Abstract
Legionella pneumophila, a facultative intracellular parasite of human alveolar macrophages and protozoa, causes Legionnaires' disease. Using mini-Tn10 mutagenesis, we previously isolated a L. pneumophila mutant that was hypersensitive to iron chelators. This mutant, NU216, and its allelic equivalent, NU216R, were also defective for intracellular infection, particularly in iron-deficient host cells. To determine whether NU216R was attenuated for virulence, we assessed its ability to cause disease in guinea pigs following intratracheal inoculation. NU216R-infected animals yielded 1,000-fold fewer bacteria from their lungs and spleen compared to wild-type-130b-infected animals that had received a 50-fold-lower dose. Moreover, NU216R-infected animals subsequently cleared the bacteria from these sites. While infection with 130b resulted in high fever, weight loss, and ruffled fur, inoculation with NU216R did not elicit any signs of disease. DNA sequence analysis revealed that the transposon insertion in NU216R lies in the first open reading frame of a two-gene operon. This open reading frame (iraA) encodes a 272-amino-acid protein that shows sequence similarity to methyltransferases. The second open reading frame (iraB) encodes a 501-amino-acid protein that is highly similar to di- and tripeptide transporters from both prokaryotes and eukaryotes. Southern hybridization analyses determined that the iraAB locus was largely limited to strains of L. pneumophila, the most pathogenic of the Legionella species. A newly derived mutant containing a targeted disruption of iraB showed reduced ability to grow under iron-depleted extracellular conditions, but it did not have an infectivity defect in the macrophage-like U937 cells. These data suggest that iraA is critical for virulence of L. pneumophila while iraB is involved in a novel method of iron acquisition which may utilize iron-loaded peptides.
Legionella pneumophila is the causative agent of Legionnaires' disease, a form of community-acquired and nosocomial pneumonia. L. pneumophila is a gram-negative facultative intracellular parasite that infects protozoa and macrophages (1, 12, 23). The organism enters alveolar macrophages by coiling or conventional phagocytosis and replicates within a phagosome that appears not to fuse with the endosomes or lysosomes (16, 17, 38). Various factors that enable L. pneumophila to productively infect protozoa and macrophages have been reported. These include the outer membrane porin (27, 43), Mip (13, 25), type II and type IV secretion systems (4a, 46, 47), type IV pili (47, 71), flagella (61; C. Dietrich, K. Heuner, J. Hacker, and B. C. Brand, Abstr. 99th Gen. Meet. Am. Soc. Microbiol. 1999, abstr. B/D-358, p. 99, 1999), a catalase-peroxidase (6), growth phase (11) and the products of the numerous dot, icm, eml, mil, and pmi loci (1, 66, 69, 73). While these factors are clearly implicated in infection, relatively few have been fully characterized at the mechanistic level and fewer still have actually been demonstrated to be critical for causing disease in animal models (13, 46, 49, 51).
Iron is a key requirement for L. pneumophila virulence and intracellular growth (22, 60, 64). Indeed, chelators that deplete the macrophage iron pool block L. pneumophila intracellular replication (8, 29, 60). Also, gamma interferon inhibits L. pneumophila growth within human monocytes by reducing the levels of intracellular iron (9, 10). The mechanisms by which L. pneumophila acquires iron, especially intracellular iron, are poorly understood. L. pneumophila can proteolytically degrade transferrin and use the released iron in steady-state, iron-limited cultures (39). However, this indirect mode of iron acquisition is unlikely to be relevant for intracellular growth, since the L. pneumophila phagosome does not contain transferrin, and the bacterium itself does not bind transferrin (15, 40, 62). L. pneumophila can bind lactoferrin, but this does not lead to iron assimilation (7). While L. pneumophila binds hemin and can utilize it as the sole iron source for extracellular growth, disruption of a gene that promotes hemin binding does not impair replication in macrophages (56). Two internal ferric reductases are thought to be important for iron assimilation by L. pneumophila, although their roles in pathogenesis have not been delineated (40, 59). It had been reported that L. pneumophila does not produce siderophores when grown in iron-deficient media (39, 45, 65). However, we have recently demonstrated that L. pneumophila does elaborate a nonhydroxamate, nonphenolate siderophore (legiobactin), but only under specific growth conditions (M. R. Liles, T. A. Scheel, and N. P. Cianciotto, submitted for publication). Curiously, we have also demonstrated the presence of a L. pneumophila homolog of a hydroxamate biosynthetic gene (37). A targeted disruption of this gene results in reduced growth within macrophages, suggesting that L. pneumophila may produce and require an additional siderophore within host cells.
We earlier reported the isolation, by mini-Tn10 mutagenesis, of various L. pneumophila (ira) mutants defective for iron acquisition and assimilation (60). Most of these mutants were hypersensitive to the iron chelator ethylenediaminediacetic acid (EDDA) but were resistant to streptonigrin, an antibiotic whose bactericidal activity is enhanced by high levels of intracellular iron (74). One of these mutants, NU216, was especially impaired for infection of macrophage-like U937 cells, showing a prolonged lag phase and a reduced growth rate (60). The impairment of NU216 in macrophages was exacerbated nearly 100-fold in the presence of the iron chelator desferrioxamine (DFX), indicating that it included a defect in intracellular iron assimilation. As a first step toward identifying the genetic basis of the phenotype of the mutant, allelic exchange was used to reintroduce the mutation into the wild-type background to confirm that the defects were actually linked to the mini-Tn10 disruption. The new strain, NU216R, was EDDA hypersensitive and showed the same phenotype in U937 cells as the original mutant. NU216 and NU216R were not impaired for extracellular growth in standard Legionella media (60).
In the present study, we further characterized the iron uptake and infectivity impairment of NU216R. First, we showed that the ira locus that was inactivated in this strain is critical for virulence in guinea pigs. Subsequent sequence analysis of the locus revealed the presence of a two-gene operon (iraAB). Additional mutant analysis indicated that iraA alone is required for intracellular infection while iraB, encoding a putative peptide transporter, may promote iron assimilation by a novel method.
MATERIALS AND METHODS
Bacterial strains, media, and plasmids.
The L. pneumophila serogroup 1 strain 130b (Wadsworth), its mini-Tn10 derivative NU216, and the reengineered allelic equivalent NU216R have been described previously (60). Additional legionellae tested for the presence of the iraAB locus are listed in Table 1. L. pneumophila strains were routinely cultured on buffered charcoal yeast extract (BCYE) agar for 2 to 3 days at 37°C (19). Antibiotics (chloramphenicol, 3 μg/ml; kanamycin, 25 μg/ml) or sucrose (5%, wt/vol) were added to the medium when appropriate. Growth curve experiments were performed in liquid buffered yeast extract medium (BYE broth) with or without the standard iron supplementation (19). A L. pneumophila plasmid library containing Sau3 AI-digested 130b DNA cloned into the BamHI site of pBR322 was maintained in Escherichia coli HB101 (36). E. coli strain NovaBlue (Novagen, Madison, Wis.) was routinely used for maintaining and propagating newly isolated plasmids. Plasmid pCDP70, containing the mutated ira locus from NU216, has been described previously (60).
TABLE 1.
Legionella strainsa used in this study
| Species | Strain | Serogroup | Implicated in Disease | iraA and iraB present |
|---|---|---|---|---|
| L. pneumophila | 130b (Wadsworth) | 1 | Yes | Yes |
| L. pneumophila | ATCC 33154 | 2 | Yes | Yes |
| L. pneumophila | ATCC 33155 | 3 | Yes | Yes |
| L. pneumophila | ATCC 33156 | 4 | Yes | Yes |
| L. pneumophila | ATCC 33216 | 5 | Yes | Yes |
| L. pneumophila | MDPHb | 6 | Yes | Yes |
| L. pneumophila | MDPH | 7 | Yes | Yes |
| L. pneumophila | MDPH | 9 | Yes | Yes |
| L. pneumophila | MDPH | 11 | Yes | Yes |
| L. pneumophila | MDPH | 12 | Yes | Yes |
| L. pneumophila | ATCC 43736 | 13 | Yes | Yes |
| L. birminghamensis | ATCC 43702 | Yes | No | |
| L. erythra | SE-32A | No | No | |
| L. feeleii | WO-44C | Yes | No | |
| L. gormanii | ATCC33297 | Yes | Yes | |
| L. hackeliae | Lansing 2 | Yes | No | |
| L. israelensis | ATCC 43119 | No | No | |
| L. jamestowniensis | ATCC 35298 | No | No | |
| L. longbeachae | ATCC 33462 | Yes | No | |
| L. micdadei | ATCC 33204 | Yes | No | |
| L. oakridgensis | ATCC 33761 | Yes | No | |
| L. parisiensis | PF-209 | Yes | No | |
| L. sainthelensi | Mt. St. Helens 4 | Yes | No | |
| L. santicrucis | ATCC 35301 | No | No | |
| L. spiritensis | ATCC 35249 | No | No |
Experimental infection of guinea pigs and U937 cells by L. pneumophila.
To assess the virulence of L. pneumophila strains, we examined their ability to cause disease in guinea pigs following intratracheal injection (13, 20, 75). The procedure for infection was described previously (46). Briefly, BCYE agar-grown bacteria were transferred to BYE broth and grown at 37°C in a shaking incubator. The bacteria were then resuspended at the desired concentration and injected into surgically exposed trachea in 0.3-ml volumes. The delivered bacterial inoculum was determined by plating on BCYE agar plates. The study used Hartley male guinea pigs weighing around 300 g. The animals were observed for signs of infection for 7 days, with daily measurements of body weight and rectal temperature. The right lower lobes of the lungs and spleen were harvested from the respective groups each day, and ground homogenates were cultured quantitatively.
Intracellular infection of macrophage-like U937 cells generally was performed as described previously (60). However, to assess the effect of intracellular iron depletion, U937 cells were treated with DFX and intracellular legionellae were quantitated at various times postinoculation (10, 29, 60). The percent reduction in recovery was calculated as 100 × (average CFU from untreated cells − average CFU from DFX-treated cells)/(average CFU from untreated wells).
Sequence and Southern analysis of the iraAB locus.
Initial sequence analysis of the ira locus mutated in NU216 was performed on subclones of pCDP70 with the transposon primer 5′-GTGACGACTGAATCCGGT. To facilitate further sequencing, we sought an additional iraAB-containing plasmid from our library of 130b DNA. To do this, we used labeled pCDP70 as a probe in colony blots that were performed with the Genius system kit (Boerhinger, Mannheim, Germany). The isolated plasmid, pCDP71, contained approximately 5 kb of Legionella DNA. The sequence of the pCDP71 insert was determined by primer walking. Sequence analysis was performed using either the DyeTerminator cycle-sequencing reaction mix or the BigDye terminator cycle-sequencing reaction mix from PE Applied Biosystems (Foster City, Calif.). Primers for sequencing and PCR were obtained from the Biotech Facility at Northwestern University Medical School, Chicago, Ill. Automated sequence analysis was performed at the Biotech Facility on an ABI Prism 373 DNA sequencer (Applied Biosystems). Sequence database searches were performed using the programs based on the BLAST algorithm (3).
To determine the distribution of the iraAB locus among legionellae, Southern hybridization analysis was performed using iraA- and iraB-specific fluorescently labeled probes and the Genius system kit. The fragments to be labeled were generated by PCR using the primers shown below in Fig. 3. Chromosomal DNA for the blots was isolated from Legionella strains as described previously (60).
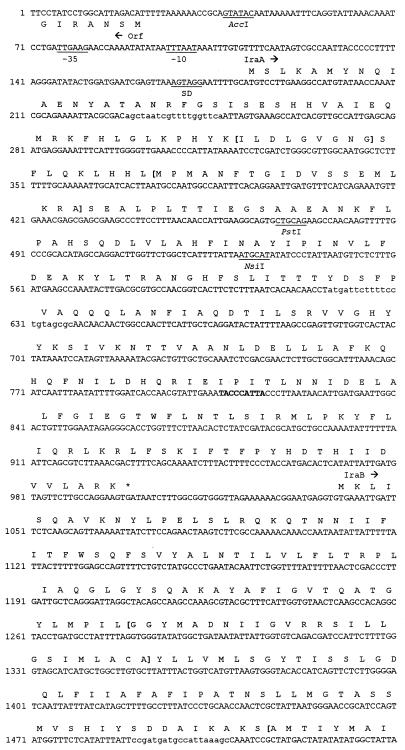
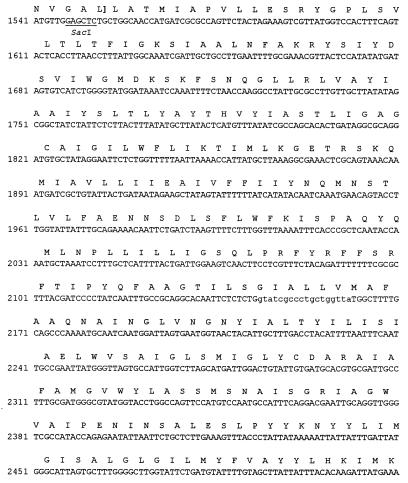
FIG. 3.
Nucleotide sequence of the L. pneumophila iraAB locus. The deduced amino acid sequences of the various ORFs and the termination codons (*) are indicated. Arrows indicate the directions of translation of the ORFs. Putative promoter sequences are indicated by the −10 and −35 designations upstream of iraA, and the Shine-Dalgarno sequence is indicated as SD. The target sequence for the miniTn10 insertion in NU216 and NU216R is indicated in bold. The two underlined sequences represent a 250-bp IR. Restriction sites used for making the constructs described in this paper are indicated. The Kmr cassette insertion in NU244 is in the SacI site. The IraA sequences corresponding to the SAM-binding consensus and the IraB sequences corresponding to the PTR consensus are enclosed within parentheses. PCR was performed using primers 5′-AGCTAATCGTTTTGGTTCA and 5′-GCGCTACAGGAAAAGAATCAT to generate iraA-specific fragments and 5′CCGATGATGCCATTAAAGC and 5′TAACCAGCAGGGCGATAC to generate iraB-specific fragments for hybridization analyses. The primer-binding regions are indicated in lowercase type. For brevity, only the beginning of the upstream ORF is shown. Double-stranded sequence data were compiled with GeneRunner (Hastings Software, Inc., Hastings, N.Y.).
Transcomplementation and directed-mutagenesis studies.
To facilitate complementation analyses of NU216R, we used pCDP71 to construct three new plasmids; pVK100 (carrying both iraA and iraB), pVK101 (carrying only iraA), and pVK102 (carrying only iraB). pVK100 was made by inserting the AccI-BstBI fragment from pCDP71 into the AccI site of the Cmr vector pSU2719 (50). pVK101 was generated by eliminating the iraB-containing SacI fragment from pVK100. To obtain pVK102, the 4.2-kb PstI-SphI fragment from pVK100 was treated with Klenow enzyme to generate blunt ends and then circularized. To attempt to generate a L. pneumophila strain with an in-frame deletion in iraA, plasmid pVK107 was generated in two steps. First, the EcoRV-SalI fragment from pCDP71 was ligated to NruI-SalI-digested pBOC20, a vector containing the counterselectable sacB (54). Subsequently, the 63-bp NsiI-PstI fragment in iraA was eliminated from pCDP74 to generate an in-frame deletion. Plasmids were introduced into L. pneumophila by electroporation (14).
To target a disruption in iraB, the PstI-containing Kmr fragment from pMB2190 was cloned into the PstI site of pBluescript II SK(+) (Stratagene, La Jolla, Calif.) to generate pVK3. The aph gene in pVK3 was determined to be in the same direction as the lacZ gene of the vector. The Kmr-containing region from pVK3 was amplified using the primer 5′-GGGAGCTCAGGTCTGCCTCGTGAAGAA and the M13 reverse primer. The SacI-digested PCR product was cloned into the SacI site of pCDP71 to generate pVK104 with a disrupted iraB gene. As the first step toward introducing the iraB mutation into L. pneumophila, the SalI fragment from pVK104 containing the inactivated iraB gene was moved into the SalI site of pBOC20. The resulting Kmr Cmr plasmid, pVK105, was electroporated into wild-type L. pneumophila. Cmr Kmr electrotransformants were then streaked onto plates containing karamycin and 5% sucrose (14). Kmr Cms sucrose-resistant colonies were selected as putative mutants containing a disruption in iraB. The identity of these mutants was confirmed by PCR analysis. PCR was performed in a 50-μl reaction mix, as recommended by the manufacturer, in the presence of 1 to 2 U of Taq polymerase (Life Technologies, Rockville, Md.). One of these iraB mutants, designated NU244, was chosen for further study.
Nucleotide sequence accession number.
The L. pneumophila iraAB locus sequence is deposited in the GenBank database at the National Center for Biotechnology Information under accession no. AF167992.
RESULTS
Cell morphology of L. pneumophila iraAB mutants.
It was observed that NU216 cells collected from 3-day-old BCYE agar plates and suspended in BYE broth did not show a correspondence between optical density at 660 nm (OD660) and CFU per milliliter that was comparable to that of 130b. Thus, NU216 cells resuspended to the same OD660 as 130b and plated onto BCYE agar plates resulted in only half the number of CFU. Gram staining revealed that the NU216 culture was far more filamentous than 130b (data not shown). To determine whether this altered cellular morphology was associated with the transposon insertion in the ira locus, we examined NU216R. The morphology of this strain was similar to that of 130b, indicating that the filamentous nature of NU216 was due to a spontaneous secondary mutation. Since NU216R possesses the iron uptake and infectivity defects of NU216 (60), we concluded that these defects are not due to any changes in cellular morphology. We therefore used NU216R for all of the studies detailed in this paper.
Pulmonary infection of guinea pigs by L. pneumophila strains.
The decreased infection of U937 cells by NU216 and NU216R suggested that the ira locus disrupted in these strains was required for virulence (60). To test this hypothesis, we evaluated the relative virulence of NU216R for guinea pigs. An initial, qualitative assessment of virulence was obtained by observing the ability of various doses of bacteria to cause fatal disease following intratracheal inoculation. Guinea pigs infected by intratracheal injection of virulent L. pneumophila develop an acute pneumonia resembling human Legionnaires' disease (13, 20, 75). Indeed, we observed that three out of four animals infected with 6 × 106 CFU of 130b succumbed to the infection within 4 days. In contrast, none of the guinea pigs infected with 7.8 × 107 CFU of NU216R had weight loss or respiratory distress, and all were alive 10 days postinoculation. Thus, the ira mutant NU216R appears to have a 50% lethal dose at least 100-fold greater than that of the wild-type strain.
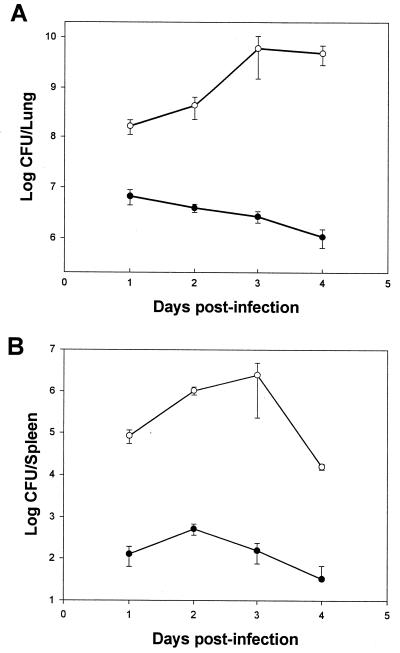
An alternative, quantitative assessment of virulence was obtained by inoculating guinea pigs with NU216R and 130b and then comparing the L. pneumophila concentrations in their lungs and the spleens on each day following infection. After the introduction of 106 CFU into the animals, the numbers of 130b increased over 100-fold in the lungs within 24 h (Fig. 1A). The numbers of wild-type bacteria in the lungs continued to increase on subsequent days, reaching a maximal value of 4.9 × 109 CFU on day 3 postinfection. In marked contrast, even when the animals were infected with a 50-fold-greater number of NU216R, this strain did not significantly increase in numbers within the lungs in the first 24 h (Fig. 1A). Furthermore, on subsequent days, the mutant bacteria steadily decreased in numbers. Thus, by day 3 postinfection, there were approximately 1,000-fold fewer CFU of NU216R than of 130b in the lungs of the infected animals. Taken together, these data indicate that this ira mutant is not able to replicate within a mammalian lung.
FIG. 1.
Replication of L. pneumophila strains within guinea pig lung and spleen. Doses of 1.0×106 CFU of 130b (open circles) or 5.2×107 CFU of NU216R (solid circles) were introduced intratracheally into guinea pigs, and the total number of bacterial CFU per lung (A) and spleen (B) were quantified in three or four animals at various times after inoculation. Each data point represents the mean ± standard error (n = 4 for days 1 and 2, and n = 3 for days 3 and 4). At every time point, there was a statistically significant difference between the numbers of wild-type and mutant bacteria (P < 0.05; Student's t test).
The wild-type bacterium disseminated to the spleen in significant numbers within the first day (Fig. 1B). Following this, the number of wild-type bacteria continued to increase for at least another day before dropping on subsequent days. This pattern of accumulation in the spleen has been observed previously (46). In contrast, 1,000-fold-less NU216R was observed in the spleen 1 day postinfection, and the number of bacteria increased only marginally on the following day and then decreased progressively. Interestingly, 1 and 2 days postinfection, the two strains showed a greater difference in numbers in the spleens compared with those in the lungs. By 10 days postinfection, the mutant bacteria were below the detectable limit (102 CFU) in both the lungs and the spleen, indicating clearance.
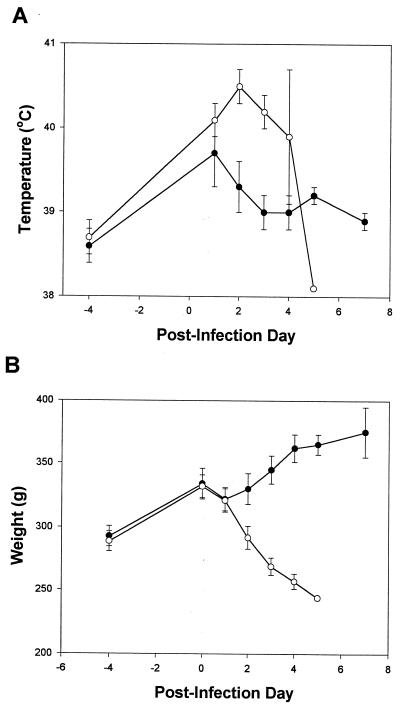
To further document the progression of the disease, the temperature and weight of the guinea pigs were monitored during the course of the clearance study. Animals in both experimental groups displayed an increase in body temperature 1 day postinfection (Fig. 2A). However, animals infected with the wild-type strain displayed higher body temperatures that continued to increase. The drastic drop in body temperature on day 5 was a reflection of their moribund state. In contrast, the temperature of NU216R-infected animals dropped at 2 days postinfection and resolved to baseline levels (Fig. 2A). As has been previously demonstrated (20, 46), animals infected with wild-type bacteria had an average weight loss of 20% by 4 days postinfection (Fig. 2B). NU216R-infected animals, on the other hand, gained weight by 12% in the same period. Taken together, the mutant's higher LD50, inability to accumulate in the lungs or spleen, and failure to mediate any signs of disease indicated that this ira locus encodes a virulence determinant(s).
FIG. 2.
Signs of L. pneumophila pulmonary infection. Rectal temperature (A) and body weight (B) of the 130b-infected (open circles) and NU216R-infected animals (solid circles) were monitored through the course of infection. Each point represents the mean ± standard error from three or more animals with the exception of one 130b-infected guinea pig on day 5. 130b-infected animals were moribund on day 5 postinfection and were sacrificed (temperature: P < 0.05 for days 2 to 4; weight: P < 0.05 for days 2 to 4 [Student's t test]).
Sequence analysis of the iraAB locus.
To characterize the genetic lesion responsible for the drastic attenuation in virulence of NU216R, we cloned and sequenced the ira locus from the L. pneumophila chromosome. To construct NU216R, we had earlier cloned an approximately 8-kb SalI fragment containing the Kmr marker and flanking DNA from NU216 (60). Using subclones of this plasmid, pCDP70, as well as pCDP71, which contains the intact ira, we sequenced this locus and mapped the transposon insertion in NU216 and NU216R. In all, 3,763 bp was sequenced from both strands (Fig. 3). This represents 815 bp upstream and 2,967 bp downstream of the transposon insertion. The base composition of this region (G+C content = 38%) was typical for L. pneumophila (47). Sequence analysis revealed that the transposon disrupted the first open reading frame (ORF) in what appears to be a two-gene operon. This orf, designated iraA, is preceded by sequences with strong homology to prokaryotic promoter sequences and an appropriately positioned Shine-Dalgarno sequence. iraA is predicted to encode a soluble 272-amino-acid protein. Although it did not show overall homology to proteins in the database, IraA contained the S-adenosylmethionine (SAM)-binding motif present in several prokaryotic and eukaryotic small-molecule methyltransferases. Recently, a consensus sequence, oL(D/E)oGsGsG-X16–21-(D/E)-X28–94-oDso (o = hydrophobic amino acids; s = small amino acid), was generated from numerous eukaryotic methyltransferases (37, 46). The IraA sequence includes the stretch ILDLGVGNG-X18-D-X87-QDTI that fits the above consensus (Fig. 3). Interestingly, a comparison of the prokaryotic methyltransferases in this family revealed a slightly different and more extended conservation of sequences surrounding the initial o(L/D)oGsGsG sequence (Fig. 4).
FIG. 4.
Partial alignment of the L. pneumophila IraA protein sequence with prokaryotic homologs. The sequences are derived from L. pneumophila (Lp) IraA, Acetobacter aceti (Aa) phosphatidylethanolamine N-methyltransferase (Pmt) (Hanada et al., unpublished), Thermotoga maritima (Tm) ubiquinone/menaquinone biosynthesis methyltransferase (UbiE) (53), Streptomyces griseus (Sg) carotenoid biosynthetic gene (CrtT) (68), Chlamydia pneumoniae (Cp) UbiE (S. Kalman, W. Mitchell, R. Marathe, C. Lammel, J. Fan, J. Olinger, J. Grimwood, R. W. Dain S, and R. S. Stephens, GenBank accession no. AEOO-1636 [unpublished results]), and Rhodobacter sphaeroides (Rs) Pmt (5). Alignments were achieved using CLUSTALW (72). In the consensus sequence, invariant residues are indicated in capitals, highly conserved residues are indicated in lowercase type, o indicates any hydrophobic amino acid, and s indicates a small amino acid. Regions corresponding to the two signature sequences unique to this class of proteins are indicated in bold type.
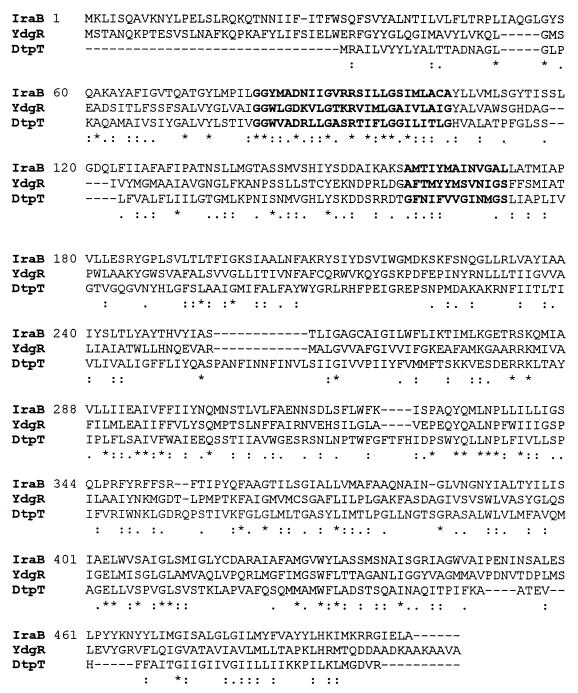
The ORF designated iraB starts with a GTG codon 35 bp downstream of the iraA termination codon (Fig. 3). The use of GTG initiation codons in L. pneumophila and elsewhere has been reported previously (36). IraB is highly similar to prokaryotic and eukaryotic members of the PTR2 family of peptide transporters (70). These integral membrane proteins are involved in the proton-dependent uptake of di- and tripeptides. IraB is most closely related to a putative 500-amino-acid protein, YdgR, from E. coli, also predicted to be a member of this family. The L. pneumophila and E. coli ORFs align along their entire length with an overall similarity of 51% and an identity of 29% (Fig. 5). The best-studied prokaryotic member of this class of proteins is the 463-amino-acid DtpT protein from Lactococcus lactis (Fig. 5) (34). Analysis using the SOSUI program predicts that IraB, like DtpT, is an integral membrane protein with 12 membrane-spanning regions (34; S. Mitaku, SOSUI, http://www.tuat.ac.jp/∼mitaku/adv_sosui/). Other members of this family which show homology to IraB include the Drosophila melanogaster oligopeptide transporter 1, the Lactobacillus helveticus di- and tripeptide transport protein, and the rat kidney oligopeptide transporter (4, 52, 67).
FIG. 5.
Complete alignment of L. pneumophila IraB with E. coli YdgR (GenBank accession no. 1742700) (2) and the Lactococcus lactis di- and tripeptide transporter, DtpT (GenBank accession no. 625863) (33). An asterisk indicates invariant residues, a colon indicates regions of high similarity, and a dot indicates regions of low similarity. Alignments were performed using Align Plus for Windows (Scientific and Educational Software, State Line, Pa.).
A large ORF (predicted to encode >450 amino acids) whose product has no significant similarity to proteins in the database is present 159 bp upstream and divergent from iraA (Fig. 3). While the function of this ORF is unknown, it is highly unlikely, based on its position and orientation relative to the transposon, to be related to the iron and/or infectivity defects in NU216R. There were no large ORFs immediately downstream of iraB (Fig. 3). Further downstream of iraB was a divergently transcribed ORF that was highly similar to the lipoic acid biogenesis gene lipA from Pelobacter carbinolicus, E. coli, and other organisms (57, 63). Lipoic acid is a coenzyme required for the activity of enzyme complexes such as pyruvate dehydrogenase (41). The 994-bp sequence between the ends of iraB and lipA contains an interesting feature, i.e., a 93-amino-acid-encoding ORF flanked by a 250-bp imperfect inverted repeat (IR) (Fig. 3). Two subrepeats of 82 bp are present within each IR, and an additional 82-bp subrepeat is present upstream and divergent from the first IR. The ORF itself is similar to sequences encoding various hypothetical “intergenic” proteins of approximately the same length (28, 44). The significance of these DNA sequences is unknown, although the IR could play a regulatory role. Thus, the sequence analyses indicated that this ira locus is bicistronic and that the impairments of NU216R were due to a loss of IraA and/or IraB.
Construction of an L. pneumophila iraB mutant.
To determine whether the iron and/or infectivity defects in NU216R were due to the loss of iraA and/or a polar effect on the expression of iraB, we attempted to complement NU216R with plasmids containing either iraA alone (pVK101), iraB alone (pVK102), iraA and iraB (pVK100), or vector alone (pSU2719). Curiously, the transformation efficiency was significantly lower with the ira-containing plasmids than with vector (data not shown). Additionally, in three separate attempts, it was observed that the strains transformed with the ira-containing plasmids took at least an extra day to appear on BCYE agar plates than did vector-transformed bacteria. Furthermore, it was observed that the iraAB-containing plasmids were not consistently maintained in these strains. This was unexpected, since in several instances we have been successful in complementing other Legionella mutants with the appropriate fragment of DNA cloned into various vectors (14, 37, 46). This suggested that overexpression of the genes from this locus was detrimental to Legionella survival. As an alternate approach to determining the relative contributions of iraA and iraB, we sought a mutant containing a nonpolar mutation in iraA. Thus, pVK107 was transformed into NU216R in order to replace the mini-Tn10-disrupted iraA with an in-frame deletion in iraA. Unfortunately, no transformants were obtained in three attempts. We finally decided to determine if a targeted disruption of iraB itself would result in the same phenotype(s) as NU216R. Using the counterselectable pVK105 for allelic exchange, we readily obtained strain NU244, which contained a Kmr cassette inserted in the middle of iraB (Fig. 3). Importantly, NU244 grew in BYE broth and on BCYE agar as well as did 130b, and so it was suitable for phenotypic analysis (see below). Incidentally, the cell morphology of this strain was similar to that of 130b. We therefore evaluated NU244 for EDDA sensitivity and infectivity to begin to determine the contribution of iraB to iron acquisition and virulence.
Sensitivity of L. pneumophila strains to iron limitation.
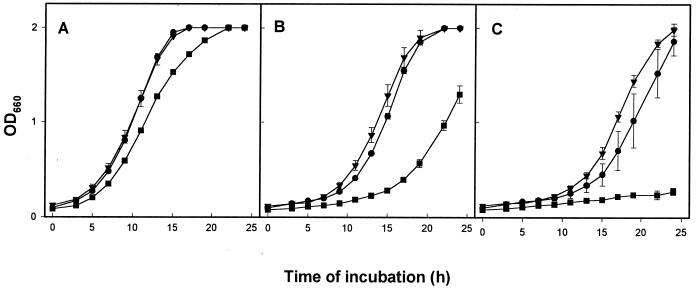
To determine if a mutation in iraB results in sensitivity to iron deprivation, strains 130b, NU216R, and NU244 were compared for their ability to grow in BYE broth containing different amounts of free iron. The three strains did not display significant differences for growth in standard BYE medium (Fig. 6A), confirming that the iraAB mutants do not have a generalized growth defect. Interestingly, however, NU244 was impaired for growth in BYE medium lacking iron supplementation, while 130b and NU216R grew as in the supplemented medium (Fig. 6B). The normal growth of 130b and NU216R in unsupplemented BYE medium is not surprising since it contains approximately 27 μM residual iron (31). The impaired growth of NU244 in unsupplemented BYE implies that it is more sensitive to iron starvation than is NU216R. To confirm this notion, we further depleted the BYE medium of iron by including 20 μM EDDA (Fig. 6C). As expected, NU244 was highly sensitive to iron depletion. To our surprise, however, NU216R had lost its hypersensitivity to EDDA since our last report (60) (Fig. 6C). In separate experiments, we observed that the original mutant, NU216, had also lost its EDDA hypersensitivity (data not shown). While this loss was observed for NU216 and NU216R on three different occasions, both strains continued to grow on karamycin and the persistence of their transposon mutations was confirmed by PCR analysis (data not shown). The impaired growth of NU244 in media lacking added iron, combined with its hypersensitivity to EDDA, indicated that iraB is involved in the assimilation of iron in extracellular conditions. Thus, we suspect that the original EDDA hypersensitivity of NU216 and NU216R was partly, if not completely, a result of a polar effect on iraB expression.
FIG. 6.
Growth of L. pneumophila strains in BYE broth containing different amounts of free iron. Strains 130b (circles), NU216R (triangles), and NU244 (squares) were inoculated from 2-day-old BCYE agar plates at equivalent OD660 to BYE broth with the standard 335 μM ferric pyrophosphate supplementation (A), BYE without iron supplementation (B), and BYE with 20 μM EDDA in the absence of iron supplementation (C). OD660 was monitored for 24 h. Each point represents the mean ± standard error from three different cultures. In panels B and C, NU244 growth was significantly different from 130b growth after 12 and 15 h of incubation, respectively (P < 0.05; Student's t test). These data are representative of two different experiments. At 30 μM EDDA, all three strains were impaired for growth (data not shown).
Intracellular infection of U937 cells by L. pneumophila strains.
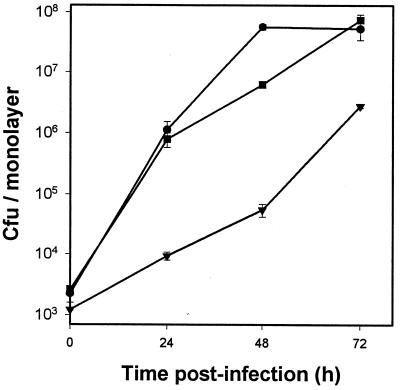
To determine whether iraB also promotes intracellular infection, we evaluated the relative ability of NU244 to infect U937 cells. Given its change in EDDA sensitivity, we were also eager to retest the capacity of NU216R to infect the macrophage-like cells. In three separate experiments (one of which is shown in Fig. 7), NU244 growth in U937 cells was comparable to that of 130b. In contrast, NU216R was as highly impaired for intracellular replication (Fig. 7), as had been reported previously (60). In other experiments, it was also observed that the original mutant, NU216, continued to be impaired for infection in U937 cells (data not shown). Thus, it appears that lesions in the linked ira genes result in distinct infectivity phenotypes; i.e., while disruption of iraA results in stable impairment of replication in macrophages, disruption of iraB does not. Nevertheless, we felt compelled to evaluate if NU244, given its growth impairment in low-iron media, would be impaired in iron-stressed macrophages. Consequently, infections were performed as above, but with U937 cells continuously maintained in the presence of various concentrations of DFX (10, 29, 60). In an initial experiment, we observed that the intracellular growth of NU244, like that of 130b, was not significantly inhibited in the presence of 10 μM DFX (Table 2). We subsequently repeated the experiment, assessing the effect of a higher concentration of DFX (15 μM) on these strains (Table 2). Again, NU244 recovery was not significantly different from that of 130b, although both strains begin to show signs of inhibition at 15 μM DFX. In a third experiment, it was observed that NU244 and the wild type behaved identically to each other whether cultured at 24, 48, or 72 h postinfection (data not shown). In the course of assessing NU244, we reexamined the DFX sensitivity of NU216R. Consistent with our earlier observation (60), NU216R was hypersensitive to intracellular iron depletion (Table 2). Thus, the growth of NU216R was inhibited by ∼73% in the presence of 10 μM and ∼88% in the presence of 15 μM DFX (P < 0.05 by Student's t test). Two conclusions can be drawn from these experiments. First, NU244, despite being very sensitive to growth in low-iron media, does not demonstrate hypersensitivity to intracellular iron depletion, indicating that iraB is critical only for extracellular iron acquisition. Second, NU216R, despite losing its EDDA inhibition phenotype in broth cultures, continued to show chelator hypersensitivity in macrophages, indicating that iraA is critical only for intracellular iron acquisition. Since NU216R was routinely maintained and passaged on laboratory media, a reversion of the sensitivity to iron-limited media without a concomitant reversion of the DFX sensitivity in macrophages is not unreasonable.
FIG. 7.
Growth of L. pneumophila strains within U937 cells. Macrophage monolayers were infected in triplicate with 130b (circles), NU216R (triangles), and NU244 (squares) from 3-day-old BCYE agar plates. Bacterial CFU per monolayer were quantified at 0, 24, 48, and 72 h after inoculation. Each data point represents the mean ± standard error for three independent monolayers. The 48-h time point for NU244 was identical to that for 130b in two independent experiments and as such does not represent a significant difference from 130b. The difference at t = 0 between 130b and NU216R is consistent with the observation that the wild-type inoculum contained proportionally more bacteria. Significant differences in recovery were observed between 130b- and NU216R-infected macrophages at all times except t = 0 h (P < 0.05; Student's t test).
TABLE 2.
Growth of L. pneumophila strains within DFX-treated U937 cells
| Expt | Strain | 105 CFU/monolayer recovered 48 h postinfectiona (mean ± SE)
|
||
|---|---|---|---|---|
| No DFX | 10 μM DFX | 15 μM DFX | ||
| I | 130b | 937 ± 55 | 813 ± 45 (P = 0.16) | ND |
| NU244 | 907 ± 106 | 753 ± 74 (P = 0.30) | ND | |
| II | 130b | 537 ± 77 | 420 ± 12 (P = 0.21) | 211 ± 0.94 (P = 0.06) |
| NU216R | 6.3 ± 0.85 | 1.8 ± 0.50 (P = 0.01) | 0.75 ± 0.26 (P = 0.003) | |
| NU244 | 583 ± 74 | 436 ± 23 (P = 0.13) | 230 ± 67 (P = 0.03) | |
U937 monolayers were incubated for 24 h in the presence of various concentrations of DFX. These cells were infected with equivalent numbers of the various L. pneumophila strains. As before (60) DFX treatment did not reduce the uptake of the legionellae (data not shown). The bacterial titers from three replicate wells were determined at 48 h. The P values (Student's t test) for the difference in recovery compared to untreated wells are indicated in parentheses.
Distribution of iraA and iraB in various Legionella species and L. pneumophila serogroups.
The L. pneumophila species has 14 serogroups, all of which have been associated with disease, while the Legionella genus currently includes 43 species, about half of which have been associated with human disease (48, 55). Given the drastic impairment of NU216R in guinea pigs, we proceeded to determine if there was a correlation between the distribution of iraAB among the various L. pneumophila serogroups and Legionella species and the disease association of the organism. To do this, the presence of iraA and iraB in various Legionella species and L. pneumophila serogroups was determined by Southern hybridizations using iraA- and iraB-specific probes (Fig. 3, Table 1). Hybridization analysis under conditions of high (10% mismatch) stringency revealed that these two ORFs were present in all 11 of the L. pneumophila serogroups tested. Of the 14 other species examined, hybridization was observed under low (30% mismatch)-stringency conditions only with DNA from L. gormanii. Thus, iraAB appears to be largely limited to L. pneumophila, the species that is mostly associated with disease.
DISCUSSION
In our earlier analysis (60), we had established that the ira mutant NU216 was defective for growth in macrophage-like U937 cells and that the infectivity defect was indeed linked to the transposon insertion. Here, we demonstrate that this mutant is also severely impaired in a guinea pig model of Legionnaires' disease. This is consistent with earlier reports showing that specific mutants defective for macrophage infection are less virulent (13, 46). Thus, while 130b established an infection in the animals, the mutant failed to elicit any signs of infection even when inoculated at 50-fold-higher levels than the wild-type strain. Consistent with this, a 1,000-fold-greater number of wild-type bacteria than of the mutant could be recovered from the animals, despite the difference in the initial inoculum. Interestingly, NU216R was more drastically impaired in the guinea pig lungs than in U937 cells, suggesting an additional role(s) for this locus in the extracellular context. Additionally, the greater difference on day 1 postinoculation between the mutant and the wild type in the spleen than in the lungs may suggest that the mutant is also defective for dissemination. The drastic impairment of NU216R following intratracheal inoculation establishes iraAB as a significant contributor to virulence, and thus one of very few L. pneumophila loci whose importance has been confirmed and extended by animal models (13, 21, 46, 49, 51).
Based on the following observations, we propose that iraAB is a two-gene operon. First, the start codon for iraB is only 35 bp downstream of the iraA termination codon. Second, there are no other ORFs in the same direction adjacent to iraA and iraB. Third, a consensus promoter and Shine-Dalgarno sequence are present upstream of iraA. The absence of an appropriately positioned Shine-Dalgarno sequence upstream of iraB may be indicative of translational coupling between iraA and iraB. Fourth, preliminary experiments suggest that the two genes are coordinately regulated. Specifically, RNA dot blot analyses revealed that both iraA and iraB transcripts are detectable in wild-type L. pneumophila isolated form U937 cells while neither transcript could be detected in broth-grown cultures (V. K. Viswanathan, P. Edelstein, C. D. Pope, and N. P. Cianciotto, Abstr. 98th Gen. Meet. Am. Soc. Microbiol. 1998, abstr. 210, p. 90, 1998). Southern hybridization analyses suggested that the iraAB genes are present in all L. pneumophila serogroups tested, as well as L. gormanii. L. gormanii is a relatively rare isolate that has been associated with disease (54). This distribution is atypical, since most other genes examined (e.g., mip, fur, and pilD) are present in all legionellae tested (36, 37, 47). Of the very few exceptions, frgA appears to be present only in L. pneumophila serogroups (37). The presence of iraAB in only two species of Legionella is reminiscent of the distribution of the hbp locus, which was found only in three Legionella species (56). It is likely that the iraAB-flanking genes lipA (Fig. 3) and hisF (located approximately 1.5 kb upstream of iraA [unpublished results]) are more extensively conserved among the various Legionella species.
Phenotypic analyses of NU216R and NU244 reveal a complex role for iraAB in L. pneumophila growth and pathogenesis. NU216R, but not NU244, is impaired for replication in macrophages, implying that IraA, but not IraB, is required for intracellular growth. The stable hypersensitivity of NU216R, but not NU244, to intracellular iron limitation indicates that the role of IraA in virulence is related to iron acquisition within macrophages. However, sequence analysis revealed that IraA is unlike any known promoter of intracellular infection or iron assimilation. Rather, IraA contains the consensus sequence shared between prokaryotic and eukaryotic small-molecule methyltransferases (35, 42). This consensus sequence is the only region of similarity among the various enzymes that use SAM as a donor to methylate a wide variety of acceptors. Indeed, mutation of the conserved residues in this region, which is thought to be the SAM-binding site, eliminates enzyme activity (35). As a group, the small-molecule methyltransferases catalyze reactions leading to the biosynthesis of compounds such as phosphatidylcholine (PC), ubiquinone, and carotenoids (5, 52, 69). Given that these compounds are involved in functions as diverse as osmoprotection, aerobic respiration, and pigmentation, respectively, it is not unreasonable to suspect that IraA could be involved in iron acquisition. The substrate for the putative IraA enzyme obviously remains to be determined. However, BLAST analyses revealed that the strongest overall match between IraA and the database is with the Acetobacter aceti phosphatidylethanolamine methyltransferase, an enzyme involved in the conversion of phosphatidylethanolamine to PC (T. Hanada, Y. Koizumi, S. Udaka, and F. Yanagida, GenBank accession no. BAA34057 [unpublished results]). Interestingly, while very few bacteria produce PC, 10 different isolates of L. pneumophila were demonstrated to have unusually high PC content, and PC was the most abundant phospholipid in almost all of them (24). Taken together, our results suggest a novel role for IraA in iron acquisition and virulence. It remains to be established if these two phenotypes are directly linked or if IraA has a function that indirectly affects both iron acquisition and virulence. Although the importance of IraA for intracellular iron acquisition is clear, it is not obvious if IraA also plays a role in extracellular iron uptake. While both broth-grown NU216 and NU216R were initially hypersensitive to EDDA, they subsequently lost this phenotype without the concomitant loss of either their transposons or infectivity defects. An unrelated second-site mutation, possibly one that enhanced iron uptake only in broth cultures, may have resulted in a partial complementation of the iraA phenotype. Alternatively, the EDDA hypersensitivity of NU216 and NU216R may not be directly due to a loss of IraA but, rather, may have resulted from a polar effect on iraB expression (see below). Loss of polarity, possibly by enhanced expression of iraB from a cryptic promoter, could explain the loss of the EDDA hypersensitivity.
The hypersensitivity of NU244 to iron starvation indicates that IraB is required for L. pneumophila extracellular iron uptake. However, IraB, like IraA, appears unlike any other iron uptake system. Several lines of evidence suggest that IraB is a homolog of the PTR2 family of peptide transporters. First, IraB shows strong similarity to di- and tripeptide transporters from various prokaryotic and eukaryotic organisms. Second, the alignment with these proteins is along the entire length of the sequence. Third, the IraB sequence AMTIYMAINVGAL (Fig. 3) is very similar to the PTR family signature sequence F-(S/T/N/M)-X-F-(V/Y/F)-h-X-I-N-h-G-(S/A)-h (where h indicates a hydrophobic residue). A second region of consensus unique to this family of proteins is also present in IraB (GGYMADNIIGVRRSILLGSIMLACA) (Fig. 3) (34). Fourth, like the PTR2 homolog DtpT from L. lactis, IraB is predicted to have 12 membrane-spanning regions (32, 34). In most microorganisms, this family represents but one of several systems of peptide transport (58). DtpT has been demonstrated to transport di- and tripeptides via a proton motive force-dependent mechanism. While it primarily transports hydrophilic and neutral peptides such as Ala-Glu, Ala-Ala, Leu-Leu, Gly-Asp, Pro-Gly, Glu-Glu, and Asp-Glu (26, 33), this protein can also mediate the uptake of peptides with two negative charges, such as Glu-Glu and Asp-Glu. It has been suggested that DtpT is required for Lactococcus to use caseins as the sole source of nitrogen (26). The role of di- and tripeptide transporters in other prokaryotes is not known, but we hypothesize that L. pneumophila utilizes IraB to import iron-loaded peptides as a source of iron. Although peptides can be components of siderophores, there have been as yet no reports of free peptides involved in iron transport in microorganisms (18). However, iron acquisition through low-molecular-weight chelates such as glycyl-l-histidyl-l-lysine has been observed in human macrophages, and cysteine-containing N-terminal peptides enhance iron uptake in Caco-2 intestinal cells (30). This study, however, does not rule out an indirect relationship between peptide transport and iron uptake.
In summary, the juxtaposition of the genes for a putative methyltransferase and a peptide transporter represents a unique situation. This is the first report of a possible link between peptide transport, a small-molecule methyltransferase, and iron acquisition. Therefore, the iraAB locus may encode a novel mechanism of iron transport that is relevant in different environments, including the mammalian lung.
ACKNOWLEDGMENTS
We thank Martha Edelstein and Jianjun Ren for their contributions to this study. We also thank Mark Liles, Virginia Aragon, Sherry Kurtz, and Tracy Aber for useful discussions. Plasmid pMB2190 was kindly provided by Brian Nichols.
This work was supported by NIH grant AI34937 awarded to N.P.C.
REFERENCES
- 1.Abu Kwaik Y. Fatal attraction of mammalian cells to Legionella pneumophila. Mol Microbiol. 1998;30:689–695. doi: 10.1046/j.1365-2958.1998.01092.x. [DOI] [PubMed] [Google Scholar]
- 2.Aiba H, Baba T, Hayashi K, Inada T, Isono K, Itoh T, Kasai H, Kashimoto K, Kimura S, Kitakawa M, Kitagawa M, Makino K, Miki T, Mizobuchi K, Mori H, Mori T, Motomera K, Nakade S, Nakamura Y, Nashimoto H, Nishio Y, Oshima T, Saito N, Sampei G, Horiuchi T. A 570-kb DNA sequence of the Escherichia coli K-12 genome corresponding to the 28.0-40.1 min region on the linkage map (supplement) DNA Res. 1996;3:435–440. doi: 10.1093/dnares/3.6.435. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amrein H, Axel R. Genes expressed in neurons of adult male Drosophila. Cell. 1997;88:459–469. doi: 10.1016/s0092-8674(00)81886-3. [DOI] [PubMed] [Google Scholar]
- 4a.Aragon, V., S. Kurtz, A. Fleiger, B. Neumeister, and N. P. Cianciotto. Secreted enzymatic activities of wild type and pilD-deficient Legionella pneumophila. Infect. Immun., in press. [DOI] [PMC free article] [PubMed]
- 5.Arondel V, Benning C, Somerville C R. Isolation and functional expression in Escherichia coli of a gene encoding phosphatidylethanolamine methyltransferase (EC 2.1.1.17) from Rhodobacter sphaeroides. J Biol Chem. 1993;268:16002–16008. [PubMed] [Google Scholar]
- 6.Bandyopadhyay P, Steinman H M. Legionella pneumophila catalase-peroxidases: cloning of the katB gene and studies of KatB function. J Bacteriol. 1998;180:5369–5374. doi: 10.1128/jb.180.20.5369-5374.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bortner C A, Arnold R R, Miller R D. Bactericidal effect of lactoferrin on Legionella pneumophila: effect of the physiological state of the organism. Can J Microbiol. 1989;35:1048–1051. doi: 10.1139/m89-174. [DOI] [PubMed] [Google Scholar]
- 8.Byrd T F, Horwitz M A. Chloroquine inhibits the intracellular multiplication of Legionella pneumophila by limiting the availability of iron. A potential new mechanism for the therapeutic effect of chloroquine against intracellular pathogens. J Clin Investig. 1991;88:351–357. doi: 10.1172/JCI115301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byrd T F, Horwitz M A. Interferon gamma-activated human monocytes downregulate transferrin receptors and inhibit the intracellular multiplication of Legionella pneumophila by limiting the availability of iron. J Clin Investig. 1989;83:1457–1465. doi: 10.1172/JCI114038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byrd T F, Horwitz M A. Lactoferrin inhibits or promotes Legionella pneumophila intracellular multiplication in nonactivated and interferon gamma-activated human monocytes depending upon its degree of iron saturation. Iron-lactoferrin and nonphysiologic iron chelates reverse monocyte activation against Legionella pneumophila. J Clin Investig. 1991;88:1103–1112. doi: 10.1172/JCI115409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byrne B, Swanson M S. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect Immun. 1998;66:3029–3034. doi: 10.1128/iai.66.7.3029-3034.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cianciotto N, Eisenstein B I, Engleberg N C, Shuman H. Genetics and molecular pathogenesis of Legionella pneumophila, an intracellular parasite of macrophages. Mol Biol Med. 1989;6:409–424. [PubMed] [Google Scholar]
- 13.Cianciotto N P, Eisenstein B I, Mody C H, Engleberg N C. A mutation in the mip gene results in an attenuation of Legionella pneumophila virulence. J Infect Dis. 1990;162:121–126. doi: 10.1093/infdis/162.1.121. [DOI] [PubMed] [Google Scholar]
- 14.Cianciotto N P, Fields B S. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc Natl Acad Sci USA. 1992;89:5188–5191. doi: 10.1073/pnas.89.11.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clemens D L, Horwitz M A. Characterization of the Mycobacterium tuberculosis phagosome and evidence that phagosomal maturation is inhibited. J Exp Med. 1995;181:257–270. doi: 10.1084/jem.181.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clemens D L, Horwitz M A. Hypoexpression of major histocompatibility complex molecules on Legionella pneumophila phagosomes and phagolysosomes. Infect Immun. 1993;61:2803–2812. doi: 10.1128/iai.61.7.2803-2812.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dowling J N, Saha A K, Glew R H. Virulence factors of the family Legionellaceae. Microbiol Rev. 1992;56:32–60. doi: 10.1128/mr.56.1.32-60.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drechsel H, Jung G. Peptide siderophores. J Pept Sci. 1998;4:147–181. doi: 10.1002/(SICI)1099-1387(199805)4:3%3C147::AID-PSC136%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 19.Edelstein P H. Improved semiselective medium for isolation of Legionella pneumophila from contaminated clinical and environmental specimens. J Clin Microbiol. 1981;14:298–303. doi: 10.1128/jcm.14.3.298-303.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edelstein P H, Calarco K, Yasui V K. Antimicrobial therapy of experimentally induced Legionnaires' disease in guinea pigs. Am Rev Respir Dis. 1984;130:849–856. doi: 10.1164/arrd.1984.130.5.849. [DOI] [PubMed] [Google Scholar]
- 21.Edelstein P H, Edelstein M A, Higa F, Falkow S. Discovery of virulence genes of Legionella pneumophila by using signature tagged mutagenesis in a guinea pig pneumonia model. Proc Natl Acad Sci USA. 1999;96:8190–8195. doi: 10.1073/pnas.96.14.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feeley J C, Gorman G W, Weaver R E, Mackel D C, Smith H W. Primary isolation media for Legionnaires' disease bacterium. J Clin Microbiol. 1978;8:320–325. doi: 10.1128/jcm.8.3.320-325.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fields B S. The molecular ecology of legionellae. Trends Microbiol. 1996;4:286–290. doi: 10.1016/0966-842x(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 24.Finnerty W R, Makula R A, Feeley J C. Cellular lipids of the Legionnaires' disease bacterium. Ann Intern Med. 1979;90:631–634. doi: 10.7326/0003-4819-90-4-631. [DOI] [PubMed] [Google Scholar]
- 25.Fischer G, Bang H, Ludwig B, Mann K, Hacker J. Mip protein of Legionella pneumophila exhibits peptidyl-prolyl-cis/trans isomerase (PPIase) activity. Mol Microbiol. 1992;6:1375–1383. doi: 10.1111/j.1365-2958.1992.tb00858.x. [DOI] [PubMed] [Google Scholar]
- 26.Foucaud C, Kunji E R, Hagting A, Richard J, Konings W N, Desmazeaud M, Poolman B. Specificity of peptide transport systems in Lactococcus lactis: evidence for a third system which transports hydrophobic di- and tripeptides. J Bacteriol. 1995;177:4652–4657. doi: 10.1128/jb.177.16.4652-4657.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gabay J E, Blake M, Niles W D, Horwitz M A. Purification of Legionella pneumophila major outer membrane protein and demonstration that it is a porin. J Bacteriol. 1985;162:85–91. doi: 10.1128/jb.162.1.85-91.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garmyn D, Ferain T, Bernard N, Hols P, Delplace B, Delcour J. Pediococcus acidilactici IdhD gene: cloning, nucleotide sequence, and transcriptional analysis. J Bacteriol. 1995;177:3427–3437. doi: 10.1128/jb.177.12.3427-3437.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gebran S J, Newton C, Yamamoto Y, Widen R, Klein T W, Friedman H. Macrophage permissiveness for Legionella pneumophila growth modulated by iron. Infect Immun. 1994;62:564–568. doi: 10.1128/iai.62.2.564-568.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glahn R P, Van Campen D R. Iron uptake is enhanced in Caco-2 cell monolayers by cysteine and reduced cysteinyl glycine. J Nutr. 1997;127:642–647. doi: 10.1093/jn/127.4.642. [DOI] [PubMed] [Google Scholar]
- 31.Goldoni P, Visca P, Pastoris M C, Valenti P, Orsi N. Growth of Legionella spp. under conditions of iron restriction. J Med Microbiol. 1991;34:113–118. doi: 10.1099/00222615-34-2-113. [DOI] [PubMed] [Google Scholar]
- 32.Hagting A, Knol J, Hasemeier B, Streutker M R, Fang G, Poolman B, Konings W N. Amplified expression, purification and functional reconstitution of the dipeptide and tripeptide transport protein of Lactococcus lactis. Eur J Biochem. 1997;247:581–587. doi: 10.1111/j.1432-1033.1997.00581.x. [DOI] [PubMed] [Google Scholar]
- 33.Hagting A, Kunji E R, Leenhouts K J, Poolman B, Konings W N. The di- and tripeptide transport protein of Lactococcus lactis. A new type of bacterial peptide transporter. J Biol Chem. 1994;269:11391–11399. [PubMed] [Google Scholar]
- 34.Hagting A, van der Velde J, Poolman B, Konings W N. Membrane topology of the di- and tripeptide transport protein of Lactococcus lactis. Biochemistry. 1997;36:6777–6785. doi: 10.1021/bi963068t. [DOI] [PubMed] [Google Scholar]
- 35.Hamahata A, Takata Y, Gomi T, Fujioka M. Probing the S-adenosylmethionine-binding site of rat guanidinoacetate methyltransferase. Effect of site-directed mutagenesis of residues that are conserved across mammalian non-nucleic acid methyltransferases. Biochem J. 1996;317:141–145. doi: 10.1042/bj3170141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hickey E K, Cianciotto N P. Cloning and sequencing of the Legionella pneumophila fur gene. Gene. 1994;143:117–121. doi: 10.1016/0378-1119(94)90615-7. [DOI] [PubMed] [Google Scholar]
- 37.Hickey E K, Cianciotto N P. An iron- and Fur-repressed Legionella pneumophila gene that promotes intracellular infection and encodes a protein with similarity to the Escherichia coli aerobactin synthetases. Infect Immun. 1997;65:133–143. doi: 10.1128/iai.65.1.133-143.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horwitz M A. Phagocytosis of the Legionnaires' disease bacterium (Legionella pneumophila) occurs by a novel mechanism: engulfment within a pseudopod coil. Cell. 1984;36:27–33. doi: 10.1016/0092-8674(84)90070-9. [DOI] [PubMed] [Google Scholar]
- 39.James B W, Mauchline W S, Dennis P J, Keevil C W. A study of iron acquisition mechanisms of Legionella pneumophila grown in chemostat culture. Curr Microbiol. 1997;34:238–243. doi: 10.1007/s002849900176. [DOI] [PubMed] [Google Scholar]
- 40.Johnson W, Varner L, Poch M. Acquisition of iron by Legionella pneumophila: role of iron reductase. Infect Immun. 1991;59:2376–2381. doi: 10.1128/iai.59.7.2376-2381.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jordan S W, Cronan J E., Jr Biosynthesis of lipoic acid and posttranslational modification with lipoic acid in Escherichia coli. Methods Enzymol. 1997;279:176–183. doi: 10.1016/s0076-6879(97)79021-9. [DOI] [PubMed] [Google Scholar]
- 42.Koonin E V. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and lambda 2 protein of reovirus. J Gen Virol. 1993;74:733–740. doi: 10.1099/0022-1317-74-4-733. [DOI] [PubMed] [Google Scholar]
- 43.Krinos C, High A S, Rodgers F G. Role of the 25 kDa major outer membrane protein of Legionella pneumophila in attachment to U-937 cells and its potential as a virulence factor for chick embryos. J Appl Microbiol. 1999;86:237–244. doi: 10.1046/j.1365-2672.1999.00667.x. [DOI] [PubMed] [Google Scholar]
- 44.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 45.Liles M R, Cianciotto N P. Absence of siderophore-like activity in Legionella pneumophila supernatants. Infect Immun. 1996;64:1873–1875. doi: 10.1128/iai.64.5.1873-1875.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liles M R, Edelstein P H, Cianciotto N P. The prepilin peptidase is required for protein secretion by and the virulence of the intracellular pathogen Legionella pneumophila. Mol Microbiol. 1999;31:959–970. doi: 10.1046/j.1365-2958.1999.01239.x. [DOI] [PubMed] [Google Scholar]
- 46a.Liles M R, Scheel T A, Cianciotto N P. Discovery of a neoclassical siderophore, legiobactin, produced by strains of Legionella pneumophila. J Bacteriol. 2000;182:749–757. doi: 10.1128/jb.182.3.749-757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liles M R, Viswanathan V K, Cianciotto N P. Identification and temperature regulation of Legionella pneumophila genes involved in type IV pilus biogenesis and type II protein secretion. Infect Immun. 1998;66:1776–1782. doi: 10.1128/iai.66.4.1776-1782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lo Presti F, Riffard S, Meugnier H, Reyrolle M, Lasne Y, Grimont P A, Grimont F, Vandenesch F, Etienne J, Fleurette J, Freney J. Legionella taurinensis sp. nov., a new species antigenically similar to Legionella spiritensis. Int J Syst Bacteriol. 1999;49:397–403. doi: 10.1099/00207713-49-2-397. [DOI] [PubMed] [Google Scholar]
- 49.Marra A, Blander S J, Horwitz M A, Shuman H A. Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc Natl Acad Sci USA. 1992;89:9607–9611. doi: 10.1073/pnas.89.20.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinez E, Bartolome B, de la Cruz F. pACYC184-derived cloning vectors containing the multiple cloning site and lacZ alpha reporter gene of pUC8/9 and pUC18/19 plasmids. Gene. 1988;68:159–162. doi: 10.1016/0378-1119(88)90608-7. [DOI] [PubMed] [Google Scholar]
- 51.Moffat J F, Edelstein P H, Regula D P, Jr, Cirillo J D, Tompkins L S. Effects of an isogenic Zn-metalloprotease-deficient mutant of Legionella pneumophila in a guinea-pig pneumonia model. Mol Microbiol. 1994;12:693–705. doi: 10.1111/j.1365-2958.1994.tb01057.x. [DOI] [PubMed] [Google Scholar]
- 52.Nakajima H, Hagting A, Kunji E R, Poolman B, Konings W N. Cloning and functional expression in Escherichia coli of the gene encoding the di- and tripeptide transport protein of Lactobacillus helveticus. Appl Environ Microbiol. 1997;63:2213–2217. doi: 10.1128/aem.63.6.2213-2217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nelson K E, Clayton R A, Gill S R, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Nelson W C, Ketchum K A, McDonald L, Utterback T R, Malek J A, Linher K D, Garrett M M, Stewart A M, Cotton M D, Pratt M S, Phillips C A, Richardson D, Heidelberg J, Sutton G G, Fleischmann R D, White O, Salzberg S L, Smith H O, Venter C J, Fraser C M. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 54.O'Connell W A, Bangsborg J M, Cianciotto N P. Characterization of a Legionella micdadei mip mutant. Infect Immun. 1995;63:2840–2845. doi: 10.1128/iai.63.8.2840-2845.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Connell W A, Dhand L, Cianciotto N P. Infection of macrophage-like cells by Legionella species that have not been associated with disease. Infect Immun. 1996;64:4381–4384. doi: 10.1128/iai.64.10.4381-4384.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O'Connell W A, Hickey E K, Cianciotto N P. A Legionella pneumophila gene that promotes hemin binding. Infect Immun. 1996;64:842–848. doi: 10.1128/iai.64.3.842-848.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oppermann F B, Steinbuchel A. Identification and molecular characterization of the aco genes encoding the Pelobacter carbinolicus acetoin dehydrogenase enzyme system. J Bacteriol. 1994;176:469–485. doi: 10.1128/jb.176.2.469-485.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Payne J W, Smith M W. Peptide transport by micro-organisms. Adv Microb Physiol. 1994;36:1–80. doi: 10.1016/s0065-2911(08)60176-9. [DOI] [PubMed] [Google Scholar]
- 59.Poch M T, Johnson W. Ferric reductases of Legionella pneumophila. Biometals. 1993;6:107–114. doi: 10.1007/BF00140111. [DOI] [PubMed] [Google Scholar]
- 60.Pope C D, O'Connell W A, Cianciotto N P. Legionella pneumophila mutants that are defective for iron acquisition and assimilation and intracellular infection. Infect Immun. 1996;64:629–636. doi: 10.1128/iai.64.2.629-636.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pruckler J M, Benson R F, Moyenuddin M, Martin W T, Fields B S. Association of flagellum expression and intracellular growth of Legionella pneumophila. Infect Immun. 1995;63:4928–4932. doi: 10.1128/iai.63.12.4928-4932.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quinn F D, Weisberg E D. Killing of Legionella pneumophila by human serum and iron-binding agents. Curr Microbiol. 1988;17:111–116. [Google Scholar]
- 63.Reed K E, Cronan J E., Jr Lipoic acid metabolism in Escherichia coli: sequencing and functional characterization of the lipA and lipB genes. J Bacteriol. 1993;175:1325–1336. doi: 10.1128/jb.175.5.1325-1336.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reeves M W, Pine L, Hutner S H, George J R, Harrell W K. Metal requirements of Legionella pneumophila. J Clin Microbiol. 1981;13:688–695. doi: 10.1128/jcm.13.4.688-695.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reeves M W, Pine L, Neilands J B, Balows A. Absence of siderophore activity in Legionella species grown in iron-deficient media. J Bacteriol. 1983;154:324–329. doi: 10.1128/jb.154.1.324-329.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roy C R, Berger K H, Isberg R R. Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol Microbiol. 1998;28:663–674. doi: 10.1046/j.1365-2958.1998.00841.x. [DOI] [PubMed] [Google Scholar]
- 67.Saito H, Terada T, Okuda M, Sasaki S, Inui K. Molecular cloning and tissue distribution of rat peptide transporter PEPT2. Biochim Biophys Acta. 1996;1280:173–177. doi: 10.1016/0005-2736(96)00024-7. [DOI] [PubMed] [Google Scholar]
- 68.Schumann G, Nurnberger H, Sandmann G, Krugel H. Activation and analysis of cryptic crt genes for carotenoid biosynthesis from Streptomyces griseus. Mol Gen Genet. 1996;252:658–666. doi: 10.1007/BF02173971. [DOI] [PubMed] [Google Scholar]
- 69.Segal G, Shuman H A. How is the intracellular fate of the Legionella pneumophila phagosome determined? Trends Microbiol. 1998;6:253–255. doi: 10.1016/s0966-842x(98)01308-0. [DOI] [PubMed] [Google Scholar]
- 70.Steiner H Y, Naider F, Becker J M. The PTR family: a new group of peptide transporters. Mol Microbiol. 1995;16:825–834. doi: 10.1111/j.1365-2958.1995.tb02310.x. [DOI] [PubMed] [Google Scholar]
- 71.Stone B J, Abu Kwaik Y. Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infect Immun. 1998;66:1768–1775. doi: 10.1128/iai.66.4.1768-1775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thompson, J. D., D. G. Higgins, and T. J. Gibson. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. [DOI] [PMC free article] [PubMed]
- 73.Vogel J P, Isberg R R. Cell biology of Legionella pneumophila. Curr Opin Microbiol. 1999;2:30–34. doi: 10.1016/s1369-5274(99)80005-8. [DOI] [PubMed] [Google Scholar]
- 74.White J R, Yeowell H N. Iron enhances the bactericidal action of streptonigrin. Biochem Biophys Res Commun. 1982;106:407–411. doi: 10.1016/0006-291x(82)91125-1. [DOI] [PubMed] [Google Scholar]
- 75.Winn W C, Jr, Davis G S, Gump D W, Craighead J E, Beaty H N. Legionnaires' pneumonia after intratracheal inoculation of guinea pigs and rats. Lab Investig. 1982;47:568–578. [PubMed] [Google Scholar]