Abstract
Antibiotic resistance is recognised as a global threat to human health by national healthcare agencies, governments and medical societies, as well as the World Health Organization. Increasing resistance to available antimicrobial agents is of concern for bacterial, fungal, viral and parasitic pathogens. One of the greatest concerns is the continuing escalation of antimicrobial resistance among Gram-negative bacteria resulting in the endemic presence of multidrug-resistant (MDR) and extremely drug-resistant (XDR) pathogens. This concern is heightened by the identification of such MDR/XDR Gram-negative bacteria in water and food sources, as colonisers of the intestine and other locations in both hospitalised patients and individuals in the community, and as agents of all types of infections. Pneumonia and other types of respiratory infections are among the most common infections caused by MDR/XDR Gram-negative bacteria and are associated with high rates of mortality. Future concerns are already heightened due to emergence of resistance to all existing antimicrobial agents developed in the past decade to treat MDR/XDR Gram-negative bacteria and a scarcity of novel agents in the developmental pipeline. This clinical scenario increases the likelihood of a future pandemic caused by MDR/XDR Gram-negative bacteria.
Short abstract
Antimicrobial resistance continues to rise among Gram-negative bacteria, leading to greater morbidity, mortality, lengths of stay and costs. The level of resistance is approaching pandemic proportions, requiring an urgent call to address this problem. https://bit.ly/3NTnDqK
Introduction
Respiratory infections are among the most common indications for hospitalisation to include admission to an intensive care unit (ICU) [1]. The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) respiratory virus pandemic is estimated to have accounted for >7 million deaths worldwide [2] and resulted in significant economic distress with the global economy contracting by 3.5% in 2020 [3]. Another “silent” pandemic that has been going on for more than three decades is the increasing prevalence of bacterial infections attributed to multidrug-resistant (MDR) and extremely drug-resistant (XDR) Gram-negative bacteria [4]. The United Nations Interagency Coordination Group on Antimicrobial Resistance Report considers bacterial antimicrobial resistance to be a major threat to human health, and a recent Wellcome Trust report suggests that nearly 300 million individuals will die over the next several decades as a direct result of antimicrobial resistance [5, 6]. Similarly, in the United States, antibiotic-resistant pathogens cause >2 million infections and 23 000 deaths per year, as reported by the United States Centers for Disease Control and Prevention [7]. Despite the introduction of novel antibiotics, the continued escalation of resistance among Gram-negative bacteria suggests that the problem of antibiotic resistance is likely to intensify in the future, leading to antimicrobial inadequacies and a potential full-blown pandemic [8].
The Extended Study on Prevalence of Infection in Intensive Care III (EPIC III) found that among the 15 165 qualifying patients, 8135 (54%) had at least one suspected or proven infection on the study day, with most being respiratory infections [1]. Moreover, antibiotic-resistant pathogens including vancomycin-resistant Enterococcus, Klebsiella species resistant to β-lactam antibiotics or carbapenem-resistant Acinetobacter species were associated with the highest risk of in-hospital death [1]. Given the importance of escalating antibiotic resistance among Gram-negative bacteria as a cause of mortality, morbidity and economic hardship, we brought together in this issue of the European Respiratory Review a multidisciplinary group of authors to discuss this possible next pandemic. The goal of this review is to provide a concise appraisal of the problem of escalating antibiotic resistance among Gram-negative bacteria as an important class of agents for respiratory infections. It is also important to understand that pandemics are not mutually exclusive, as evident by the SARS-CoV-2 pandemic which contributed to outbreaks of MDR/XDR Gram-negative infections including pneumonia [9, 10]. Therefore, as a medical community we must maintain a state of preparedness in order to deal with future pandemics, to which antibiotic resistant Gram-negative bacteria will likely contribute, if not cause outright.
Antibiotic resistant Gram-negative pulmonary infections: key organisms and their epidemiology
According to recent estimates, antibiotic-resistant infections were associated with nearly 5 million deaths globally in 2019 alone [11]. The most common site of infection among these was the lower respiratory tract, which accounted for 1.5 million of the deaths and >75 000 000 disability-adjusted life-years [11]. Although not delineated according to infectious syndrome by the authors, the Gram-negative pathogens associated with the most deaths included Escherichia coli, Klebsiella pneumoniae, Acinetobacter baumannii and Pseudomonas aeruginosa, each associated with >250 000 deaths in 2019, although incidence of the pathogens and death rates varied substantially by region [11].
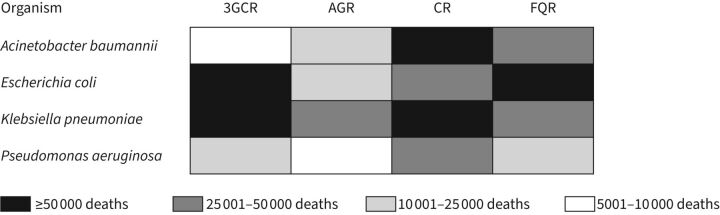
Among individual pathogen–antimicrobial combinations, in 2019, >50 000 deaths each were directly attributable to third-generation cephalosporin-resistant K. pneumoniae and E. coli, carbapenem-resistant A. baumannii and K. pneumoniae, and fluoroquinolone-resistant E. coli (figure 1) [11]. The deaths associated with and attributable to antimicrobial resistance as estimated by the Antimicrobial Resistance Collaborators [11] were derived from a variety of data sources from across the world. However, these estimates were derived from 2019 data among patients who became ill before the onset of the SARS-CoV-2 pandemic. Studies showed that even before the SARS-CoV-2 pandemic, cases of pneumonia due to antimicrobial-resistant Gram-negative bacteria were on the rise in many regions throughout the world [12]. 3 years into the pandemic, we know that patients hospitalised with SARS-CoV-2 receive antimicrobials at very high rates (≥75% in some instances), a situation which will only exacerbate the global antimicrobial resistance crisis that was burgeoning pre-pandemic [13]. In a vicious positive-feedback cycle, patients with SARS-CoV-2 are known to have higher rates of ventilator-associated pneumonia (VAP) [14], a situation which will lead to increased levels of broad-spectrum antimicrobial use, and subsequent development of antimicrobial resistance. In addition, despite the high numbers of antimicrobial-resistant lower respiratory infections found in the aforementioned study [11], this may be an underestimate, as causative pathogens in pneumonia are identified in only ∼30–60% of patients [15–18].
FIGURE 1.
Global deaths in 2019 directly attributable to Gram-negative antimicrobial resistance by pathogen–antimicrobial. Data from [11]. 3GCR: third-generation cephalosporin resistance; AGR: aminoglycoside resistance; CR: carbapenem resistance; FQR: fluoroquinolone resistance.
In the study by the Antimicrobial Resistance Collaborators, methicillin-resistant Staphylococcus aureus (MRSA) and drug-resistant E. coli were the two most common organisms causing mortality [11]. For patients with pneumonia, there is good evidence that MRSA nare swabs have a high negative predictive value and can be used in antimicrobial stewardship efforts to stop unnecessary anti-MRSA antimicrobial use [19]. Unfortunately, there is no easy analogue/stand-in to assist in ruling out drug-resistant Gram-negative pneumonia. Some efforts have attempted to understand the relationship between intestinal colonisation with antimicrobial-resistant Gram-negatives, including those producing extended-spectrum β-lactamases and/or carbapenemases, and the development of future infection, but operationalising such practices has proven challenging and may lead to overtreatment with novel antimicrobials, which must be preserved to prevent development of new resistance [20–23]. Even before the SARS-CoV-2 pandemic, the match between prescribed antimicrobials and recovered pathogens was poor, with MRSA and Pseudomonas being treated far more often than they were detected [24].
At the individual level, patients with antimicrobial-resistant Gram-negative pneumonia are more likely to have been treated previously with antibiotics, be bed-bound, have longer lengths of hospitalisation, have a previous microbiology history of resistant organisms, and to have certain acute and chronic medical comorbidities, with variation by hospital case-mix and region [16, 24–29]. Understanding the risk factors for infection with antibiotic-resistant organisms is crucial for effective empiric treatment decision making. One helpful tool is the “PES” score aimed at identifying antibiotic-resistant Pseudomonas, Enterobacterales and MRSA in patients with pneumonia [30] (table 1).
TABLE 1.
The “PES” score to assess the risk of pneumonia due to Pseudomonas aeruginosa, Enterobacteriaceae with extended-spectrum β-lactamases and methicillin-resistant Staphylococcus aureus pathogens
| Age, years | |
| <40 | 0 |
| 40–65 | 1 |
| >65 | 2 |
| Male | 1 |
| Previous antibiotic use | 2 |
| Chronic respiratory disorder | 2 |
| Chronic renal disease | 3 |
| At emergency | |
| Consciousness impairment | 2 |
| Fever | −1 |
Risk scores for infection are low: ≤1, moderate: 2–4, high: ≥5.
With continued antimicrobial selection pressure, additional mechanisms of antimicrobial resistance are likely to evolve, as has happened with each new antimicrobial that has been developed and used clinically. Identifying risk factors for the development of antimicrobial resistance and the prospective use of algorithms to predict which hospitalised patients will go on to develop antimicrobial resistance may be helpful in certain instances and/or localities, but rapid detection of pathogens and their antimicrobial resistance profiles is likely to be more sensitive for resistance detection [31]. The implementation of rapid molecular diagnostics, especially for pneumonia and bloodstream infections, may be a key step forward in minimising antimicrobial overuse.
Overview of main resistance mechanisms
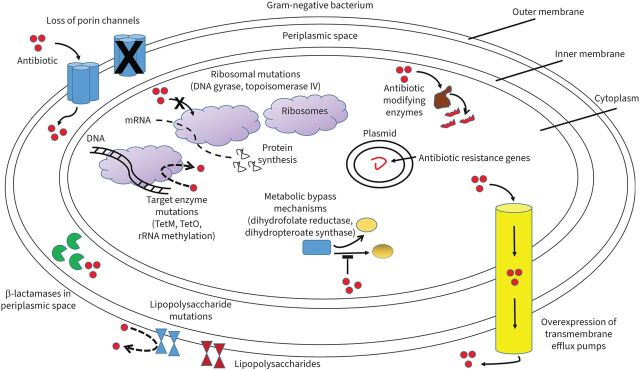
Resistance mechanisms contributing to the development of multidrug-resistant Gram-negative organisms include enzymatic inactivation, efflux pumps, porin mutations and target site modifications [32] (figure 2). Mutations leading to resistance may be intrinsic to the organism or acquired via plasmid-mediated transmission [33]. Each plasmid-mediated conjugation may contain multiple resistance determinants, thus conferring a multidrug-resistant phenotype [32]. Table 2 summarises the relevant resistance mechanisms discussed in this section, in addition to organisms commonly harbouring such mechanisms, the phenotypic result and potential therapeutic approaches.
FIGURE 2.
Mechanisms of antibiotic resistance in Gram-negative bacteria. Shown are loss of porin channels which reduce antibiotic movement across the bacterial membrane; β-lactamases in the periplasmic space inactivating β-lactams; increased transmembrane efflux pump expression expelling antibiotics from within the bacteria; antibiotic-modifying enzymes altering antibiotics so they cannot interact with end targets; antibiotic target and ribosomal mutations interfering with antibiotic actions; metabolic bypass mechanisms allowing alternative enzyme pathways bypassing antibiotic inhibitory effects; and lipopolysaccharide mutations limiting specific antibiotics such as polymyxins from disrupting the cell membrane.
TABLE 2.
Antibiotic mechanisms of resistance of Gram-negative organisms
| Resistance mechanism | Pathogens commonly harbouring mechanism | Antibiotic classes impacted by mechanism | Therapeutic options to overcome mechanism |
| Enzymatic inactivation | |||
| ESBL | Enterobacterales, P. aeruginosa, A. baumannii, S. maltophilia | Penicillins, cephalosporins (all), aztreonam | Carbapenems non-BL based on susceptibility testing |
| Amp-C | Inducible: E. cloacae, C. freundii, K. aerogenes Stable derepressed: E. coli, A. baumannii, Shigella spp. Plasmid-mediated: K. pneumoniae, E. coli, Salmonella spp. |
Strong inducers: cephamycins, aminopenicillins, first-generation cephalosporins Weak inducers: piperacillin-tazobactam, third-generation cephalosporins, aztreonam |
Cefepime Carbapenem non-BL based on susceptibility testing |
| Carbapenemases | |||
| KPC | Enterobacterales | Penicillins, cephalosporins (all), carbapenems, aztreonam | Newer-generation BL/BLI Cefiderocol non-BL based on susceptibility testing |
| MBL (NDM, VIM, IMP) | P. aeruginosa, S. maltophilia, Enterobacterales | Penicillins, cephalosporins (all), carbapenems | Ceftazidime/avibactam PLUS aztreonam cefiderocol non-BL based on susceptibility testing |
| OXA-48 | Acinetobacter, Enterobacterales | Penicillins, cephalosporins (narrow), carbapenems | Ceftazidime/avibactam Cefiderocol non-BL based on susceptibility testing |
| Other inactivating enzymes | |||
| AME | Enterobacterales, Acinetobacter, Pseudomonas, S. maltophilia | Aminoglycosides | Based on susceptibility testing |
| Porin mutations | |||
| OprD protein | Pseudomonas, Acinetobacter | Aminoglycosides, carbapenems | Based on susceptibility testing |
| Omp proteins | Enterobacterales, Acinetobacter | Aminoglycosides, carbapenems, tigecycline | |
| Efflux pumps | |||
| MexAB-OprM | Pseudomonas | β-lactams including carbapenems, macrolides, fluoroquinolones, tetracyclines, TMP/SMX | Based on susceptibility testing |
| MexXY-OprM | Pseudomonas | Aminoglycosides | |
| MexCD-OprJ | Pseudomonas | β-lactams including carbapenems | |
| Target site modification | |||
| 16s rRNA mutation or methylation 30S ribosomal mutation | Pseudomonas, Enterobacterales | Aminoglycosides | Based on susceptibility testing |
| Topoisomerase IV or DNA gyrase mutation | Pseudomonas, Enterobacterales, Acinetobacter, S. maltophilia | Fluoroquinolones | |
| 23S rRNA mutation or methylation 50S ribosomal mutation | Enterobacterales | Macrolides | |
| RNA polymerase mutation | Enterobacterales | Rifampin | |
| DHFR overproduction or DHPS mutation | Enterobacterales | TMP/SMX | |
| Lipid A neutralisation | Enterobacterales, Pseudomonas, Acinetobacter | Colistin | |
| PBP mutation | Pseudomonas | β-lactams including carbapenems |
ESBL: extended-spectrum β-lactamases; KPC: Klebsiella pneumoniae carbapenemases; MBL: metallo-β-lactamases; NDM: New Delhi MBL; VIM: Verona integron-encoded MBL; IMP: imipenem's MBL; OXA: OXA-β-lactamases; AME: aminoglycoside-modifying enzymes; DHFR: dihydrofolate reductase; DHPS: dihydropteroate synthase; PBP: penicillin-binding protein; P. aeruginosa: Pseudomonas aeruginosa; A. baumannii: Acinetobacter baumannii; S. maltophilia: Stenotrophomonas maltophilia; BL: β-lactam; E. cloacae: Enterobacter cloacae; C. freundii: Citrobacter freundii; K. aerogenes: Klebsiella aerogenes; E. coli: Escherichia coli; K. pneumoniae: Klebsiella pneumoniae; BLI: β-lactam inhibitor; TMP: trimethoprim; SMX: sulfamethoxazole.
β-Lactamases
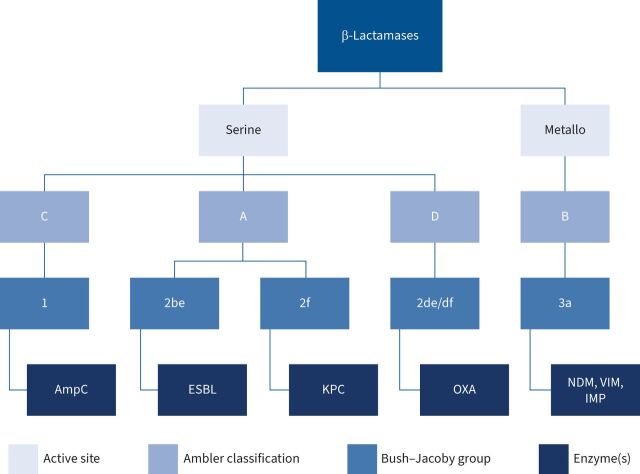
β-Lactamases are categorised molecularly with the Ambler classification based on the enzymatic active site structure and amino acid sequences, dividing them into classes A (narrow and extended-spectrum β-lactamases), C (AmpC cephalosporinases) and D (oxacillinases), which all utilise serine for hydrolysis, and B (metallo-β-lactamases), which utilise zinc ions for hydrolysis [34]. Functionally, β-lactamases are categorised into Bush–Jacoby groups, based upon their hydrolysis and inhibition profiles [34–36]. Phenotypically, β-lactamases include extended-spectrum β-lactamases (ESBLs), AmpC cephalosporinases and carbapenemsases [36]. A summary of β-lactamase classification is displayed in figure 3.
FIGURE 3.
Molecular, functional and phenotypic classification of β-lactamase enzymes. ESBL: extended-spectrum β-lactamase; KPC: Klebsiella pneumoniae carbapenemases; OXA: OXA-β-lactamases; NDM: New Delhi metallo-β-lactamase; VIM: Verona integron-encoded metallo-β-lactamase; IMP: imipenem's metallo-β-lactamase.
ESBLs
ESBLs confer resistance to penicillins, cephalosporins including extended-spectrum (third and fourth generations) and monobactams, and are categorised as Ambler class A and Bush–Jacoby group 2 [35, 37]. This group of β-lactamases is genetically diverse, with the most common mutations including TEM, SHV and CTX-M isolated in Enterobacterales species [32, 38]. CTX-M-15 is the most common variant worldwide, but there are nearly 500 recognised variants of the various mutations [35]. Contemporary ESBLs are reliably inhibited by newer β-lactamase inhibitors, including avibactam, relebactam and vaborbactam; however, older-generation β-lactamase inhibitors including clavulanate, sulbactam and tazobactam may succumb to the inoculum effect or may be rendered ineffective against isolates harbouring inhibitor-resistant β-lactamases [35, 39, 40]. OXA-type ESBLs (Ambler class D) are less common, but include derivations of OXA-10 and OXA-2, commonly detected in isolates of P. aeruginosa [35].
AmpC
AmpC β-lactamases are classified as Ambler class C and Bush–Jacoby group 1 and are often isolated in Enterobacterales species [32, 41]. These enzymes result from either the inducible or stable derepression of ampC regulatory protein production or from transmission of plasmid-mediated ampC genes [42, 43]. Susceptibility testing of an organism with stable derepressed AmpC production will demonstrate resistance to third-generation cephalosporins and cephamycins, while inducible AmpC producing organisms will initially test susceptible to third-generation cephalosporins. These inducible organisms become resistant upon exposure to certain β-lactams, which causes disabling of the negative regulator protein, AmpR, resulting in increasing production of AmpC [42]. Potent inducers of AmpC production include the aminopenicillins, first-generation cephalosporins, and cephamycins, while weak inducers include piperacillin-tazobactam, aztreonam, and third-generation cephalosporins, all of which may be hydrolysed in varying levels of AmpC production. Cefepime and carbapenems are inducers of AmpC production, but withstand its hydrolysis [42].
Carbapenemases
Carbapenemases include β-lactamases from Ambler class A (K. pneumoniae carbapenemases (KPC)), class B (New Delhi metallo-β-lactamases (NDM), Verona integron-encoded metallo-β-lactamases (VIM) and imipenem's metallo-β-lactamases (IMP)) and class D (OXA-48 variants) [36]. Carbapenemase enzymes are isolated in Enterobacterales, Pseudomonas and Acinetobacter species [44].
KPCs are the most clinically relevant of the class A carbapenemases and have been identified across the globe [44–46]. Despite their nomenclature, these genes are found on transferable plasmids and are isolated in other Enterobacterales species, in addition to K. pneumoniae [47].
The three subclasses of metallo-β-lactamases (MBLs) (NDM, VIM and IMP) render resistance to carbapenems via hydrolysis utilising zinc, and they are not inhibited by available β-lactamase inhibitors [44, 48].
OXA-β-lactamases, specifically OXA-48 variants, have been isolated in Enterobacterales species. Because of their heterogeneity, the global prevalence of OXA-48 β-lactamases may be underestimated [44].
In addition to carbapenems, most carbapenemases also hydrolyse penicillins and cephalosporins, while only KPCs hydrolyse monobactams [45, 46]. Similar to ESBLs, KPCs are generally inhibited by traditional β-lactamase inhibitors, while OXA carbapenemases are not [45, 46]. KPCs are additionally susceptible to all of the newer-generation β-lactamase inhibitors. OXA carbapenemases are not reliably inhibited by vaborbactam, but demonstrate variable susceptibility to avibactam and relebactam [49]. Gram-negative organisms may also produce aminoglycoside-modifying enzymes (AMEs), including acetyltransferases, nucleotidyltransferases and phosphotransferases, all of which inactivate aminoglycosides [50, 51]. Less commonly, fluoroquinolone-, macrolide- and rifampin-inactivating enzymes may be produced by various Enterobacterales, Pseudomonas and Acinetobacter species [33].
Target site modification
Although alterations in penicillin-binding proteins are commonly observed with Gram-positive organisms, Pseudomonas species may harbour such mechanisms, leading to reduced β-lactam susceptibility [33]. Modification of fluoroquinolone target proteins occurs by mutations of gyrA/gyrB and parC/parE genes [32, 52], while aminoglycoside binding site alteration occurs by mutation of the 30S ribosomal subunit or by production of 16s rRNA methyltransferases via organisms harbouring armA and various rmt genes [50, 51]. Additionally, resistance to colistin is observed with alterations in the negative charge of the bacterial cell membrane, which prevents binding and insertion of the positively charged drug at its target site [33].
Efflux pumps
In nonfermenting Gram-negative bacteria, such as Pseudomonas species, resistance to β-lactams and fluoroquinolones is often caused by overproduction of the MexAB-OprM efflux pump, whereas overproduction of MexXY-OprM transporters results in resistance to aminoglycosides [32, 52, 53]. This overproduction is mediated by mutations in the nalB, nfxB and nfxC genes [52].
Porin mutations
Porin channels play an important role in antibiotic uptake into Gram-negative bacteria [32]. Mutations resulting in inactivation or decreased expression of porin proteins, most notably OprD, leads to decreased permeability of several antibiotic classes including carbapenems, aminoglycosides and fluoroquinolones [32, 51, 52].
Gram-negative bacteria can harbour multiple resistance mechanisms that manifest clinically as a multidrug resistance phenotype [32]. Multiple β-lactamases may coexist, such as the combination of carbapenemases with an AmpC or ESBL, which, along with the presence of an AME-, ESBL- and AmpC-producing organism also having porin mutations results in a slowed rate of bacterial penetration, facilitating enzymatic hydrolysis of β-lactams, including carbapenems [44]. Additionally, the presence of such β-lactamases in combination with various drug efflux pumps or alterations in target sites can also confer resistance to other classes of antibiotics, and thus the manifestation of multidrug resistant organisms [44]. The growing prevalence and global impact of such multidrug resistance is of great concern.
Impact of Gram-negative antibiotic-resistant pulmonary infection on outcomes
There is a consensus that timely and appropriate antibiotic treatment, defined as an antibiotic regimen with in vitro activity against the causative pathogens, is a necessary first step to optimise the outcomes of patients with serious infections, especially in the ICU setting [54–58]. The guiding principle of timely appropriate antibiotic treatment of serious infections is endorsed by the most recent 2021 Surviving Sepsis Campaign International Guidelines for the management of sepsis and septic shock with a strong recommendation [59]. Kumar et al. [58] demonstrated that for every hour's delay until appropriate antibiotic administration, crude mortality increased by more than 10%. Vazquez-Guillamet et al. [60] studied >1000 subjects with septic shock and calculated that appropriate therapy enhanced the likelihood of survival at least three-fold. More importantly, this converted into a number needed to treat (NNT) to save one life of only 4, and the prevalence-adjusted pathogen-specific NNT to prevent one patient death was lowest for infections caused by MDR bacteria (NNT=20) [60]. Thus, both appropriate antibiotic selection and their timely administration are necessary to optimise patient outcomes.
Bassetti et al. [61] summarised the data regarding appropriate early therapy of serious infections in a meta-analysis of 114 studies of appropriate therapy in severe bacterial infections including pneumonia. Appropriate initial antibiotic therapy not only significantly reduced in-hospital mortality (OR 0.44, 95% CI 0.38–0.50), but also reduced length of stay by >2.5 days [61]. Delayed appropriate antibiotic therapy has also been shown to be associated with greater mortality in VAP [62]. Moreover, several nosocomial pneumonia registration trials for agency drug approval have demonstrated that when inactive antibiotic therapy is administered to patients, primarily inactive due to inadequate dosing or antibiotic exposure, mortality is increased [63–66]. These studies confirm the importance of delivering an appropriate and adequately dosed antibiotic regimen to patients with bacterial lung infections in order to improve patient outcomes, especially hospital survival.
The strongest evidence supporting early appropriate antibiotic therapy for severe infections comes from a randomised prospective trial examining this issue. The MERINO trial compared piperacillin-tazobactam to meropenem in patients with bloodstream infection caused by ceftriaxone-nonsusceptible E. coli or K. pneumoniae; in essence, it was a trial of appropriate versus inappropriate therapy [57]. The source of infections was primarily urinary tract and intra-abdominal with <5% being pneumonia. Noninferiority of the piperacillin-tazobactam arm could not be established, with 12.3% of 187 patients randomised to piperacillin-tazobactam dying at 30 days compared with 3.7% of 191 patients randomised to meropenem [57]. Microbiological failures were also more common among patients randomised to receive pipercillin-tazobactam when infected with Amp-C overexpressing Gram-negative bacteria [67].
MDR and XDR Gram-negative bacterial infections are more likely to initially receive inappropriate antibiotic treatment, resulting in worse outcomes. Zilberberg et al. [68] studied 1064 patients with Gram-negative bacteraemia, of whom 351 (29.2%) did not survive hospitalisation. Nonsurvivors were significantly more likely to have infection with an MDR isolate and to have received inappropriate initial antibiotic therapy. A multivariate analysis demonstrated that presence of infection with an MDR isolate was strongly associated with the receipt of inappropriate initial antibiotic therapy (adjusted OR 13.05, 95% CI 7.00–24.31). The same group of investigators also evaluated 1423 patients from United States hospitals with pneumonia or sepsis due to A. baumannii [69]. Harbouring MDR A. baumannii increased the risk of receiving inappropriate initial antibiotic therapy more than five-fold, and inappropriate initial antibiotic therapy nearly doubled hospital mortality. More recently, Martinez-Nadal et al. [70] evaluated 1615 episodes of bacteraemia associated with neutropenia, of which 394 (24%) received inappropriate initial antibiotic therapy. Patients with MDR Gram-negative bacteria, accounting for 221 (14%) of all isolates, were more likely to receive inappropriate initial antibiotic therapy (39% versus 7%, p<0.001). Overall mortality was also higher in patients with Gram-negative bacteraemia who received inappropriate initial antibiotic therapy (36% versus 24%, p=0.004). Thus, it can be seen that the presence of antibiotic resistance among Gram-negative bacteria is associated with greater initial administration of inappropriate antibiotic therapy, which in turn leads to greater mortality.
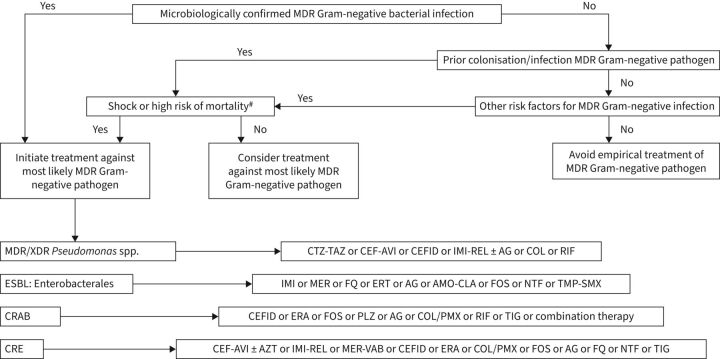
The problem of antimicrobial resistance appears to be escalating. Data from the National Healthcare Safety Network from 2015 to 2017 show that among P. aeruginosa isolates in ICU patients, 26.3% were resistant to carbapenems, 26.5% were resistant to extended-spectrum cephalosporins and 27.1% were resistant to fluoroquinolones, with 18.6% classified as MDR [71]. Increasing resistance has led clinicians to employ antimicrobial “cocktails” in the hopes of salvaging patients with MDR/XDR infections [72, 73]. Verona integron-encoded metallo-β-lactamase-positive P. aeruginosa (VIM-PA) has emerged as one of the most resistant Gram-negative pathogens, along with carbapenem-resistant A. baumannii. In a study from the Netherlands, VIM-PA bacteraemia was associated with greater mortality compared to carbapenems-susceptible infection [74]. Thus, it is evident that increasing future infections with MDR/XDR pathogens will be associated with greater morbidity and mortality, particularly in respiratory infections. Figure 4 offers an approach to the clinical management of antibiotic-resistant Gram-negative infections in the hospital setting.
FIGURE 4.
Clinical approach to the treatment of microbiologically confirmed or suspected multidrug-resistant (MDR) Gram-negative bacterial infection. XDR: extremely drug-resistant; CTZ-TAZ: ceftolozane/tazobactam; CEF-AVI: centazidime-avibactam; CEFID: cefiderocol; IMI-REL: imipenem-cilastatin; AG: aminoglycoside; RIF: rifampicin; ESBL: extended-spectrum β-lactamase producing; MER: meropenem; FQ: fluoroquinolone; ERT: ertapenem; AMO-CLA: amoxicillin-clavulanate; FOS: fosfomycin; NTF: nitrofurantoin; TMP-SMX: trimethoprim-sulfamethoxazole; CRAB: carbapenem-resistant Acinetobacter baumannii; ERA: eravacycline; PLZ: plazomicin; COL-PMX: colistin-polymyxin B; TIG: tigecycline; CRE: carbapenem-resistant Enterobacterales; AZT: aztreonam; MER-VAB: meropenem-vaborbactam. #: high risk of mortality is considered >15%.
Novel and pipeline antibiotics
Several antibiotics have been introduced to combat Gram-negative resistance and expand coverage against MDR Gram-negative infections, including respiratory tract infections (table 3) [75]. Their precise role in empirical and definitive treatment is beyond the scope of this review; however, well-written reviews and guidelines highlight the importance of pertinent risk factors, current and prior culture and susceptibility data, local resistance rates, available clinical and/or in vitro data and other factors [75–77]. Moreover, the use of novel antibiotics as combination therapy provides a unique approach to the treatment of MDR Gram-negative infections. For example, ceftazidime/avibactam has broad activity against serine β-lactamases; however, it is hydrolysed by MBL. In contrast, aztreonam is resistant to MBL hydrolysis, but susceptible to serine β-lactamases. Available in vitro and in vivo data have demonstrated synergistic activity with this combination against MBL-producing Gram-negative organisms [78–81].
TABLE 3.
Novel and pipeline antibiotic treatment options for Gram-negative respiratory infections
| Drug class and indication(s) # | Notable activity¶ | Development phase | Comments | |
| Novel antibiotics | ||||
| Ceftolozane/ tazobactam | BL/BLI HABP/VABP | ESBL-E, MDR P. aeruginosa | FDA/EMA approved | Verify ESBL-E No CRE activity |
| Ceftazidime/ avibactam | BL/BLI HABP/VABP | MDR P. aeruginosa, CRE (class A, KPC), AmpC and ESBL-E | FDA/EMA approved | No MBL activity Not reliable against A. baumannii Limited anaerobic activity |
| Imipenem/ cilastatin/ relebactam | BL/BLI HABP/VABP | CRE (KPC), ESBL-E, MDR P. aeruginosa | FDA/EMA approved | No additional activity against A. baumannii, compared to imipenem No MBL activity Anaerobe and MSSA activity |
| Meropenem/ vaborbactam | BL/BLI HABP/VABP | CRE (KPC), ESBL-E, non-MDR P. aeruginosa and non-MDR A. baumannii | FDA/EMA approved | No additional activity against MDR P. aeruginosa or MDR A. baumannii compared to meropenem No MBL activity Anaerobe and MSSA activity |
| Cefiderocol | BL HABP/VABP | CRE (KPC and MBL), ESBL-E, MDR A. baumannii and P. aeruginosa | FDA/EMA approved | Inactive against Gram-positives and anaerobes |
| Delafloxacin | Fluoroquinolone CABP |
ESBL-E | FDA/EMA approved | Limited P. aeruginosa activity Broad Gram-positive and atypical coverage |
| Eravacycline | Fluorocycline No indications |
CRE (KPC and MBL), EBSL-E, MDR A. baumannii | FDA/EMA approved | Limited clinical efficacy data against MDR infections Often lower MICs than tigecycline Low rates of and in vitro activity against C. difficile |
| Omadacycline | Tetracycline CABP | ESBL-E, KPC, MBL, MDR A. baumannii | FDA/EMA approved | No Pseudomonas or Proteus spp. activity Broad Gram-positive and atypical coverage |
| Plazomicin | Aminoglycoside No indications |
CRE (KPC and MBL), ESBL-E | FDA/EMA approved | Variable P. aeruginosa activity No A. baumannii activity Limited safety and efficacy data Consider in combination therapy regimens |
| Pipeline antibiotics | ||||
| Arbekacin | Aminoglycoside | Aminoglycoside-inactivating Gram-positive and Gram-negative pathogens | Phase III | Highly active against MRSA and shows activity against MDR-P. aeruginosa and -A. baumannii |
| Aztreonam/ avibactam | Monobactam/BLI | ESBL, KPC, MBL, AmpC, OXA-48 | Phase III | No increased activity to P. aeruginosa compared to aztreonam monotherapy No A. baumannii activity |
| Cefepime/ enmetazobactam | BL/BLI | ESBL, AmpC, limited evidence of KPC and OXA-48 | Phase III | No increased activity to P. aeruginosa compared to cefepime monotherapy No class B β-lactamase activity |
| Cefepime/ taniborbactam | BL/BLI | ESBL, KPC, class B β-lactamases (VIM, NDM, SPM-1, GIM-1), AmpC, OXA-48 | Phase III | Adds activity to cefepime against CRE and carbapenem-resistant P. aeruginosa |
| Cefepime/ zidebactam | BL/BLI | ESBL, KPC, MBLs, AmpC, OXA-48 | Phase III | Active against carbapenem-resistant P. aeruginosa Limited Acinetobacter spp. activity |
| Murepavadin | OMPTA | Highly active against MDR P. aeruginosa | Phase III | Showed activity against colistin, ceftolozane/tazobactam and tobramycin nonsusceptible P. aeruginosa |
| Sulopenem | Carbapenem | ESBL, AmpC | Phase III | Does not provide additional activity to current carbapenems |
BL: β-lactam; BLI: β-lactamase inhibitor; HABP: hospital-acquired bacterial pneumonia; VABP: ventilator-associated bacterial pneumonia; ESBL-E: extended spectrum β-lactamase Enterobacterales; MDR: multidrug-resistant; P. aeruginosa: Pseudomonas aeruginosa; FDA: United States Food and Drug Administration; EMA: European Medicines Agency; CRE: carbapenem-resistant Enterobacteriaceae; KPC: Klebsiella pneumoniae carbapenemase; MBL: metallo-β-lactamase; A. baumannii: Acinetobacter baumannii; MSSA: methicillin-sensitive Staphylococcus aureus; CABP: community-acquired bacterial pneumonia; MIC: minimum inhibitory concentration; C difficile: Clostridium difficile; OXA: OXA-β-lactamases; VIM: Verona integron-encoded metallo-β-lactamases; NDM: New Delhi metallo-β-lactamases. #: EMA or FDA; ¶: inclusive of in vitro and in vivo data, which may not correlate with clinical efficacy.
Pipeline antibiotics will provide extended Gram-negative coverage and may become useful options for the treatment of MDR Gram-negative respiratory infections (table 3). For example, the addition of the novel β-lactamase inhibitor zidebactam to the β-lactam cefepime improves activity against KPC, ESBL, OXA-48 and MBLs [82, 83]. The expanded spectrum provided by and mechanism of β-lactam/β-lactamase inhibitor combinations differs among agents [49, 82]. Avoidance of aminoglycoside-inactivating enzymes affords the novel synthetic aminoglycoside, arbekacin, improved activity against MDR Gram-negatives and Gram-positives [84]. Murepavadin highlights a novel drug class of outer membrane protein targeting antibiotics by binding to the lipopolysaccharide transport protein D present on the outer membrane of MDR P. aeruginosa [85]. Initially encouraging phase I and II data have been stifled by the termination of the phase III PRISM-MDR trial due to safety concerns surrounding adjunctive use of murepavadin in patients with MDR P. aeruginosa VAP [86]. These data emphasise the need for ongoing study of all novel antimicrobials for MDR Gram-negative respiratory infections.
Monoclonal antibodies
The success of monoclonal antibodies (mAbs) for treatment and/or prevention of various diseases has reinvigorated consideration in treatment and prevention of bacterial infections. Antibacterial mAbs may disrupt bacterial infections via several mechanisms; however, targeting polysaccharides, proteins or toxins associated with proliferation, adhesion, host immunity protection, host cell damage and/or increased virulence are most common (table 4) [87, 88]. Encouraging results in prevention or treatment of Clostridioides difficile, inhalational anthrax and MRSA infections have been reported; however, data in Gram-negative respiratory infections remain preliminary [89–91].
TABLE 4.
Monoclonal antibody treatment options for Gram-negative respiratory infections
| Target | Gram-negative target(s) | Development phase | Studied place in therapy | |
| A1102 | LPS-O-antigen D-galactan-III | K. pneumoniae | Pre-clinical | Prevention |
| A1124 | LPS-O-antigen o25b | E. coli | Pre-clinical | Prevention |
| AR-101 | LPS 011 exopolysaccharide | P. aeruginosa | Phase II | Treatment |
| AR-105 | Alginate exopolysaccharide | P. aeruginosa | Phase II | Prevention and treatment |
| Anti-Hyr1 | Hyr1 peptide 5 | A. baumannii and K. pneumoniae | Pre-clinical | Prevention |
| KB001-A | Pcrv protein | P. aeruginosa | Phase II/III | Prevention |
| MEDI3902 | Pcrv protein and Psl exopolysaccharide | P. aeruginosa | Phase II | Prevention |
LPS: lipopolysaccharide; K. pneumoniae: Klebsiella pneumoniae; E. coli: Escherichia coli; P. aeruginosa: Pseudomonas aeruginosa; A. baumannii: Acinetobacter baumannii.
The mAb KB001-A targets the PcrV protein on P. aeruginosa, reducing levels of toxin transport and lung injury/inflammation in animal models [92]. In cystic fibrosis patients, KB001-A was not associated with decreased time to need for antibiotics, but increased forced expiratory volume in 1 s and reduced inflammatory markers [92]. A randomised phase II trial of mechanically ventilated patients colonised with P. aeruginosa reported no difference in incidence of treatment-related adverse events, but reduced incidence of P. aeruginosa pneumonia development in patients receiving KB001-A [93]. The EVADE trial is assessing the ability of MEDI3902, a bivalent/multi-target mAb, to prevent P. aeruginosa infections in patients mechanically ventilated colonised with P. aeruginosa [94]. Panobacumab and AR-105 target polysaccharides produced by P. aeruginosa and are under investigation as adjuncts with standard-of-care antibiotics for nosocomial P. aeruginosa infections [95, 96]. Continued advancements in molecular modelling and bioinformatics are likely to yield additional targets for pathogens associated with MDR Gram-negative respiratory infections [97].
Antibacterial mAbs show great promise for the treatment and prevention of Gram-negative bacterial respiratory infections. Noteworthy advantages include target specificity, reduced risk of resistance emergence, minimal disturbance of gut microbiota and unique pharmacokinetic profiles. Still, limitations related to target specificity, cost, antibody development, administration, availability and clinical data hamper use in routine clinical practice [87]. Results of ongoing clinical trials are awaited to properly assess their efficacy, safety and appropriate place in therapy.
Bacteriophage therapy
Bacteriophages (phages) are the most abundant viruses/organisms on Earth and can infect and replicate within bacterial cells, causing lysis [98]. Leveraging phages to target pathogenic bacteria has seen a resurgence due to threats of antimicrobial resistance and advancements in molecular engineering (table 5). Bacteriophages offer several presumed advantages related to their specificity and ubiquity, including minimal effects on microbiota, safety profile, administration flexibility, and self-limiting properties. However, there is a paucity of data in these regards, combined with unknowns surrounding monitoring, toxin release, antibody production and inevitable development of resistance. Considerations of the unique interplay with bacteria, antibiotics and the immune system are necessary to optimise delivery and production, minimise or leverage resistance and understand their safety.
TABLE 5.
Bacteriophage therapies for Gram-negative respiratory infections
| Phage components | Bacterial target | Development phase | Administration route | |
| AB-PA01 cocktail | Pa193, Pa204, Pa222, Pa223 | P. aeruginosa | Pre-clinical | Intravenous ± nebulised |
| AB-PA01 m1 | Pa193, Pa204, Pa222, Pa223, Pa176 | P. aeruginosa | Pre-clinical | Intravenous ± nebulised |
| Acinetobacter therapies | 2ϕ ± ϕAb124 | A. baumannii | Pre-clinical | Topical or nebulised |
| Achromobacter cocktail | Not specified | A. xylosoxidans | Pre-clinical | Oral + nebulised |
| Navy Phage cocktail 1 | Paϕ1, PaSKWϕ17, PaSKWϕ22 | P. aeruginosa | Pre-clinical | Intravenous ± nebulised |
| Navy Phage cocktail 2 | PaATFϕ1 and PaATFϕ3 | P. aeruginosa | Pre-clinical | Intravenous |
| Adaptive Therapeutics phage | BdPF16phi4281 | B. dolosa | Pre-clinical | Intravenous |
P. aeruginosa: Pseudomonas aeruginosa; A. baumannii: Acinetobacter baumannii; A. xylosoxidans: Achromobacter xylosoxidans; B. dolosa; Burkholderia dolosa.
Clinical evidence for phages is limited to case series and reports. A systematic review highlighted the potential of phages in difficult-to-treat respiratory infections [99]. One such case described success of adjunctive phage therapy for the treatment of a P. aeruginosa VAP and bronchopleural fistula after thoracotomy, resulting in both clinical improvement and bacterial eradication at 6-month follow-up [100]. Phage therapy has exceptional potential for patients with MDR Gram-negative respiratory infections; however, well-designed clinical trials are still necessary.
Oligonucleotides
Antimicrobial oligonucleotides are synthetic nucleic acid sequences able to silence genes vital to bacterial survival (table 6) [98]. In vivo and in vitro data suggest that oligonucleotides may serve as a future treatment modality of severe Gram-negative respiratory infections [98]. Oligonucleotides targeting the genes MexB, blaNDM-1, and CTX-M-15 have resulted in improved minimum inhibitory concentration (MIC) profiles of common antimicrobials [101, 102]. Like mAbs and phages, oligonucleotides offer a target-specific method with potentially longer-lasting effects and reduced resistance development; however, limitations surrounding their adequate delivery, use with antibiotics and unestablished safety and efficacy profile highlight the need for continued study.
TABLE 6.
Oligonucleotides for Gram-negative respiratory infections
| Gene silenced and typical function | Silencing clinical result | Development phase | Bacterial target | |
| CTX-M-15 ASO |
blaCTX−M-15 Encodes resistance to third-generation cephalosporins |
Reduced MIC to third-generation cephalosporins | Pre-clinical | E. coli; other Gram-negatives |
| MexB siRNA |
mexB Encodes efflux pump component |
Restored activity of resistant antibiotics | Pre-clinical | E. coli; other Gram-negatives |
| NDM-1 ASO |
blaNDM-1 Encodes carbapenemases |
Restored activity of resistant antibiotics | Pre-clinical | P. aeruginosa; other Gram-negatives |
NDM: New Delhi metallo-β-lactamases; MIC: minimum inhibitory concentration; E. coli: Escherichia coli; P. aeruginosa: Pseudomonas aeruginosa.
Role of stewardship in curbing MDR spread
Antimicrobial stewardship programmes (ASPs) are an essential tool in helping prevent the spread of MDR respiratory infections, and involve a multidisciplinary approach to ensure that patients receive appropriate initial antibiotic therapy, appropriate dosing and route of administration, as well as sufficient duration of antimicrobial therapy [103, 104]. The goal of ASPs is to improve clinical outcomes of patients, decrease costs associated with antimicrobial use and control the spread of resistant organisms [105–107]. It is increasingly recognised that ASPs are an essential part of management of hospitalised patients, especially those in the ICU, and its use is recommended by the Infectious Diseases Society of America and international sepsis guidelines [59, 104, 108].
The ICU is an essential area for ASPs, due to the frequent use of antibiotics, high levels of MDR organisms and implications for poor clinical outcomes in patients treated with inappropriate antibiotics [105–107, 109]. Despite more than half of patients in the ICU having suspected or confirmed infection, antibiotic use is in the ICU is still frequently overly broad or too narrow [1, 106]. The consequence of overly broad antimicrobial use in the ICU leads to the development of antimicrobial-resistant organisms, which is associated with increased mortality [1, 110, 111]. Infection with an MDR organism is also associated with increased all-cause mortality and increased likelihood of hospital readmission [112].
Respiratory infections in critically ill patients represent an important area for antimicrobial stewardship, and pneumonia is the most common infection found in the ICU [1]. Initial antibiotic therapy that is inappropriately narrow has been shown to be associated with an increased risk of mortality, while initial antibiotic therapy that is inappropriately broad-spectrum has also been associated with increased risk of mortality [113]. Appropriate duration of antibiotic therapy is also an important aspect of ASPs, as unnecessarily prolonged courses of antibiotics are associated with adverse patient outcomes and the development of MDR organisms [111, 113]. Teshome et al. [111] showed that each additional day of broad-spectrum antibiotic use directed against P. aeruginosa was associated with an increased risk of development of MDR infection. The benefits of shorter courses of antibiotics for pneumonia were shown in a systematic review and meta-analysis of >1000 patients with hospital-acquired pneumonia (HAP) and VAP found that a shorter course of antibiotics resulted in increased 28-day antibiotic-free days as well as a reduction in recurrent VAP due to MDR organisms [114].
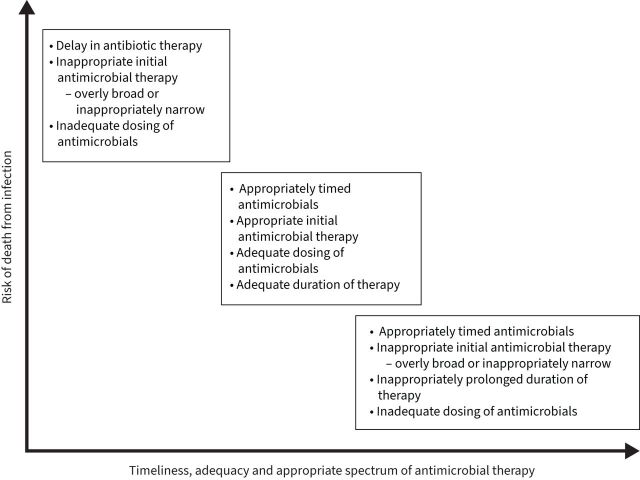
In a retrospective cohort analysis of >17 000 patients admitted to the hospital with sepsis and positive cultures, Rhee et al. [113] found that overly broad empirical antibiotic therapy was associated with increased mortality, with an estimated 20% increase in the odds of death. The adverse effects of overly broad antibiotic therapy were also seen in a retrospective study of over 1900 patients admitted to the hospital with pneumonia, which showed that the use of broad-spectrum antibiotics in community-onset pneumonia was associated with longer hospital stays, increased rates of Clostridium difficile infection and increased mortality [115]. Unnecessarily broad antibiotic therapy has the unfortunate consequence of disrupting the microbiome, which refers to the numerous bacteria, fungi and viruses that exist in a symbiotic relationship with a human body [116, 117]. The use of broad-spectrum antibiotics has been shown to alter the beneficial bacteria that make up the microbiome, leading to dysbiosis and placing the patient at risk for colonisation with MDR organisms and subsequent opportunistic infections [116]. ASPs can be beneficial in preserving the host microbiome by limiting unnecessary antimicrobial use and limiting length of therapy to an appropriate duration. The importance of appropriate timing, duration and spectrum of antimicrobial activity is detailed in figure 5.
FIGURE 5.
The importance of appropriate timing, duration and spectrum of antimicrobial activity.
The success of an ASP is dependent on several key elements and must include a multidisciplinary approach that utilises the expertise of ICU physicians, infectious disease physicians, pharmacists and microbiologists [104, 106]. Several of the key elements that are recommended by the United States Centers for Disease Control and Prevention for a successful ASP are detailed in table 7 [104]. Prospective audit and feedback of antibiotic use is an effective aspect of an ASP, which functions as a review of broad-spectrum antibiotic therapy leading to antibiotic de-escalation when appropriate [103, 104]. It has been shown to be associated with a reduction in use of broad-spectrum antibiotics in critically ill patients, reduction in resistance of Gram-negative organisms, with no adverse consequences on mortality or ICU length of stay [103, 108, 118].
TABLE 7.
Key components of an effective antimicrobial stewardship programme
| Description | Outcomes | |
| Pre-authorisation | Prescribers are required to gain approval prior to use of certain antimicrobials | Ensures appropriate antibiotic selection and dosing Prevents unnecessary initiation of antibiotics |
| Prospective audit and feedback | External review of antibiotics after initiation, with suggestions from experts on optimal use | Reduction in unnecessary use of broad-spectrum antibiotics |
| Facility-specific treatment guidelines | Specific treatment guidelines for common conditions, such as community-acquired pneumonia, UTI and surgical prophylaxis, taking into account national guidelines and local susceptibilities and formulary options | Simplifies antibiotic prescribing for common conditions Allows for consensus between stewardship team and prescribers |
| Antibiotic time-outs | Provider-led reassessment of the continuing need for antibiotics after additional clinical information has returned | Improves the appropriateness of antibiotic use Encourages antibiotic de-escalation when appropriate |
| Pharmacy-based interventions | Prescribing interventions by pharmacy to optimise antibiotic use May be incorporated into electronic health record |
Dose optimisation, dose adjustments based on renal function, changing intravenous antibiotics to oral formulation |
| Rapid diagnostics | Early pathogen identification using technology such as PCR to identify bacterial infection prior to awaiting culture results | Earlier identification of causative organism can lead to antibiotic de-escalation and optimisation of antibiotics |
| Clinical decision support system | Incorporation of clinical and patient-specific data into electronic health record to provide providers with relevant information prior to prescribing antibiotics | Decreased overall antibiotic use and improved use of antibiotics in patients with sepsis |
UTI: urinary tract infection
Clinical decision support systems are another important part of an effective ASP, which are designed to incorporate numerous clinical and patient-specific data in order to provide more information to clinicians so that they can make more appropriate decisions regarding antimicrobial therapy. Previous studies have shown that clinical support tools that make antibiotic recommendations using patient data and antibiogram information can decrease overall use of antibiotics [119] as well as improve initial antibiotic therapy in patients with sepsis [120].
Rapid diagnostic testing to identify a source of infection remains an area of need for ASPs in the ICU. Earlier identification of causative organisms, as well as rapid diagnostics that are sensitive in ruling out specific organisms that are treated with broad-spectrum antimicrobials can lead to rapid de-escalation of antibiotics [103]. A randomised controlled trial of 45 mechanically ventilated patients with suspected pneumonia showed that testing with a PCR of bronchoalveolar lavage fluid for MRSA resulted in significant reduction in vancomycin and linezolid use when MRSA was not detected compared to the control group [121]. PCR-based assays for detection of Gram-negative organisms remains a challenging area, but recent studies have demonstrated good sensitivity of PCR-based assays in detecting respiratory pathogens, including detection of potential resistant organisms, which could lead to improvements in antimicrobial prescribing [122, 123].
One alternative to empiric broad-spectrum antibiotics for critically ill patients with a suspected HAP/VAP is to utilise surveillance cultures to guide initial antimicrobial therapy. If MDR pathogens are found on surveillance cultures, initial antibiotic therapy can be directed towards that pathogen, while more narrowed antibiotics could be used in a patient without MDR colonisation. Multiple studies have investigated the use of surveillance cultures, with varying degrees of sensitivity and specificity, depending on the sites of surveillance as well as the frequency of surveillance cultures [124]. A prospective observational cohort study investigated the role of surveillance cultures for identifying MDR P. aeruginosa by performing daily tracheal aspirates on patients mechanically ventilated for >48 h, and found 75 patients with newly colonised Pseudomonas, 27% of whom had MDR P. aeruginosa and 45% of whom went on to develop VAP, which the authors argued emphasised the importance of early detection of MDR colonisation to guide antibiotic therapy [125]. A systematic review and meta-analysis investigated whether surveillance cultures of lower respiratory tract specimens accurately predicted VAP organisms, and they found a pooled sensitivity of 75% and specificity of 92% in culture-positive VAP, showing that surveillance cultures may be able to accurately predict MDR VAP [126]. Cultures may need to be taken more than twice weekly, which contributes to the main limitation of the surveillance strategy, which is its high cost [124, 126].
Selective oropharyngeal decontamination (SOD) and selective digestive decontamination (SDD) are additional methods used to reduce the burden of MDR pathogens in ICUs. SOD/SDD refers to prophylactic treatment with antibiotics, often involving a paste in the oropharynx, enteral suspension of antimicrobials and an intravenous antibiotic, typically early on after ICU admission [127]. There have been numerous studies looking into the effect of SOD/SDD on development of MDR infections and ICU outcomes, with previous studies in ICUs with low levels of antibiotic resistance showing benefits of SOD/SDD, including lower levels of development of MDR pathogens and improved mortality [127, 128]. The use of SOD/SDD in ICUs with high endemic levels of MDR pathogens is more controversial, although a recent prospective observational cohort study found that implementation of an SDD programme in an ICU in Spain with high endemic levels of bacterial resistance led to a reduction of infections caused by MDR organisms, while also being associated with lower rates of VAP and bloodstream infections [129]. The most recent guidelines from the European Respiratory Society suggest utilising SOD but not SDD in areas with low levels of antimicrobial resistance for patients who are mechanically ventilated for >48 h [130].
There have been several studies demonstrating the benefits of ASPs in reducing the development of MDR organisms. A large systematic review and meta-analysis of 26 studies investigating the effects of implementation of an ASP on hospitalised patients showed a pooled percentage change of total antibiotic use of −19%, which increased to −39% in ICU patients [131]. Additionally, implementation of an ASP was associated with a reduction of infection with MDR organisms such as MRSA, carbapenem-resistant P. aeruginosa and ESBL Klebsiella [131]. A separate systematic review and meta-analysis included 32 studies that investigated development of infection and colonisation with MDR organisms, finding that ASP led to a 51% reduction in the development of an infection or colonisation with an MDR Gram-negative bacteria, a 48% reduction in ESBL Gram-negative bacteria and a 37% reduction in MRSA infection and colonisation [132].
Conclusions
The rising tide of antibiotic resistance leading to endemic spread and infection with MDR/XDR Gram-negative bacteria is a cause for great alarm. Greater research and clinical efforts now and in the future need to be directed towards improved methods for the diagnosis, prevention and treatment of MDR/XDR Gram-negative bacteria, as well as respiratory infections attributed to these pathogens. Only in this way can we prevent a future pandemic of infection attributed to MDR/XDR respiratory infections. The importance of this goal is highlighted by the future possibility of having converging respiratory infection pandemics, one caused by a respiratory virus and the other by MDR/XDR Gram-negative bacteria.
Footnotes
Number 4 in the Series “Respiratory infections” Edited by Antoni Torres and Michael S. Niederman
This article has an editorial commentary: https://doi.org/10.1183/16000617.0150-2022
Provenance: Commissioned article, peer reviewed.
Previous articles in this series: No. 1: Kumar K, Daley CL, Griffith DE, et al. Management of Mycobacterium avium complex and Mycobacterium abscessus pulmonary disease: therapeutic advances and emerging treatments. Eur Respir Rev 2022; 31: 210212. No. 2: Cilloniz C, Luna CM, Hurtado JC, et al. Respiratory viruses: their importance and lessons learned from COVID-19. Eur Respir Rev 2022; 31: 220051. No. 3: Cavallazzi R, Ramirez JA. How and when to manage respiratory infections out of hospital. Eur Respir Rev 2022; 31: 220092.
Author contributions: M.H. Kollef, D. Reynolds, J.P. Burnham, M. McCabe, V. Yuenger, S.T. Micek, K. Betthauser and C. Vazquez Guillamet each made substantial contributions to the conception and design of the work, participated in acquisition, analysis, and interpretation of data, and have drafted the work or substantively revised it. All authors have approved the submitted version and any substantially modified version that involves the author's contribution to the study, and all authors have agreed both to be personally accountable for the author's own contributions and to ensure that questions related to the accuracy or integrity of any part of the work, even ones in which the author was not personally involved, are appropriately investigated, resolved, and the resolution documented in the literature.
Conflict of interest: M.H Kollef's work is supported by the Barnes-Jewish Hospital Foundation. The remaining authors declare that they have no competing interests.
References
- 1.Vincent JL, Sakr Y, Singer M, et al. . Prevalence and outcomes of infection among patients in intensive care units in 2017. JAMA 2020; 323: 1478–1487. doi: 10.1001/jama.2020.2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Institute for Health Metrics and Evaluation (IHME) . COVID-19 has caused 6.9 million deaths globally, more than double what official reports show. www.healthdata.org/news-release/covid-19-has-caused-69-million-deaths-globally-more-double-what-official-reports-show Date last accessed: 12 April 2022. Date last updated: 6 May 2021.
- 3.Yeyati EL, Filipinni F. Social and Economic Impact of COVID-19. www.brookings.edu/wp-content/uploads/2021/06/Social-and-economic-impact-COVID.pdf Date last accessed: 12 April 2022. Date last updated: 1 June 2021.
- 4.Magiorakos AP, Srinivasan A, Carey RB, et al. . Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012; 18: 268–281. doi: 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 5.United Nations Interagency Coordination Group on Antimicrobial Resistance . No Time to Wait: Securing the Future from Drug-Resistant Infections. www.who.int/docs/default-source/documents/no-time-to-wait-securing-the-future-from-drug-resistant-infections-en.pdfsfvrsn=5b424d7_6 Date last accessed: 12 April 2022. Date last updated: 1 April 2019.
- 6.Wellcome Trust, United Kingdom Department of Health . 2014 Review on Antimicrobial Resistance. Available from: https://amr-review.org/ Date last accessed: 12 April 2022. Date last updated: 31 July 2014.
- 7.Centers for Disease Control and prevention . Antibiotic Resistance: A Global Threat. www.cdc.gov/drugresistance/solutions-initiative/stories/ar-global-threat.html. Date last accessed: 12 April 2022. Date last updated: 18 February 2020.
- 8.Talbot GH, Jezek A, Murray BE, et al. . The Infectious Diseases Society of America's 10×'20 Initiative (10 new systemic antibacterial agents US Food and Drug Administration approved by 2020): is 20×'20 a possibility? Clin Infect Dis 2019; 69: 1–11. doi: 10.1093/cid/ciz089 [DOI] [PubMed] [Google Scholar]
- 9.Shinohara DR, Dos Santos Saalfeld SM, Martinez HV, et al. . Outbreak of endemic carbapenem-resistant Acinetobacter baumannii in a coronavirus disease 2019 (COVID-19)-specific intensive care unit. Infect Control Hosp Epidemiol 2022; 43: 815–817. doi: 10.1017/ice.2021.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez S, Innes GK, Walters MS, et al. . Increase in hospital-acquired carbapenem-resistant Acinetobacter baumannii infection and colonization in an acute care hospital during a surge in COVID-19 admissions – New Jersey, February–July 2020. MMWR Morb Mortal Wkly Rep 2020; 69: 1827–1831. doi: 10.15585/mmwr.mm6948e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antimicrobial Resistance Collaborators . Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 2022; 399: 629–655. doi: 10.1016/S0140-6736(21)02724-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sader HS, Castanheira M, Arends SJR, et al. . Geographical and temporal variation in the frequency and antimicrobial susceptibility of bacteria isolated from patients hospitalized with bacterial pneumonia: results from 20 years of the SENTRY Antimicrobial Surveillance Program (1997–2016). J Antimicrob Chemother 2019; 74: 1595–1606. doi: 10.1093/jac/dkz074 [DOI] [PubMed] [Google Scholar]
- 13.Langford BJ, So M, Raybardhan S, et al. . Antibiotic prescribing in patients with COVID-19: rapid review and meta-analysis. Clin Microbiol Infect 2021; 27: 520–531. doi: 10.1016/j.cmi.2020.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vacheron CH, Lepape A, Savey A, et al. . Increased incidence of ventilator-acquired pneumonia in coronavirus disease 2019 patients: a multicentric cohort study. Crit Care Med 2022; 50: 449–459. doi: 10.1097/CCM.0000000000005297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher K, Trupka T, Micek ST, et al. . A prospective one-year microbiologic survey of combined pneumonia and respiratory failure. Surg Infect 2017; 18: 827–833. doi: 10.1089/sur.2017.111 [DOI] [PubMed] [Google Scholar]
- 16.Barreto JV, Dias CC, Cardoso T. Risk factors for community-onset pneumonia caused by drug-resistant pathogens: a prospective cohort study. Eur J Intern Med 2022; 96: 66–73. doi: 10.1016/j.ejim.2021.10.005 [DOI] [PubMed] [Google Scholar]
- 17.Webber DM, Wallace MA, Burnham CA, et al. . Evaluation of the BioFire FilmArray pneumonia panel for detection of viral and bacterial pathogens in lower respiratory tract specimens in the setting of a tertiary care academic medical center. J Clin Microbiol 2020; 58: e00343-20. doi: 10.1128/JCM.00343-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrol ED, Mankhambo LA, Guiver M, et al. . PCR improves diagnostic yield from lung aspiration in Malawian children with radiologically confirmed pneumonia. PLoS One 2011; 6: e21042. doi: 10.1371/journal.pone.0021042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parente DM, Cunha CB, Mylonakis E, et al. . The clinical utility of methicillin-resistant Staphylococcus aureus (MRSA) nasal screening to rule out MRSA pneumonia: a diagnostic meta-analysis with antimicrobial stewardship implications. Clin Infect Dis 2018; 67: 1–7. doi: 10.1093/cid/ciy024 [DOI] [PubMed] [Google Scholar]
- 20.Torres I, Huntley D, Tormo M, et al. . Multi-body-site colonization screening cultures for predicting multi-drug resistant Gram-negative and Gram-positive bacteremia in hematological patients. BMC Infect Dis 2022; 22: 172. doi: 10.1186/s12879-022-07154-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang R, Han JH, Lautenbach E, et al. . Clinical prediction tool for extended-spectrum beta-lactamase-producing enterobacterales as the etiology of a bloodstream infection in solid organ transplant recipients. Transpl Infect Dis 2021; 23: e13599. doi: 10.1111/tid.13599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peterlin L, Žagar M, Lejko Zupanc T, et al. . Should the patients colonized with extended-spectrum beta-lactamase-producing Gram-negative bacilli (E-GNB) coming to hospital from the community with pneumonia get anti-E-GNB active empirical treatment? J Chemother 2017; 29: 287–291. doi: 10.1080/1120009X.2016.1263173 [DOI] [PubMed] [Google Scholar]
- 23.Mascitti H, Duran C, Nemo EM, et al. . Factors associated with bacteraemia due to multidrug-resistant organisms among bacteraemic patients with multidrug-resistant organism carriage: a case control study. Antimicrob Resist Infect Control 2018; 7: 116. doi: 10.1186/s13756-018-0412-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones BE, Brown KA, Jones MM, et al. . Variation in empiric coverage versus detection of methicillin-resistant Staphylococcus aureus and Pseudomonas aeruginosa in hospitalizations for community-onset pneumonia across 128 US Veterans Affairs Medical Centers. Infect Control Hosp Epidemiol 2017; 38: 937–944. doi: 10.1017/ice.2017.98 [DOI] [PubMed] [Google Scholar]
- 25.Costa RD, Baptista JP, Freitas R, et al. . Hospital-acquired pneumonia in a multipurpose intensive care unit: one-year prospective study. Acta Med Port 2019; 32: 746–753. doi: 10.20344/amp.11607 [DOI] [PubMed] [Google Scholar]
- 26.Patro S, Sarangi G, Das P, et al. . Bacteriological profile of ventilator-associated pneumonia in a tertiary care hospital. Indian J Pathol Microbiol 2018; 61: 375–379. doi: 10.4103/IJPM.IJPM_487_16 [DOI] [PubMed] [Google Scholar]
- 27.Fernández-Martínez NF, Carcel-Fernández S, De la Fuente-Martos C, et al. . Risk factors for multidrug-resistant Gram-negative bacteria carriage upon admission to the intensive care unit. Int J Environ Res Public Health 2022; 19: 1039. doi: 10.3390/ijerph19031039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zilberberg MD, Nathanson BH, Sulham K, et al. . Carbapenem resistance, inappropriate empiric treatment and outcomes among patients hospitalized with Enterobacteriaceae urinary tract infection, pneumonia and sepsis. BMC Infect Dis 2017; 17: 279. doi: 10.1186/s12879-017-2383-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zilberberg MD, Nathanson BH, Sulham K, et al. . A novel algorithm to analyze epidemiology and outcomes of carbapenem resistance among patients with hospital-acquired and ventilator-associated pneumonia: a retrospective cohort study. Chest 2019; 155: 1119–1130. doi: 10.1016/j.chest.2018.12.024 [DOI] [PubMed] [Google Scholar]
- 30.Prina E, Ranzani OT, Polverino E, et al. . Risk factors associated with potentially antibiotic-resistant pathogens in community-acquired pneumonia. Ann Am Thorac Soc 2015; 12: 153–160. doi: 10.1513/AnnalsATS.201407-305OC [DOI] [PubMed] [Google Scholar]
- 31.Timbrook TT, Morton JB, McConeghy KW, et al. . The effect of molecular rapid diagnostic testing on clinical outcomes in bloodstream infections: a systematic review and meta-analysis. Clin Infect Dis 2017; 64: 15–23. doi: 10.1093/cid/ciw649 [DOI] [PubMed] [Google Scholar]
- 32.Ruppé E, Woerther PL, Barbier F. Mechanisms of antimicrobial resistance in Gram-negative bacilli. Ann Intensive Care 2015; 5: 61. doi: 10.1186/s13613-015-0061-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kakoullis L, Papachristodoulou E, Chra P, et al. . Mechanisms of resistance in important Gram-positive and Gram-negative pathogens and novel antibiotic solutions. Antibiotics 2021; 10: 415. doi: 10.3390/antibiotics10040415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bush K, Jacoby GA. Updated functional classification of β-lactamases. Antimicrob Agents Chemother 2010; 54: 969–976. doi: 10.1128/AAC.01009-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castanheira M, Simner PJ, Bradford PA. Extended-spectrum β-lactamases: an update on their characteristics, epidemiology and detection. JAC Antimicrob Resist 2021; 3: dlab092. doi: 10.1093/jacamr/dlab092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawa T, Kooguchi K, Moriyama K. Molecular diversity of extended-spectrum β-lactamases and carbapenemases, and antimicrobial resistance. J Intensive Care 2020; 8: 13. doi: 10.1186/s40560-020-0429-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamma PD, Aitken SL, Bonomo RA, et al. . IDSA Guidance on the Treatment of Antimicrobial-Resistant Gram-Negative Infections: Version 1.0. 2020. Available from: https://www.idsociety.org/practice-guideline/amr-guidance/# [Google Scholar]
- 38.Ur Rahman S, Ali T, Ali I, et al. . The growing genetic and functional diversity of extended spectrum beta-lactamases. Biomed Res Int 2018; 2018: 9519718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Docobo-Pérez F, López-Cerero L, López-Rojas R, et al. . Inoculum effect on the efficacies of amoxicillin-clavulanate, piperacillin-tazobactam, and imipenem against extended-spectrum β-lactamase (ESBL)-producing and non-ESBL-producing Escherichia coli in an experimental murine sepsis model. Antimicrob Agents Chemother 2013; 57: 2109–2113. doi: 10.1128/AAC.02190-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harada Y, Morinaga Y, Kaku N, et al. . In vitro and in vivo activities of piperacillin-tazobactam and meropenem at different inoculum sizes of ESBL-producing Klebsiella pneumoniae. Clin Microbiol Infect 2014; 20: O831–O839. doi: 10.1111/1469-0691.12677 [DOI] [PubMed] [Google Scholar]
- 41.Jacoby GA. AmpC β-lactamases. Clin Microbiol Rev 2009; 22: 161–182. doi: 10.1128/CMR.00036-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamma PD, Doi Y, Bonomo RA, et al. . A primer on AmpC β-lactamases: necessary knowledge for an increasingly multidrug-resistant world. Clin Infect Dis 2019; 69: 1446–1455. doi: 10.1093/cid/ciz173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamma PD, Aitken SLL, Bonomo RA, et al. . IDSA guidance on the treatment of antimicrobial-resistant Gram-negative infections: version 2.0. 2022. Available from: https://www.idsociety.org/practice-guideline/amr-guidance-2.0/ [Google Scholar]
- 44.Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis 2017; 215: S28–S36. doi: 10.1093/infdis/jiw282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med 2012; 18: 263–272. doi: 10.1016/j.molmed.2012.03.003 [DOI] [PubMed] [Google Scholar]
- 46.Nordmann P, Poirel L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect 2014; 20: 821–830. doi: 10.1111/1469-0691.12719 [DOI] [PubMed] [Google Scholar]
- 47.Diene SM, Rolain JM. Carbapenemase genes and genetic platforms in Gram-negative bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter species. Clin Microbiol Infect 2014; 20: 831–838. doi: 10.1111/1469-0691.12655 [DOI] [PubMed] [Google Scholar]
- 48.Tilahun M, Kassa Y, Gedefie A, et al. . Emerging carbapenem-resistant Enterobacteriaceae infection, its epidemiology and novel treatment options: a review. Infect Drug Resist 2021; 14: 4363–4374. doi: 10.2147/IDR.S337611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yahav D, Ciske CG, Grāmantniece A, et al. . New β-lactam-β-lactamase inhibitor combinations. Clin Microbiol Rev 2021; 34: e00115-20. doi: 10.1128/CMR.00115-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doi Y, Wachino J, Arakawa Y. Aminoglycoside resistance: the emergence of acquired 16s ribosomal RNA methyltransferases. Infect Dis Clin North Am 2016; 30: 523–537. doi: 10.1016/j.idc.2016.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramirez MS, Tolmasky ME. Aminoglycoside modifying enzymes. Drug Resist Updat 2010; 13: 151–171. doi: 10.1016/j.drup.2010.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Behzadi P, Baráth Z, Gajdács M. It's not easy being green: a narrative review on the microbiology, virulence and therapeutic prospects of multidrug-resistant Pseudomonas aeruginosa . Antibiotics 2021; 10: 42. doi: 10.3390/antibiotics10010042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishino K, Yamasaki S, Nakashima R, et al. . Function and inhibitory mechanisms of multidrug efflux pumps. Front Microbiol 2021; 12: 737288. doi: 10.3389/fmicb.2021.737288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kollef MH, Sherman G, Ward S, et al. . Inadequate antimicrobial treatment of infections: a risk factor for hospital mortality among critically ill patients. Chest 1999; 115: 462–474. doi: 10.1378/chest.115.2.462 [DOI] [PubMed] [Google Scholar]
- 55.Lodise TP Jr, Patel N, Kwa A, et al. . Predictors of 30-day mortality among patients with Pseudomonas aeruginosa bloodstream infections: impact of delayed appropriate antibiotic selection. Antimicrob Agents Chemother 2007; 51: 3510–3515. doi: 10.1128/AAC.00338-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kollef M, Micek S, Hampton N, et al. . Septic shock attributed to Candida infection: importance of empiric therapy and source control. Clin Infect Dis 2012; 54: 1739–1746. doi: 10.1093/cid/cis305 [DOI] [PubMed] [Google Scholar]
- 57.Harris PNA, Tambyah PA, Lye DC, et al. . Effect of piperacillin-tazobactam vs meropenem on 30-day mortality for patients with E. coli or Klebsiella pneumoniae bloodstream infection and ceftriaxone resistance: a randomized clinical trial. JAMA 2018; 320: 984–994. doi: 10.1001/jama.2018.12163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar A, Roberts D, Wood KE, et al. . Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 2006; 34: 1589–1596. doi: 10.1097/01.CCM.0000217961.75225.E9 [DOI] [PubMed] [Google Scholar]
- 59.Evans L, Rhodes A, Alhazzani W, et al. . Surviving Sepsis campaign: international guidelines for management of sepsis and septic shock: 2021. Crit Care Med 2021; 49: e1063–e1143. doi: 10.1097/CCM.0000000000005337 [DOI] [PubMed] [Google Scholar]
- 60.Vazquez-Guillamet MC, Scolari M, Zilberberg MD, et al. . Using the number needed to treat to assess appropriate antimicrobial therapy as a determinant of outcome in severe sepsis and septic shock. Crit Care Med 2014; 42: 2342–2329. doi: 10.1097/CCM.0000000000000516 [DOI] [PubMed] [Google Scholar]
- 61.Bassetti M, Rello J, Blasi F, et al. . Systematic review of the impact of appropriate versus inappropriate initial antibiotic therapy on outcomes of patients with severe bacterial infections. Int J Antimicrob Agents 2020; 56: 106184. doi: 10.1016/j.ijantimicag.2020.106184 [DOI] [PubMed] [Google Scholar]
- 62.Iregui M, Ward S, Sherman G, et al. . Clinical importance of delays in the initiation of appropriate antibiotic treatment for ventilator-associated pneumonia. Chest 2002; 122: 262–268. doi: 10.1378/chest.122.1.262 [DOI] [PubMed] [Google Scholar]
- 63.Awad SS, Rodriguez AH, Chuang YC, et al. . A phase 3 randomized double-blind comparison of ceftobiprole medocaril versus ceftazidime plus linezolid for the treatment of hospital-acquired pneumonia. Clin Infect Dis 2014; 59: 51–61. doi: 10.1093/cid/ciu219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kollef MH, Chastre J, Clavel M, et al. . A randomized trial of 7-day doripenem versus 10-day imipenem-cilastatin for ventilator-associated pneumonia. Crit Care 2012; 16: R218. doi: 10.1186/cc11862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Freire AT, Melnyk V, Kim MJ, et al. . Comparison of tigecycline with imipenem/cilastatin for the treatment of hospital-acquired pneumonia. Diagn Microbiol Infect Dis 2010; 68: 140–151. doi: 10.1016/j.diagmicrobio.2010.05.012 [DOI] [PubMed] [Google Scholar]
- 66.Kollef MH, Micek ST. Limitations of registration trials for nosocomial pneumonia. Clin Infect Dis 2021; 73: e4549–e4551. doi: 10.1093/cid/ciaa926 [DOI] [PubMed] [Google Scholar]
- 67.Stewart AG, Paterson DL, Young B, et al. . Meropenem versus piperacillin-tazobactam for definitive treatment of bloodstream infections caused by AmpC β-lactamase-producing Enterobacter spp, Citrobacter freundii, Morganella morganii, Providencia spp, or Serratia marcescens: a pilot multicenter randomized controlled trial (MERINO-2). Open Forum Infect Dis 2021; 8: ofab387. doi: 10.1093/ofid/ofab387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zilberberg MD, Shorr AF, Micek ST, et al. . Multi-drug resistance, inappropriate initial antibiotic therapy and mortality in Gram-negative sepsis and septic shock: a retrospective cohort study. Crit Care 2014; 18: 596. doi: 10.1186/s13054-014-0596-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zilberberg MD, Nathanson BH, Sulham K, et al. . Multidrug resistance, inappropriate empiric therapy, and hospital mortality in Acinetobacter baumannii pneumonia and sepsis. Crit Care 2016; 20: 221. doi: 10.1186/s13054-016-1392-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martinez-Nadal G, Puerta-Alcalde P, Gudiol C, et al. . Inappropriate empirical antibiotic treatment in high-risk neutropenic patients with bacteremia in the era of multidrug resistance. Clin Infect Dis 2020; 70: 1068–1074. doi: 10.1093/cid/ciz319 [DOI] [PubMed] [Google Scholar]
- 71.Reynolds D, Kollef M. The epidemiology and pathogenesis and treatment of Pseudomonas aeruginosa infections: an update. Drugs 2021; 81: 2117–2213. doi: 10.1007/s40265-021-01635-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alanezi G, Almulhem A, Aldriwesh M, et al. . A triple antimicrobial regimen for multidrug-resistant Klebsiella pneumonia in a neonatal intensive care unit outbreak: a case series. J Infect Public Health 2022; 15: 138–141. doi: 10.1016/j.jiph.2021.10.008 [DOI] [PubMed] [Google Scholar]
- 73.Pelaez-Bejarano A, Sánchez-Del Moral R, Montero-Pérez O, et al. . Successful treatment of Verona integron-encoded metallo-β-lactamase-producing Klebsiella pneumoniae infection using the combination of ceftazidime/avibactam and aztreonam. Eur J Hosp Pharm 2022; 29: 113–115. doi: 10.1136/ejhpharm-2021-002772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Persoon MC, Voor In't Holt AF, Wielders CCH, et al. . Mortality associated with carbapenem-susceptible and Verona Integron-encoded Metallo-β-lactamase-positive Pseudomonas aeruginosa bacteremia. Antimicrob Resist Infect Control 2020; 9: 25. doi: 10.1186/s13756-020-0682-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bassetti M, Garau J. Current and future perspectives in the treatment of multidrug-resistant Gram-negative infections. J Antimicrob Chemother 2021; 76: Suppl. 4, iv23–iv37. doi: 10.1093/jac/dkab352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kalil AC, Metersky ML, Klompas M, et al. . Management of adults with hospital-acquired and ventilator-associated pneumonia: 2016 clinical practice guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin Infect Dis 2016; 63: e61–e111. doi: 10.1093/cid/ciw353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tamma PD, Aitken SL, Bonomo RA, et al. . Infectious Diseases Society of America guidance on the treatment of extended-spectrum β-lactamase producing Enterobacterales (ESBL-E), carbapenem-resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with difficult-to-treat resistance (DTR-P. aeruginosa). Clin Infect Dis 2021; 72: e169–e183. doi: 10.1093/cid/ciaa1478 [DOI] [PubMed] [Google Scholar]
- 78.Marshall S, Hujer AM, Rojas LJ, et al. . Can ceftazidime-avibactam and aztreonam overcome β-lactam resistance conferred by metallo-β-lactamases in Enterobacteriaceae? Antimicrob Agents Chemother 2017; 61: e02243-16. doi: 10.1128/AAC.02243-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Davido B, Fellous L, Lawrence C, et al. . Ceftazidime-avibactam and aztreonam, an interesting strategy to overcome β-lactam resistance conferred by metallo-β-lactamases in Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother 2017; 61: e01008-17. doi: 10.1128/AAC.01008-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wenzler E, Deraedt MF, Harrington AT, et al. . Synergistic activity of ceftazidime-avibactam and aztreonam against serine and metallo-β-lactamase-producing Gram-negative pathogens. Diagn Microbiol Infect Dis 2017; 88: 352–354. doi: 10.1016/j.diagmicrobio.2017.05.009 [DOI] [PubMed] [Google Scholar]
- 81.Lee M, Abbey T, Biagi M, et al. . Activity of aztreonam in combination with ceftazidime-avibactam against serine- and metallo-β-lactamase-producing Pseudomonas aeruginosa. Diagn Microbiol Infect Dis 2021; 99: 115227. doi: 10.1016/j.diagmicrobio.2020.115227 [DOI] [PubMed] [Google Scholar]
- 82.Isler B, Harris P, Stewart AG, et al. . An update on cefepime and its future role in combination with novel β-lactamase inhibitors for MDR Enterobacterales and Pseudomonas aeruginosa. J Antimicrob Chemother 2021; 76: 550–560. doi: 10.1093/jac/dkaa511 [DOI] [PubMed] [Google Scholar]
- 83.Mushtaq S, Garello P, Vickers A, et al. . Activity of cefepime/zidebactam (WCK 5222) against ‘problem’ antibiotic-resistant Gram-negative bacteria sent to a national reference laboratory. J Antimicrob Chemother 2021; 76: 1511–1522. doi: 10.1093/jac/dkab067 [DOI] [PubMed] [Google Scholar]
- 84.Matsumoto T. Arbekacin: another novel agent for treating infections due to methicillin-resistant Staphylococcus aureus and multidrug-resistant Gram-negative pathogens. Clin Pharmacol 2014; 6: 139–148. doi: 10.2147/CPAA.S44377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sader HS, Flamm RK, Dale GE, et al. . Murepavadin activity tested against contemporary (2016–17) clinical isolates of XDR Pseudomonas aeruginosa. J Antimicrob Chemother 2018; 73: 2400–2404. doi: 10.1093/jac/dky227 [DOI] [PubMed] [Google Scholar]
- 86.Polyphor . A multicenter, open-label, randomized, active-controlled, parallel group, pivotal study to investigate the efficacy, safety and tolerability, and pharmacokinetics of murepavadin combined with one anti-pseudomonal antibiotic versus two anti-pseudomonal antibiotics in adult subjects with ventilator-associated bacterial pneumonia suspected or confirmed to be due to Pseudomonas aeruginosa (PRISM-MDR). NCT03409679. 2019. https://clinicaltrials.gov/ct2/show/NCT03409679 Date last accessed: 28 March 2022. Date last updated: 29 August 2019.
- 87.Motley MP, Banerjee K, Fries BC. Monoclonal antibody-based therapies for bacterial infections. Curr Opin Infect Dis 2019; 32: 210–216. doi: 10.1097/QCO.0000000000000539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zurawski DV, McLendon MK. Monoclonal antibodies as an antibacterial approach against bacterial pathogens. Antibiotics 2020; 9: 155. doi: 10.3390/antibiotics9040155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wilcox MH, Gerding DN, Poxton IR, et al. . Bezlotoxumab for prevention of recurrent Clostridium difficile infection. N Engl J Med 2017; 376: 305–317. doi: 10.1056/NEJMoa1602615 [DOI] [PubMed] [Google Scholar]
- 90.Mazumdar S. Raxibacumab. MAbs 2009; 1: 531–538. doi: 10.4161/mabs.1.6.10195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.François B, Jafri HS, Chastre J, et al. . Efficacy and safety of suvratoxumab for prevention of Staphylococcus aureus ventilator-associated pneumonia (SAATELLITE): a multicentre, randomised, double-blind, placebo-controlled, parallel-group, phase 2 pilot trial. Lancet Infect Dis 2021; 21: 1313–1323. doi: 10.1016/S1473-3099(20)30995-6 [DOI] [PubMed] [Google Scholar]
- 92.Jain R, Beckett VV, Konstan MW, et al. . KB001-A, a novel anti-inflammatory, found to be safe and well-tolerated in cystic fibrosis patients infected with Pseudomonas aeruginosa. J Cyst Fibros 2018; 17: 484–491. doi: 10.1016/j.jcf.2017.12.006 [DOI] [PubMed] [Google Scholar]
- 93.François B, Luyt CE, Dugard A, et al. . Safety and pharmacokinetics of an anti-PcrV PEGylated monoclonal antibody fragment in mechanically ventilated patients colonized with Pseudomonas aeruginosa: a randomized, double-blind, placebo-controlled trial. Crit Care Med 2012; 40: 2320–2326. doi: 10.1097/CCM.0b013e31825334f6 [DOI] [PubMed] [Google Scholar]
- 94.MedImmune . A phase 2 proof-of-concept study to evaluate the efficacy and safety of MEDI3902 in mechanically ventilated patients for the prevention of nosocomial pneumonia caused by Pseudomonas aeruginosa. NCT02696902. https://clinicaltrials.gov/ct2/show/NCT02696902. Date last accessed: 28 March 2022. Date last updated: 4 February 2021.
- 95.Que YA, Lazar H, Wolff M, et al. . Assessment of panobacumab as adjunctive immunotherapy for the treatment of nosocomial Pseudomonas aeruginosa pneumonia. Eur J Clin Microbiol Infect Dis 2014; 33: 1861–1867. doi: 10.1007/s10096-014-2156-1 [DOI] [PubMed] [Google Scholar]
- 96.Aridis Pharmaceuticals . Placebo-controlled, double-blind, randomized study of Aerucin® as adjunct therapy to antibiotics in the treatment of P. aeruginosa pneumonia. NCT03027609. https://clinicaltrials.gov/ct2/show/NCT03027609. Date last accessed: 28 March 2022. Date last updated: 17 March 2022.
- 97.Youssef EG, Zhang L, Alkhazraji S, et al. . Monoclonal IgM antibodies targeting Candida albicans Hyr1 provide cross-kingdom protection against Gram-negative bacteria. Front Immunol 2020; 11: 76. doi: 10.3389/fimmu.2020.00076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Streicher LM. Exploring the future of infectious disease treatment in a post-antibiotic era: a comparative review of alternative therapeutics. J Glob Antimicrob Resist 2021; 24: 285–295. doi: 10.1016/j.jgar.2020.12.025 [DOI] [PubMed] [Google Scholar]
- 99.Uyttebroek S, Chen B, Onsea J, et al. . Safety and efficacy of phage therapy in difficult-to-treat infections: a systematic review. Lancet Infect Dis 2022; 22: E208–E220. 10.1016/S1473-3099(21)00612-5 [DOI] [PubMed] [Google Scholar]
- 100.Maddocks S, Fabijan AP, Ho J, et al. . Bacteriophage therapy of ventilator-associated pneumonia and empyema caused by Pseudomonas aeruginosa. Am J Respir Crit Care Med 2019; 200: 1179–1181. doi: 10.1164/rccm.201904-0839LE [DOI] [PubMed] [Google Scholar]
- 101.Gong FY, Zhang DY, Zhang JG, et al. . siRNA-mediated gene silencing of MexB from the MexA-MexB-OprM efflux pump in Pseudomonas aeruginosa. BMB Rep 2014; 47: 203–238. doi: 10.5483/BMBRep.2014.47.4.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sully EK, Geller BL, Li L, et al. . Peptide-conjugated phosphorodiamidate morpholino oligomer (PPMO) restores carbapenem susceptibility to NDM-1-positive pathogens in vitro and in vivo. J Antimicrob Chemother 2017; 72: 782–790. doi: 10.1093/jac/dkw476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dellit TH, Owens RC, McGowan JE, et al. . Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America guidelines for developing an institutional program to enhance antimicrobial stewardship. Clin Infect Dis 2007; 44: 159–177. doi: 10.1086/510393 [DOI] [PubMed] [Google Scholar]
- 104.Centers for Disease Control and Prevention . Core Elements of Hospital Antibiotic Stewardship Programs. Atlanta, US Department of Health and Human Services, 2019. Available from: www.cdc.gov/antibiotic-use/healthcare/pdfs/hospital-core-elements-H.pdf [Google Scholar]
- 105.Lawrence KL, Kollef MH. Antimicrobial stewardship in the intensive care unit: advances and obstacles. Am J Respir Crit Care Med 2009; 179: 434–438. doi: 10.1164/rccm.200809-1394CP [DOI] [PubMed] [Google Scholar]
- 106.Wunderink RG, Srinivasan A, Barie PS, et al. . Antibiotic stewardship in the intensive care unit. An official American Thoracic Society workshop report in collaboration with the AACN, CHEST, CDC, and SCCM. Ann Am Thorac Soc 2020; 17: 531–540. doi: 10.1513/AnnalsATS.202003-188ST [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pickens CI, Wunderink RG. Principles and practice of antibiotic stewardship in the ICU. Chest 2019; 156: 163–171. doi: 10.1016/j.chest.2019.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Barlam TF, Cosgrove SE, Abbo LM, et al. . Implementing an antibiotic stewardship program: guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clin Infect Dis 2016; 62: e51–e77. doi: 10.1093/cid/ciw118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kollef MH, Micek ST. Antimicrobial stewardship programs: mandatory for all ICUs. Crit Care 2012; 16: 179. doi: 10.1186/cc11853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tamma PD, Avdic E, Li DX, et al. . Association of adverse events with antibiotic use in hospitalized patients. JAMA Intern Med 2017; 177: 1308–1315. doi: 10.1001/jamainternmed.2017.1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Teshome BF, Vouri SM, Hampton N, et al. . Duration of exposure to antipseudomonal β-lactam antibiotics in the critically ill and development of new resistance. Pharmacotherapy 2019; 39: 261–270. doi: 10.1002/phar.2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Barrasa-Villar JI, Aibar-Remón C, Prieto-Andrés P, et al. . Impact on morbidity, mortality, and length of stay of hospital-acquired infections by resistant microorganisms. Clin Infect Dis 2017; 65: 644–652. doi: 10.1093/cid/cix411 [DOI] [PubMed] [Google Scholar]
- 113.Rhee C, Kadri SS, Dekker JP, et al. . Prevalence of antibiotic-resistant pathogens in culture-proven sepsis and outcomes associated with inadequate and broad-spectrum empiric antibiotic use. JAMA Netw Open 2020; 3: e202899. doi: 10.1001/jamanetworkopen.2020.2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pugh R, Grant C, Cooke RP, et al. . Short-course versus prolonged-course antibiotic therapy for hospital-acquired pneumonia in critically ill adults. Cochrane Database Syst Rev 2015; 8: CD007577. doi: 0.1002/14651858.CD007577.pub3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Webb BJ, Sorensen J, Jephson A, et al. . Broad-spectrum antibiotic use and poor outcomes in community-onset pneumonia: a cohort study. Eur Respir J 2019; 54: 1900057. doi: 10.1183/13993003.00057-2019 [DOI] [PubMed] [Google Scholar]
- 116.Wuethrich I, Pelzer BW, Khodamoradi Y, et al. . The role of the human gut microbiota in colonization and infection with multidrug-resistant bacteria. Gut Microbes 2021; 13: 1–13. doi: 10.1080/19490976.2021.1911279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kollef MH, Torres A, Shorr AF, et al. . Nosocomial infection. Crit Care Med 2021; 49: 169–187. doi: 10.1097/CCM.0000000000004783 [DOI] [PubMed] [Google Scholar]
- 118.Elligsen M, Walker SAN, Pinto R, et al. . Audit and feedback to reduce broad-spectrum antibiotic use among intensive care unit patients: a controlled interrupted time series analysis. Infect Control Hosp Epidemiol 2012; 33: 354–361. doi: 10.1086/664757 [DOI] [PubMed] [Google Scholar]
- 119.Buising KL, Thursky KA, Robertson MB, et al. . Electronic antibiotic stewardship – reduced consumption of broad-spectrum antibiotics using a computerized antimicrobial approval system in a hospital setting. J Antimicrob Chemother 2008; 62: 608–616. doi: 10.1093/jac/dkn218 [DOI] [PubMed] [Google Scholar]
- 120.Thiel SW, Asghar MF, Micek ST, et al. . Hospital-wide impact of a standardized order set for the management of bacteremic severe sepsis. Crit Care Med 2009; 37: 819–824. doi: 10.1097/CCM.0b013e318196206b [DOI] [PubMed] [Google Scholar]
- 121.Paonessa JR, Shah RD, Pickens CI, et al. . Rapid detection of methicillin-resistant Staphylococcus aureus in BAL: a pilot randomized controlled trial. Chest 2019; 155: 999–1007. doi: 10.1016/j.chest.2019.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Buchan BW, Windham S, Balada-Llasat JM, et al. . Practical comparison of the BioFire FilmArray pneumonia panel to routine diagnostic methods and potential impact on antimicrobial stewardship in adult hospitalized patients with lower respiratory tract infections. J Clin Microbiol 2020; 58: e00135-20. doi: 10.1128/JCM.00135-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Peiffer-Smadja N, Bouadma L, Mathy V, et al. . Performance and impact of a multiplex PCR in ICU patients with ventilator-associated pneumonia or ventilated hospital-acquired pneumonia. Crit Care 2020; 24: 366. doi: 10.1186/s13054-020-03067-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Blot S, Depuydt P, Vogelaers D. Maximizing rates of empiric appropriate antibiotic therapy with minimized use of broad-spectrum agents: are surveillance cultures the key? Intensive Care Med 2008; 34: 2130–2103. doi: 10.1007/s00134-008-1249-7 [DOI] [PubMed] [Google Scholar]
- 125.Yang K, Zhuo H, Guglielmo J, et al. . Multidrug-resistant Pseudomonas aeruginosa ventilator-associated pneumonia: the role of endotracheal aspirate surveillance cultures. Ann Pharmacother 2009; 43: 28–35. doi: 10.1345/aph.1L210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Brusselaers N, Labeau S, Vogelaers D, et al. . Value of lower respiratory tract surveillance cultures to predict bacterial pathogens in ventilator-associated pneumonia: systematic review and diagnostic test accuracy meta-analysis. Intensive Care Med 2013; 39: 365–375. doi: 10.1007/s00134-012-2759-x [DOI] [PubMed] [Google Scholar]
- 127.Plantinga NL, Bonten MJM. Selective decontamination and antibiotic resistance in ICUs. Crit Care 2015; 19: 259. doi: 10.1186/s13054-015-0967-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Price R, MacLennan G, Glen J, et al. . Selective digestive or oropharyngeal decontamination and topical oropharyngeal chlorhexidine for prevention of death in general intensive care: systematic review and network meta-analysis. BMJ 2014; 348: g2197. doi: 10.1136/bmj.g2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sánchez-Ramírez C, Hípola-Escalada S, Miriam Cabrera-Santana M, et al. . Long-term use of selective digestive decontamination in an ICU highly endemic for bacterial resistance. Crit Care 2018; 22: 141. doi: 10.1186/s13054-018-2057-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Torres A, Niederman MS, Chastre J, et al. . International ERS/ESICM/ESCMID/ALAT guidelines for the management of hospital-acquired pneumonia and ventilator-associated pneumonia: guidelines for the management of hospital-acquired pneumonia (HAP)/ventilator-associated pneumonia (VAP) of the European Respiratory Society (ERS), European Society of Intensive Care Medicine (ESICM), European Society of Clinical Microbiology and Infectious Diseases (ESCMID) and Asociación Latinoamericana del Tórax (ALAT). Eur Respir J 2017; 50: 1700582. doi: 10.1183/13993003.00582-2017 [DOI] [PubMed] [Google Scholar]
- 131.Karanika S, Paudel S, Grigoras C, et al. . Systematic review and meta-analysis of clinical and economic outcomes from the implementation of hospital-based antimicrobial stewardship programs. Antimicrob Agents Chemother 2016; 60: 4840–4852. doi: 10.1128/AAC.00825-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Baur D, Gladstone BP, Burket F, et al. . Effect of antibiotic stewardship on the incidence of infection and colonisation with antibiotic-resistant bacteria and Clostridium difficile infection: a systematic review and meta-analysis. Lancet Infect Dis 2017; 17: 990–1001. doi: 10.1016/S1473-3099(17)30325-0 [DOI] [PubMed] [Google Scholar]