or amgydalofugal projections to the hypothalamus is consistent with such a role. Similarly, MC3R null mice show enhanced sensitivity to administration of liraglutide, suggesting a possible role for MC3R in modulating the impact of ascending interosensory information from the brainstem (Sweeney et al., 2021). Equally significant is the fact that MC3R is expressed in nearly all AgRP neurons of the ARH (Sweeney et al., 2021), and through both ascending and descending projections from these cells, it may modulate the activity of target regions involved in mediating the sensory valence of hunger signals (Betley et al., 2015, 2016; Y. Chen et al., 2016; Girardet et al., 2018). Thus, the breadth and diversity of functional neural systems that either contain a high density of MC3R neurons or are innervated by them suggests an expanded functional role for MC3R signaling in behavioral regulation. In addition, MC3R neuronal labeling is sexually dimorphic in several brain regions, suggesting that MC3R signaling likely impacts physiological responses to melanocortins differently in males and females. Thus, the organization of neurons that express MC3R demonstrates the capacity to impact a diverse array of functionally distinct neural systems by modulating the integration of a variety of sensory cues and may do so differently in males and females.
4.1 |. MC3R in the hypothalamus
The MC3R has been proposed to play an important role in rheostatic control of energy balance (Ghamari-Langroudi et al., 2018), and the hypothalamus is an important site for the integration of sensory, cognitive, and behavioral state information that likely impacts this adaptive regulatory function (Risold et al., 1997; Swanson, 2000; Thompson & Swanson, 2003). MC3R neurons and fibers are found in all major hypothalamic regions, including each major nucleus in the medial zone of the hypothalamus, which collectively comprise the behavior control column (Swanson, 2000). In addition, both labeled cells and fibers are well represented in periventricular parts of the hypothalamus, which include functionally defined zones containing releasing hormone neurons (neuroendocrine motor zone) or that control autonomic nervous system activity (hypothalamic visceromotor pattern generator) (Thompson & Swanson, 2003). Similarly, labeled neurons and fibers in the lateral zone of the hypothalamus (Simerly, 2015) provide a substrate for the regulation of consummatory behavior and direct connections with the cerebral cortex. The only hypothalamic nucleus where MC3R labeling is noticeably sparse or absent is the suprachiasmatic nucleus, a nodal component of neural circuits controlling circadian rhythmicity (Lu et al., 2001; Mohawk et al., 2012; Watts et al., 1987). This finding is consistent with the previous observation that circadian rhythms in food intake and locomotor activity are largely intact in MC3R null mice (Renquist et al., 2011). However, MC3R does modulate entrainment to meal intake, but this occurs independently of the suprachiasmatic nucleus (G. M. Sutton et al., 2008). Within the behavioral control column, MC3R labeling was most pronounced in the ventromedial hypothalamic nucleus, anterior hypothalamic nucleus, and descending division of the paraventricular hypothalamic nucleus. These areas are critical nodes for the control of reproductive, defensive, and ingestive behaviors and thus imply that MC3R may participate in modulating these behaviors (Canteras et al., 1994; Risold et al., 1994; Simerly, 1998; Swanson, 2000). The dorsomedial hypothalamus exhibits dense MC3R labeling and influences defensive and ingestive behaviors, as well as autonomic responses (Simonds et al., 2014; Thompson & Swanson, 2003).
As noted previously, MC3R expression is abundant in the arcuate nucleus and has been colocalized to a number of important cell types (Lam et al., 2021; Sweeney et al., 2021). Notably, the neuroendocrine division of the paraventricular hypothalamic nucleus (Markakis & Swanson, 1997; Swanson, 2000) appears to receive particularly strong inputs from MC3R neurons because fiber labeling is considerably higher than neuronal labeling, indicating that this area is primarily a target of MC3R neurons. Altogether, the anatomical evidence presented here suggests that MC3R is well positioned to impact a variety of motivated behaviors that are essential to survival. In addition to intrahypothalamic connections between regions that contain high densities of MC3R neurons, the hypothalamus receives multimodal sensory information through descending projections from the cerebral hemispheres, as well as ascending projections from the brainstem (Swanson, 2000). MC3R cells and fibers are found in several of these sensory pathways and may represent additional access points for melanocortin-mediated modulation of motivated behavior. The discussion below of motor neural networks that control motivated behavior will follow the neuroanatomical organization of brain circuitry described in detail by Swanson (2000).
4.2 |. Descending pathways to the hypothalamus
4.2.1 |. Cortical and septohippocampal inputs
The infralimbic and prelimbic cortical areas contained the highest densities of MC3R neurons, which likely provide direct inputs to the medial and lateral zones of the hypothalamus (Risold et al., 1997; Saper, 2000). Whether these prefrontal MC3R projections to the hypothalamus convey polymodal sensory information related to experience or cognitive aspects of feeding behavior remains to be investigated. Labeled neurons were also noted in other cortical regions, including the anterior cingulate area, insula, gustatory area, primary somatosensory area, and ventral retrosplenial area, suggesting that MC3R signaling may impact multiple aspects of polymodal sensory processing. The high density of MC3R neurons in the subiculum contributes labeled axons that course through the medial corticohypothalamic tract to innervate structures located primarily in the medial zone of the hypothalamus (Canteras & Swanson, 1992; Kishi et al., 2000; Swanson & Cowan, 1975). The subiculum sends dense projections to the lateral septal nucleus, which also contains numerous MC3R neurons and provides topographically organized projections to several hypothalamic nuclei involved in neuroendocrine and autonomic regulation, including the medial preoptic, anterior, ventromedial, dorsal premammillary, ventral premammillary, and medial mammillary nuclei (Risold & Swanson, 1997; Swanson & Cowan, 1979). Thus, on purely anatomical grounds, it is possible that MC3R signaling impacts the activity of several well-characterized pathways conveying multimodal sensory information from the cerebral cortex to discrete functional domains of the hypothalamus.
4.2.2 |. Inputs from amygdala and bed nuclei of the stria terminalis
The majority of labeled MC3R neurons in the amygdala were localized to three nuclei, each with distinct patterns of projections (Canteras et al., 1995; Petrovich et al., 1996; Petrovich & Swanson, 1997; Swanson & Petrovich, 1998). The highest density of MC3R neurons was located in the posterodorsal part of the medial nucleus of the amygdala, which is heavily innervated by the accessory olfactory bulb and projects strongly to the principal nucleus of the BST. This information is conveyed to the hypothalamus from the principal nucleus through strong, sexually dimorphic projections to periventricular and medial components of the hypothalamus that have a significant impact on reproduction (Gu et al., 2003). This latter observation may be relevant to an apparent role for MC3R in puberty (Lam et al., 2021). The principal nucleus of the BST contains the most MC3R neurons of any part of the BST, but whether these cells provide return projections to the posterodorsal part of the medial amygdalar nucleus, which contains a high density of labeled axons, requires verification. The basomedial nucleus and lateral zone of the posterior part of the cortical nucleus receive inputs from the main olfactory system, and each contains MC3R neurons that project to dorsal and ventral components of the anterior division of the BST, which contain very high densities of labeled axons but few MC3R cell bodies. Many MC3R neurons are located in the central amygdaloid nucleus (lateral part), which projects to the anterolateral area of the BST and plays an important role in multiple aspects of consummatory behavior and autonomic regulation (Cai et al., 2014; Dong et al., 2001; Hulsman et al., 2021; Izadi & Radahmadi, 2021; Kafami & Nasimi, 2016; Petrovich & Swanson, 1997; Saha, 2005). Notably, amygdalar cell groups that do not project directly to the BST (lateral and anterior basolateral nuclei) appear to lack MC3R neurons.
4.2.3 |. Ascending pathways to the hypothalamus
Although the nucleus of the solitary tract (NTS) is a critical pathway that transmits viscerosensory information to hypothalamic regions containing high densities of MC3R neurons, only a few MC3R cells are present within the NTS itself. However, the parabrachial nucleus (PB) is the major relay for ascending visceral information from the NTS, and MC3R cells are abundant in this important nucleus. The PB relays viscerosensory and nociceptive information through direct projections to the dorsomedial, anteroventral preoptic, lateral preoptic, median preoptic, ventromedial, lateral, paraventricular, and arcuate nuclei of the hypothalamus (Fulwiler & Saper, 1984; Saper & Loewy, 1980). Furthermore, the PB can also indirectly influence the activity of hypothalamic nuclei through its projections to the bed nuclei of the stria terminalis and central nucleus of the amygdala (J. Y. Chen et al., 2018; Jaramillo et al., 2020, 2021; Moga et al., 1989; Saper & Loewy, 1980; Ye & Veinante, 2019). MC3R neurons in the PB are primarily localized to its lateral subnucleus, which may therefore participate in the processing and transfer of viscerosensory and nociceptive information to hypothalamic nuclei known to respond to these stimuli (Andermann & Lowell, 2017; Campos et al., 2018; J. Y. Chen et al., 2018; Malick et al., 2001).
4.3 |. Hypothalamic output
The hypothalamus integrates sensory, cognitive, and behavioral state information and regulates multiple behaviors and physiological responses through its descending projections to brainstem and spinal motor systems. The hypothalamus also provides ascending projections directly from the lateral hypothalamus to the cerebral cortex or indirectly via projections to rostromedial components of the dorsal thalamus, such as the paraventricular thalamic nucleus (Risold et al., 1997). Accordingly, we observed heavy MC3R axonal labeling in brainstem regions important for motor control, including the nucleus of the solitary tract, dorsal motor nucleus of the vagus nerve, and area postrema. Dense labeling of MC3R fibers and cell bodies is also present in the paraventricular nucleus and nucleus of reuniens, which are important sites for conveying information regarding behavioral state from the hypothalamus to the cortex.
4.4 |. Sexually dimorphic MC3R neuronal expression
Until recently, MC3R expression had been examined primarily in male rodents. Increased Mc3r mRNA abundance is observed in the ARH of male mice compared to female mice, and the opposite is seen in the AVPV (Sweeney et al., 2021). Our data in MC3R-Cre::tdTom mice reported here confirm these observations. Apparent sex differences in the density of MC3R labeling in the BSTpr and ventral premammillary region indicate that these populations of MC3R neurons may also be sexually dimorphic. The greater number of neurons in the BSTpr of females is surprising because there are over twice as many neurons in the BSTpr of males, and previously reported sex differences favored males (Simerly, 2002). MC3R labeling patterns exhibit considerable overlap with the distribution of androgen and estrogen receptors (Simerly et al., 1990), indicating that neurons in certain regions may coexpress MC3R and sex steroid receptors. In the arcuate nucleus of the hypothalamus, approximately 50% of Mc3r neurons coexpress estrogen receptor 1 (unpublished results). Whether the differences observed in MC3R expression are due to differences in the expression of the Mc3r gene or a different complement of neurons between the sexes remains to be determined. Thus, anatomical evidence suggests that MC3R signaling may lead to different behavioral and functional outcomes in males and females and that sex steroid receptors may mediate this sexual differentiation (Simerly et al., 1997). Deletion of MC3R increases dopamine content in the ventral tegmental area (VTA) and decreases sucrose intake and preference only in females (Lippert et al., 2014). Additionally, activating MC3R neurons in the VTA decreases feeding in females but not males (Dunigan et al., 2021). Females lacking MC3R also exhibit reductions in locomotor activity and decreased corticosterone, whereas males display hyperinsulinemia (A. S. Chen et al., 2000). Recent evidence also indicates that male and female MC3R KO mice respond differently to the effects of behavioral stressors on food intake (Sweeney et al., 2021). Only male MC3R KO mice display enhanced anorexia following restraint, but females exhibit a more pronounced anorexic response to the novelty-suppressed feeding test (Sweeney et al., 2021). Future studies designed to link observed anatomical and functional differences in melanocortin signaling between males and females are warranted.
5 |. CONCLUSIONS
Collectively, a detailed evaluation of the distribution of neurons that express MC3R reveals a brain-wide organization with the potential to modify the activity of convergent sensory systems known to regulate the expression of a wide variety of motivated behaviors and associated physiological responses. This organization is especially apparent in the 3D reconstructions of whole-brain data sets collected with light-sheet imaging. We found these datasets to enhance the discovery of labeling patterns that are difficult to discern in conventional 2D representations (e.g., projections of confocal images or single sections on a wide field microscope), and the speed and fluidity of such interactive interrogations promoted the discovery of new anatomical relationships. Striking features of these data sets are the large numbers of MC3R neurons located in telencephalic regions, especially the cerebral cortex, hippocampal formation and amygdala, and their anatomical relationship to hypothalamic regions containing dense clusters of MC3R neurons. By targeting fluorescence to axons and terminals with the synaptophysin-tdTomato labeling strategy, we were able to visualize major afferent tracts connecting these regions in their entirety, and this information may prove useful for planning future experimental interventions or manipulations, such as those associated with lesions or imaging experiments. Equally important is the enhanced throughput provided by utilization of a semiautomated method for capturing quantitative estimates of neuronal density in discrete regions throughout the brain. Although no computer-assisted brain registration pipeline is error proof, the relative estimates presented here closely match our qualitative and quantitative sampling. Future studies designed to identify the functional properties of MC3R neural systems visualized here should continue to enhance our understanding of how melanocortin signaling impacts not only energy balance but also an expanding range of other essential survival behaviors.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Simerly Lab for comments and discussion on early versions of this manuscript. This work was supported by NIH grants R01DK106476 (Richard B. Simerly), R01DK126715 (Roger D. Cone), and F32DK123879 (Michelle N. Bedenbaugh).
Abbreviations:
- AAA
anterior amygdalar area
- ACAd
anterior cingulate area, dorsal part
- ACAv
anterior cingulate area, ventral part
- ACB
nucleus accumbens
- aco
anterior commissure, olfactory limb
- AG
gyrus ambiens
- AHN
anterior hypothalamic nucleus
- AId
agranular insular area, dorsal part
- AIp
agranular insular area, posterior part
- AIv
agranular insular area, ventral part
- AP
area postrema
- APN
anterior pretectal nucleus
- ARH
arcuate nucleus hypothalamus
- AUDd
dorsal auditory areas
- AUDv
ventral auditory areas
- AVPV
anteroventral periventricular nucleus hypothalamus
- BLA
basolateral nucleus amygdala
- BLAp
basolateral nucleus amygdala, posterior part
- BMA
basomedial nucleus amygdala
- BMAp
basomedial nucleus amygdala, posterior part
- BSTal
bed nuclei stria terminalis, anterior division, anterolateral area
- BSTam
bed nuclei stria terminalis, anterior division, anteromedial area
- BSTov
bed nuclei stria terminalis, anterior division, oval nucleus
- c
central canal
- CA1
field CA1, Ammon’s horn
- CA3
field CA3, Ammon’s horn
- cc
corpus callosum
- CEA
central nucleus amygdala
- CEAm
central nucleus amygdala, medial part
- CM
central medial nucleus thalamus
- COApl
cortical nucleus amygdala, posterior part, lateral zone
- COApm
cortical nucleus amygdala, posterior part, medial zone
- CP
caudoputamen
- CUN
cuneiform nucleus
- DG
dentate gyrus
- DMH
dorsomedial nucleus hypothalamus
- DMX
dorsal motor nucleus of the vagus nerve
- fa
corpus callosum, anterior forceps
- fr
fasciculus retroflexus
- fx
columns of the fornix
- GP
globus pallidus
- GU
gustatory area
- HPF
hippocampal formation
- IAM
interanteromedial nucleus thalamus
- ICc
inferior colliculus, central nucleus
- ICd
inferior colliculus, dorsal nucleus
- ICe
inferior colliculus, external nucleus
- ILA
infralimbic area
- IMD
intermediodorsal nucleus thalamus
- int
internal capsule
- IO
inferior olivary complex
- IPN
interpeduncular nucleus
- IRN
intermediate reticular nucleus
- LA
lateral nucleus amygdala
- LD
lateral dorsal nucleus thalamus
- LDT
Laterodorsal tegmental nucleus
- LGv
lateral geniculate complex, ventral part
- LHA
lateral hypothalamic area
- LM
lateral mammillary nucleus
- LP
lateral posterior nucleus thalamus
- LPO
lateral preoptic area
- LSr
lateral septal nucleus, rostral part
- LSv
lateral septal nucleus, ventral part
- m
subiculum, ventral part, molecular layer
- MARN
magnocellular reticular nucleus
- MD
mediodorsal nucleus thalamus
- MDRNd
medullary reticular nucleus, dorsal part
- MDRNv
medullary reticular nucleus, ventral part
- ME
median eminence
- MEPO
median preoptic nucleus
- MGd
medial geniculate complex, dorsal part
- MGm
medial geniculate complex, medial part
- ml
medial lemniscus
- MM
medial mammillary nucleus
- mo
molecular area
- MOp
primary motor area
- MOs
secondary motor area
- MPO
medial preoptic area
- MRN
mesencephalic reticular nucleus
- MS
medial septal nucleus
- mtt
mammillothalamic tract
- NDB
diagonal band nucleus
- NLOT
Nucleus of the lateral olfactory tract
- NTS
nucleus of the solitary tract
- och
optic chiasm
- OP
olivary pretectal nucleus
- opt
optic tract
- PA
posterior nucleus amygdala
- PAG
periaqueductal gray
- PB
parabrachial nucleus
- PBIv
parabrachial nucleus, lateral division, ventral lateral part
- PBl
parabrachial nucleus, lateral division
- PBlc
parabrachial nucleus, lateral division, central lateral part
- PBle
parabrachial nucleus, lateral division, external layer
- PBmm
parabrachial nucleus, medial division, medial part
- PCG
pontine central gray
- PH
posterior hypothalamic nucleus
- PL
prelimbic area
- POL
posterior limiting nucleus thalamus
- PP
peripeduncular nucleus
- PRN
pontine reticular nucleus
- PRNc
pontine reticular nucleus, caudal part
- PTLp
posterior parietal association areas
- PVH
paraventricular nucleus hypothalamus
- PVT
paraventricular nucleus thalamus
- py
pyramid
- RE
nucleus reuniens
- RSPagl
retrosplenial area, agranular lateral part
- RSPd
retrosplenial area, dorsal part
- RSPv
retrosplenial area, ventral part
- SCH
suprachiasmatic nucleus
- SCm
superior colliculus, motor-related
- scp
superior cerebellar peduncles
- SCs
superior colliculus, sensory related
- SFO
subfornical organ
- SGN
suprageniculate nucleus
- SI
substantia innominata
- SN
substantia nigra
- so
supraoptic nucleus
- sp
subiculum, ventral part, pyramidal layer
- SPFp
subparafascicular nucleus thalamus, parvicellular part
- SPVC
spinal nucleus of the trigeminal, caudal part
- SPVl
spinal nucleus of the trigeminal, interpolar part
- sr
subiculum, ventral part, stratum radiatum
- SSp
primary somatosensory area
- SSs
supplementary somatosensory area
- SUBd
subiculum, dorsal part
- SUBv
subiculum, ventral part
- SUM
supramammillary nucleus
- V3
third ventricle
- V4
fourth ventricle
- VAL
ventral anterior-lateral complex of the thalamus
- VISam
anteromedial visual area
- VL
lateral ventricle
- VPM
ventral posteromedial nucleus of the thalamus
- VTA
ventral tegmental area
- XII
twelfth cranial nerve
- ZI
zona incerta
Footnotes
CONFLICT OF INTEREST
Roger D. Cone is on the SAB and an equity holder in Courage Therapeutics. Patrick Sweeney is an equity holder in Courage Therapeutics. Other authors declare no conflict of interest.
DATA AVAILABILITY STATEMENT
Data related to the current study are available from the corresponding author upon reasonable request.
REFERENCES
- Andermann ML, & Lowell BB (2017). Toward a wiring diagram understanding of appetite control. Neuron, 95, 757–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begriche K,Levasseur PR,Zhang J,Rossi J,Skorupa D,Solt LA,Young B, Burris TP, Marks DL, Mynatt RL, & Butler AA (2011). Genetic dissection of the functions of the melanocortin-3 receptor, a seven-transmembrane G-protein-coupled receptor, suggests roles for central and peripheral receptors in energy homeostasis. Journal of Biological Chemistry, 286, 40771–40781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betley JN, Xu S, Cao ZFH, Gong R, Magnus CJ, Yu Y, & Sternson SM (2015). Neurons for hunger and thirst transmit a negative-valence teaching signal. Nature, 521, 180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddinger JE, Lazarenko RM, Scott MM, & Simerly R. (2020). Leptin suppresses development of GLP-1 inputs to the paraventricular nucleus of the hypothalamus. eLife, 9, e59857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bria A, Bernaschi M, Guarrasi M, & Iannello G. (2019). Exploiting multilevel parallelism for stitching very large microscopy images. Frontiers in Neuroinformatics, 13, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett CJ, Li C, Webber E, Tsaousidou E, Xue SY, Brüning JC, & Krashes MJ (2016). Hunger-driven motivational state competition. Neuron, 92, 187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler AA (2006). The melanocortin system and energy balance. Peptides, 27, 281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler AA, & Cone RD (2002). The melanocortin receptors: Lessons from knockout models. Neuropeptides, 36, 77–84. [DOI] [PubMed] [Google Scholar]
- Butler AA, Kesteson RA, Khong K, Cullen MJ, Pelleymounter MA, Dekoning J, Baetscher M, & Cone RD (2000). A unique metalolic sysdrone causes obesity in the melanocortin-3 receptor-deficient mouse. Endocrinology, 141, 3518–3521. [DOI] [PubMed] [Google Scholar]
- Butler AA, Marks DL, Fan W, Kuhn CM, Bartolome M, & Cone RD (2001). Melanocortin-4 receptor is required for acute homeostatic responses to increased dietary fat. Nature Neuroscience, 4, 605–611. [DOI] [PubMed] [Google Scholar]
- Cai H, Haubensak W, Anthony TE, & Anderson DJ (2014). Central amygdala PKC-δ+ neurons mediate the influence of multiple anorexigenic signals. Nature Neuroscience, 17, 1240–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos CA, Bowen AJ, Roman CW, & Palmiter RD (2018). Encoding of danger by parabrachial CGRP neurons. Nature, 555, 617–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canteras NS, Simerly RB, & Swanson LW (1994). Organization of projections from the ventromedial nucleus of the hypothalamus: A Phaseolus vulgaris-leucoagglutinin study in the rat. Journal of Comparative Neurology, 348, 41–79. [DOI] [PubMed] [Google Scholar]
- Canteras NS, Simerly RB, & Swanson LW (1995). Organization of projections from the medial nucleus of the amygdala: A PHAL study in the rat. Journal of Comparative Neurology, 360, 213–245. [DOI] [PubMed] [Google Scholar]
- Canteras NS, & Swanson LW (1992). Projections of the ventral subiculum to the amygdala, septum, and hypothalamus: A PHAL anterograde tract-tracing study in the rat. Journal of Comparative Neurology, 324, 180–194. [DOI] [PubMed] [Google Scholar]
- Catania A, Gatti S, Colombo G, & Lipton JM (2004). Targeting melanocortin receptors as a novel strategy to control inflammation. Pharmacological Reviews, 56, 1–29. [DOI] [PubMed] [Google Scholar]
- Chandramohan G, Durham N, Sinha S, Norris K, & Vaziri ND (2009). Role of γ melanocyte-stimulating hormone–renal melanocortin 3 receptor system in blood pressure regulation in salt-resistant and salt-sensitive rats. Metabolism, 58, 1424–1429. [DOI] [PubMed] [Google Scholar]
- Chen AS, Marsh DJ, Trumbauer ME, Frazier EG, Guan X-M, Yu H, Rosenblum CI, Vongs A, Feng Y, Cao L, Metzger JM, Strack AM, Camacho RE, Mellin TN, Nunes CN, Min W, Fisher J, Gopal-Truter S, MacIntyre DE, … Ploeg L. H. T. V. d. (2000). Inactivation of the mouse melanocortin-3 receptor results in increased fat mass and reduced lean body mass. Nature Genetics, 26, 97–102. [DOI] [PubMed] [Google Scholar]
- Chen JY, Campos CA, Jarvie BC, & Palmiter RD (2018). Parabrachial CGRP neurons establish and sustain aversive taste memories. Neuron, 100, 891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Lin Y-C, Zimmerman CA, Essner RA, & Knight ZA (2016). Hunger neurons drive feeding through a sustained, positive reinforcement signal. eLife, 5, e18640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone RD (2006). Studies on the physiological functions of the melanocortin system. Endocrine Reviews, 27, 736–749. [DOI] [PubMed] [Google Scholar]
- Dean KM, Roudot P, Welf ES, Danuser G, & Fiolka R. (2015). Deconvolution-free subcellular imaging with axially swept light sheet microscopy. Biophysical Journal, 108, 2807–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H-W, Petrovich GD, & Swanson LW (2001). Topography of projections from amygdala to bed nuclei of the stria terminalis. Brain Research Reviews, 38, 192–246. [DOI] [PubMed] [Google Scholar]
- Dunigan AI, Olson DP, & Roseberry AG (2021). VTA MC3R neurons control feeding in an activity- and sex-dependent manner in mice. Neuropharmacology, 197, 108746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellacott KLJ, Murphy JG, Marks DL, & Cone RD (2007). Obesity-induced inflammation in white adipose tissue is attenuated by loss of melanocortin-3 receptor signaling. Endocrinology, 148, 6186–6194. [DOI] [PubMed] [Google Scholar]
- Fan W, Dinulescu DM, Butler AA, Zhou J, Marks DL, & Cone RD (2000). The central melanocortin system can directly regulate serum insulin levels. Endocrinology, 141, 3072–3079. [DOI] [PubMed] [Google Scholar]
- Fan W, Ellacott KLJ, Halatchev IG, Takahashi K, Yu P, & Cone RD (2004). Cholecystokinin-mediated suppression of feeding involves the brainstem melanocortin system. Nature Neuroscience, 7, 335–336. [DOI] [PubMed] [Google Scholar]
- Fulwiler CE, & Saper CB (1984). Subnuclear organization of the efferent connections of the parabrachial nucleus in the rat. Brain Research Reviews, 7, 229–259. [DOI] [PubMed] [Google Scholar]
- Gantz I, Konda Y, Tashiro T, Shimoto Y, Miwa H, Munzert G, Watson SJ, DelValle J, & Yamada T. (1993). Molecular cloning of a novel melanocortin receptor. Journal of Biological Chemistry, 268, 8246–50. [PubMed] [Google Scholar]
- Garfield AS, Lam DD, Marston OJ, Przydzial MJ, & Heisler LK (2009). Role of central melanocortin pathways in energy homeostasis. Trends in Endocrinology & Metabolism, 20, 203–215. [DOI] [PubMed] [Google Scholar]
- Getting SJ, Riffo-Vasquez Y, Pitchford S, Kaneva M, Grieco P, Page CP, Perretti M, & Spina D. (2008). A role for MC3R in modulating lung inflammation. Pulmonary Pharmacology & Therapeutics, 21, 866–873. [DOI] [PubMed] [Google Scholar]
- Ghamari-Langroudi M, Cakir I, Lippert RN, Sweeney P, Litt MJ, Ellacott KLJ, & Cone RD (2018). Regulation of energy rheostasis by the melanocortin-3 receptor. Science Advances, 4, eaat0866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardet C, Marks DL, & Butler AA (2018). Melanocortin-3 receptors expressed on agouti-related peptide neurons inhibit feeding behavior in female mice. Obesity, 26, 1849–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu G, Cornea A, & Simerly RB (2003). Sexual differentiation of projections from the principal nucleus of the bed nuclei of the stria terminalis. Journal of Comparative Neurology, 460, 542–562. [DOI] [PubMed] [Google Scholar]
- Hahn JD, & Swanson LW (2012). Connections of the lateral hypothalamic area juxtadorsomedial region in the male rat. Journal of Comparative Neurology, 520, 1831–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedde PN, & Gratton E. (2018). Selective plane illumination microscopy with a light sheet of uniform thickness formed by an electrically tunable lens. Microscopy Research and Technique, 81, 924–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsman AM, Terburg D, Roelofs K, & Klumpers F. (2021). Roles of the bed nucleus of the stria terminalis and amygdala in fear reactions. Handbook of clinical neurology, (vol.) 179, pp. 419–432. Elsevier. [DOI] [PubMed] [Google Scholar]
- Huszar D, Lynch CA, Fairchild-Huntress V, Dunmore JH, Fang Q, Berkemeier LR, Gu W, Kesterson RA, Boston BA, Cone RD, Smith FJ, Campfield LA, Burn P, & Lee F. (1997). Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell, 88, 131–141. [DOI] [PubMed] [Google Scholar]
- Hutton LA, Gu G, & Simerly RB (1998). Development of a sexually dimorphic projection from the bed nuclei of the stria terminalis to the anteroventral periventricular nucleus in the rat. Journal of Neuroscience, 18, 3003–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izadi MS, & Radahmadi M. (2021). Overview of the central amygdala role in feeding behaviour. British Journal of Nutrition, 217, 953–960. [DOI] [PubMed] [Google Scholar]
- Jaramillo AA, Brown JA, & Winder DG (2021). Danger and distress: Parabrachial-extended amygdala circuits. Neuropharmacology, 198, 108757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo AA, Williford KM, Marshall C, Winder DG, & Centanni SW (2020). BNST transient activity associates with approach behavior in a stressful environment and is modulated by the parabrachial nucleus. Neurobiology Stress, 13, 100247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafami M, & Nasimi A. (2016). Contribution of amygdala to the pressor response elicited by microinjection of angiotensin II into the bed nucleus of the stria terminalis. Brain Research Bulletin, 127, 202–207. [DOI] [PubMed] [Google Scholar]
- Kamitakahara A, Xu B, & Simerly R. (2016). Ventromedial hypothalamic expression of Bdnf is required to establish normal patterns of afferent GABAergic connectivity and responses to hypoglycemia. Molecular Metabolism, 5, 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Small CJ, Russell SH, Morgan DGA, Abbott CR, AlAhmed SH, Hay DL, Ghatei MA, Smith DM, & Bloom SR (2002). Effects of melanocortin receptor ligands on thyrotropin-releasing hormone release: Evidence for the differential roles of melanocortin 3 and 4 receptors. Journal of Neuroendocrinology, 14, 276–282. [DOI] [PubMed] [Google Scholar]
- Kishi T, Tsumori T, Ono K, Yokota S, Ishino H, & Yasui Y. (2000). Topographical organization of projections from the subiculum to the hypothalamus in the rat. Journal of Comparative Neurology, 419, 205–222. [DOI] [PubMed] [Google Scholar]
- Lam BYH, Williamson A, Finer S, Day FR, Tadross JA, Soares AG, Wade K, Sweeney P, Bedenbaugh MN, Porter DT, Melvin A, Ellacott KLJ, Lippert RN, Buller S, Rosmaninho-Salgado J, Dowsett GKC, Ridley KE, Xu Z, Cimino I, … O’Rahilly S. (2021). MC3R links nutritional state to childhood growth and the timing of puberty. Nature, 599, 436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen T-M, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, … Wohnoutka PE (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature, 445, 168–176. [DOI] [PubMed] [Google Scholar]
- Lippert RN, Ellacott KLJ, & Cone RD (2014). Gender-specific roles for the melanocortin-3 receptor in the regulation of the mesolimbic dopamine system in mice. Endocrinology, 155, 1718–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Zhang Y-H, Chou TC, Gaus SE, Elmquist JK, Shiromani P, & Saper CB (2001). Contrasting effects of ibotenate lesions of the paraventricular nucleus and subparaventricular zone on sleep–wake cycle and temperature regulation. Journal of Neuroscience, 21, 4864–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malick A, Jakubowski M, Elmquist JK, Saper CB, & Burstein R. (2001). A neurohistochemical blueprint for pain-induced loss of appetite. Proceedings of the National Academy of Sciences, 98, 9930–9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markakis EA, & Swanson LW (1997). Spatiotemporal patterns of secretomotor neuron generation in the parvicellular neuroendocrine. Brain Research Reviews, 24, 255–291. [DOI] [PubMed] [Google Scholar]
- Marks DL, Butler AA, Turner R, Brookhart G, & Cone RD (2003). Differential role of melanocortin receptor subtypes in cachexia. Endocrinology, 144, 1513–1523. [DOI] [PubMed] [Google Scholar]
- Marks DL, & Cone RD (2003). The Role of the melanocortin-3 receptor in cachexia. Annals of the New York Academy of Sciences, 994, 258–266. [DOI] [PubMed] [Google Scholar]
- Mavrikaki M, Girardet C, Kern A, Brantley AF, Miller CA, Macarthur H, Marks DL, & Butler AA (2016). Melanocortin-3 receptors in the limbic system mediate feeding-related motivational responses during weight loss. Molecular Metabolism, 5, 566–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mioni C, Giuliani D, Cainazzo MM, Leone S, Iannone C, Bazzani C, Grieco P, Novellino E, Tomasi A, Bertolini A, & Guarini S. (2003). Further evidence that melanocortins prevent myocardial reperfusion injury by activating melanocortin MC3 receptors. European Journal of Pharmacology, 477, 227–234. [DOI] [PubMed] [Google Scholar]
- Moga MM, Saper CB, & Gray TS (1989). Bed nucleus of the stria terminalis: Cytoarchitecture, immunohistochemistry, and projection to the parabrachial nucleus in the rat. Journal of Comparative Neurology, 283, 315–332. [DOI] [PubMed] [Google Scholar]
- Mohawk JA, Green CB, & Takahashi JS (2012). Central and peripheral circadian clocks in mammals. Neuroscience, 35, 445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens C, Keijzer M, Vries KD, Scheurink A, & Dijk GV (2005). Effects of high-fat diets with different carbohydrate-to-protein ratios on energy homeostasis in rats with impaired brain melanocortin receptor activity. American Journal of Physiology-Regulatory, Integrative and Comparative Physiology, 289, R156–R163. [DOI] [PubMed] [Google Scholar]
- Mountjoy KG, Mortrud MT, Low MJ, Simerly RB, & Cone RD (1994). Localization of the melanocortin-4 receptor (MC4-R) in neuroendocrine and autonomic control circuits in the brain. Molecular Endocrinology, 8, 1298–1308. [DOI] [PubMed] [Google Scholar]
- Pandit R, Omrani A, Luijendijk MCM, Vrind V. A. J. d., Rozen AJV, Ophuis R, Garner K, Kallo I, Ghanem A, Liposits Z, Conzelmann KK, Vanderschuren L, Fleur S. E. l., & Adan RAH (2016). Melanocortin 3 receptor signaling in midbrain dopamine neurons increases the motivation for food reward. Neuropsychopharmacol, 41, 2241–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y-G, Sohn CH, Chen R, McCue M, Yun DH, Drummond GT, Ku T, Evans NB, Oak HC, Trieu W, Choi H, Jin X, Lilascharoen V, Wang J, Truttmann MC, Qi HW, Ploegh HL, Golub TR, Chen S-C, … Chung K. (2019). Protection of tissue physicochemical properties using polyfunctional crosslinkers. Nature Biotechnology, 37, 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei H, Patterson CM, Sutton AK, Burnett KH, Myers MG, & Olson DP (2019). Lateral hypothalamic Mc3R-expressing neurons modulate locomotor activity, energy expenditure, and adiposity in male mice. Endocrinology, 160, 343–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovich GD, Risold PY, & Swanson LW (1996). Organization of projections from the basomedial nucleus of the amygdala: A PHAL study in the rat. Journal of Comparative Neurology, 374, 387–420. [DOI] [PubMed] [Google Scholar]
- Petrovich GD, & Swanson LW (1997). Projections from the lateral part of the central amygdalar nucleus to the postulated fear conditioning circuit. Brain Research, 763, 247–254. [DOI] [PubMed] [Google Scholar]
- Polston EK, Gu G, & Simerly RB (2004). Neurons in the principal nucleus of the bed nuclei of the stria terminalis provide sexually dimorphic GABAergic input to the anteroventral periventricular nucleus of the hypothalamus. Neuroscience, 123, 793–803. [DOI] [PubMed] [Google Scholar]
- Polston EK, & Simerly RB (2003). Sex-specific patterns of galanin, cholecystokinin, and substance P expression in neurons of the principal bed nucleus of the stria terminalis are differentially reflected within three efferent preoptic pathways in the juvenile rat. Journal of Comparative Neurology, 465, 551–559. [DOI] [PubMed] [Google Scholar]
- Renquist BJ, Lippert RN, Sebag JA, Ellacott KLJ, & Cone RD (2011). Physiological roles of the melanocortin MC3 receptor. European Journal of Pharmacology, 660, 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risold PY, Canteras NS, & Swanson LW (1994). Organization of projections from the anterior hypothalamic nucleus: A Phaseolus vulgaris-leucoagglutinin study in the rat. Journal of Comparative Neurology, 348, 1–40. [DOI] [PubMed] [Google Scholar]
- Risold PY, & Swanson LW (1997). Connections of the rat lateral septal complex. Brain Research Reviews, 24, 115–195. [DOI] [PubMed] [Google Scholar]
- Risold PY, Thompson RH, & Swanson LW (1997). The structural organization of connections between hypothalamus and cerebral. Brain Research Reviews, 24, 197–254. [DOI] [PubMed] [Google Scholar]
- Roselli-Rehfuss L, Mountjoy KG, Robbins LS, Mortrud MT, Low MJ, Tatro JB, Entwistle ML, Simerly RB, & Cone RD (1993). Identification of a receptor for gamma melanotropin and other proopiomelanocortin peptides in the hypothalamus and limbic system. Proceedings of the National Academy of Sciences, 90, 8856–8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S. (2005). Role of the central nucleus of the amygdala in the control of blood pressure: Descending pathways to medullary cardiovascular nuclei. Clinical and Experimental Pharmacology and Physiology, 32, 450–456. [DOI] [PubMed] [Google Scholar]
- Saper CB (2000). Hypothalamic connections with the cerebral cortex. Progress in Brain Research, 126, 39–48. [DOI] [PubMed] [Google Scholar]
- Saper CB, & Loewy AD (1980). Efferent connections of the parabrachial nucleus in the rat. Brain Research, 197, 291–317. [DOI] [PubMed] [Google Scholar]
- Shanmugarajah L, Dunigan AI, Frantz KJ, & Roseberry AG (2017). Altered sucrose self-administration following injection of melanocortin receptor agonists and antagonists into the ventral tegmental area. Psychopharmacology, 234, 1683–1692. [DOI] [PubMed] [Google Scholar]
- Simerly RB (1998). Organization and regulation of sexually dimorphic neuroendocrine pathways. Behavioural Brain Research, 92, 195–203. [DOI] [PubMed] [Google Scholar]
- Simerly RB (2002). Wired for reproduction: Organization and development of sexually dimorphic circuits in the mammalian forebrain. Annual Review of Neuroscience, 25, 507–536. [DOI] [PubMed] [Google Scholar]
- Simerly RB (2005). Wired on hormones: Endocrine regulation of hypothalamic development. Current Opinion in Neurobiology, 15, 81–85. [DOI] [PubMed] [Google Scholar]
- Simerly RB (2015). The rat nervous system (4th ed.). Elsevier. [Google Scholar]
- Simerly RB, Swanson LW, Chang C, & Muramatsu M. (1990). Distribution of androgen and estrogen receptor mRNA-containing cells in the rat brain: An in situ hybridization study. Journal of Comparative Neurology, 294, 76–95. [DOI] [PubMed] [Google Scholar]
- Simerly RB, Zee MC, Pendleton JW, Lubahn DB, & Korach KS (1997). Estrogen receptor-dependent sexual differentiation of dopaminergic neurons in the preoptic region of the mouse. Proceedings of the National Academy of Sciences, 94, 14077–14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonds SE, Pryor JT, Ravussin E, Greenway FL, Dileone R, Allen AM, Bassi J, Elmquist JK, Keogh JM, Henning E, Myers MG, Licinio J, Brown RD, Enriori PJ, O’Rahilly S, Sternson SM, Grove KL, Spanswick DC, Farooqi IS, & Cowley MA (2014). Leptin mediates the increase in blood pressure associated with obesity. Cell, 159, 1404–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton AK, Goforth PB, Gonzalez IE, Dell’Orco J, Pei H, Myers MG, & Olson DP (2021). Melanocortin 3 receptor-expressing neurons in the ventromedial hypothalamus promote glucose disposal. Proceedings of the National Academy of Sciences, 118, e2103090118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton GM, Perez-Tilve D, Nogueiras R, Fang J, Kim JK, Cone RD, Gimble JM, Tschöp MH, & Butler AA (2008). The melanocortin-3 receptor is required for entrainment to meal intake. Journal of Neuroscience, 28, 12946–12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton GM, Trevaskis JL, Hulver MW, McMillan RP, Markward NJ, Babin MJ, Meyer EA, & Butler AA (2006). Diet-genotype interactions in the development of the obese, insulin-resistant phenotype of C57BL/6J mice lacking melanocortin-3 or −4 receptors. Endocrinology, 147, 2183–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney J, Kamentsky L, Evans NB, Xie K, Park Y. - G., Drummond, G., Yun, D. H., & Chung, K. (2019). Scalable image processing techniques for quantitative analysis of volumetric biological images from light-sheet microscopy. 10.1101/576595 [DOI] [Google Scholar]
- Swanson LW, & Cowan W. (1975). Hippocampo-hypothalamic connections: Origin in subicular cortex, not ammon’s horn. Science, 189, 303–304. [DOI] [PubMed] [Google Scholar]
- Swanson LW (2000). Cerebral hemisphere regulation of motivated behavior. Brain Research, 886, 113–164. [DOI] [PubMed] [Google Scholar]
- Swanson LW, & Cowan WM (1979). The connections of the septal region in the rat. Journal of Comparative Neurology, 186, 621–655. [DOI] [PubMed] [Google Scholar]
- Swanson LW, & Petrovich GD (1998). What is the amygdala? Trends in Neuroscience (Tins), 21, 323–331. [DOI] [PubMed] [Google Scholar]
- Sweeney P, Bedenbaugh MN, Maldonado J, Pan P, Fowler K, Williams SY, Gimenez LE, Ghamari-Langroudi M, Downing G, Gui Y, Hadley CK, Joy ST, Mapp AK, Simerly RB, & Cone RD (2021). The melanocortin-3 receptor is a pharmacological target for the regulation of anorexia. Science Translational Medicine, 13, eabd6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RH, & Swanson LW (2003). Structural characterization of a hypothalamic visceromotor pattern generator network. Brain Research Reviews, 41, 153–202. [DOI] [PubMed] [Google Scholar]
- Versteeg DHG, Bergen PV, Adan RAH, & Wildt DJD (1998). Melanocortins and cardiovascular regulation. European Journal of Pharmacology, 360, 1–14. [DOI] [PubMed] [Google Scholar]
- Watts AG, Swanson LW, & Sanchez-Watts G. (1987). Efferent projections of the suprachiasmatic nucleus: I. Studies using anterograde transport of Phaseolus vulgaris leucoagglutinin in the rat. Journal of Comparative Neurology, 258, 204–229. [DOI] [PubMed] [Google Scholar]
- West KS, Lu C, Olson DP, & Roseberry AG (2019). Alpha-melanocyte stimulating hormone increases the activity of melanocortin-3 receptor-expressing neurons in the ventral tegmental area. Journal of Physiology, 597, 3217–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, & Tao Y-X (2016). Mutations in Melanocortin-3 receptor gene and human obesity. Progress in Molecular Biology and Translational Science, 140, 97–129. [DOI] [PubMed] [Google Scholar]
- Ye J, & Veinante P. (2019). Cell-type specific parallel circuits in the bed nucleus of the stria terminalis and the central nucleus of the amygdala of the mouse. Brain Structure and Function, 224, 1067–1095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data related to the current study are available from the corresponding author upon reasonable request.