Abstract
Viridans group streptococci (VS) from the oral cavity entering the bloodstream may initiate infective endocarditis (IE). We aimed to identify genes expressed in response to a pH increase from slightly acidic (pH 6.2) to neutral (pH 7.3) as encountered by VS entering the bloodstream from the oral cavity. Using a recently developed promoter-screening vector, we isolated five promoter fragments from the genomic DNA of Streptococcus gordonii CH1 responding to this stimulus. No common regulatory sequences were identified in these promoter fragments that could account for the coordinate expression of the corresponding genes. One of the isolated fragments contained the promoter region and 5′ end of a gene highly homologous to the methionine sulfoxide reductase gene (msrA) of various bacterial and eukaryotic species. This gene has been found to be activated in S. gordonii strain V288 in a rabbit model of IE (A. O. Kiliç, M. C. Herzberg, M. W. Meyer, X. Zhao, and L. Tao, Plasmid 42:67–72, 1999). We isolated and characterized the msrA gene of S. gordonii CH1 and constructed a chromosomal insertion mutant. This mutant was more sensitive to hydrogen peroxide, suggesting a role for the streptococcal MsrA in protecting against oxidative stress. Moreover, MsrA appeared to be important for the growth of S. gordonii CH1 under aerobic and anaerobic conditions. Both these properties of MsrA may contribute to the ability of S. gordonii to cause IE.
Viridans group streptococci (VS), which colonize the teeth and oral mucosal surfaces of humans, are isolated from 40 to 60% of patients with native valve infective endocarditis (IE) (33). In one of the early steps in the development of IE, VS from the oral cavity gain access to the bloodstream, causing a transient bacteremia. Subsequently, VS may adhere to a preformed cardiac vegetation, a meshwork of platelets and fibrin present on endocardial lesions (9). Several surface components of VS are thought to be involved in their adherence to the vegetations, like FimA of Streptococcus parasanguis (3, 34) and extracellular polysaccharides of various VS species (4, 28, 30). The adherent bacteria are able to multiply rapidly within the vegetation (5, 9).
After VS enter the bloodstream, their adaptation to this new environment presumably involves the expression of genes, induced upon sensing of signals from the changed environment. One of these signals may be a change in the pH. Many bacteria are known to respond to pH changes. Most investigations have focused on adaptive responses to a decrease in pH. Acidification induces expression of specific genes in several bacterial pathogens, like Salmonella enterica serovar Typhimurium (20) and Vibrio cholerae (6), and upregulates the expression of the major stress protein DnaK in Streptococcus mutans, a member of the VS group (15). However, when VS enter the bloodstream, the bacteria experience an increase in pH from slightly acidic (6.0 to 6.5) (25) in the dental plaque to near neutral (7.3) in blood. As this stimulus is possibly involved in the induction of VS genes that might play a role in the colonization of the vegetation by VS, and therefore is involved in the pathogenesis of IE, we isolated promoters whose activities were upregulated by this pH increase. One of the isolated fragments contained part of an msrA homolog, a gene whose expression was recently found to be induced in Streptococcus gordonii V288 in the experimental rabbit model of IE (16). We therefore cloned and further characterized this putative S. gordonii virulence gene.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. gordonii strain CH1, also referred to as strain Challis (37), and its msrA insertion mutant MM1 (this study) were cultured in Todd-Hewitt (TH) broth (Oxoid, Basingstoke, Hampshire, England) or on TH agar at 37°C in a 5% CO2 atmosphere. TH broth and TH agar plates were supplemented with 5 μg of erythromycin or chloramphenicol per ml or 500 μg of spectinomycin per ml when required. Escherichia coli strains DH5α (Gibco-BRL, Breda, The Netherlands), BHB2600 (13), and Top10F′ (Invitrogen, Groningen, The Netherlands) were cultured in Luria-Bertani medium or on Luria-Bertani agar. When required, 100 μg of erythromycin or ampicillin per ml and 10 μg of chloramphenicol per ml were added.
DNA isolation, DNA manipulations, and bacterial transformation.
Plasmid DNA was isolated from E. coli using the Wizard Plus SV miniprep DNA purification system (Promega Corporation, Madison, Wis.), and from S. gordonii CH1 as described previously (36). Streptococcal chromosomal DNA was isolated using the Puregene Chromosomal DNA isolation kit for gram-positive bacteria and yeast (Gentra Systems Inc., Minneapolis, Minn.), with some minor modifications. Lysozyme (Sigma Chemical Co., St. Louis, Mo.) and mutanolysin (Sigma) were added to the lysis mixture of the DNA isolation kit at final concentrations of 5 mg/ml and 20 U/ml, respectively, and the period of incubation to obtain protoplasts was extended to 2 h at 37°C. Routine DNA manipulations were performed as described by Sambrook et al. (29), and enzymes were purchased from Boehringer GmbH (Mannheim, Germany). Transformation of E. coli was done by standard electroporation (7). S. gordonii CH1 was transformed using an optimized electroporation protocol for VS (34a).
Genomic DNA library and selection of neutral-pH-inducible promoters.
The construction of the novel broad-host-range selection vector pMM223 (GenBank accession no. AF076212) and of a genomic expression library of S. gordonii CH1 in this vector will be described elsewhere (34a). Briefly, genomic DNA of strain CH1 was digested with Sau3A, and fragments were ligated into the BglII site of pMM223. Recombinant plasmids, containing chromosomal fragments 100 to 1,000 bp in size, were introduced into the homologous host by electroporation. Transformants were pooled from the transformation plates to constitute the S. gordonii expression library. This library contained approximately 105 independent clones, statistically representing the entire genome (29).
To isolate neutral-pH-inducible promoter fragments from this library, 25 μl containing 2.5 × 105 CFU was plated onto TH agar (pH 7.3) supplemented with 5 μg of erythromycin per ml for plasmid maintenance and 500 μg of spectinomycin per ml for selection of active streptococcal promoters. After incubation at 37°C for 36 h, colonies resistant to erythromycin as well as to spectinomycin were plated onto TH agar (pH 6.2), again supplemented with erythromycin and spectinomycin to identify colonies susceptible to spectinomycin at this lower pH. As a control for the viability of the isolated S. gordonii clones, these were also restreaked onto TH agar plates (pH 6.2) supplemented with erythromycin only and onto other plates (pH 7.3) with erythromycin and spectinomycin. From clones that failed to grow on the pH 6.2 agar, but which did grow on the pH 7.3 agar in the presence of spectinomycin, the cloned chromosomal fragments were amplified.
PCR amplification and DNA sequence analysis.
Cloned chromosomal DNA fragments from selected S. gordonii CH1 strains were amplified from crude bacterial lysates by PCR (14), using primers AV9 (5′-ATGTCACTAGTCTCTACAAC-3′) and AV4 (5′-AATTGGATCCCGGGTTTTTTTATAATTTTTTTAATCTG-3′) and Taq DNA polymerase (Promega Corporation). Amplicons were purified using the High Pure PCR product purification kit (Boehringer) and sequenced by PCR-mediated Taq Dye Deoxy terminator cycle sequencing (Perkin-Elmer, Foster City, Calif.) on an Applied Biosystems (San Jose, Calif.) model 373 DNA sequencer. Primer AV9 or primer AV19 (5′-CTCCTCACTATTTTGATTAG-3′), annealing upstream and downstream of the unique BglII site of pMM223, respectively, were used to sequence the cloned fragments. The sequences obtained were analyzed using the BLAST program (1). For the identification of possible common sequence features, the CLUSTAL program was used (12).
Measurement of in vitro growth rate.
To determine the relative activity of isolated neutral-pH-inducible promoter fragments, the growth rates in the presence and absence of spectinomycin of the clones carrying these fragments were determined at both pH 6.2 and pH 7.3 (Vriesema et al., submitted). In short, a single colony of each clone was grown at 37°C in TH supplemented with erythromycin for plasmid maintenance. After overnight incubation, the cultures were diluted 100-fold in fresh medium containing erythromycin and spectinomycin or erythromycin alone. Growth was monitored by measuring the optical density at 620 nm over time, and the mid-log-phase doubling time (t1/2) was determined. The relative promoter activity at each pH was expressed as the ratio of growth in the presence and absence of spectinomycin [t1/2(+spec)/t1/2(−spec)].
Isolation and characterization of the streptococcal msrA gene.
Chromosomal DNA of S. gordonii CH1 was digested to completion with HindIII, and the resulting fragments were self-ligated. Using primers AV40 (5′-CAAGCCCCAGAAACACCCGC-3′) and AV41 (5′-CAGTGGGATACGCCAATGGAC-3′), corresponding to the complement of nucleotides 23 to 42 and to nucleotides 83 to 103 of the identified streptococcal msrA homolog, respectively (see Results), a fragment of approximately 3.5 kb was amplified. The purified amplicon was ligated into the PCR cloning vector pCR2.1 (Invitrogen) and introduced into E. coli TOP10F′ cells. Part of this fragment, containing the 3′ end of the msrA gene, was subcloned as a 2.0-kb EcoRI fragment into pUC19 (39), generating pMM1226. After digestion with BamHI and SphI, subclones with fragments of decreasing sizes were created by exonuclease III (Boehringer) digestion according to standard procedures (29). Individual fragments were sequenced using the universal M13(−21) and M13(Reverse) primers, and the sequence of the streptococcal msrA homolog was compiled.
Primer extension assay.
Total RNA was extracted from S. gordonii CH1 using the RNeasy Mini Kit (Qiagen GmbH, Hilden, Germany). Ten micrograms of total cell RNA was used in the primer extension reactions. The RNA was incubated for 5 min at 65°C with 0.2 pmol of primer AV40 in hybridization buffer (70 mM Tris-HCl [pH 8.3], 14 mM MgCl2, 14 mM dithiothreitol) in a final volume of 14 μl. The mixture was gradually cooled to room temperature, and the volume was adjusted to 20 μl by the addition of dATP, dGTP, and dCTP to a final concentration of 100 μM and dCTP to a final concentration of 10 μM. To this mixture, 15 μCi of [α-32P]dCTP with a specific activity of 3,000 Ci/mmol was added. cDNA was synthesized by the addition of 12.5 U of avian myoblastosis virus reverse transcriptase (Boehringer) and incubation at 42°C for 30 min. The reaction was terminated by the addition of 5 μl of sequencing loading buffer. In addition, a sequence reaction was performed with the same primer, using the T7 Sequenase version 2.0 DNA sequencing kit (Amersham Life Science, Inc., Cleveland, Ohio) and [α-35S]dATP. The primer extension reaction was electrophoresed on a 6% polyacrylamide–7 M urea gel, parallel to the sequence reaction which served as a marker for determination of the size of the synthesized cDNA.
Construction of an S. gordonii CH1 msrA insertion mutant.
The complete msrA gene, including its putative promoter sequence, was amplified from the S. gordonii CH1 chromosomal DNA using the Expand long-template PCR kit (Boehringer) and primers AV45 (5′-AATTACTAGTGAAATGAAGAATATGGCTGGGTTGAGAAG-3′) and AV46 (5′-ATATACTAGTGCCAACGCTCAGCAAAAAAGGCCTG-3′). The amplicon obtained, approximately 1.1 kb, was cloned into pCR2.1, creating vector pMM1227. The erythromycin resistance gene of the broad-host-range vector pMG36e (32) was isolated as a 1.0-kb EcoRI-NsiI fragment, and the sticky ends were filled in using Klenow fragment enzyme polymerase (Boehringer). This fragment was ligated into the unique HindII site of the msrA gene in pMM1227, and the resulting vector, pMM1228, was linearized with BglII. This linear plasmid DNA was introduced into S. gordonii CH1 by electroporation, and erythromycin-resistant clones were selected on agar plates. Chromosomal integration of the msrA copy carrying the inserted erythromycin resistance gene was confirmed by Southern blotting. To complement the mutation, the msrA gene was obtained as an EcoRI fragment from pMM1227 and ligated into the unique EcoRI site of the broad-host-range vector pNZ124 (27), resulting in plasmid pMM1229. After this construct was introduced into the insertion mutant and into the wild type, colonies resistant to erythromycin and chloramphenicol were selected on TH agar plates.
Southern blotting.
Southern blots were prepared according to standard procedures (29) using Zeta-probe membranes (Bio-Rad, Hercules, Calif.). The 1.1-kb amplified msrA gene was used as the homologous DNA probe. The DNA probe was random-primed labeled with digoxigenin-11-dUTP using the DIG system for filter hybridization (Boehringer). Hybridization was done in DIG Easy Hyb hybridization solution (Boehringer) at 60°C, and DIG-labeled nucleic acids were visualized with anti-DIG-horseradish peroxidase and CSPD (Boehringer) as described by the manufacturer.
Hydrogen peroxide inhibition assay.
To test the susceptibility of bacteria to H2O2, a disk inhibition assay was performed, essentially as described by Moskovitz et al. (23). Bacteria were grown to stationary phase in TH broth. One milliliter of the bacterial suspension was added to 5 ml of liquid TH agar at 42°C and poured onto TH agar plates. A 1.3-cm-diameter filter disk (Whatman Scientific Ltd., Maidstone, United Kingdom) was placed on the plate and impregnated with either 20 μl of H2O or 20 μl of a 30% H2O2 solution. The plates were incubated overnight at 37°C.
Nucleotide sequence accession numbers.
The nucleotide sequence of the promoter fragment SGPP1224 has been assigned GenBank accession no. AF153501. The complete nucleotide sequence of the msrA gene from S. gordonii CH1 and the partial sequences of a pyrD homolog and a putative open reading frame have been assigned GenBank accession no. AF128264.
RESULTS
Isolation of pH-regulated promoters from S. gordonii.
A genomic library of S. gordonii CH1 was used for the selection of neutral-pH-inducible promoters. A total of 146 spectinomycin-resistant colonies apparently carrying an active promoter fragment grew on TH agar plates (pH 7.3) supplemented with erythromycin (5 μg/ml) and spectinomycin (500 μg/ml). The relatively limited number of spectinomycin-resistant clones was presumably due to the high antibiotic concentration used for selection. Two of the spectinomycin-resistant clones (CH1 pMM1223 and CH1 pMM1224) showed no growth on spectinomycin-containing TH plates (pH 6.2), and the growth of three other clones (CH1 pMM1221, CH1 pMM1222, and CH1 pMM1225) was strongly reduced on these plates. All five clones grew well on the two control plates. The growth of the other 141 spectinomycin-resistant clones did not show any difference on any of the three plates. This indicated that the five selected S. gordonii CH1 clones had lower promoter activities at pH 6.2 than at pH 7.3.
Identification and characterization of the pH-regulated promoters.
To identify the promoters of the five selected strains, the cloned genomic fragments were amplified by PCR and sequenced completely. Four of five promoter fragments showed sequence homology to known entries in the EMBL, GenBank, and DDBJ databases (Table 1).
TABLE 1.
Identified sequence homologies for the isolated neutral-pH-inducible promoter fragments from S. gordonii CH1
| Strain | Promoter | Accession no.a | Database matchb |
|---|---|---|---|
| CH1 pMM1221 | SGP1221 | AJ236900 | cysK of B. subtilis (P37887) |
| CH1 pMM1222 | SGP1222c | AF128264 | msrA of S. pneumoniae (U41735) |
| CH1 pMM1223 | SGP1223 | AF127175 | hydA of C. acetobutylicum (U15277) |
| CH1 pMM1224 | SGP1224 | AF153501 | No homology |
GenBank database.
Accession numbers for EMBL, GenBank, and DDBJ databases are in parentheses.
This promoter fragment is identical to SGP1225.
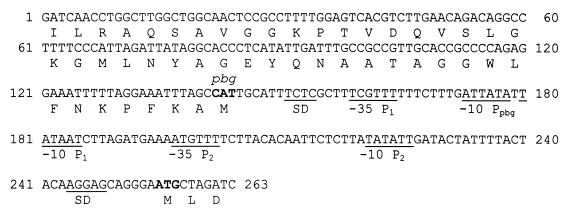
SGP1221 showed homology to the 5′ end of cysK from Bacillus subtilis, as well as to cysK homologs in several other bacterial species (Mycobacterium, E. coli, and serovar Typhimurium). SGP1223 showed limited similarity to the promoter region of the hydA gene of Clostridium acetobutylicum ATCC 824. We had already isolated these promoter fragments in previous studies, using other experimental settings (35, 36). The sequence within SGP1224 (Fig. 1) presumably responsible for the expression of the promoterless spectinomycin gene of pMM223 did not have similarity to known sequences. Upstream and in the inverse orientation an open reading frame was located, the translated amino acid sequence of which was homologous to the N-terminal region of the 6-phosphate-beta-glucosidase of several bacterial species, including B. subtilis and E. coli. Several regions were identified in this fragment that could act as promoters driving either the expression of the promoterless spectinomycin gene of pMM223 or that of the oppositely oriented phospho-beta-glucosidase (pbg)-like gene (Fig. 1).
FIG. 1.
Complete nucleotide sequence of the promoter fragment SGP1224. Putative −35 and −10 promoter regions and Shine-Dalgarno sequences (SD) are underlined. P1 and P2 are possible promoter stretches driving expression of the promoterless spectinomycin gene, and Ppbg is a putative promoter driving expression of the inversely oriented pbg-like gene. Translational start sites (ATG) are printed in boldface, and partial open reading frames are shown.
SGP1222 and SGP1225 appeared to be identical genomic-DNA fragments. The sequence was highly homologous to the 5′ end of the methionine sulfoxide reductase (msrA) gene from different bacterial and eukaryotic organisms. The translated sequence of this fragment showed strong identity to the N terminus of the MsrA protein of Streptococcus pneumoniae (Swissprot database accession no. P35593). The upstream sequence was a possible open reading frame with over 85% identity at the protein level to the dihydroorotate dehydrogenase (PyrD) of Streptococcus thermophilus ST11 (EMBL database accession no. Y12213), an enzyme involved in the de novo biosynthesis of pyrimidine.
The inducibility of the selected clones was confirmed by determination of the ratio of the growth rates in liquid medium in the presence and in the absence of spectinomycin. All clones showed a reduction in this growth rate ratio at pH 6.2 (Table 2), although the difference was much less pronounced than on solid medium. Although the activities of all promoters were upregulated by an increase in the pH, no general structure was identified in the sequences of the promoter fragments that might account for this regulation.
TABLE 2.
Activity at pH 6.2 and 7.3 of a constitutive and pH-regulated promoters isolated from S. gordonii CH1, recorded as the ratio of growth in medium with and without Spa
| Strain | Activity
|
|
|---|---|---|
| pH 6.2 | pH 7.3 | |
| CH1 pMM240 | 0.94 ± 0.01 | 0.98 ± 0.01 |
| CH1 pMM1221 | 0.90 ± 0.03 | 0.99 ± 0.01 |
| CH1 pMM1222 | 0.73 ± 0.09 | 0.93 ± 0.03 |
| CH1 pMM1223 | 0.68 ± 0.05 | 0.86 ± 0.06 |
| CH1 pMM1224 | 0.62 ± 0.05 | 0.82 ± 0.10 |
Growth was monitored by measuring the absorbance at 620 nm (A620) over time, and the mid-log-phase doubling time (t1/2) was determined. Relative promoter activity at the different pH values is expressed as the ratio of growth in the presence and absence of spectinomycin at each pH tested [t1/2(+spec)/ t1/2(−spec)]. pMM240 contains a constitutive promoter of S. gordonii CH1.
Isolation and characterization of the msrA gene from S. gordonii CH1.
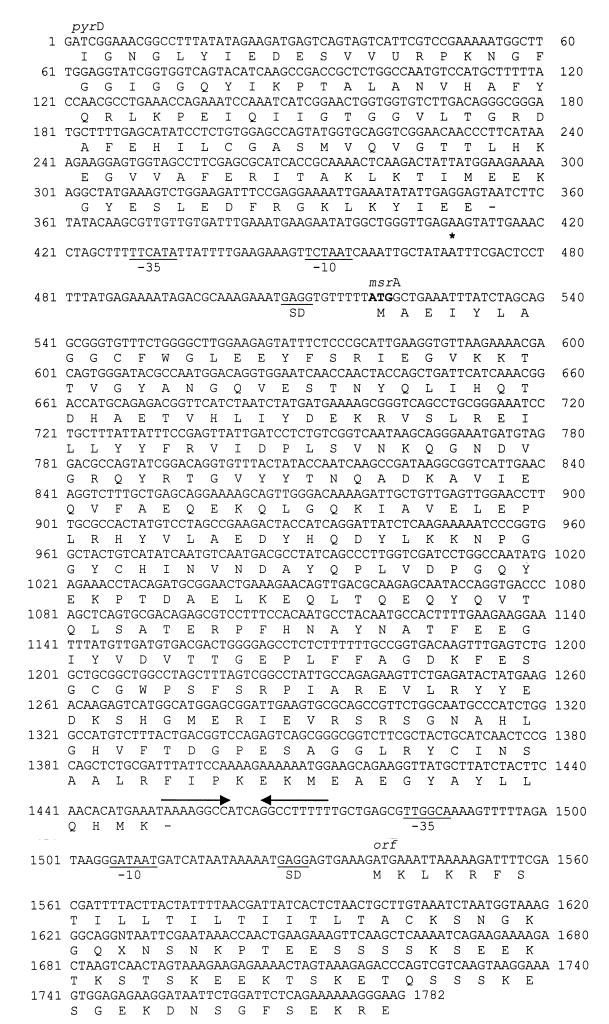
As the activity of the msrA promoter homolog of S. gordonii V288 was recently found to be induced in the experimental rabbit model of IE (16), we further characterized the corresponding S. gordonii CH1 msrA homolog. The 3′ end of the CH1 msrA gene was amplified by inside-out PCR on a self-ligated HindIII digest of chromosomal DNA using primer pair AV40-AV41. After subcloning and exonuclease III treatment of the 3.5-kb amplicon, a final fragment of approximately 1.2 kb was sequenced. This sequence contained the 3′ end of msrA and overlapped the sequence of the SGP1222 promoter fragment, which allowed the assembly of a total sequence of 1,782 nucleotides (Fig. 2).
FIG. 2.
Complete nucleotide sequence of the msrA gene from S. gordonii CH1 and partial sequences of a pyrD homolog and of a putative open reading frame, located upstream and downstream of the streptococcal msrA, respectively. Putative −35 and −10 promoter hexamers and Shine-Dalgarno sequences (SD) are underlined, and the transcriptional start site of msrA is indicated with an asterisk. Inverted repeats, which might form a transcriptional termination stem-loop, are indicated with arrows. Translational start sites (ATG) are printed in boldface, and the translated amino acid sequences of msrA and of the partial open reading frames upstream and downstream of msrA are shown. Nucleotides 1 to 747 represent the sequence of the isolated promoter fragments SGP1222 and SGP1225.
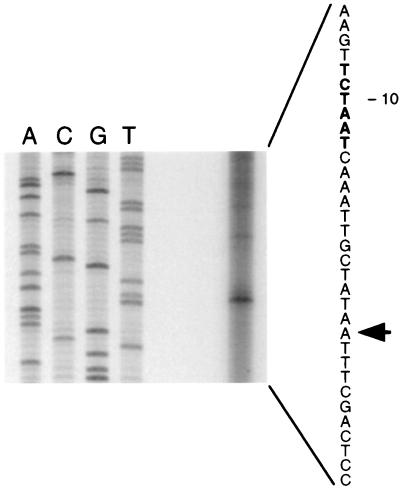
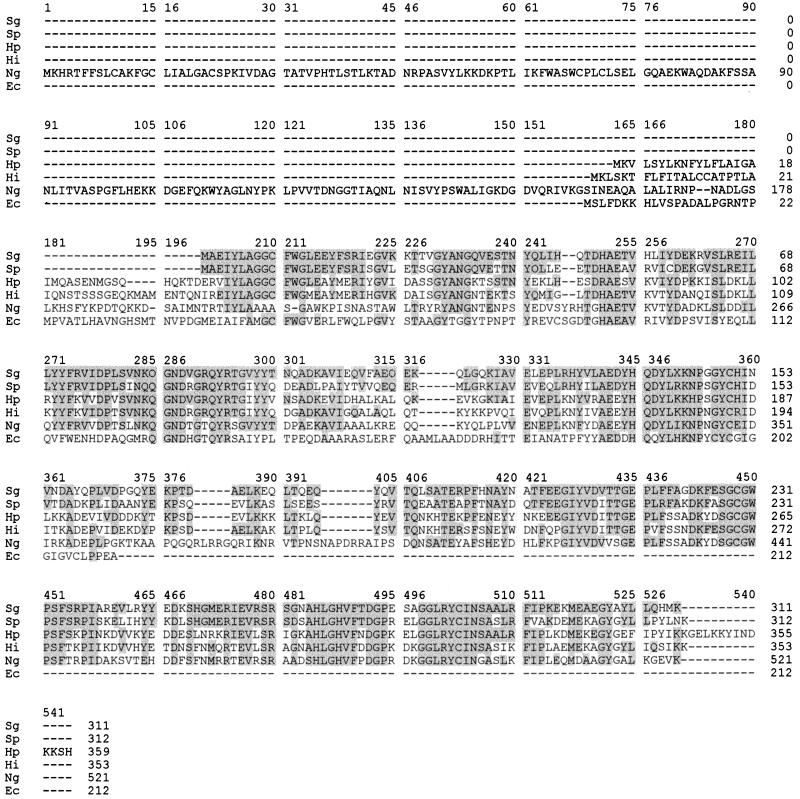
The S. gordonii CH1 msrA gene consisted of 933 nucleotides. A potential ribosome binding site was found 8 nucleotides upstream of the ATG translation start. Primer extension analysis revealed the transcription initiation site located 50 nucleotides upstream of the translation start site of the gene (Fig. 3). Preceding this transcription start site, putative −35 and −10 regions were identified. At the end of the gene, inverted repeats, capable of forming a terminator stem-loop structure with a free energy of −11.4 kcal, were identified. The S. gordonii CH1 msrA gene encodes a putative protein of 311 amino acids with a predicted molecular mass of 35.7 kDa and a pI of 5.35. Comparison of the translated amino acid sequence to entries in the databases revealed strong homology throughout the protein to other MsrA homologs (Fig. 4). There was 68 and 72.6% identity at the DNA and protein levels, respectively, to the methionine sulfoxide reductase of S. pneumoniae. Upstream of the pneumococcal msrA sequence, so-called BOX elements are present that are possibly involved in regulation of gene expression (17). No such structures were detected upstream of the translational start site of the msrA gene of S. gordonii CH1.
FIG. 3.
Determination of the transcription start site of S. gordonii CH1 msrA. The transcription start site is indicated with an arrow, and the putative −10 region in the coding strand is presented in boldface.
FIG. 4.
Amino acid sequence alignment of MsrA of S. gordonii (Sg) with MsrA proteins of S. pneumoniae (Sp), Helicobacter pylori (Hp), Haemophilus influenzae (Hi), and E. coli (Ec), and with the homologous PilB of N. gonorrhoeae (Ng). Amino acid sequence alignment was performed with the CLUSTAL program. The shaded boxes enclose residues of the MsrA protein from S. gordonii CH1 that are found at identical positions within one or more of the other MsrA sequences or within N. gonorrhoeae PilB.
Downstream of the msrA gene, another possible open reading frame was identified (Fig. 2). A putative ribosome binding site and −35 and −10 promoter regions were present in the intergenic region preceding this open reading frame. The open reading frame and its translated amino acid sequence did not have homology to any known sequences in the databases.
Effect of msrA mutation on sensitivity to oxidative stress and on growth.
An msrA mutant of S. gordonii CH1 was constructed by insertion of an erythromycin resistance marker. Erythromycin-resistant clones were tested for successful integration by Southern blotting. Strain MM1 was found to have the erythromycin resistance gene inserted into the msrA gene, resulting in an increase in size by 1.0 kb of the chromosomal fragment hybridizing with the msrA probe (Fig. 5).
FIG. 5.

Southern blot of S. gordonii strain CH1 and its msrA insertion mutant MM1. The hybridizing fragment in the wild-type strain is increased in size in the mutant strain by 1.0 kb, due to the inserted erythromycin resistance gene.
As MsrA is known to play a role in protection against oxidative damage in other bacterial and eukaryotic species (21–24, 38), sensitivity to oxidative stress of the S. gordonii CH1 msrA mutant was tested using an H2O2 disk inhibition assay (23). Growth of the mutant strain was more strongly reduced than that observed for the parent strain when the disk was impregnated with 30% H2O2. Complementation of the mutation by introduction of an intact copy of the msrA gene on a low-copy-number plasmid into the mutant strain MM1 decreased the inhibition zone to that observed with the wild type (Table 3). No growth inhibition was observed for either strain when the disk was impregnated with water. These data strongly indicate that the absence of a functional msrA gene renders S. gordonii more susceptible to H2O2 stress.
TABLE 3.
Effect of H2O2 treatment on the growth of S. gordonii CH1 and its msrA insertion mutant S. gordonii MM1
| Genotype | H2O2a | Area of growth inhibition (cm2)b |
|---|---|---|
| msrA+ | − | 0 |
| msrA::Emr | − | 0 |
| msrA+ | + | 7.8 |
| msrA::Emr | + | 11.2 |
| msrA+ (pMM1229; msrA+) | + | 7.8 |
| msrA::Emr (pMM1229; msrA+) | + | 7.8 |
In control experiments, the disks were impregnated with 20 μl of H2O instead of H2O2. +, H2O2; −, H2O.
The amount of growth inhibition is expressed as the area of the clear zone minus the area of the disk, as no growth was observed under the control conditions. The values are the averages of at least three experiments.
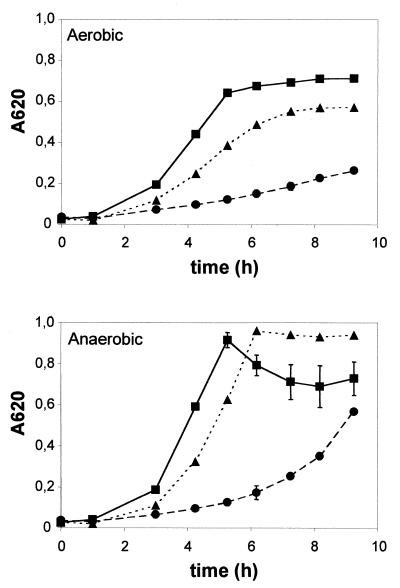
Next, the growth rate in TH broth of the different strains was assessed, in order to define a possible influence of the absence of a functional MsrA on bacterial multiplication. Growth of the mutant MM1 was strongly reduced compared to that of the wild-type strain CH1 when it was cultured at 37°C under either aerobic or anaerobic conditions. The growth rate of the msrA-complemented mutant was almost identical to that of the wild-type strain, CH1 (Fig. 6). These results imply a function of the streptococcal MsrA homolog in bacterial multiplication, in addition to its role in protection against oxidative stress.
FIG. 6.
Growth of S. gordonii CH1 (squares), its msrA mutant MM1 (circles), and the complemented mutant (MM1 pMM1229; triangles) under aerobic (top) and anaerobic (bottom) conditions in TH medium. The values are the averages of three experiments, and the standard error of the mean is indicated for each value.
DISCUSSION
In this study we found that a slight increase in the environmental pH, as observed when VS from the oral cavity gain access to the bloodstream, induces or upregulates the expression of specific genes. Indeed, five clones containing a promoter whose activity was upregulated when the pH was raised from 6.2 to 7.3 were isolated from an S. gordonii CH1 expression library. No common regulatory sequences that might be involved in a coordinate pH-regulated gene expression could be identified in the sequenced promoter regions. Another example of response by VS to an increase in pH is the intracellular thrombin-like activity of Streptococcus sanguis, which is reduced at acidic pH and is increased upon alkalification of the medium (18). Induction of gene expression upon increase of the environmental pH might, therefore, be a general response mechanism within the VS in order to survive when the bacteria translocate from the oral cavity to the blood.
One of the isolated pH-regulated promoter fragments, SGP1221, showed homology at both the DNA and protein levels to the cysteine synthase of B. subtilis. We have also identified this promoter fragment (EMBL database accession no. AJ236900) in recent screening experiments for constitutively active promoters from S. gordonii CH1 (35). In those experiments we used agar plates at pH 7.8, which explains the isolation of this promoter. In B. subtilis, CysK is expressed under normal laboratory conditions, but expression levels can be up- or downregulated by different environmental stimuli, e.g., cold shock, heat shock, and salt stress (11). In S. gordonii the level of expression of this gene is regulated by variation in the external pH, a stimulus which might also regulate expression of the B. subtilis cysK gene.
Fragment SGP1223 showed limited homology to the promoter region of the hydA gene from C. acetobutylicum ATCC 824. Expression of this gene in C. acetobutylicum is known to be transcriptionally regulated by the environmental pH (10). SGP1223 was identical to a neutral-pH-inducible promoter fragment we had identified earlier (Vriesema et al., submitted), indicating reproducibility of the screening system.
One neutral-pH-inducible promoter fragment was isolated twice from the genomic DNA library (SGP1222 and SGP1225). The fragment showed homology to the msrA gene found in many prokaryotic (E. coli, S. pneumoniae, and Neisseria gonorrhoeae) (23, 38) and eukaryotic (Saccharomyces cerevisiae, rat, and human) species (21, 22, 24). This gene encodes methionine sulfoxide reductase, a protein involved in the reduction of oxidized proteins. The sulfur groups of methionine residues are highly sensitive to oxidation by oxygen radicals, and oxidized proteins are in general not functional. Reduction of oxidized methionine residues by MsrA restores the protein function, thus decreasing the need for de novo protein synthesis (8). A second function recently suggested for MsrA is its involvement in the stabilization of adhesins. Mutation in E. coli msrA decreased fimbria-mediated mannose-dependent agglutination of erythrocytes, and mutation of S. pneumoniae msrA caused decreased binding to specific glycoconjugate-containing receptors on vascular endothelial and lung cells (38). Finally, the methionine sulfoxide reductase might also be involved in signal transduction, as it is highly homologous to PilB of N. gonorrhoeae (38), the sensor component of the PilAB two-component regulator system (31). However, such a function could not be identified for the MsrA from S. pneumoniae (26).
The promoter of the msrA gene from S. gordonii V288 is activated in vivo in a rabbit model of endocarditis (16). In addition, methionine sulfoxide reductase has been demonstrated to be of importance for the survival of Staphylococcus aureus in a murine bacteremia model (19). Although an S. aureus msrA deletion mutant was not attenuated in its virulence in this model, in mixed infections the wild-type was almost solely reisolated (19). This indicates that the MsrA protein is beneficial for bacterial survival in this host.
MsrA of S. gordonii CH1 appeared to be involved in protection against oxidative stress, as growth of the msrA mutant strain MM1 on solid medium in the presence of H2O2 was much more reduced than the growth of wild-type CH1. This may well be of great importance for survival in vivo, as blood-borne bacteria are challenged by oxidative radicals produced by polymorphonuclear leukocytes and other cells of the host immune system (2).
In addition, MsrA was required for maximal growth, under both aerobic and anaerobic conditions. The observed growth reduction of the S. gordonii mutant under aerobic conditions was not caused by an increased sensitivity to oxidative damage, as a similar difference in growth rate between the wild-type and the mutant strain was observed when they were cultured under anaerobic conditions. Complementation of the mutation almost completely restored growth to wild-type levels. In contrast, in E. coli, mutation of msrA did not affect growth (23). It seems that MsrA of S. gordonii CH1, in addition to having a function in protection against oxidative damage, plays an important role in bacterial growth. This phenomenon might also explain the above-mentioned survival benefit of wild-type S. aureus in mixed infections with its msrA mutant in the murine bacteremia model (19). In addition, MsrA will probably prove to be of importance in IE, as rapid bacterial multiplication is a major characteristic of VS in the development of this disease (5, 9).
ACKNOWLEDGMENTS
We thank Jan Kok (Department of Genetics, University of Groningen, Haren, The Netherlands) for plasmid pMG36e and Richard van Kranenburg (NIZO, Ede, The Netherlands) for plasmid pNZ124. In addition, we are grateful to Bianca Klasens for technical assistance, Wim van Est and Eelco Roos for excellent photographic work, and Martine van Vugt for critical reading of the manuscript.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E F, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Beaman L, Beaman B L. The role of oxygen and its derivatives in microbial pathogenesis and host defense. Annu Rev Microbiol. 1984;38:27–48. doi: 10.1146/annurev.mi.38.100184.000331. [DOI] [PubMed] [Google Scholar]
- 3.Burnette-Curley D, Wells V, Viscount H, Munro C L, Fenno J C, Fives-Taylor P, Macrina F L. FimA, a major virulence factor associated with Streptococcus parasanguis endocarditis. Infect Immun. 1995;63:4669–4674. doi: 10.1128/iai.63.12.4669-4674.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dall L H, Herndon B L. Association of cell-adherent glycocalyx and endocarditis production by viridans group streptococci. J Clin Microbiol. 1990;28:1698–1700. doi: 10.1128/jcm.28.8.1698-1700.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dankert J, van der Werff J, Zaat S A J, Joldersma W, Klein D, Hess J. Involvement of bactericidal factors from thrombin stimulated platelets in clearance of adherent viridans streptococci in experimental infective endocarditis. Infect Immun. 1995;63:663–671. doi: 10.1128/iai.63.2.663-671.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiRita V J, Mekalanos J J. Genetic regulation of bacterial virulence. Annu Rev Genet. 1989;23:455–482. doi: 10.1146/annurev.ge.23.120189.002323. [DOI] [PubMed] [Google Scholar]
- 7.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dowson C G, Barcus V, King S, Pickerill P, Whatmore A, Yeo M. Horizontal gene transfer and the evolution of resistance and virulence determinants in Streptococcus. J Appl Microbiol. 1997;83(Suppl.):42S–51S. doi: 10.1046/j.1365-2672.83.s1.5.x. [DOI] [PubMed] [Google Scholar]
- 9.Durack D T, Beeson P B. Experimental bacterial endocarditis. I. Colonization of a sterile vegetation. Br J Exp Pathol. 1972;53:44–49. [PMC free article] [PubMed] [Google Scholar]
- 10.Gorwa M-F, Croux C, Soucaille P. Molecular characterization and transcriptional analysis of the putative hydrogenase gene of Clostridium acetobutylicum ATCC 824. J Bacteriol. 1996;178:2668–2675. doi: 10.1128/jb.178.9.2668-2675.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graumann P, Schröder K, Schmid R, Marahiel M A. Cold shock stress-induced proteins in Bacillus subtilis. J Bacteriol. 1996;178:4611–4619. doi: 10.1128/jb.178.15.4611-4619.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higgins D G, Sharp P M. Fast and sensitive multiple sequence alignments on a microcomputer. Comput Appl Biosci. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- 13.Hohn B. In vitro packaging of λ and cosmid DNA. Methods Enzymol. 1979;68:299–309. doi: 10.1016/0076-6879(79)68021-7. [DOI] [PubMed] [Google Scholar]
- 14.Hynes W L, Ferretti J J, Gilmore M S, Segarra R A. PCR amplification of streptococcal DNA using crude cell lysates. FEMS Microbiol Lett. 1992;94:139–142. doi: 10.1016/0378-1097(92)90597-h. [DOI] [PubMed] [Google Scholar]
- 15.Jayaraman G C, Penders J E, Burne R A. Transcriptional analysis of the Streptococcus mutans hcrA, grpE and dnaK genes and regulation of expression in response to heat shock and environmental acidification. Mol Microbiol. 1997;25:329–341. doi: 10.1046/j.1365-2958.1997.4671835.x. [DOI] [PubMed] [Google Scholar]
- 16.Kiliç A O, Herzberg M C, Meyer M W, Zhao X, Tao L. Streptococcal reporter gene-fusion vector for identification of in vivo expressed genes. Plasmid. 1999;42:67–72. doi: 10.1006/plas.1999.1408. [DOI] [PubMed] [Google Scholar]
- 17.Martin B, Humbert O, Camara M, Guenzi E, Walker J, Mitchell T, Andrew P, Prudhomme M, Alloing G, Hakenbeck R, Morrison D A, Boulnois G J, Claverys J-P. A highly conserved repeated DNA element located in the chromosome of Streptococcus pneumoniae. Nucleic Acids Res. 1992;20:3479–3483. doi: 10.1093/nar/20.13.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayo J A, Harty D W S, Knox K W. Modulation of glycosidase and protease activities by chemostat growth conditions in an endocarditis strain of Streptococcus sanguis. Oral Microbiol Immunol. 1995;10:342–348. doi: 10.1111/j.1399-302x.1995.tb00164.x. [DOI] [PubMed] [Google Scholar]
- 19.Mei J M, Nourbakhsh F, Ford C W, Holden D W, Achen M G. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol Microbiol. 1997;26:399–407. doi: 10.1046/j.1365-2958.1997.5911966.x. [DOI] [PubMed] [Google Scholar]
- 20.Miller S I, Krukal A M, Mekalanos J J. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci USA. 1991;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moskovitz J, Berlett B S, Poston J M, Stadtman E R. The yeast peptide-methionine sulfoxide reductase functions as an antioxidant in vivo. Proc Natl Acad Sci USA. 1997;94:9585–9589. doi: 10.1073/pnas.94.18.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moskovitz J, Jenkins N A, Gilbert D J, Copeland N G, Jursky F, Weissbach H, Brot N. Chromosomal localization of the mammalian peptide-methionine sulfoxide reductase gene and its differential expression in various tissues. Proc Natl Acad Sci USA. 1996;93:3205–3208. doi: 10.1073/pnas.93.8.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moskovitz J, Rahman M A, Strassman J, Yancey S O, Kushner S R, Brot N, Weissbach H. Escherichia coli peptide methionine sulfoxide reductase gene: regulation of expression and role in protecting against oxidative damage. J Bacteriol. 1995;177:502–507. doi: 10.1128/jb.177.3.502-507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moskovitz J, Weissbach H, Brot N. Cloning and expression of a mammalian gene involved in the reduction of methionine sulfoxide residues in proteins. Proc Natl Acad Sci USA. 1996;93:2095–2099. doi: 10.1073/pnas.93.5.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nolte W A. Defense mechanisms of the mouth. In: Nolte W A, editor. Oral microbiology. St. Louis, Mo: The C.V. Mosby Company; 1982. pp. 245–260. [Google Scholar]
- 26.Pearce B J, Yin Y B, Masure H R. Genetic identification of exported proteins in Streptococcus pneumoniae. Mol Microbiol. 1993;9:1037–1050. doi: 10.1111/j.1365-2958.1993.tb01233.x. [DOI] [PubMed] [Google Scholar]
- 27.Platteeuw C, Simons G, De Vos W M. Use of the Escherichia coli β-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl Environ Microbiol. 1993;60:587–593. doi: 10.1128/aem.60.2.587-593.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramirez-Ronda C H. Adherence of glucan-positive and glucan-negative streptococcal strains to normal and damaged heart valves. J Clin Investig. 1978;62:805–814. doi: 10.1172/JCI109192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 30.Scheld W M, Valone J A, Sande M A. Bacterial adherence in the pathogenesis of endocarditis. Interaction of bacterial dextran, platelets, and fibrin. J Clin Investig. 1978;61:1394–1404. doi: 10.1172/JCI109057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taha M-K, Dupuy B, Saurin W, So M, Marchal C. Control of pilus expression in Neisseria gonorrhoeae as an original system in the family of two-component regulators. Mol Microbiol. 1991;5:137–148. doi: 10.1111/j.1365-2958.1991.tb01834.x. [DOI] [PubMed] [Google Scholar]
- 32.van de Guchte M, van der Vossen J M B M, Kok J, Venema G. Construction of a lactococcal expression vector: expression of hen egg white lysozyme in Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1989;55:224–228. doi: 10.1128/aem.55.1.224-228.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van der Meer J T M, van Vianen W, van Leeuwen W B, Valkenburg H A, Thompson J, Michel M F. Distribution, antibiotic susceptibility and tolerance of bacterial isolates in culture-positive cases of endocarditis in The Netherlands. Eur J Clin Microbiol Infect Dis. 1991;10:728–734. doi: 10.1007/BF01972497. [DOI] [PubMed] [Google Scholar]
- 34.Viscount H B, Munro C L, Burnette-Curley D, Peterson D L, Macrina F L. Immunization with FimA protects against Streptococcus parasanguis endocarditis in rats. Infect Immun. 1997;65:994–1002. doi: 10.1128/iai.65.3.994-1002.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Vriesema, A. J. M., R. Brinkman, J. Kok, J. Dankert, and S. A. J. Zaat. Broad-host-range shuttle vectors for the screening of regulated promoter activity in viridans group streptococci: isolation of a pH-regulated promoter. Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 35.Vriesema A J M, Dankert J, Zaat S A J. Isolation and characterization of promoter regions from Streptococcus gordonii CH1. Curr Microbiol. 1999;39:321–326. doi: 10.1007/s002849900466. [DOI] [PubMed] [Google Scholar]
- 36.Vriesema A J M, Zaat S A J, Dankert J. A simple procedure for isolation of cloning vectors and endogenous plasmids from viridans group streptococci and Staphylococcus aureus. Appl Environ Microbiol. 1996;62:3527–3529. doi: 10.1128/aem.62.9.3527-3529.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wells, V. D., C. L. Munro, M. C. Sulavik, D. B. Clewell, and F. L. Macrina. Infectivity of a glucan synthesis-defective mutant of Streptococcus gordonii (Challis) in a rat endocarditis model. FEMS Microbiol. Lett. 112:301–306. [DOI] [PubMed]
- 38.Wizemann T M, Moskovitz J, Pearce B J, Cundell D R, Arvidson C G, So M, Weissbach H, Brot N, Masure H R. Peptide methionine sulfoxide reductase contributes to the maintenance of adhesins in three major pathogens. Proc Natl Acad Sci USA. 1996;93:7985–7990. doi: 10.1073/pnas.93.15.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–109. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]