Abstract
The gut microbiome can affect various aspects of both behavior and physiology, including exercise ability, but effects on voluntary exercise have rarely been studied. We studied females from a selection experiment in which 4 replicate High Runner (HR) lines of mice are bred for voluntary exercise and compared with 4 non-selected control (C) lines. HR and C mice differ in several traits that likely interact with the gut microbiome, including higher daily running distance, body temperatures when running, spontaneous physical activity when housed without wheels, and food consumption. After two weeks of wheel access to reach a stable plateau in daily running, mice were administered broad-spectrum antibiotics for 10 days. Antibiotic treatment caused a significant reduction in daily wheel-running distance in the HR mice (−21%) but not in the C mice. Antibiotics did not affect body mass or food consumption in either HR or C mice, and we did not observe sickness behavior. Wheel running by HR mice did not recover during the 12 days following cessation of antibiotics. The decreased wheel-running in HR but not C mice, with no apparent negative side effects of antibiotics, suggests that the HR microbiome is an important component of their high-running phenotype.
Keywords: Antibiotics, Behavior, Exercise, Gut microbiota, Host-microbe interaction, Wheel running
1. Introduction
Mammals have evolved in a world dominated by bacteria, and their gut microbiome has become an essential component of the host’s body (McFall-Ngai et al., 2013; Milani et al., 2020). The gut microbiome is crucial for numerous aspects of host biology, including digestion, metabolism, and immune function (Dominguez-Bello et al., 2019; Gilbert et al., 2018; Kohl and Carey, 2016). Considerable evidence indicates that mammalian hosts and their gut microbiomes have coevolved (Ley et al., 2008; Zaneveld et al., 2008), but the microbiome is also influenced by numerous environmental factors on both acute and chronic time scales (Koskella and Bergelson, 2020).
Numerous studies demonstrate that environmental alterations to the normal gut microbiome can affect the behavior and physiology of mammalian hosts (Cryan and Dinan, 2014; Fontaine and Kohl, 2020). These effects on the host may occur through multiple mechanisms, including afferent and efferent signals between the gastrointestinal tract and the brain (via the vagus nerve), that may be influenced by the gut microbiome (Fülling et al., 2019). In addition, bacterial-derived metabolites can be important energy sources for the host (Krautkramer et al., 2021; Rowland et al., 2018). Furthermore, gut microbes can influence the host through their effect on hormonal and immunological pathways (Clarke et al., 2014; Eshleman and Alenghat, 2021).
The influence of gut microbes on behavior has been demonstrated primarily by studies of microbiome depletion via antibiotic treatment and transplants into germ-free mice (Bercik et al., 2011; Leclercq et al., 2017; Luo et al., 2018; Marin et al., 2017; Sudo et al., 2004; Tochitani et al., 2016). For example, BALB/c mice have lower levels of exploratory behavior than NIH Swiss Webster mice under both specific-pathogen free conditions and in the germ-free state (Bercik et al., 2011). Reciprocal transplants into germ-free BALB/c and NIH Swiss mice resulted in “transplanted behavior.” Germ-free NIH Swiss mice colonized with BALB/c microbiota had decreased exploratory behavior in a step-down test, and germ-free BALB/c mice colonized with NIH Swiss microbiota had increased exploratory behavior, compared to baseline (Bercik et al., 2011).
Voluntary exercise behavior, defined as locomotion that is not motivated by any external factor or required for survival (Garland et al., 2011), plays a key role in mammalian health (Hawley et al., 2014; Heinonen et al., 2014; Silverman and Deuster, 2014). Although many studies have shown that exercise can affect the gut microbiome (Campbell and Wisniewski, 2017; Mailing et al., 2019; Mohr et al., 2020), how the microbiome affects exercise behavior is poorly understood (Crowson and McClave, 2020; Hughes, 2020; Oyanagi et al., 2018). The expression of voluntary exercise, as with all voluntary behaviors, will depend on intrinsic motivational state, but may also be limited by physical abilities (e.g. see Garland et al., 2011; Good et al., 2015; Jaromin et al., 2019; Kelly et al., 2014; Lightfoot et al., 2018).
One way that the gut microbiome might affect motivation is by altering signals from the mammalian gastrointestinal tract to the brain (Fülling et al., 2019). The gut microbiome has also been suggested to regulate reward processes in the brain (García-Cabrerizo et al., 2021). Therefore, given that wheel-running can be a self-rewarding, motivated behavior in rodents (Garland et al., 2011; Novak et al., 2012; Sherwin, 1998), changes in the gut microbiome community have the potential to impact motivation to exercise.
One mechanism by which the microbiome might affect exercise ability is through bacterial-derived metabolites (Bindels and Delzenne, 2013; Grosicki et al., 2018; Ticinesi et al., 2017). Bacterial symbionts produce short-chain fatty acids (SCFAs) from the fermentation of carbohydrates in the gastrointestinal tract, which can travel through circulation and affect skeletal muscle performance (Przewłócka et al., 2020). Butyrate, one of the three most abundant SCFAs, is associated with increased epithelial cell wall integrity and increased glucose uptake in skeletal muscle (Ticinesi et al., 2019). In addition to altering the gut microbiome community, antibiotics have been shown to decrease luminal butyrate concentration and decrease serum glucose levels in C57BL/6 mice, resulting in a shift to glucose instead of butyrate as the gut epithelial cell energy source (Zarrinpar et al., 2018). Alterations in host energy substrate availability could affect exercise behavior. Furthermore, as noted in the Discussion, two studies have demonstrated that antibiotic treatment affects treadmill endurance capacity and muscle physiology in mice (Nay et al., 2019; Okamoto et al., 2019).
In rodent models, voluntary exercise is generally measured as voluntary wheel-running behavior (Garland et al., 2011). Although a few studies have examined the effects of antibiotics on mouse activity in an open-field arena (Ceylani et al., 2018; Diaz Heijtz et al., 2011), this type of test has little to do with voluntary exercise behavior or spontaneous physical activity in home cages (Careau et al., 2012; Novak et al., 2012; Zombeck et al., 2011). To our knowledge, only one study has tested effects of the microbiome on voluntary wheel running. Male C57BL/6N mice were given 1 week of antibiotic treatment, followed by 7 weeks of a high-fat diet, then transplanted with cecal contents from either sedentary mice or mice with 12 weeks of wheel access, and finally provided with 7 days of wheel access (Oyanagi et al., 2018). Mice transplanted with microbiomes from mice with exercise exposure had higher levels of wheel running compared to the mice colonized with microbiomes from sedentary mice.
The purpose of the present study was to test for effects of eliminating the gut microbiome on voluntary exercise behavior. To increase statistical power (see below), we used mice from an ongoing artificial selection experiment that breeds for high voluntary wheel running over multiple generations. Mice from the 4 replicate High Runner (HR) and 4 non-selected Control (C) lines have several key physiological and behavioral differences, suggesting their gut microbes are likely to be different under baseline conditions and might also respond differently to antibiotics. Over 89 generations of selective breeding, the HR mice have evolved to run ~3-fold more revolutions per day than C mice (Cadney et al., 2021a; Cadney et al., 2021b; Careau et al., 2013; Copes et al., 2015; McNamara et al., 2021; Swallow et al., 1998). In addition to increased physical activity, HR mice have higher food consumption (for their body size), higher body temperature when active, higher activity levels when housed without wheels, and altered hormone levels (Copes et al., 2015; Malisch et al., 2009; Swallow et al., 2009), all of which could affect the gut environment acutely, as well as alter the selective regime experienced by symbiotic microbes.
As the HR mice run near their physiological limits (Claghorn et al., 2017; Girard et al., 2001; Rezende et al., 2005), dysbiosis of their gut microbiome and possible associated reduction in energy substrate availability has the potential to adversely affect wheel-running behavior. In addition, HR mice appear to have higher motivation for wheel running than C mice (e.g. see Belke and Garland, 2007; Garland et al., 2016; Rhodes et al., 2005; Saul et al., 2017), which could be affected by antibiotic treatment. In a previous study, we found differences in the gut microbiome between adult HR and C mice (McNamara et al., 2021). As discussed in our previous paper, the HR microbiome might have coevolved after many generations of selective breeding and/or it might reflect acute effects of exercise. In either case, we hypothesized that reduction of the gut microbiome would have a larger adverse effect on the amount of wheel running in HR mice than in C mice.
2. Materials and Methods
All procedures were approved by the Institutional Animal Care and Use Committee of the University of California, Riverside.
2.1. Experimental design
Mice were sampled from generation 89 of an ongoing experiment in which 4 replicate High Runner (HR) lines are selectively bred for voluntary wheel-running behavior and compared with 4 replicate non-selected Control (C) lines. Briefly, the selection experiment began in 1993 with 224 outbred, genetically variable Hsd:ICR strain mice (Swallow et al., 1998). Mice were randomly mated for 2 generations and then randomly assigned into 4 replicate HR lines bred for high wheel running and 4 replicate C lines. Each generation, the highest-running HR female and male from each of 10 families are chosen as breeders, based on the average revolutions on days 5 and 6 of a 6-day period of wheel access as young adults (Swallow et al., 1998). The wheels are Wahman-type activity wheels with 1.12 m-circumference, 10-cm wide running surface of 10-mm metal mesh, with clear Plexiglas walls. Breeders in the C lines are chosen without regard to wheel running. Mice are paired outside their family but within their line, and sibling matings are never allowed. Photoperiod is 12:12 L:D and temperature is ~22°C. All mice receive Standard Laboratory Rodent Diet (SD) from Harlan Teklad (W-8604), which contains 24.3% kJ from protein, 4% kJ from fat, and 40.2% from carbohydrate. Pregnant dams are given Harlan Teklad Lab mouse breeder diet [S-2335] 7004 through weaning.
For the present experiment, 12 to 16 females (sampled from as many different families as possible) from each of the 4 replicate HR and 4 C lines were weaned at 21 days of age. We chose female mice, rather than both sexes, to avoid aggressive behavior in males when cohoused (Kappel et al., 2017). Mice were housed individually for < 1 week, then randomly cohoused in groups of four (including mixing of the HR and C lines in an attempt to homogenize the gut microbiome, as mice are coprophagic (Laukens et al., 2016)). Each week, mice were again randomized into clean cages, and this occurred three times (Fig. 1).
Figure 1.
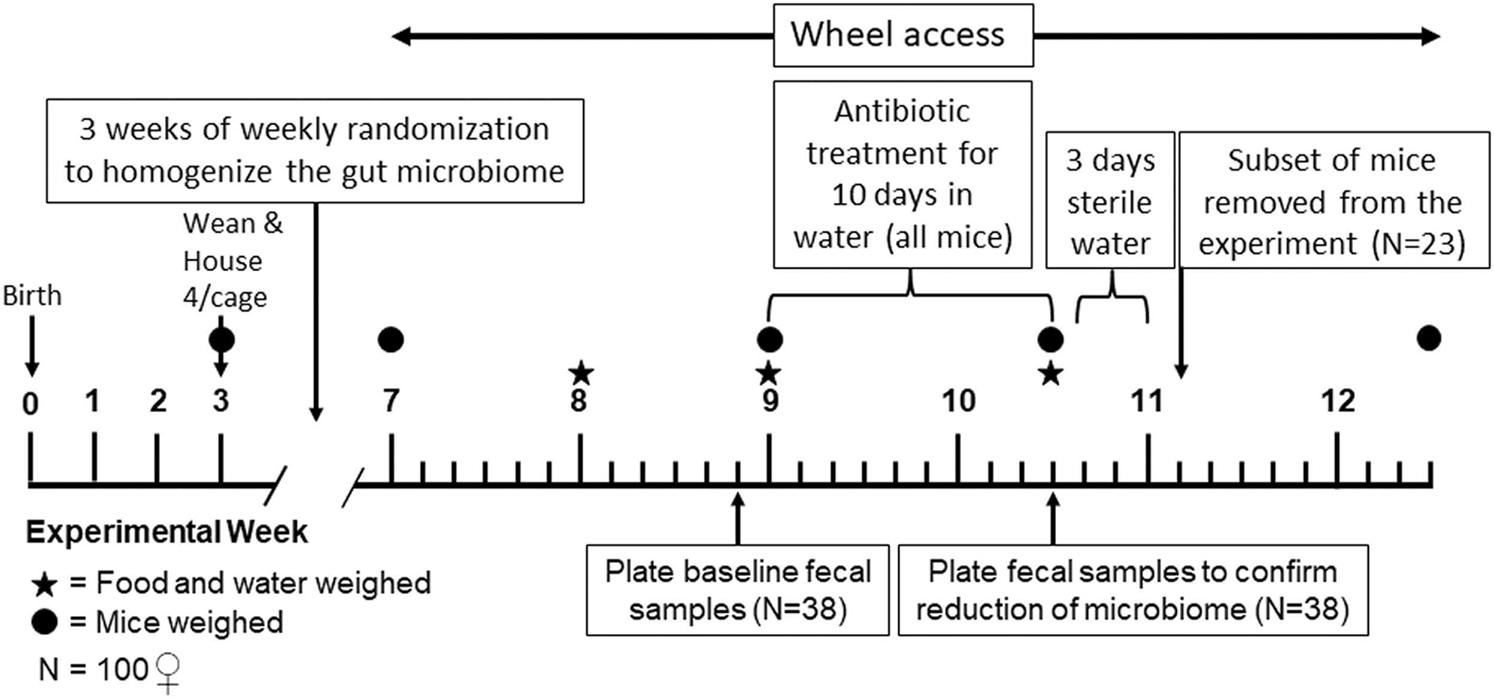
Experimental Timeline.
At ~7 weeks of age, mice were weighed and placed into individual cages with wheels (same as used in the routine selective breeding protocol) for two weeks to allow daily running distances to reach a stable plateau (Copes et al., 2015; Thompson et al., 2018). Numerous previous studies in the HR mice have shown that the wheel running plateau remains for several weeks (Dumke et al., 2001; Meek et al., 2012; Swallow et al., 1999). After two weeks, mice were weighed, fecal samples were collected, and all mice were given a broad-spectrum antibiotic cocktail in the drinking water to greatly reduce the gut microbiome. The antibiotic cocktail contained 1 g/L ampicillin, 1 g/L neomycin, 125 mg/L vancomycin, and 2.5 g/L Splenda to increase cocktail palatability (similar to Ichinohe et al., 2011; Rakoff-Nahoum et al., 2004; Reikvam et al., 2011). The antibiotic treatment lasted for 10 days; body mass, antibiotic water consumption, and food consumption were measured at the start and end of treatment. After completion of the antibiotic treatment, 23 mice were randomly removed for a separate experiment, another fecal sample was collected, and the mice were given sterile water for 3 days to washout the antibiotics. Mice were then provided with tap water for 9 days to allow the gut microbiome to naturally “recover” (either from environmental sources or through bacteria present in the gut). Wheel access continued for 12 days after the cessation of antibiotic treatment (Fig. 1).
2.2. Wheel Running
As noted above, mice were housed individually in home cages with attached wheels. Sensors attached to the wheel record the daily number of revolutions in each 1-minute interval during a 23 hr measurement period (Swallow et al., 1998). Following our previous studies, we calculated the total number of revolutions per day (distance), the number of 1-minute intervals with at least one revolution (duration of activity), the mean running speed (revolutions/interval), and the maximum number of revolutions observed for any 1-minute interval. Before and after housing mice with wheels, we measured wheel freeness by recording the number of revolutions per wheel until it reaches a stop after accelerating each wheel to a constant speed (Copes et al., 2015).
2.3. Spontaneous physical activity
As in previous studies, each home cage was fitted with an infrared sensor (Talon TL-Xpress-A; Crow Electronics, Fort Lee, NJ) connected to a digital I/O board (ICS 2313; ICS Electronics, Pleasanton, CA), and a Macintosh computer with custom software (Copes et al., 2015). The sensors have an ~90° field of view with a reset time of 1–2 seconds if no motion is detected. The sensor software takes readings ~3 times per second, recording movement detection as a “1” and no movement detection as a “0”. For each minute a mean value (0–1) is computed, and the software saves mean values every 10 minutes. To calculate the total home-cage activity, or “spontaneous physical activity” (Acosta et al., 2015; Garland et al., 2011), we summed the total activity recorded in a 23 hr measurement period. We also tallied the number of 1-minute intervals during which motion was detected (duration of activity) and used this to estimate the mean intensity of activity, i.e., the average amount of activity per minute when any home-cage activity was occurring (Copes et al., 2015). We also measured sensor sensitivity with a heated curling iron passed back and forth in front of the sensor 10 times over a period of 10 s.
2.4. Food and Water Consumption
Average daily water consumption during the initial wheel access (seven days) and during antibiotic treatment (ten days) was calculated by weighing the water bottle at the start and end of the period. We had 4 control water bottles to account for water bottle leakage. Average daily food consumption was calculated in a similar fashion to water consumption, by weighing food hoppers with due allowance for wastage (Koteja et al., 2003).
2.5. Fecal Sampling and Plating
To confirm reduction of the gut microbiome, we determined the number of colony-forming units (CFUs) for a subset of mice from a baseline sample (N=38) collected after two weeks of wheel access and a post-antibiotic treatment fecal sample from the same subset of mice. Mice were placed into clean, but not completely aseptic, individual cages until defecation. Fecal pellet mass was determined by weighing sterile tubes before and after collection of the pellet. Pellets were suspended in 500 μL sterile phosphate-buffered saline (PBS) using a BeadBeater for 30 seconds at 1,400 rpm. 5 μL of the fecal suspension was plated on Luria Bertani media in a serial dilution to 10−6 (two per mouse) and incubated both aerobically and anaerobically at 37°C. After a 24 hr incubation, the colonies were counted and the colony-forming units calculated by dividing the number of colonies per mL plated by the total dilution factor.
2.6. Statistical Analyses
We used linear mixed models to determine the effects of genetic background (High Runner versus Control linetype), antibiotic treatment (pre-antibiotic versus post-antibiotic), and their interaction on wheel running, spontaneous physical activity, body mass, water consumption, and food consumption. Following numerous previous studies of these mice, the effect of linetype was tested relative to the variance among replicate lines, which are a nested random effect within linetype, with 1 and 6 d.f. (SAS Procedure Mixed with repeated measures, SAS Institute, Cary, NC, USA) (e.g., Acosta et al., 2015; Swallow et al., 1998). The effect of antibiotics and the antibiotic × linetype interaction were also tested with 1 and 6 d.f. Outliers were removed when standardized residuals were greater than approximately 3 and we accepted nominal statistical significance at p<0.05. Data are presented as untransformed least squares means and standard errors. Such covariates as body mass, wheel freeness (results not shown), and home-cage sensor sensitivity (results not shown) were included in the models where appropriate. For food and water consumption statistical tests were run both with and without the amount of wheel running as a covariate (e.g., see Copes et al., 2015; Hiramatsu and Garland, 2018).
A measure of effect size (in this case, Pearson’s r) was calculated for all main and interactive effects. We chose Pearson’s r because of (1) it’s ease of use (in this case requiring only an F-statistic and d.f.) and (2) it’s ease of interpretation (r family effect sizes are measures of the strength of association). For a brief primer on effect sizes and why to include them, see Rosenthal (1994), Sullivan and Feinn (2012), and Lakens (2013).
3. Results
3.1. Depletion of the gut microbiome
Prior to antibiotic treatment, plates averaged 4.18 × 106 aerobic colony-forming units, with no statistical difference between High Runner (N=18) and Control (N=20) mice (ANOVA, F1,6=0.32, p=0.5905). Ten days of antibiotic treatment reduced the colonies to non-detectible amounts for both linetypes. Anaerobic plates were not usable due to technical issues.
3.2. Wheel running
In a repeated-measures analysis, once a wheel-running plateau was attained (Fig. 2A: days 11–13), HR mice ran ~3.5-fold more than C mice, then antibiotic treatment reduced the number of revolutions run per day (days 22–24) in only the HR mice (Fig. 3A: linetype × antibiotic treatment interaction F1,6=15.60, p=0.0075; differences of least squares means p=0.0023 for HR mice and p=0.2071 for C mice). Antibiotic treatment reduced the number of minutes mice ran per day (Fig. 3B) for both linetypes (F1,6=60.79, p=0.0002; differences of least squares means p=0.0018 for HR mice and p=0.0012 for C mice). The linetype × antibiotic interaction approached significance for average running speed and had a large effect size (Fig. 3C: F1,6=4.92, p=0.0683, r=0.6712), with a slight tendency for a decreased running speed in the HR lines, and increased running speed in the C lines (differences of least squares means p=0.1925 for HR mice and p=0.1209 for C mice). However, HR mice continued to run faster than C mice, both before and during antibiotics (Fig. 3C and 3D).
Figure 2.
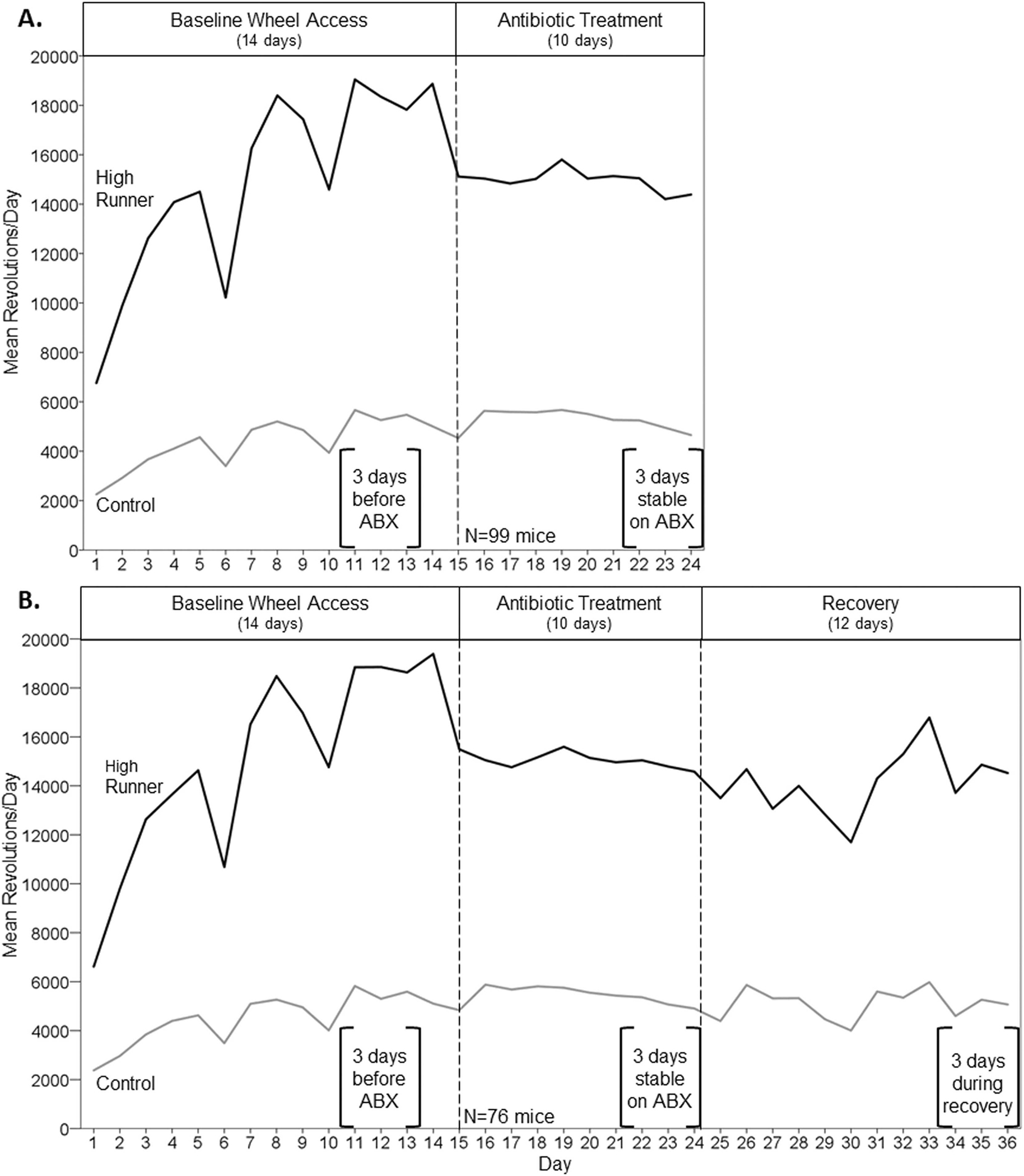
Average daily wheel running (revolutions per 23 hours) across the course of the experiment. Values were calculated by taking the simple means of the 4 HR lines and the 4 C lines for each individual day, and then averaging those values to obtain the average for the HR and C lines. Data were truncated on days 8, 14, and 25 (~20 hours) when the water bottles were being changed. Brackets indicate days that were used in statistical analyses. See Supplemental Figure 2 for wheel running mean revolutions per day for each of the 4 HR and 4 C lines. A. N=99 mice, through day 24 of the experiment. Days 11–13 before antibiotics were compared with days 22–24 during antibiotics. B. N=76 mice, excluding a random subset that were removed on day 28 for a separate experiment. The microbiome was allowed to naturally recover after antibiotic treatment (days 25–36). Days 34–36 were used to indicate wheel running in recovery.
Figure 3.
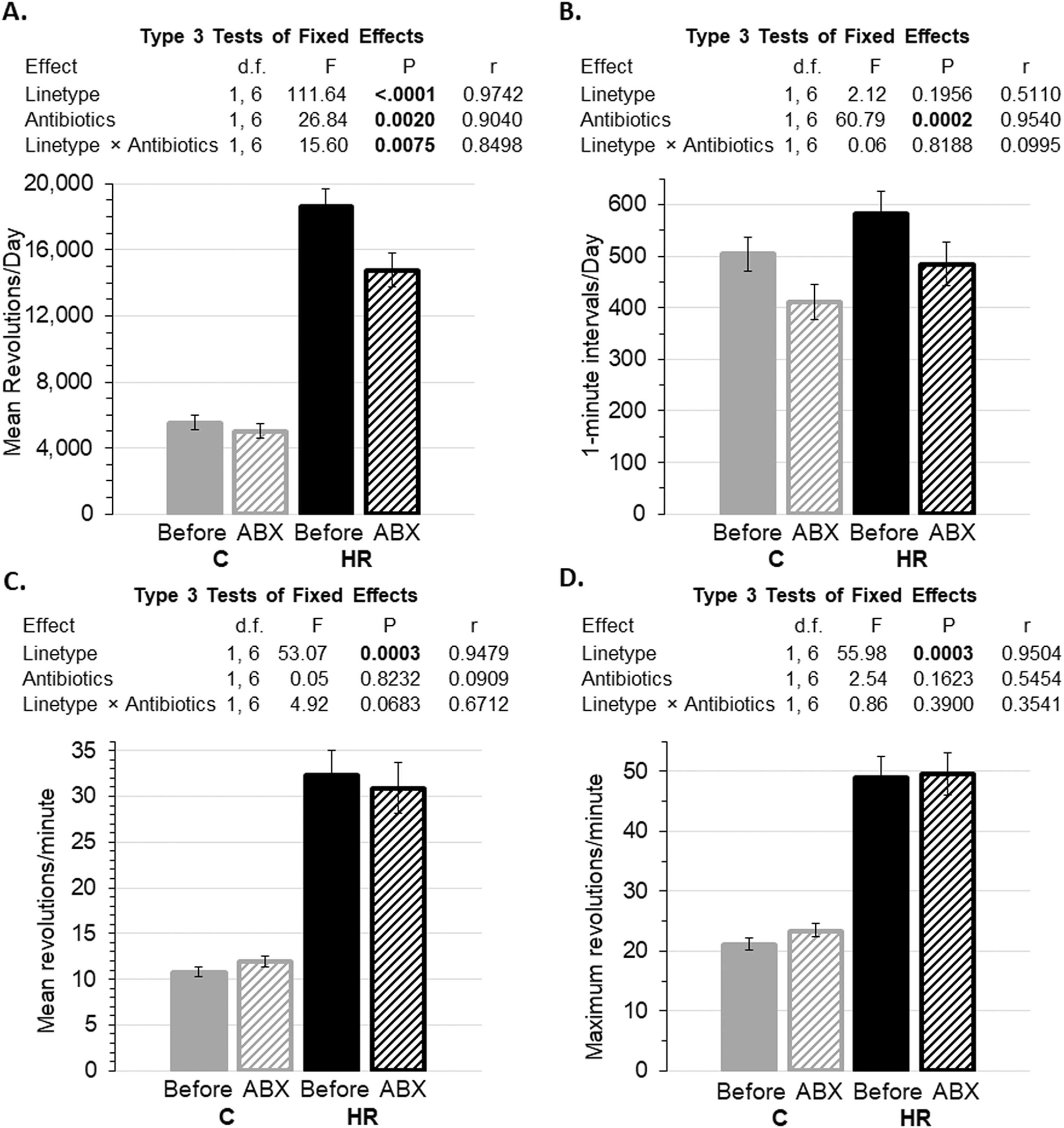
Wheel running before (averages for days 11–13) and during (averages for days 22–24) antibiotics (ABX; see Figure 2). Data are presented as least squares means ± standard errors and type 3 tests of fixed effects from repeated-measures analyses using values for individual days in SAS Procedure Mixed. N=92 mice and 543 observations after removal of statistical outliers (see Methods). Pearson’s r is a measure of effect size (see Methods). A. Revolutions per day. Antibiotic treatment reduced the revolutions per day (days 22–24) in only the HR mice (linetype × antibiotic treatment interaction, p=0.0075; differences of least squares means p=0.0023 for HR mice and p=0.2071 for C mice). B. Number of 1-minute intervals with at least one revolution. Antibiotic treatment reduced the number of minutes mice run per day for both linetypes (p=0.0002; differences of least squares means p=0.0018 for HR mice and p=0.0012 for C mice). C. Revolutions per minute. Antibiotics had no statistical effect on average speed. D. Maximum revolutions per minute. Antibiotics had no statistical effect on maximum running speed.
After antibiotic treatment stopped, mice remained in home cages with wheel access and regular food and drinking water for 12 days (Fig. 2B). Halting antibiotics had no statistical effect on the daily running distance, number of minutes mice ran per day, average speed, or maximum running speed (Fig. 4). In particular, the daily wheel-running distance of the HR mice did not recover to levels before antibiotics (Fig. 4A: differences of least squares means p=0.0004 for HR mice before antibiotics versus after recovery). During the recovery period, the HR mice ran ~2.7-fold more revolutions per day than C mice (Fig. 4A), which was mainly related to their faster running (Fig. 4C and 4D).
Figure 4.
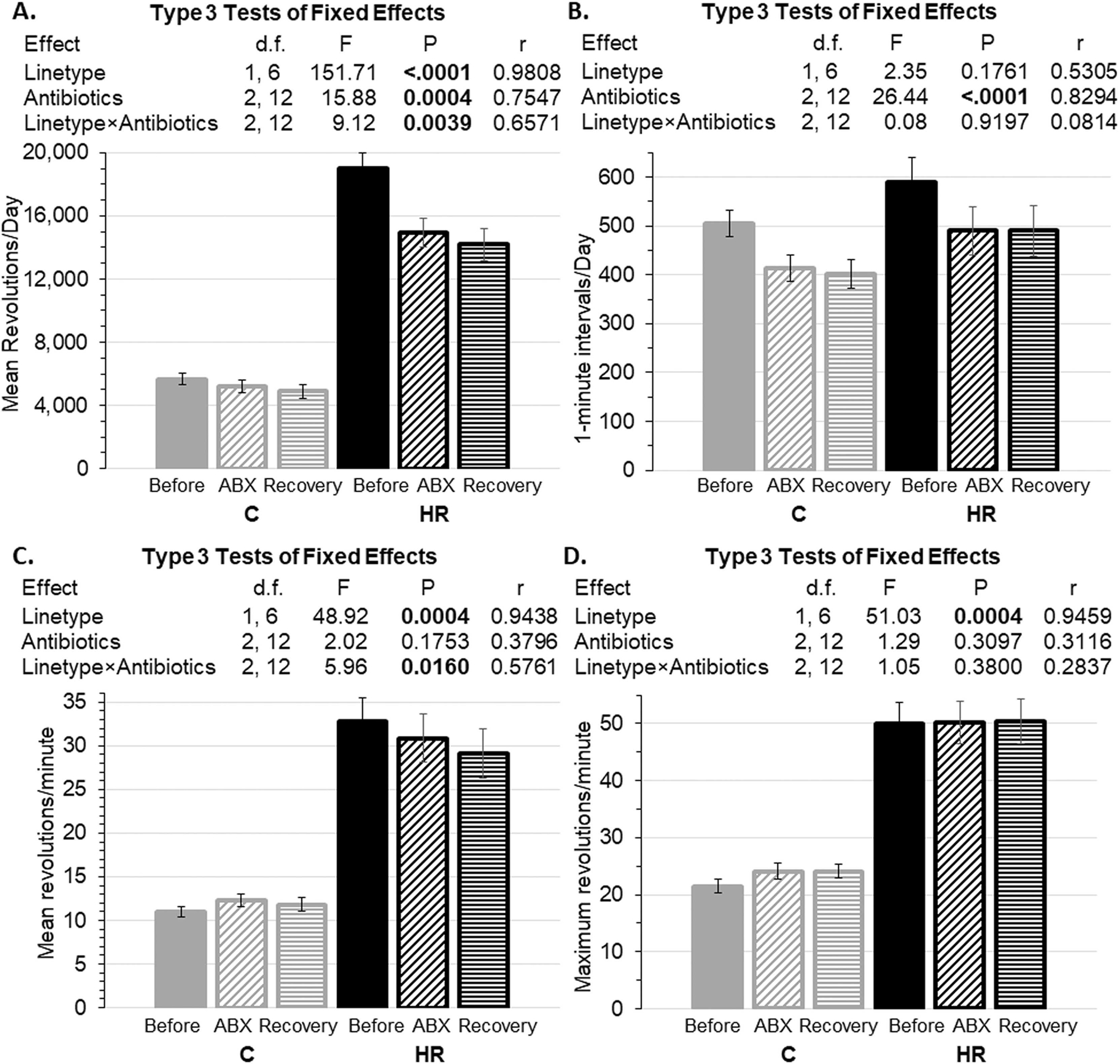
Wheel running before antibiotics (averages for days 11–13), during antibiotics (averages for days 22–24), and after recovery (days 34–36) (see Figure 2). Data are presented as least squares means ± standard error from repeated-measures analyses using values for individual days in SAS Procedure Mixed. N=73 mice and 639 observations after removal of statistical outliers (see Methods). Pearson’s r is a measure of effect size (see Methods). A. Revolution per day. B. Number of 1-minute intervals with at least one revolution. C. Revolutions per minute. D. Maximum revolutions per minute. Overall, mice did not recover to pre-treatment levels of running (see Figures 2 and 3) within the time frame of this experiment.
3.3. Spontaneous physical activity measured in home cages
In a repeated-measures analysis using the same experimental days before (days 11–13) and during (days 22–24) antibiotic treatment as above, antibiotic treatment tended to decrease total cage activity in C mice, although the interaction did not reach significance (Supplemental Fig. 1A: F1,6=3.55, p=0.1084, r=0.6097; differences of least squares means p=0.0576 for C mice and p=0.4914 for HR mice). This decrease in total cage activity for C mice was partially explained by a trend for reduced activity duration in C mice but not in HR mice (Supplemental Fig. 1B: F1,6=3.79, p=0.0996, r=0.6222; differences of least squares means p=0.0063 and p=0.5649, respectively). Antibiotics tended to increase mean intensity of activity (total/minutes) in HR mice, but not in C mice (Supplemental Fig. 1C: F1,6=2.85, p=0.1425; differences of least squares means p=0.7159 for C mice and p=0.1166 for HR mice). Data were not available for the period of recovery from antibiotics.
3.4. Body mass
Consistent with previous studies (Kelly et al., 2017; Meek et al., 2009; Thompson et al., 2018), HR females tended to weigh less than C females for the duration of the experiment (Fig. 5), but the difference never reached statistical significance (all p>0.2000). Separate repeated-measures analysis of mass before versus after 10 days of antibiotics indicated mice were larger after antibiotic treatment (F 1,6= 22.68, p=0.0031). Analysis of mass after 10 days of antibiotics versus after 12 days of recovery indicated no significant difference (F 1,6= 0.01, p=0.9321).
Figure 5.
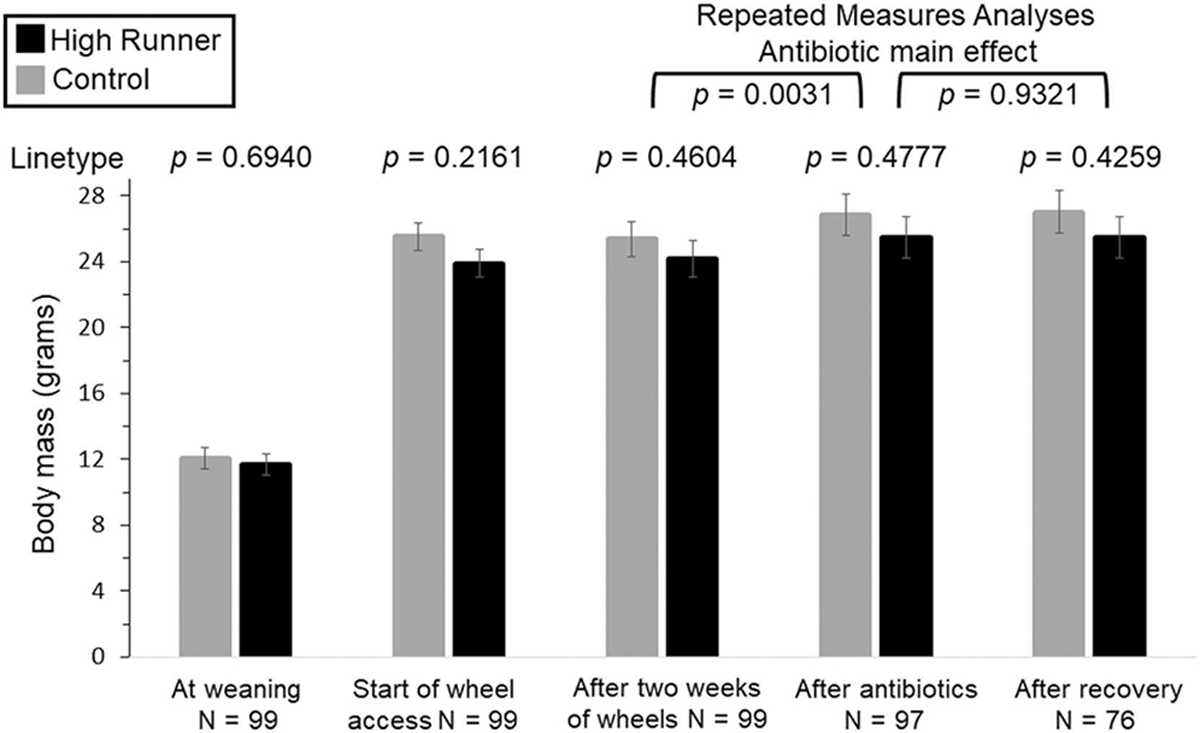
Body mass measured at various timepoints. Analyses were first done separately for each time point (not repeated-measures) and values in the figure are least squares means ± standard errors from those analyses. p-values above each pair of bars are for the linetype comparison at each time point. In addition, separate repeated-measures analysis of mass before versus after 10 days of antibiotics indicated mice were larger after antibiotic treatment (treatment p=0.0031, linetype p=0.4646, interaction p=0.8329). A similar analysis of mass after antibiotics versus after 12 days of recovery indicated no effect of recovery (treatment p=0.9321, linetype p=0.4780, interaction p=0.6612).
3.5. Food and water consumption
With body mass as a covariate, antibiotics had no effect on food consumption for C mice, but HR mice consumed less food during antibiotic treatment (days 15–24) than pre-antibiotics (days 8–14) (Fig. 6A: linetype × antibiotic treatment interaction F 1,6 = 6.24, p=0.0467). HR mice also consumed significantly more food than C mice during both the pre-antibiotic and antibiotic periods (F 1,6 = 9.15, p=0.0232). When the amount of wheel running (revs/day) was added as a covariate, it was a highly significant predictor of food consumption (Fig. 6B, F1,166=61.23, p<0.0001) and the effects of linetype (p=0.9365), as well as the linetype × antibiotic interaction, became non-significant (p=0.2854).
Figure 6.
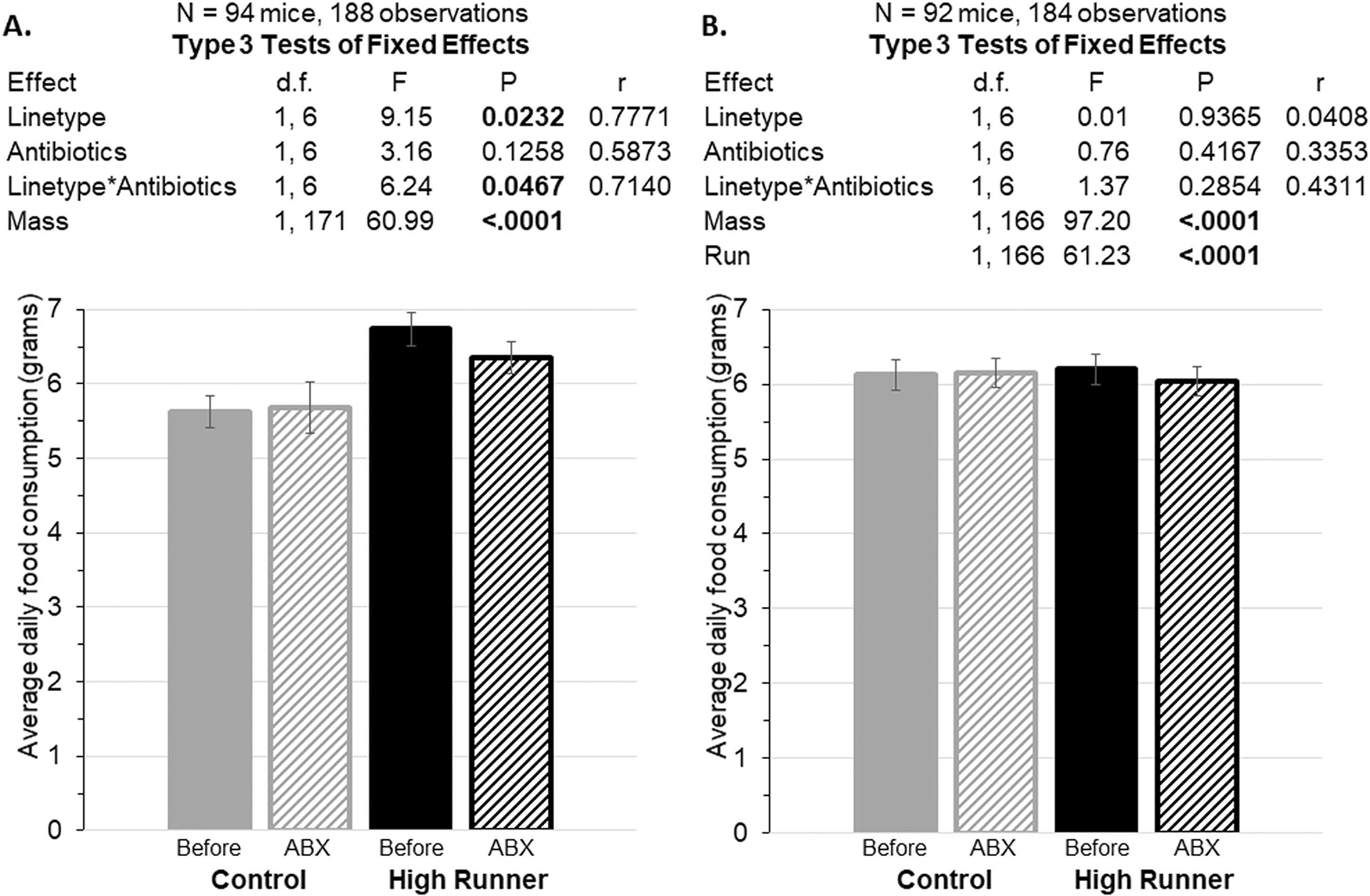
Average daily food consumption during the baseline period (days 8–14) compared to during the antibiotic (ABX) treatment period (days 15–24), with body mass as a covariate. Data are presented as least squares mean ± standard errors from repeated-measures analyses in SAS Procedure Mixed. Pearson’s r is a measure of effect size (see Methods). A. Antibiotics decreased food consumption in the HR mice (repeated-measures analysis interaction p=0.0467). B. Food consumption with wheel running as an additional covariate. The interaction between linetype and antibiotics is no longer significant.
Accounting for body mass, both linetypes drank significantly more of the antibiotic water during experimental days 15–24 compared to regular water during days 8–14 (F 1,6 = 24.76, p=0.0025; Fig. 7A). With wheel running as an additional covariate (Fig. 7B, F1,166=11.38, p=0.0009), the effect of antibiotics was still highly significant (F1,6=33.46, p=0.0012).
Figure 7.
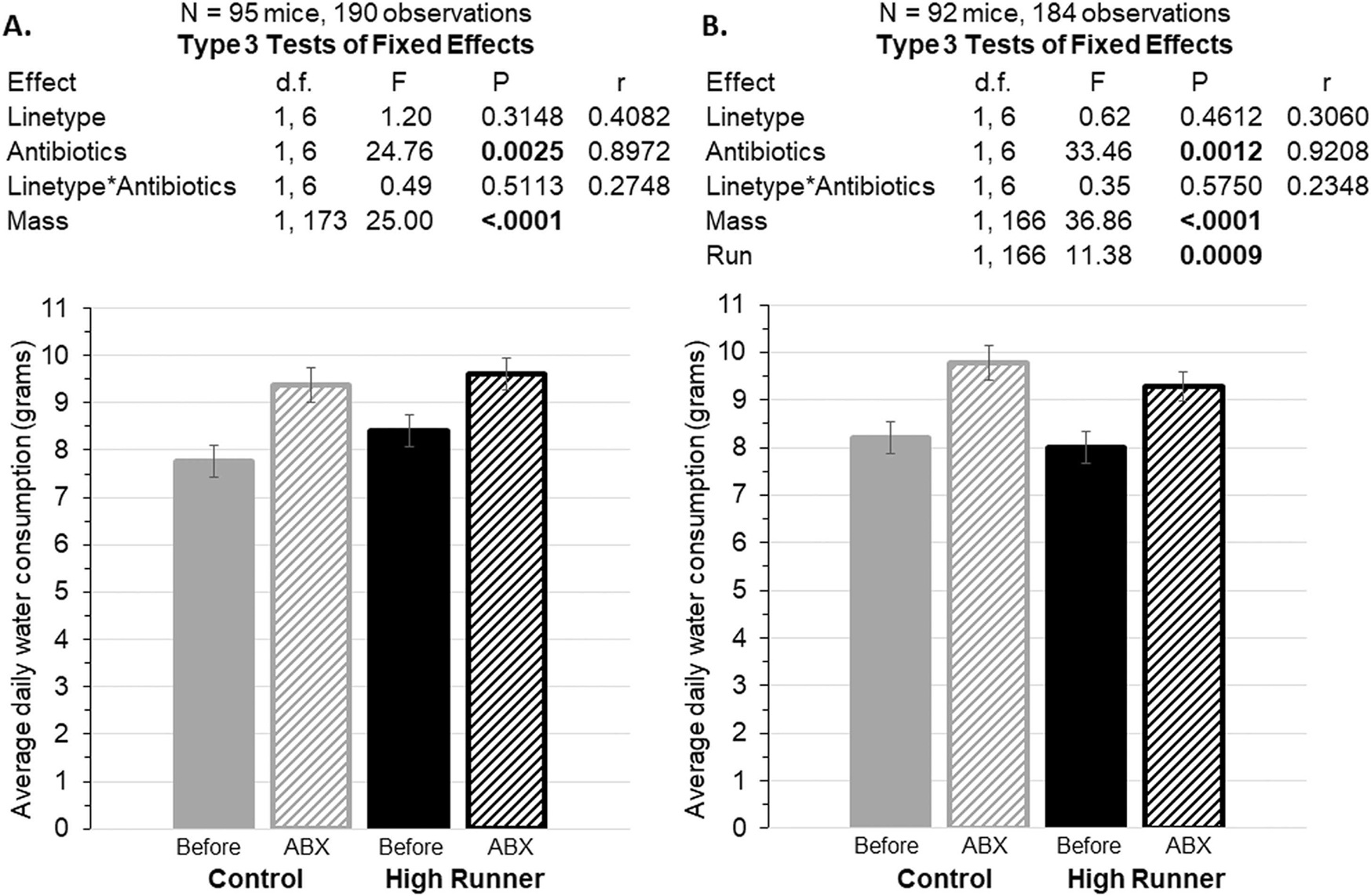
Average daily water consumption during the baseline period (days 8–14) compared to during the antibiotic (ABX) treatment period (days 15–24), with body mass as a covariate. Data are presented as least squares means ± standard errors from repeated-measures analyses in SAS Procedure Mixed. Pearson’s r is a measure of effect size (see Methods). A. Water consumption increased while on antibiotics. B. When included as an additional covariate, wheel running was a significant positive predictor, but water consumption was still higher while on antibiotics. This result differs from food consumption (Figure 6).
4. Discussion
The antibiotic treatment we administered greatly reduced the gut microbiome in both High Runner and Control lines, based on aerobic plating. Although we were not able to show the effects of antibiotics on anaerobic bacteria, previous antibiotic cocktails that included ampicillin, streptomycin, and neomycin in combination with other antibiotics have shown a successful decrease or depletion of anaerobic colonies (Carvalho et al., 2012; Castro-Mejía et al., 2018; Reikvam et al., 2011). Ten days of antibiotic treatment significantly reduced daily wheel-running distance in selectively bred HR lines of mice, but not in their non-selected C counterparts (Fig. 3). Moreover, mice from the HR lines did not recover to pre-antibiotic treatment levels of wheel running during 12 days of recovery (Fig. 4). Based on body mass, food consumption, and behavioral observations, antibiotic treatment did not appear to cause sickness behavior (Fig. 5, 6).
4.1. Antibiotic treatment alters wheel-running behavior
Broad-spectrum antibiotic treatment significantly reduced the distance of daily wheel running by the HR lines (−21%), but had no statistical effect on C lines (Fig. 3A). At a more granular level, antibiotics significantly reduced the duration of daily running in both HR and C lines (Fig. 3B), but in the latter the average running speed increased slightly (Fig. 3C), such that daily running distance was not significantly affected. Mice from the HR lines did not recover to pre-antibiotic treatment levels of wheel running during 12 days of recovery (Fig. 4).
Reducing the gut microbiome could have altered the motivation and/or ability for sustained, aerobically supported exercise, both of which are higher in HR mice relative to C mice (e.g. see Cadney et al., 2021b; Hiramatsu et al., 2017; Meek et al., 2009; Singleton and Garland, 2019). Previous research has shown that antibiotic treatment can affect exercise ability in mice. Three weeks of antibiotic treatment in male mice of the inbred C57BL/6 strain decreased treadmill endurance capacity, as measured by time to exhaustion during a submaximal test, with no significant effect on body mass, food consumption or water consumption (Nay et al., 2019). Antibiotics also decreased fatigue resistance in an ex vivo contractile test of extensor digitorum longus muscle and glycogen content of the gastrocnemius. Eleven days of natural reseeding with soiled bedding from the control group normalized muscle performance and glycogen levels. These results demonstrate the importance of bacterial species on host optimal skeletal muscle function. In a separate study of male C57BL/6J mice (Okamoto et al., 2019), two weeks of antibiotic treatment reduced treadmill running time, decreased tibialis anterior muscle mass, reduced fecal bacterial density, decreased fecal short-chain fatty acid (SCFA) concentration, and decreased concentration of acetate in plasma. Given that acetate is the dominant SCFA in circulation, the authors speculated it could have the largest influence on exercise tolerance, as any change in acetate concentration would change the potential energy source for muscles during exercise. Continuous administration of acetate for one week increased treadmill running time after the initial impairment of endurance capacity caused by two weeks of antibiotic treatment. Future studies of the HR mice could introduce SCFAs into circulation to test whether this would counter the negative effects of microbiome ablation on wheel running and/or exercise ability.
As noted in the Introduction, the microbiome has been shown to affect various aspects of behavior, and some of the effects likely occur through changes in motivation. More specifically, the gut microbiome has been suggested to play a role in reward circuits (García-Cabrerizo et al., 2021). For example, ileum infusion of antibiotics for 25 days in piglets decreased concentrations of hypothalamic (1.13-fold) and circulating dopamine (1.18-fold) (Gao et al., 2018). The mechanism(s) for antibiotic treatment to modulate brain function and behavior may be in part from influence of antibiotics upon amino acids being absorbed from the gut. Once in circulation, large neutral amino acid transporters import essential amino acids across the blood-brain barrier to where they become precursors to serotonin and catecholamines, including dopamine (Zaragozá, 2020). A variety of lines of evidence indicate that the motivation/reward system of the HR mice is altered (e.g. see Belke and Garland, 2007; Garland et al., 2016; Rhodes et al., 2005; Saul et al., 2017). Several studies indicate alterations in dopamine signaling in the HR mice (Rhodes et al., 2005; Saul et al., 2017), which could suggest that reduction of the gut microbiome by antibiotic treatment may have altered dopamine signaling in a way that reduced motivation for wheel running. Future studies of the HR mice could use pharmacological manipulation of reward pathways (e.g. Keeney et al., 2012; Rhodes and Garland, 2003; Rhodes et al., 2005) in an attempt to counteract the negative effect of antibiotic treatment on wheel running.
Separating motivation from ability when examining voluntary exercise is challenging. Our data for daily wheel-running distance can be separated into components that estimate the duration of running versus average running speed. We speculate that duration of activity is more affected by motivation, whereas average (and maximum) running speed is more reflective of ability (see also Kay, 2017; Swallow et al., 2009). Antibiotic administration had stronger negative effects on the duration of activity than on speed (Fig. 3). These results suggest that future studies should explore possible effects of microbiome manipulation on brain motivation and reward systems.
4.2. Lack of recovery in wheel-running after cessation of antibiotics
Wheel-running behavior of HR mice did not recover to pre-antibiotic levels even 12 days after cessation of antibiotic treatment (Fig. 4). However, it is important to note that we did not attempt to reseed the gut microbiome community after cessation of antibiotics. In a small pilot study with 7-week old female inbred C57BL/6 strain mice, 10 days of antibiotic treatment caused an immediate reduction in the richness of the gut microbiome and altered the overall community structure (Laubitz et al., 2021). The gut microbiome was then allowed to naturally recover with no reseeding of the community. Twenty days after the cessation of antibiotic treatment, 31 genera primarily belonging to the Firmicutes phylum had gone extinct, although gut microbiome diversity and community structure had recovered. In a separate study, “humanized” germ-free mice (transplanted with human microbes) were given daily streptomycin treatment for 5 days (Ng et al., 2019). During the first day of antibiotic treatment there was an initial drastic drop in the aerobic and anaerobic colony-forming units and a decrease in the number of observed species. The initial drop in diversity began to recover during days 2–5 of the antibiotic treatment. Nine days after the cessation of antibiotics the gut microbiome diversity failed to recover to pre-treatment levels, largely driven by a decrease in the Bacteroidetes phylum, while the CFUs returned to normal levels during day 2 of the antibiotic treatment (Ng et al., 2019).
Given the results of Laubitz et al. (2021) and Ng et al. (2019), and in the absence of reseeding, if wheel-running behavior were to recover, then it likely would have taken longer than the 12 days we allowed. In future experiments, we plan to re-seed the HR microbiome with soiled bedding from HR and/or C mice that had not been administered antibiotics, and also allow a longer period of time for possible recovery. In addition, transplant experiments will be used to further explore the contribution of the HR microbiome to their high motivation and/or ability for exercise.
4.3. High variability in wheel-running behavior
Mice in general, including HR and C mice, have high day-to-day variation in wheel running behavior (e.g., see Fig. 4 in Acosta et al., 2017). Indeed, this variability has been the subject of previous papers (e.g., see Biro et al., 2018; Eisenmann et al., 2009). In the present study of females, some variation would be related to the estrus cycle, but that should not account for consistent drops on days 6 and 10 that occurred for both HR and C mice (Fig. 2). The cause of those drops is unknown, but could be related to elevated noise levels in the vivarium and increases in humidity when cages are cleaned in a room down the hall. Importantly, the wheel-testing rooms were always left undisturbed, except when we entered to download data once each day.
At first glance, Fig. 2 also suggests that variability seems to have decreased in both HR and C lines when antibiotics were administered. However, whereas Figure 2 shows all of the days of data for wheel running, we only used days 11–13 before antibiotics, days 22–24 during antibiotics, and days 34–36 during recovery for statistical comparisons. Considering only those days, the variability does not appear much different across the three treatment phases; moreover, the mixed model analyses provided no clear evidence for differences in variability in relation to treatment phase.
4.4. Absence of sickness behavior
Protocols in which a broad-spectrum antibiotic cocktail was given to mice in water bottles have sometimes been shown to cause health problems, even within five days (Knoop et al., 2016; Reikvam et al., 2011). For example, BALB/c mice refused to consume the antibiotic cocktail after 4 days, which resulted in a weight loss of greater than 20% compared to baseline (Reikvam et al., 2011). In the present study, antibiotics (with Splenda) did not cause reduced water consumption. In addition, we observed no decrease in body mass (Fig. 5). In fact, body mass increased with antibiotic treatment in both linetypes (Fig. 5). Given that the mice we used in our experiment were only 9 weeks old when antibiotic treatment began, the mice were likely still growing (e.g., see Fig. 4 in Bronikowski et al., 2006), or at least adding fat mass. Furthermore, we observed no decrease in food consumption when analyzed with wheel running as a covariate (Fig. 6B). Finally, daily checks, which included checking for diarrheal symptoms and for any mice clearly unwell or lying prone, did not reveal any obvious signs of sickness behavior (e.g., see Downs et al., 2012).
4.5. Food consumption
Antibiotics decreased food consumption in HR but not C mice (Fig. 6A), similar to the effect on daily wheel-running distance (Fig. 3A). Decreased voluntary exercise would reduce energy requirements for HR mice (e.g., see Copes et al., 2015; Hiramatsu and Garland, 2018). When the amount of wheel running (revs/day) was used as a covariate, there was no apparent change in food consumption of HR mice before and during antibiotic treatment and no difference between the HR and C lines (Fig. 6B). Together, these results suggest that both the higher food consumption of HR mice in general (e.g., see Copes et al., 2015), and their decreased food consumption during antibiotic treatment, are a function of their wheel-running behavior.
4.6. Water consumption
With body mass as a covariate, both HR and C mice had a significant increase in water consumption during antibiotic treatment when compared to pre-antibiotic values (Fig. 7). Adding the number of revolutions run per day as a covariate did not change this pattern. One possible explanation for an increase in water consumption during antibiotic treatment is that mice liked the taste of the Splenda in the antibiotic cocktail, leading them to consume more of the antibiotic cocktail as compared with ordinary tap water. One study using the same artificially selected mouse model found that both HR and C mice always drank more water with Splenda (and with other sweeteners) as compared with plain tap water (Thompson et al., 2018). However, antibiotic-induced kidney injury could also explain the increased water consumption while on antibiotic treatment (Sinha Ray et al., 2016; Yang et al., 2019).
4.7. Home-cage activity
The effects of antibiotic treatment on home-cage activity are somewhat puzzling, as only the Control mice seem to have been affected. More specifically, C mice on antibiotics had reduced home-cage activity, caused by a reduction in the number of active minutes in their home cage (Supplemental Fig. 1). These results suggest that if antibiotics are affecting motivation to run (see Section 4.1 above), then they may also be affecting motivation for physical activity in general. But this interpretation fails to explain the lack of an antibiotic effect on home-cage activity in the HR mice.
4.8. Limitations
Constraints on available resources did not allow us to include HR and C groups housed with wheel and provided with water supplemented with artificial sweetener but not antibiotics. Such groups would have served as an additional “control” for the effects of artificial sweeteners per se. However, previously we showed that artificial sweeteners alone did not affect wheel running in either HR or C mice (Thompson et al., 2018). Therefore, we are confident that the effects observed in the present study can be attributed to antibiotics.
We were also unable to sequence the gut microbiome community, which is highly sensitive to both antibiotic and exercise treatments (Blaser, 2016; Mach and Fuster-Botella, 2017; Rosa et al., 2018). Culturing the majority of bacterial species in the gut microbiome is a challenge (see review Lagkouvardos et al., 2017), but we were able to show that 10 days of treatment with a broad-spectrum antibiotic cocktail eliminated aerobic CFUs under these particular conditions. We could not demonstrate the effectiveness of the antibiotic cocktail on the anaerobic community due to technical issues with the plating. As the gut microbiome is primarily composed of anaerobic bacteria, plating of these CFUs would be important in future studies. We were also unable to plate samples after the 12-day recovery period, which we would expect to recover to near baseline levels. Nevertheless, we were able to show a significant change in the wheel-running of HR mice caused by a broad-spectrum antibiotic cocktail. Here, we did not attempt to determine the mechanistic basis of the apparent effect of gut microbiome depletion on wheel running. Future studies involving reseeding of the microbiome, in addition to transplantation of HR and C microbes, are warranted. Also of interest would be studies of standard inbred strains to test for covariation between wheel-running behavior and the microbiome complement (e.g. see Careau et al., 2012; Carmody et al., 2015; Garland et al., 2016; Kay et al., 2019; Org et al., 2015).
Supplementary Material
Supplemental Figure 1. Home-cage activity before (averages for days 11–13) and during (averages for days 22–24) antibiotics (ABX). Note that all mice had wheel access during the entire period. Due to equipment failures, data were not available for the period of recovery from antibiotics. Data are presented as least squares means ± standard errors from repeated-measures analyses of individual daily values in SAS Procedure Mixed. Pearson’s r is a measure of effect size (see Methods). A. Although the interaction did not reach statistical significance, C mice on antibiotics tended to have decreased total cage activity during antibiotic treatment (differences of least squares means p=0.0576 for C mice and p=0.4914 for HR mice). B. Although the interaction did not reach statistical significance, antibiotics reduced activity duration in C mice but not in HR mice (differences of least squares means p=0.0063 and p=0.5649, respectively). C. Antibiotics tended to increase mean intensity of activity (total/minutes) in HR mice, but not in C mice (differences of least squares means p=0.7159 for C mice and p=0.1166 for HR mice).
Supplemental Figure 2. Wheel running mean revolutions per day (23 hours) for each of the 4 HR and 4 C lines. Data were truncated on days 8, 14, and 25 (~20 hours) when the water bottles were being changed. Boxes indicate days that were used in statistical analyses. A. N=99 mice, through day 24 of the experiment. Days 11–13 before antibiotics were compared with days 22–24 during antibiotics. B. N=76 mice, excluding a subset that were removed on day 28 for a separate experiment. The microbiome was allowed to naturally recover after antibiotic treatment (days 25–36). Days 34–36 were used to indicate wheel running in recovery.
Acknowledgments
This work was supported by a US NSF grant to T.G. (DEB-1655362) and a US NIH grant to A.H. (R35GM124724).
Footnotes
Declaration of Interests
The authors declare no competing or financial interests.
REFERENCES
- Acosta W, Meek TH, Schutz H, Dlugosz EM, Vu KT and Garland T Jr.. (2015). Effects of early-onset voluntary exercise on adult physical activity and associated phenotypes in mice. Physiol. Behav. 149, 279–286. [DOI] [PubMed] [Google Scholar]
- Acosta W, Meek TH, Schutz H, Dlugosz EM and Garland T Jr. (2017). Preference for Western diet coadapts in High Runner mice and affects voluntary exercise and spontaneous physical activity in a genotype-dependent manner. Behav. Processes 135, 56–65. [DOI] [PubMed] [Google Scholar]
- Belke TW and Garland T Jr.. (2007). A brief opportunity to run does not function as a reinforcer for mice selected for high daily wheel-running rates. J. Exp. Anal. Behav. 88, 199–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercik P, Denou E, Collins J, Jackson W, Lu J, Jury J, Deng Y, Blennerhassett P, Macri J, McCoy KD, et al. (2011). The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology 141, 599–609.e3. [DOI] [PubMed] [Google Scholar]
- Bindels LB and Delzenne NM (2013). Muscle wasting: The gut microbiota as a new therapeutic target? Int. J. Biochem. Cell Biol. 45, 2186–2190. [DOI] [PubMed] [Google Scholar]
- Biro PA, Garland T Jr., Beckmann C, Ujvari B, Thomas and Post JR (2018). Metabolic scope as a proximate constraint on individual behavioral variation: effects on personality, plasticity, and predictability. Am. Nat. 192, 142–154. [DOI] [PubMed] [Google Scholar]
- Blaser MJ (2016). Antibiotic use and its consequences for the normal microbiome. Science 352, 544–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronikowski AM, Morgan TJ, Garland T Jr. and Carter PA (2006). The evolution of aging and age-related physical decline in mice selectively bred for high voluntary exercise. Evol. Int. J. Org. Evol. 60, 1494–1508. [PubMed] [Google Scholar]
- Cadney MD, Schwartz NE, McNamara MP, Schmill MP, Castro AA, Hillis DA and Garland T Jr. (2021a). Cross-fostering selectively bred High Runner mice affects adult body mass but not voluntary exercise. Physiol. Behav. In revision. [DOI] [PubMed] [Google Scholar]
- Cadney MD, Hiramatsu L, Thompson Z, Zhao M, Kay JC, Singleton JM, Albuquerque R. L. de, Schmill MP, Saltzman W and Garland T Jr. (2021b). Effects of early-life exposure to Western diet and voluntary exercise on adult activity levels, exercise physiology, and associated traits in selectively bred High Runner mice. Physiol. Behav. 234, 113389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell SC and Wisniewski PJ (2017). Exercise is a novel promoter of intestinal health and microbial diversity. Exerc. Sport Sci. Rev. 45, 41–47. [DOI] [PubMed] [Google Scholar]
- Careau V, Bininda-Emonds ORP, Ordonez G and Garland T Jr. (2012). Are voluntary wheel running and open-field behavior correlated in mice? Different answers from comparative and artificial selection approaches. Behav. Genet. 42, 830–844. [DOI] [PubMed] [Google Scholar]
- Careau V, Wolak ME, Carter PA and Garland T Jr. (2013). Limits to behavioral evolution: the quantitative genetics of a complex trait under directional selection: quantitative genetics of a selection limit. Evolution 67, 3102–3119. [DOI] [PubMed] [Google Scholar]
- Carmody RN, Gerber GK, Luevano JM, Gatti DM, Somes L, Svenson KL and Turnbaugh PJ (2015). Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 17, 72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho BM, Guadagnini D, Tsukumo DML, Schenka AA, Latuf-Filho P, Vassallo J, Dias JC, Kubota LT, Carvalheira JBC and Saad MJA (2012). Modulation of gut microbiota by antibiotics improves insulin signalling in high-fat fed mice. Diabetologia 55, 2823–2834. [DOI] [PubMed] [Google Scholar]
- Castro-Mejía JL, Jakesevic M, Fabricius NF, Krych Ł, Nielsen DS, Kot W, Bendtsen KM, Vogensen FK, Hansen CHF and Hansen AK (2018). Gut microbiota recovery and immune response in ampicillin-treated mice. Res. Vet. Sci. 118, 357–364. [DOI] [PubMed] [Google Scholar]
- Ceylani T, Jakubowska-Doğru E, Gurbanov R, Teker HT and Gozen AG (2018). The effects of repeated antibiotic administration to juvenile BALB/c mice on the microbiota status and animal behavior at the adult age. Heliyon 4, e00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claghorn GC, Thompson Z, Wi K, Van L and Garland T Jr. (2017). Caffeine stimulates voluntary wheel running in mice without increasing aerobic capacity. Physiol. Behav. 170, 133–140. [DOI] [PubMed] [Google Scholar]
- Clarke G, Stilling RM, Kennedy PJ, Stanton C, Cryan JF and Dinan TG (2014). Minireview: gut microbiota: the neglected endocrine organ. Mol. Endocrinol. 28, 1221–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copes LE, Schutz H, Dlugosz EM, Acosta W, Chappell MA and Garland T Jr. (2015). Effects of voluntary exercise on spontaneous physical activity and food consumption in mice: Results from an artificial selection experiment. Physiol. Behav. 149, 86–94. [DOI] [PubMed] [Google Scholar]
- Crowson MM and McClave SA (2020). Does the intestinal microbiome impact athletic performance? Curr. Gastroenterol. Rep. 22, 53. [DOI] [PubMed] [Google Scholar]
- Cryan JF and Dinan TG (2014). Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 15, 701–712. [DOI] [PubMed] [Google Scholar]
- Diaz Heijtz R, Wang S, Anuar F, Qian Y, Björkholm B, Samuelsson A, Hibberd ML, Forssberg H and Pettersson S (2011). Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. U. S. A. 108, 3047–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Godoy-Vitorino F, Knight R and Blaser MJ (2019). Role of the microbiome in human development. Gut 68, 1108–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs CJ, Schutz H, Meek TH, Dlugosz EM, Acosta W, de Wolski KS, Malisch JL, Hayes JP and Garland T Jr. (2012). Within-lifetime trade-offs but evolutionary freedom for hormonal and immunological traits: evidence from mice bred for high voluntary exercise. J. Exp. Biol. 215, 1651–1661. [DOI] [PubMed] [Google Scholar]
- Dumke CL, Rhodes JS, Garland T Jr., Maslowski E, Swallow JG, Wetter AC and Cartee GD (2001). Genetic selection of mice for high voluntary wheel running: effect on skeletal muscle glucose uptake. J. Appl. Physiol. 91, 1289–1297. [DOI] [PubMed] [Google Scholar]
- Eisenmann JC, Wickel EE, Kelly SA, Middleton KM and Garland T Jr. (2009). Day-to-day variability in voluntary wheel running among genetically differentiated lines of mice that vary in activity level. Eur. J. Appl. Physiol. 106, 613–619. [DOI] [PubMed] [Google Scholar]
- Eshleman EM and Alenghat T (2021). Epithelial sensing of microbiota-derived signals. Genes Immun. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine SS and Kohl KD (2020). Optimal integration between host physiology and functions of the gut microbiome. Philos. Trans. R. Soc. B Biol. Sci. 375, 20190594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fülling C, Dinan TG and Cryan JF (2019). Gut microbe to brain signaling: what happens in vagus…. Neuron 101, 998–1002. [DOI] [PubMed] [Google Scholar]
- Gao K, Pi Y, Mu C-L, Peng Y, Huang Z and Zhu W-Y (2018). Antibiotics-induced modulation of large intestinal microbiota altered aromatic amino acid profile and expression of neurotransmitters in the hypothalamus of piglets. J. Neurochem. 146, 219–234. [DOI] [PubMed] [Google Scholar]
- García-Cabrerizo R, Carbia C, Riordan KJ, Schellekens H and Cryan JF (2021). Microbiota-gut-brain axis as a regulator of reward processes. J. Neurochem. 157, 1495–1524. [DOI] [PubMed] [Google Scholar]
- Garland T Jr., Schutz H, Chappell MA, Keeney BK, Meek TH, Copes LE, Acosta W, Drenowatz C, Maciel RC, Dijk G. van, et al. (2011). The biological control of voluntary exercise, spontaneous physical activity and daily energy expenditure in relation to obesity: human and rodent perspectives. J. Exp. Biol. 214, 206–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland T Jr., Zhao M and Saltzman W (2016). Hormones and the evolution of complex traits: insights from artificial selection on behavior. Integr. Comp. Biol. 56, 207–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV and Knight R (2018). Current understanding of the human microbiome. Nat. Med. 24, 392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard I, McAleer MW, Rhodes JS and Garland T Jr. (2001). Selection for high voluntary wheel-running increases speed and intermittency in house mice (Mus domesticus). J. Exp. Biol. 204, 4311–4320. [DOI] [PubMed] [Google Scholar]
- Good DJ, Li M and Deater-Deckard K (2015). A genetic basis for motivated exercise. Exerc. Sport Sci. Rev. 43, 231–237. [DOI] [PubMed] [Google Scholar]
- Grosicki GJ, Fielding RA and Lustgarten MS (2018). Gut microbiota contribute to age-related changes in skeletal muscle size, composition, and function: biological basis for a gut-muscle axis. Calcif. Tissue Int. 102, 433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley JA, Hargreaves M, Joyner MJ and Zierath JR (2014). Integrative biology of exercise. Cell 159, 738–749. [DOI] [PubMed] [Google Scholar]
- Heinonen I, Kalliokoski KK, Hannukainen JC, Duncker DJ, Nuutila P and Knuuti J (2014). Organ-specific physiological responses to acute physical exercise and long-term training in humans. Physiology 29, 421–436. [DOI] [PubMed] [Google Scholar]
- Hiramatsu L and Garland T Jr. (2018). Mice selectively bred for high voluntary wheel-running behavior conserve more fat despite increased exercise. Physiol. Behav. 194, 1–8. [DOI] [PubMed] [Google Scholar]
- Hiramatsu L, Kay J, Thompson Z, Singleton J, Claghorn G, de Albuquerque RL, Ho B, Ho B, Sanchez G and Garland T Jr. (2017). Maternal exposure to Western diet affects adult body composition and voluntary wheel running in a genotype-specific manner in mice. Physiol. Behav. 179, 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes RL (2020). A review of the role of the gut microbiome in personalized sports nutrition. Front. Nutr. 6, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS and Iwasaki A (2011). Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. U. S. A. 108, 5354–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaromin E, Sadowska ET and Koteja P (2019). The effect of monoamines reuptake inhibitors on aerobic exercise performance in bank voles from a selection experiment. Curr. Zool. 65, 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappel S, Hawkins P and Mendl M (2017). To group or not to group? Good practice for housing male laboratory mice. Animals 7, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay J,C (2017). Inherited and phenotypically plastic characteristics of cardiac and skeletal muscle physiology in mice selectively bred for high voluntary wheel running. [Google Scholar]
- Kay JC, Claghorn GC, Thompson Z, Hampton TG and Garland T Jr. (2019). Electrocardiograms of mice selectively bred for high levels of voluntary exercise: Effects of short-term exercise training and the mini-muscle phenotype. Physiol. Behav. 199, 322–332. [DOI] [PubMed] [Google Scholar]
- Keeney BK, Meek TH, Middleton KM, Holness LF and Garland T Jr. (2012). Sex differences in cannabinoid receptor-1 (CB1) pharmacology in mice selectively bred for high voluntary wheel-running behavior. Pharmacol. Biochem. Behav. 101, 528–537. [DOI] [PubMed] [Google Scholar]
- Kelly SA, Nehrenberg DL, Hua K, Garland T Jr. and Pomp D (2014). Quantitative genomics of voluntary exercise in mice: transcriptional analysis and mapping of expression QTL in muscle. Physiol. Genomics 46, 593–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SA, Gomes FR, Kolb EM, Malisch JL and Garland T Jr. (2017). Effects of activity, genetic selection and their interaction on muscle metabolic capacities and organ masses in mice. J. Exp. Biol. 220, 1038–1047. [DOI] [PubMed] [Google Scholar]
- Knoop KA, McDonald KG, Kulkarni DH and Newberry RD (2016). Antibiotics promote inflammation through the translocation of native commensal colonic bacteria. Gut 65, 1100–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl KD and Carey HV (2016). A place for host-microbe symbiosis in the comparative physiologist’s toolbox. J. Exp. Biol. 219, 3496–3504. [DOI] [PubMed] [Google Scholar]
- Koskella B and Bergelson J (2020). The study of host–microbiome (co)evolution across levels of selection. Philos. Trans. R. Soc. B Biol. Sci. 375, 20190604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koteja P, Carter PA, Swallow JG and Garland T Jr. (2003). Food wasting by house mice: variation among individuals, families, and genetic lines. Physiol. Behav. 80, 375–383. [DOI] [PubMed] [Google Scholar]
- Krautkramer KA, Fan J and Bäckhed F (2021). Gut microbial metabolites as multi-kingdom intermediates. Nat. Rev. Microbiol. 19, 77–94. [DOI] [PubMed] [Google Scholar]
- Lagkouvardos I, Overmann J and Clavel T (2017). Cultured microbes represent a substantial fraction of the human and mouse gut microbiota. Gut Microbes 8, 493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakens D (2013). Calculating and reporting effect sizes to facilitate cumulative science: a practical primer for t-tests and ANOVAs. Front. Psychol. 4,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubitz D, Typpo K, Midura-Kiela M, Brown C, Barberán A, Ghishan FK and Kiela PR (2021). Dynamics of gut microbiota recovery after antibiotic exposure in young and old mice (a pilot study). Microorganisms 9, 647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laukens D, Brinkman BM, Raes J, De Vos M and Vandenabeele P (2016). Heterogeneity of the gut microbiome in mice: guidelines for optimizing experimental design. FEMS Microbiol. Rev. 40, 117–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq S, Mian FM, Stanisz AM, Bindels LB, Cambier E, Ben-Amram H, Koren O, Forsythe P and Bienenstock J (2017). Low-dose penicillin in early life induces long-term changes in murine gut microbiota, brain cytokines and behavior. Nat. Commun. 8, 15062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R and Gordon JI (2008). Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 6, 776–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot JT, De Geus EJC, Booth FW, Bray MS, Den Hoed M, Kaprio J, Kelly SA, Pomp D, Saul MC, Thomis MA, et al. (2018). Biological/genetic regulation of physical activity level: consensus from genbiopac. Med. Sci. Sports Exerc. 50, 863–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Zeng B, Zeng L, Du X, Li B, Huo R, Liu L, Wang H, Dong M, Pan J, et al. (2018). Gut microbiota regulates mouse behaviors through glucocorticoid receptor pathway genes in the hippocampus. Transl. Psychiatry 8,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach N and Fuster-Botella D (2017). Endurance exercise and gut microbiota: A review. J. Sport Health Sci. 6, 179–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailing LJ, Allen JM, Buford TW, Fields CJ and Woods JA (2019). Exercise and the gut microbiome: a review of the evidence, potential mechanisms, and implications for human health. Exerc. Sport Sci. Rev. 47, 75–85. [DOI] [PubMed] [Google Scholar]
- Malisch JL, Breuner CW, Kolb EM, Wada H, Hannon RM, Chappell MA, Middleton KM and Garland T Jr. (2009). Behavioral despair and home-cage activity in mice with chronically elevated baseline corticosterone concentrations. Behav. Genet. 39, 192–201. [DOI] [PubMed] [Google Scholar]
- Marin IA, Goertz JE, Ren T, Rich SS, Onengut-Gumuscu S, Farber E, Wu M, Overall CC, Kipnis J and Gaultier A (2017). Microbiota alteration is associated with the development of stress-induced despair behavior. Sci. Rep. 7, 43859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. 110, 3229–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara MP, Singleton JM, Cadney MD, Ruegger PM, Borneman J and Garland T Jr. (2021). Early-life effects of juvenile Western diet and exercise on adult gut microbiome composition in mice. J. Exp. Biol. 224, jeb239699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek TH, Lonquich BP, Hannon RM and Garland T Jr. (2009). Endurance capacity of mice selectively bred for high voluntary wheel running. J. Exp. Biol. 212, 2908–2917. [DOI] [PubMed] [Google Scholar]
- Meek TH, Dlugosz EM, Vu KT and Garland T Jr. (2012). Effects of leptin treatment and Western diet on wheel running in selectively bred high runner mice. Physiol. Behav. 106, 252–258. [DOI] [PubMed] [Google Scholar]
- Milani C, Alessandri G, Mancabelli L, Mangifesta M, Lugli GA, Viappiani A, Longhi G, Anzalone R, Duranti S, Turroni F, et al. (2020). Multi-omics approaches to decipher the impact of diet and host physiology on the mammalian gut microbiome. Appl. Environ. Microbiol. 86, e01864–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr AE, Jäger R, Carpenter KC, Kerksick CM, Purpura M, Townsend JR, West NP, Black K, Gleeson M, Pyne DB, et al. (2020). The athletic gut microbiota. J. Int. Soc. Sports Nutr. 17, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nay K, Jollet M, Goustard B, Baati N, Vernus B, Pontones M, Lefeuvre-Orfila L, Bendavid C, Rué O, Mariadassou M, et al. (2019). Gut bacteria are critical for optimal muscle function: a potential link with glucose homeostasis. Am. J. Physiol. Endocrinol. Metab. 317, E158–E171. [DOI] [PubMed] [Google Scholar]
- Ng KM, Aranda-Díaz A, Tropini C, Frankel MR, Van Treuren W, O’Laughlin CT, Merrill BD, Yu FB, Pruss KM, Oliveira RA, et al. (2019). Recovery of the gut microbiota after antibiotics depends on host diet, community context, and environmental reservoirs. Cell Host Microbe 26, 650–665.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak CM, Burghardt PR and Levine JA (2012). The use of a running wheel to measure activity in rodents: Relationship to energy balance, general activity, and reward. Neurosci. Biobehav. Rev. 36, 1001–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T, Morino K, Ugi S, Nakagawa F, Lemecha M, Ida S, Ohashi N, Sato D, Fujita Y and Maegawa H (2019). Microbiome potentiates endurance exercise through intestinal acetate production. Am. J. Physiol.-Endocrinol. Metab. 316, E956–E966. [DOI] [PubMed] [Google Scholar]
- Org E, Parks BW, Joo JWJ, Emert B, Schwartzman W, Kang EY, Mehrabian M, Pan C, Knight R, Gunsalus R, et al. (2015). Genetic and environmental control of host-gut microbiota interactions. Genome Res. 25, 1558–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyanagi E, Uchida M, Kremenik MJ and Yano H (2018). Altered gut microbiota by voluntary exercise induces high physical activity in high-fat diet mice. J. Phys. Fit. Sports Med. 7, 81–85. [Google Scholar]
- Przewłócka K, Folwarski M, Kaźmierczak-Siedlecka K, Skonieczna-Żydecka K and Kaczor JJ (2020). Gut-muscle axis exists and may affect skeletal muscle adaptation to training. Nutrients 12, 1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S and Medzhitov R (2004). Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241. [DOI] [PubMed] [Google Scholar]
- Reikvam DH, Erofeev A, Sandvik A, Grcic V, Jahnsen FL, Gaustad P, McCoy KD, Macpherson AJ, Meza-Zepeda LA and Johansen F-E (2011). Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PLoS ONE 6,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezende EL, Chappell MA, Gomes FR, Malisch JL and Garland T Jr. (2005). Maximal metabolic rates during voluntary exercise, forced exercise, and cold exposure in house mice selectively bred for high wheel-running. J. Exp. Biol. 208, 2447–2458. [DOI] [PubMed] [Google Scholar]
- Rhodes JS and Garland T Jr. (2003). Differential sensitivity to acute administration of Ritalin, apormorphine, SCH 23390, but not raclopride in mice selectively bred for hyperactive wheel-running behavior. Psychopharmacology (Berl.) 167, 242–250. [DOI] [PubMed] [Google Scholar]
- Rhodes JS, Gammie SC and Garland T Jr. (2005). Neurobiology of mice selected for high voluntary wheel-running activity. Integr. Comp. Biol. 45, 438–455. [DOI] [PubMed] [Google Scholar]
- Rosa CP, Brancaglion GA, Miyauchi-Tavares TM, Corsetti PP and de Almeida LA (2018). Antibiotic-induced dysbiosis effects on the murine gastrointestinal tract and their systemic repercussions. Life Sci. 207, 480–491. [DOI] [PubMed] [Google Scholar]
- Rosenthal R (1994). Parametric measures of effect size. In The handbook of research synthesis, pp. 231–244. New York, NY, US: Russell Sage Foundation. [Google Scholar]
- Rowland I, Gibson G, Heinken A, Scott K, Swann J, Thiele I and Tuohy K (2018). Gut microbiota functions: metabolism of nutrients and other food components. Eur. J. Nutr. 57, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul MC, Majdak P, Perez S, Reilly M, Garland T Jr. and Rhodes JS (2017). High motivation for exercise is associated with altered chromatin regulators of monoamine receptor gene expression in the striatum of selectively bred mice. Genes Brain Behav. 16, 328–341. [DOI] [PubMed] [Google Scholar]
- Sherwin CM (1998). Voluntary wheel running: a review and novel interpretation. Anim. Behav. 56, 11–27. [DOI] [PubMed] [Google Scholar]
- Silverman MN and Deuster PA (2014). Biological mechanisms underlying the role of physical fitness in health and resilience. Interface Focus 4, 20140040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton JM and Garland T Jr. (2019). Influence of corticosterone on growth, home-cage activity, wheel running, and aerobic capacity in house mice selectively bred for high voluntary wheel-running behavior. Physiol. Behav. 198, 27–41. [DOI] [PubMed] [Google Scholar]
- Sinha Ray A, Haikal A, Hammoud KA and Yu ASL (2016). Vancomycin and the risk of aki: a systematic review and meta-analysis. Clin. J. Am. Soc. Nephrol. CJASN 11, 2132–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu X-N, Kubo C and Koga Y (2004). Postnatal microbial colonization programs the hypothalamic–pituitary–adrenal system for stress response in mice. J. Physiol. 558, 263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan GM and Feinn R (2012). Using effect size—or why the p value is not enough. J. Grad. Med. Educ. 4, 279–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swallow JG, Carter PA and Garland T Jr. (1998). Artificial selection for increased wheel-running behavior in house mice. Behav. Genet. 28, 227–237. [DOI] [PubMed] [Google Scholar]
- Swallow JG, Koteja P, Carter PA and Garland T Jr. (1999). Artificial selection for increased wheel-running activity in house mice results in decreased body mass at maturity. J. Exp. Biol. 202, 2513–2520. [DOI] [PubMed] [Google Scholar]
- Swallow JG, Hayes JP, Koteja P and Garland T Jr. (2009). Selection experiments and experimental evolution of performance and physiology. Exp. Evol. Concepts Methods Appl. Sel. Exp. 301–351. [Google Scholar]
- Thompson Z, Kolb EM and Garland T Jr. (2018). High-runner mice have reduced incentive salience for a sweet-taste reward when housed with wheel access. Behav. Processes 146, 46–53. [DOI] [PubMed] [Google Scholar]
- Ticinesi A, Lauretani F, Milani C, Nouvenne A, Tana C, Del Rio D, Maggio M, Ventura M and Meschi T (2017). Aging gut microbiota at the cross-road between nutrition, physical frailty, and sarcopenia: is there a gut–muscle axis? Nutrients 9, 1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticinesi A, Lauretani F, Tana C, Nouvenne A, Ridolo E and Meschi T (2019). Exercise and immune system as modulators of intestinal microbiome: implications for the gut-muscle axis hypothesis. 25, 84–95. [PubMed] [Google Scholar]
- Tochitani S, Ikeno T, Ito T, Sakurai A, Yamauchi T and Matsuzaki H (2016). Administration of non-absorbable antibiotics to pregnant mice to perturb the maternal gut microbiota is associated with alterations in offspring behavior. PLOS ONE 11, e0138293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Zhong H, Xu C and Xu G (2019). Spotlights on antibiotic-induced acute kidney injury: the evidence to date. Iran. J. Kidney Dis. 13, 10–20. [PubMed] [Google Scholar]
- Zaneveld J, Turnbaugh PJ, Lozupone C, Ley RE, Hamady M, Gordon JI and Knight R (2008). Host-bacterial coevolution and the search for new drug targets. Curr. Opin. Chem. Biol. 12, 109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragozá R (2020). Transport of amino acids across the blood-brain barrier. Front. Physiol. 11, 973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinpar A, Chaix A, Xu ZZ, Chang MW, Marotz CA, Saghatelian A, Knight R and Panda S (2018). Antibiotic-induced microbiome depletion alters metabolic homeostasis by affecting gut signaling and colonic metabolism., Antibiotic-induced microbiome depletion alters metabolic homeostasis by affecting gut signaling and colonic metabolism. Nat. Commun. Nat. Commun. 9, 2872–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zombeck JA, DeYoung EK, Brzezinska WJ and Rhodes JS (2011). Selective breeding for increased home cage physical activity in collaborative cross and hsd:icr mice. Behav. Genet. 41, 571–582. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Home-cage activity before (averages for days 11–13) and during (averages for days 22–24) antibiotics (ABX). Note that all mice had wheel access during the entire period. Due to equipment failures, data were not available for the period of recovery from antibiotics. Data are presented as least squares means ± standard errors from repeated-measures analyses of individual daily values in SAS Procedure Mixed. Pearson’s r is a measure of effect size (see Methods). A. Although the interaction did not reach statistical significance, C mice on antibiotics tended to have decreased total cage activity during antibiotic treatment (differences of least squares means p=0.0576 for C mice and p=0.4914 for HR mice). B. Although the interaction did not reach statistical significance, antibiotics reduced activity duration in C mice but not in HR mice (differences of least squares means p=0.0063 and p=0.5649, respectively). C. Antibiotics tended to increase mean intensity of activity (total/minutes) in HR mice, but not in C mice (differences of least squares means p=0.7159 for C mice and p=0.1166 for HR mice).
Supplemental Figure 2. Wheel running mean revolutions per day (23 hours) for each of the 4 HR and 4 C lines. Data were truncated on days 8, 14, and 25 (~20 hours) when the water bottles were being changed. Boxes indicate days that were used in statistical analyses. A. N=99 mice, through day 24 of the experiment. Days 11–13 before antibiotics were compared with days 22–24 during antibiotics. B. N=76 mice, excluding a subset that were removed on day 28 for a separate experiment. The microbiome was allowed to naturally recover after antibiotic treatment (days 25–36). Days 34–36 were used to indicate wheel running in recovery.


