Abstract
In the lipopolysaccharides of Escherichia coli there are five distinct core oligosaccharide (core OS) structures, designated K-12 and R1 to R4. The objective of this work was to determine the prevalences of these core OS types within the species. Unique sequences in the waa (core OS biosynthesis) gene operon were used to develop a PCR-based system that facilitated unequivocal determination of the core OS types in isolates of E. coli. This system was applied to the 72 isolates in the E. coli ECOR collection, a compilation of isolates that is considered to be broadly representative of the genetic diversity of the species. Fifty (69.4%) of the ECOR isolates contained the R1 core OS, 8 (11.1%) were representatives of R2, 8 (11.1%) were R3, 2 (2.8%) were R4, and only 4 (5.6%) were K-12. R1 is the only core OS type found in all four major phylogenetic groups (A, B1, B2, and D) in the ECOR collection. Virulent extraintestinal pathogenic E. coli isolates tend to be closely related to group B2 and, to a lesser extent, group D isolates. All of the ECOR representatives from the B2 and D groups had the R1 core OS. In contrast, commensal E. coli isolates are more closely related to group A, which contains isolates representing each of the five core OS structures. R3 was the only core OS type found in 38 verotoxigenic E. coli (VTEC) isolates from humans and cattle belonging to the common enterohemorrhagic E. coli serogroups O157, O111, and O26. Although isolates from other VTEC serogroups showed more core OS diversity, the R3 type (83.1% of all VTEC isolates) was still predominant. When non-VTEC commensal isolates from cattle were analyzed, it was found that most possessed the R1 core OS type.
The lipopolysaccharides (LPSs) of Escherichia coli consist of (i) a hydrophobic lipid A component that forms the outer leaflet of the outer membrane, (ii) a phosphorylated, nonrepetitive hetero-oligosaccharide known as the core oligosaccharide (core OS), and (iii) a polysaccharide (O-PS) that extends from the cell surface and that forms the O antigen detected in serotyping (37). The smooth LPS (S-LPS) molecules found in most clinical isolates of E. coli are composed of this three-part structure, whereas rough LPS (R-LPS) lacks the O antigen and can have a truncated core OS. The extent of structural diversity in E. coli LPS molecules ranges from the highly conserved lipid A to the extreme variations reflected in more than 170 known O antigens (19). The core OS is conceptually divided into inner and outer core regions. The inner core is composed primarily of l-glycero-d-manno-heptose (heptose) and 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo) residues, and this part of the core OS is phosphorylated in E. coli. The principal features of the inner core OS structure are conserved among members of the Enterobacteriaceae, presumably reflecting its essential role in outer-membrane stability (16). The inner core OS structure carries additional (often nonstoichiometric) glycosyl substituents, but these vary according to core OS type (16, 18). The outer region of the core OS displays more variation, and differences in this region distinguish the five core OS types in E. coli: R1, R2, R3, R4, and K-12. While all of the outer core OSs share a structural theme, with a (hexose)3 carbohydrate backbone and two side chain residues, the order of hexoses in the backbone and the nature, position, and linkage of the side chain residues can all vary (Fig. 1). The structures for the R1 and R4 outer core OSs are highly similar, differing in only a single β-linked residue. Members of the genus Shigella are often proposed to be members of E. coli, and the R1 core OS has been found in the LPSs of Shigella sonnei and in some isolates of Shigella flexneri (9, 22, 25). The terminal part of the R2 core OS resembles the corresponding region from Salmonella enterica serovar Typhimurium in both structure and function, whereas the outer core backbone and substitution of GlcI are identical to corresponding features of the K-12 core structure (14). There are also structural similarities between the core OS terminus in the R3 type and that in a recently reported second core type in the genus Salmonella (31).
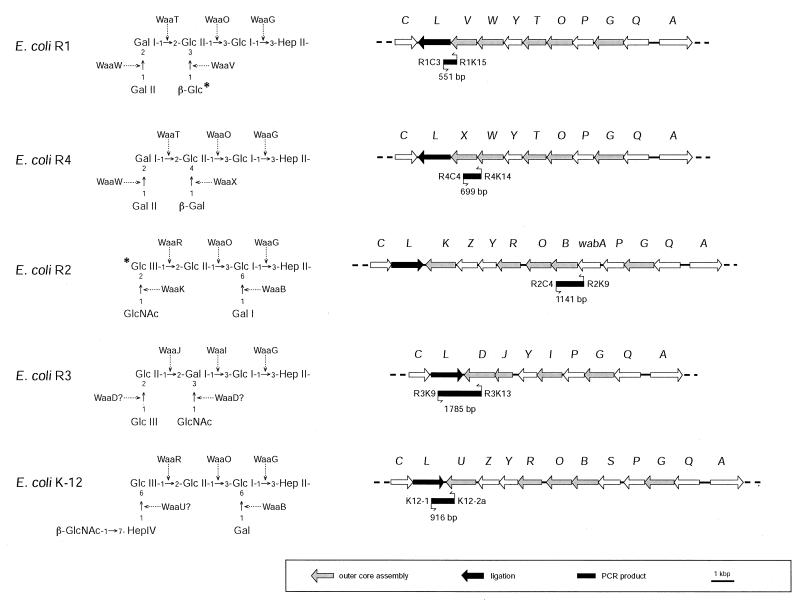
FIG. 1.
Structures of the five known outer core OSs from the LPSs of E. coli and genetic organization of the central waaQ operon from each of the waa (core OS biosynthesis) loci. HepII is the last residue of the inner core OS. The asterisks indicate the points of attachment of O antigen, but this has only been determined experimentally for the R1 and R2 core OSs. The gene products responsible for each residue of the core OS are indicated. Details can be found elsewhere (16). PCR products diagnostic for each core OS type are shown below the relevant physical maps. The primer sequences are given in Table 1.
Comparative sequence analyses have been performed for the chromosomal waa (formerly rfa) loci responsible for core OS biosynthesis in isolates representing the five known E. coli core OS types (reviewed in reference 16). These data, together with structural determinations of core OSs from defined mutants, have helped resolve the functions of many of the outer core OS biosynthesis gene products and specifically those involved in the determination of the core OS type. Core OS distributions in commensal E. coli (fecal) isolates and in isolates from septicemia and urinary tract infections (UTIs) have been examined by serological methods (2, 10). Although the R1 core OS type predominates in these analyses, it is not clear that these results are reflective of the broader species, and the objective of our study was to address this issue. Using characteristic waa sequences unique to each core OS type, we developed a PCR-based LPS core typing system for E. coli isolates. The application of this system is described here.
(This study was presented in part at the 5th meeting of the International Endotoxin Society, Santa Fe, N. Mex., September 1998 [abstr. 128], and at the 100th annual meeting of the American Society for Microbiology, Chicago, Ill., June 1999 [abstr. B/D41]).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The E. coli core OS prototypes used in this study were (i) strain F470 (R1 core), an R-LPS derivative of O8:K27 (41); (ii) F632 (R2), an R-LPS derivative of O100:K?(B):H2 (13); (iii) F653 (R3), an R-LPS derivative of O111:K58− (47); (iv) F2513 (R4), an R-LPS derivative of O14:K7 (40); and (v) W3110 (CGSC4466), a representative isolate of E. coli K-12. H. Brade (Forschungszentrum, Borstel, Germany) generously provided the R1, R2, R3, and R4 strains. Strain 1502 is a prototype of the putative “R1R4 mixed core type” (2) (see below) and was kindly provided by B. J. Appelmelk. The following isolates belonging to the O16 serogroup were obtained from F. Scheutz (Statens Seruminstitut, Copenhagen, Denmark): C462-74 (O16:K1:H48), C233-87 (O16:K2:H6), C523-76 (O16:K38:H20), C301-86 (O16:K100), C1812-80 (O16:K1:H4), C945-96 (O16:K1:H−), and C451-93 (O16:K1:H6). The 72 E. coli isolates that constitute the ECOR collection were generously provided by H. Ochman (University of Rochester). To examine the relationship between core OS type and verotoxigenic (VT-positive) phenotype, 85 E. coli isolates from the collection held at the Health Canada Guelph Laboratory were investigated. Fifty-six were human and animal verotoxigenic E. coli (VTEC) isolates representing the predominant enterohemorrhagic E. coli (EHEC) serogroups O26, O111, and O157 (23). To examine the broader relationships, several VTEC isolates from five other serogroups associated with bloody diarrhea (i.e., O55, O91, O103, O113, and O145 [11, 20, 35]) and from serotypes rarely (or never) identified among human VTEC were examined (1, 11). To determine whether the core OS distribution differed among VTEC isolates and VT-negative commensal E. coli isolates of the same serotypes, 20 isolates from cattle or meats were tested. These included 11 isolates of E. coli O157 of various H types and 9 isolates belonging to other known VTEC serogroups. The VT status of the field isolates and the presence of genes encoding the EHEC hemolysin and intimin in non-VTEC isolates were established by published PCR-based protocols (33, 38). The serotypes of E. coli isolates were established by standard serological methods (8) at the Health Canada Guelph Laboratory. Bacteria were grown on Luria-Bertani agar incubated at 37°C.
PCR amplification of core type-specific fragments from the chromosomal waa loci.
Oligonucleotide primers for sequencing and for PCR amplification of DNA products characteristic of R1, R2, R3, R4, and K-12 core OS types were designed from the nucleotide sequences of genes in the chromosomal waa region from strains F470, F632, F653, and F2513 and from E. coli K-12, respectively (Table 1). Primers were synthesized using a Perkin-Elmer 394 DNA synthesizer at the Guelph Molecular Supercentre (University of Guelph). A mixture of all 10 primers was used in each PCR amplification. Chromosomal DNA PCR templates were purified from fresh overnight bacterial cultures by using InstaGene matrix (Bio-Rad). Amplification reactions were performed by using a Perkin-Elmer Gene Amp PCR system 2400 thermocycler. PCR conditions included an initial denaturation step at 94°C for 1 min; each cycle after this step had a denaturing step at 94°C for 20 s, an annealing step at 50°C for 30 s, and a polymerization step at 72°C for 2 min 15 s. After 35 amplification cycles, a final polymerization step was performed at 72°C for 2 min. Amplification was carried out using Taq polymerase from Roche Diagnostics. PCR amplification products were separated by electrophoresis on 0.8% agarose gels and visualized with ethidium bromide. PCR amplification products were prepared for sequencing by using QIAquick PCR purification columns (Qiagen). Automated sequencing of PCR products was carried out at the Guelph Molecular Supercentre using a Perkin-Elmer ABI 377 DNA sequencing system. DNA sequencing was used to confirm the identities of the fragments from core OS prototype strains and from random isolates of previously undetermined core types. Random strains were also examined by hybridization using waa-specific gene probes to ensure the validity of the PCR data.
TABLE 1.
Oligonucleotide primers used in this study
| Primer | Sequencea (5′–3′) | GenBank accession no.b |
|---|---|---|
| R1C3 | GGGATGCGAACAGAATTAGT | AF019746 |
| R1K15 | TTCCTGGCAAGAGAGATAAG | AF019746 |
| R2C4 | GATCGACGCCGGAATTTTTT | AF019375 |
| R2K9 | AGCTCCATCATCAAGTGAGA | AF019375 |
| R3C2 | GGCCAAAACACTATCTCTCA | AF019745 |
| R3K13 | GTGCCTAGTTTATACTTGAA | AF019745 |
| R4C4 | TGCCATACTTTATTCATCA | AF019747 |
| R4K14 | TGGAATGATGTGGCGTTTAT | AF019747 |
| K12-1 | TTCGCCATTTCGTGCTACTT | X62530, M80599, M86935, AE000440, U00096, M86305, U00039, M95398 |
| K12-2a | TAATGATAATTGGAATGCTGC | X62530, M80599, M86935, AE000440, U00096, M86305, U00039, M95398 |
The locations of the primer sequences and the amplified fragments are shown in Fig. 1.
GenBank accession numbers are for the complete waa loci.
RESULTS
Diagnostic PCR for determination of the known E. coli LPS core types.
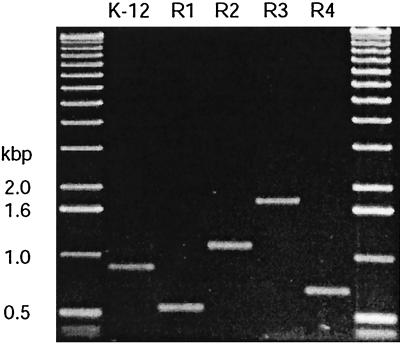
Each of the E. coli core OSs shows unique structural features in its outer core regions, and the genes determining these features have been established (Fig. 1) (reviewed in reference 16). These analyses provided unique DNA sequences that could be exploited for core OS diagnostics. The R1 and R4 core OS structures are very similar, and the majority of the corresponding waa locus sequences are essentially identical (15). However, the structures differ in the nature and linkage of the β-linked hexose residue attached to GlcII, reflecting differences in the activities of the waaV (R1) and waaX (R4) gene products. Although the waaL gene encodes an O-antigen ligase required for addition of O-PS to the completed R1 or R4 core OSs, the nature of the linkage site varies, and this is reflected in the different primary sequences of the waaL gene products (15). The R1 primer set is dependent on the waaV gene encoding the unique glycosyltransferase for the branch β-(1→2)-linked Glc residue that forms the attachment site for O-PS in this core type and sequences in the R1-specific waaL. For the R4 core type, one primer is based on sequences for waaW, a gene encoding the transferase for the terminal α-(1→2)-linked Gal residue that is conserved in core types R1 and R4. Specificity of the R4 primer set is conferred by using a second primer based on sequences from waaX, the gene encoding the unique β-(1→4)-linked Gal transferase, which generates the only structural difference between the outer core OSs of types R1 and R4 (15). Detection of the R2 core type uses one primer based on sequences in the gene encoding the transferase for the α-(1→6)-linked side chain Gal residue (waaB). The second primer site is located within wabA (formerly called waaS). The role of the wabA gene product is not resolved, but it is thought to be involved in an R2 type-specific modification of the inner core region (14, 16). The R3 core type is detected by primers located in the R3-specific waaL gene and a unique gene, waaD, whose precise role in outer core OS biosynthesis is not yet established (16). The primer set for the K-12 core type is based on sequences for the K-12-specific ligase gene (waaL) and the waaU gene that is thought to encode the transferase that adds the α-(1→6)-linked Hep residue in the outer core (14). Oligonucleotide primer pairs were designed such that easily resolved amplification products, with sizes ranging from 550 bp to 1.7 kbp, identify each core OS type (Fig. 2).
FIG. 2.
Agarose gel showing the electrophoretic mobilities of PCR products obtained from the prototype strains representing each core OS type.
Although structural analyses have identified only five distinct core types in the LPSs of E. coli, the existence of an additional core type was proposed following a survey of core OS type distribution using type-specific polyclonal rabbit antisera (2). Appelmelk et al. found that some bacteremic isolates (13 isolates or 9.4% of the tested isolates) reacted with both R1 and R4 sera, suggesting an R1R4 mixed core type (2). DNA from a prototype R1R4 mixed core type (strain 1502) was examined by PCR. Diagnostic fragments for the R1 type were obtained, with no evidence of R4-specific amplification products. To confirm this result, additional R4-specific primer sets were tested, and again no amplification products were obtained (E. Frirdich, K. Amor, and C. Whitfield, unpublished data). Thus, the waa locus from strain 1502 is apparently similar to R1 and lacks the sequences and genetic organization that define the R4 core type. To test the possibility that R4-specific genes might be located at an additional unlinked locus in a standard R1 genetic background, chromosomal DNA was digested with restriction enzymes and examined by Southern hybridization using an internal probe from the waaX gene of R4 (i.e., the gene whose product defines the R4 core OS type). A positive reaction was obtained with DNA from the R4 prototype (strain F2513), but none was detected at either high or low stringency with strain 1502 (Frirdich et al., unpublished data). We conclude that the R1R4 mixed core type isolate is actually an R1 core type isolate.
Distribution of core types in the ECOR collection.
Previous studies involving serological examination of core types have focused on isolates from (i) blood cultures (2); (ii) a collection including isolates from bacteremia or UTIs, together with fecal isolates from asymptomatic individuals (10); and (iii) VTEC isolates (6). However, such isolates would not necessarily provide an unbiased evaluation of core types in natural populations since these isolates come from niches where they would require specific virulence determinants. Furthermore, E. coli is a highly clonal species (50), and the phylogenetic relationships among the various isolates examined with respect to core OS type were not determined.
The 72 isolates of the ECOR collection originate from diverse geographic locations, and they differ in their characteristic patterns in multilocus enzyme electrophoresis (MLEE) analysis of 38 “housekeeping” enzymes (30). The ECOR collection of isolates is considered to be broadly representative of the genetic diversity within the E. coli species, and these isolates are therefore often used as a framework for describing clonal relationships in E. coli. We examined the ECOR collection to obtain a better view of the distribution of core OS types. The O serotype of each isolate in the ECOR collection was determined, and their core OS types were unequivocally assigned by PCR analysis. Fifty of the 72 isolates (69.4%) had the R1 core OS. Of the remainder, 8 (11.1%), 8 (11.1%), 2 (2.8%), and 4 (5.6%) isolates represented core OS types R2, R3, R4, and K-12, respectively. The results are shown in the context of the phylogenetic tree in Fig. 3.
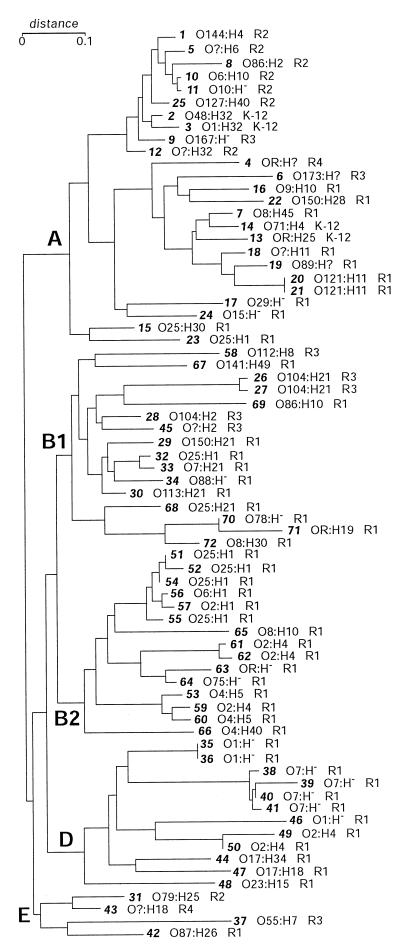
FIG. 3.
Phylogenetic tree of the ECOR isolates (17) showing the distribution of the five core OS types. The number of the ECOR isolate is given in boldface italics. For each isolate the determined O:H serotype and core OS type are listed. OR, R-LPS; ?, antigen not determined. The serotyping results reported here showed some differences to those reported elsewhere (T. S. Whittam, http: //www.bio.psu.edu/People/Faculty/Whittam/Lab). In many cases the discrepancies reflect a nontypeable reaction in one set of data or the other and may reflect subtle differences in growth conditions, protocol, or antisera. In the analysis reported here, there were significantly less nontypeable antigens. In some cases, the serotypes varied between the two studies. To verify the data reported here, two independently held ECOR collections were examined and each of the isolates giving different serotypes was retested.
Four major phylogenetic groups (A, B1, B2, and D) were identified in the ECOR phylogenetic tree by neighbor-joining analysis of MLEE using 38 loci (17) (Fig. 3). Four additional isolates (ECOR31, -43, -37, and -42) form a minor group (sometimes termed group E). Group A isolates (ECOR1 to -25) are thought to represent a distinct lineage comprising E. coli K-12 and isolates related to it (17). All five core OS types occur within group A, and the limited number of ECOR isolates with the K-12 core OS are confined to this group. The minor group (group E) also shows diversity in core OS type, with single isolates representing types R1, R2, R3, and R4. In contrast, the remaining groups show much less core OS diversity. Group B1 isolates contain primarily R1 (11 of 16; 68.8%) and R3 (5 of 16; 31.2%) core OSs. All of the isolates representing groups B2 and D have type R1 core OSs.
Distribution of the K-12 core OS type.
E. coli isolates with the K-12 core type occur relatively infrequently among natural populations of E. coli. The K-12 core OS type is confined to group A and is found in a total of four isolates (ECOR2, -3, -13, and -14) possessing serotypes O48, O1, OR, and O71, respectively. Common laboratory E. coli K-12 derivatives have R-LPS with the K-12 core OS, but analysis of the rfb (O-antigen biosynthesis) locus has shown that the original serotype of the K-12 lineage was O16 (24, 45). No members of the O16 serogroup were identified in the ECOR collection by serotyping (Fig. 3) so, to determine whether there was any correlation between the O16 antigen and the K-12 core OS, we examined several serogroup O16 isolates by PCR. Within this limited number of isolates, only one (serotype O16:K1:H4, strain C1812-80) with the K-12 core OS type was identified; the remaining five strains possessed R1 core OSs. As seen with some serotypes examined in the study by Gibb et al. (10), the same O antigen can occur in strains with different core OS types.
Prevalence of the R3 core type among VTEC isolates.
Many studies on the development of cross-reactive and cross-protective antibodies against E. coli core OSs have utilized LPS from strain J5 as an immunogen. The J5 strain is an R-LPS mutant that contains a truncated R3 core OS structure (12, 28). The parent of strain J5 belonged to serogroup O111, a common VT-positive serogroup of EHEC. To examine a possible correlation between the R3 core and O111 antigen, the core OS type of 15 field isolates of serogroup O111 was investigated. These isolates represent two different serotypes based on H antigens (O111:H8 and O111:H−). All proved to be type R3 (Table 2). To investigate any association between the R3 core OS type and the VT-positive genotype, the analysis was expanded to include further VTEC isolates representing other serogroups. Overall, 54 of 65 (83.1%) VTEC isolates had the R3 core OS type. All 48 VT-positive human and animal isolates representing EHEC serogroups O26, O55, O91, O103, and O157 were core type R3 (Table 2). Furthermore, the R3 core OS was the most frequent among the selected animal VTEC isolates belonging to serotypes rarely associated with human illness (66.7%) (Table 2). Of the four other core OS types, R1 was the most common among the VTEC isolates tested (8 of 65; 12.3%) and R1 predominated in serogroups O113 and O145. The frequencies of core types R2, R4, and K-12 were 0 (0%), 1 (1.5%), and 2 of 65 (3.1%), respectively.
TABLE 2.
Core OS types in VTEC and non-VTEC isolates
| Group | Serotype | No. of isolates tested with core type:
|
|||||
|---|---|---|---|---|---|---|---|
| R1 | R2 | R3 | R4 | K-12 | Total no. | ||
| VTEC associated with human disease (human and animal isolates) | O26:H11 | 5 | 5 | ||||
| O55:H7 | 1 | 1 | |||||
| O91:H21 | 4 | 4 | |||||
| O103:H2 | 4 | 4 | |||||
| O111:H− | 12 | 12 | |||||
| O111:H8 | 3 | 3 | |||||
| O113:H4 | 1 | 1 | |||||
| O113:H21 | 3 | 3 | |||||
| O145:H− | 3 | 1 | 4 | ||||
| O145:H8 | 1 | 1 | |||||
| O157:H7 | 14 | 14 | |||||
| O157:H− | 4 | 4 | |||||
| Total (%) | 7 (12.5) | 0 (0) | 48 (85.7) | 0 (0) | 1 (1.8) | 56 (100) | |
| VTEC not associated with human disease (animal isolates) | O6:H34 | 1 | 1 | ||||
| O40:H8 | 1 | 1 | |||||
| O49:H− | 1 | 1 | |||||
| O74:H52 | 1 | 1 | |||||
| O82:H8 | 1 | 1 | |||||
| O85:H− | 1 | 1 | |||||
| O112:H2 | 1 | 1 | |||||
| O171:H2 | 1 | 1 | |||||
| O172:H− | 1 | 1 | |||||
| Total (%) | 1 (11.1) | 0 (0) | 6 (66.7) | 1 (11.1) | 1 (11.1) | 9 (100) | |
| All VTEC | Total (%) | 8 (12.3) | 0 (0) | 54 (83.1) | 1 (1.5) | 2 (3.1) | 65 (100) |
| Non-VTEC of VTEC serogroups isolated from animals and meats | O2:H5 | 1 | 1 | ||||
| O2:H10 | 1 | 1 | |||||
| O6:H4 | 1 | 1 | |||||
| O8:H21 | 1 | 1 | |||||
| O15:H45 | 1 | 1 | |||||
| O64:H7 | 1 | 1 | |||||
| O112:H8 | 1 | 1 | |||||
| O117:H9 | 1 | 1 | |||||
| O156:H25a | 1 | 1 | |||||
| O157:H7a | 2 | 2 | |||||
| O157:H12 | 1 | 1 | |||||
| O157:H16 | 1 | 1 | |||||
| O157:H19 | 1 | 1 | |||||
| O157:H25 | 1 | 1 | |||||
| O157:H38 | 1 | 1 | |||||
| O157:H45 | 1 | 1 | |||||
| O157:H− | 1 | 2 | 3 | ||||
| Total (%) | 10 (50.0) | 2 (10.0) | 8 (40.0) | 20 (100.0) | |||
Isolates of O156:H25 and O157:H7 were probably VTEC isolates that had lost their toxin genes.
Also included in our survey were a variety of non-VTEC commensal isolates from cattle and meats belonging to known VTEC serogroups. These were examined to determine whether the R3 core type was favored in commensal bovine E. coli having the same O antigens as known human and/or bovine VTEC. Of 20 isolates, 10 (50%) were type R1, 2 (10%) were R2, and 8 (40%) were R3 (Table 3). Five of the eight isolates with the R3 core type belonged to recognized EHEC serotypes (O157:H7, O157:H−) or to O156:H25, another virulent VTEC serotype. Since VTEC isolates may readily lose their VT genes (21), these and the 15 other non-VTEC isolates were tested by multiplex PCR for genes encoding two other virulence factors of EHEC, the EHEC hemolysin and intimin (33). Two VT-negative isolates of E. coli O157:H7 and one VT-negative isolate of E. coli O156:H25 carried genes for both factors (K. Ziebell, R. P. Johnson, and C. Whitfield, unpublished data). The presence of both virulence factors in isolates of well-recognized virulent VTEC serotypes strongly suggests that they were originally VT positive but had lost their VT genes. Therefore, they should be excluded from the non-VTEC group. In this case, the frequency of the R3 core type in non-VTEC bovine isolates would be reduced to 5 of 17 (29.4%) and the R1 core type would be more prevalent (10 of 17; 58.8%). Hence, the predominance of the R1 core type appears to be restored in VT-negative bovine E. coli.
TABLE 3.
Comparison of results from core OS typing studies
| Collection | Reference | No. (%) of isolates with core type:
|
Total no. | ||||
|---|---|---|---|---|---|---|---|
| R1 | R2 | R3 | R4 | K-12 | |||
| ECOR, all isolates | This study | 50 (69.4) | 8 (11.1) | 8 (11.1) | 2 (2.8) | 4 (5.6) | 72 |
| ECOR group A | This study | 11 (44) | 7 (28) | 2 (8) | 1 (4) | 4 (16) | 25 |
| ECOR group B1 | This study | 11 (68.8) | 5 (31.2) | 16 | |||
| ECOR group B2 | This study | 15 (100) | 15 | ||||
| ECOR group D | This study | 12 (100) | 12 | ||||
| ECOR minor group (group E) | This study | 1 (25) | 1 (25) | 1 (25) | 1 (25) | 4 | |
| VTEC | This study | 8 (12.3) | 54 (83.1) | 1 (1.5) | 2 (3.1) | 65 | |
| VTEC | 6 | 28 (100) | 28 | ||||
| Non-VTEC commensals from cattle and meats | This studya | 10 (50.0) | 2 (10.0) | 8 (40.0) | 20 | ||
| This study, adjustedb | 10 (58.8) | 2 (11.8) | 5 (29.4) | 17 | |||
| Human fecal isolates | 10 | 11 (52.4) | 4 (19) | 2 (9.5) | 21 | ||
| Bacteremic isolates | 1 | 94 (68) | 9 (6.5) | 12 (8.7) | 7 (5.1) | 3 (2.2) | 138 |
| Septicemic isolates | 10 | 48 (60.8) | 6 (7.6) | 12 (15.2) | 79 | ||
| UTI isolates | 10 | 64 (80) | 4 (5) | 4 (5) | 80 | ||
| Gibb et al. study, total | 10 | 123 (68.3) | 14 (7.8) | 18 (10) | 180 | ||
Includes three isolates that are probably VTEC isolates that have lost the VT genes.
Excludes three isolates that probably have lost their VT genes.
DISCUSSION
There has been a long-standing interest in immune responses against LPS from the perspective of immunotherapeutic approaches to neutralizing endotoxin activity and vaccine development. Clearly, a detailed understanding of the prevalence of core OS types is a prerequisite for such approaches, and this has driven previous studies. Core OS distributions in commensal E. coli (fecal) isolates and in isolates from septicemia and UTIs have been assessed (2, 10). Although the R1 core OS type predominated in these analyses (Table 3), it was not clear that these results were reflective of the broader species, and the objective of our study was to address this issue. Our PCR data indicate the prevalence of the R1 core OS in natural populations of E. coli and provide a phylogenetic basis for its distribution in specific groups of clinical isolates.
The number of E. coli isolates for which the core OS types have been definitively shown by structural determination is limited (reviewed in reference 18). Therefore the PCR-based core OS typing system reported here was validated using the same collection of prototype strains used to establish previous serological tests (2, 10). With the exception of core OS type R2, the sequences used for the PCR primers focused on genes essential for characteristic structures that complete the core OS and that are essential for addition of O antigen (16). The majority of the isolates produced an O antigen, providing independent evidence that the PCR test is based on genes that are expressed. The PCR-based system gave an unequivocal result for the core OS types of all tested isolates. In contrast, studies with core-specific monoclonal antibodies (MAbs) yielded a significant number of isolates for which core OS types could not be determined. These nontypeable strains are potentially all representatives of core OS type R4 or K-12, as there were no MAbs that specifically recognized these core OS types available (10). Furthermore, the precise LPS epitopes recognized by the MAbs are unknown, so the molecular basis for their core OS type determination is unclear.
The basis of the core OS specificity for a set of polyclonal antisera is also unknown although these reagents could identify representatives of each of the five E. coli core OS types (2). Using these polyclonal sera, Appelmelk et al. found that some bacteremic isolates (13 isolates or 9.4% of the tested isolates) reacted with both R1 and R4 sera, suggesting an R1R4 mixed core type (2). Several possible explanations for this result were proposed. These isolates either (i) express both R1 and R4 LPS molecules, (ii) have a single LPS molecule containing both R1 and R4 epitopes, or (iii) produce a novel core type that is chemically distinct from both R1 and R4 but that contains cross-reactive epitopes. These possibilities can be addressed in the light of more recent data describing the molecular basis for the various core types. Structural differences in the outer core OS from the R1 and R4 core OSs reside in unique β-linked side branches (Fig. 1). These require type-specific glycosyltransferases WaaV (R1) and WaaX (R4) (15). However, an R1R4 mixed core type could not result simply from coexpression of the waaX and waaV genes because the WaaV enzyme adds the glycosyl residue that defines the linkage site for O-PS in the R1 core OS. It is assumed by analogy that the corresponding β-linked (Gal) residue in the R4 core OS added by WaaX also forms its O-PS ligation site, although this has not been confirmed by experimentation. As the ligation sites differ, an R1R4 mixed core type would require coexpression of the R1- and R4-specific ligase gene (waaL) products, as well as both waaV and waaX. In the PCR-based analysis presented here, a prototype R1R4 isolate (strain 1502) was found to have the waa locus of the R1 core OS type and the waa genes essential for formation of the R4 core OS type were not detected. The reaction of strain 1502 (and others) with both R1 and R4 antisera could reflect a cross-reactive epitope in the inner core region of the LPS. Inner-core modifications are often nonstoichiometric, and some vary from isolate to isolate (18).
Despite the overall similarity in the prevalences of core OS types in the ECOR collection and previously studied pathogenic isolates (2, 10) (Table 3), several reasons make it difficult to directly compare the numerical values. For example, the Applemelk et al. study (2) likely underreports R1 due to the R1R4 mixed core type (see above). In the Gibb et al. study (10), no core determination could be made for 14% of the isolates due to the lack of MAbs for these core OS types, as indicated above.
E. coli is considered to be a highly clonal species (50). However, the extent of genetic variation within any group of E. coli isolates depends on their sources. For example, whereas the commensal microflora is multiclonal (49), specific clones (or groups of clones) are reflected in isolates from intestinal infections such as those with EHEC (26, 51) and enteropathogenic E. coli (EPEC) (32), as well as in isolates from extraintestinal infections such as septicemia (27, 43). This is presumably due the requirements for specific sets of virulence determinants. In studies of enteroaggregative E. coli isolates (7), chromosomal markers and some plasmid-encoded genes were found to have a heterogeneous clonal distribution. However, all isolates shared a horizontally disseminated member of a conserved family of virulence plasmids that may confer the characteristic aggregative adherence phenotype. In a recent analysis, UTI EPEC, EHEC, enteroinvasive E. coli, and enterotoxigenic E. coli isolates were examined by MLEE and their phylogeny was compared to the phylogeny of the ECOR collection (36). Clusters containing one or more of the pathogenic derivatives were distributed among the four major established phylogenetic groups in the ECOR collection.
Genome sizes in E. coli range from 4.6 to 5.5 Mb (3, 4). E. coli isolates showing the largest genomes are found in phylogenetic groups B2 and D, while those from isolates in group A (including E. coli K-12; 4.639 Mb) are relatively smaller (3, 4). Specific virulence genes are known to be associated with extraintestinal infections, and DNA insertions carrying virulence genes are distributed throughout the larger genomes of isolates from sepsis and UTI (39). In surveys of the distribution of known virulence genes and loci from extraintestinal pathogens, including the kps (group 2 capsule synthesis), pap (P-pilus biogenesis), sfa (S-pilus biogenesis), and hly (α-hemolysin formation) loci, the highest concentration of these virulence determinants occurred in group B2 (5, 34). Most group B2 isolates in the ECOR collection are primarily from humans and other primates (42), and, as might be expected, group B2 isolates were found to be the most virulent in a mouse lethality model (34). Smaller numbers of virulent isolates are found in groups A, B1, and D, where distribution of the examined virulence markers is limited. Commensals were mainly found in groups A and B1. The broad distribution of the R1 core type in the ECOR collection and its particular prevalence in phylogenetic groups B2 and D therefore provide an explanation for its high incidence in bacteremic and UTI isolates (Table 3). Small numbers of septicemic and UTI isolates of E. coli were found by serological studies to contain the K-12 and R4 core OS types (2, 10), and these isolates are likely to be related to isolates in either group A or the minor group. Within the ECOR collection are several isolates originally isolated from UTI patients (ECOR11, -14, -40, -50, -56, -60, -62, and -64 [36]). Six of these isolates are members of phylogenetic groups B2 and D and are now known to contain the R1 core OS type. The remaining two isolates, ECOR11 (core type R2) and ECOR14 (K-12), belong to group A. Herzer et al. found seven isolates from UTIs or septicemia that were related to ECOR isolates in groups B1 and D (17). The R1 type would be expected to predominate among these isolates.
It remains to be established whether the structural organization of LPS containing the R1 core type confers some selective advantage, either in the virulence of extraintestinal pathogenic E. coli or in facilitating the acquisition of virulence genes by such isolates through horizontal gene transfer. The attachment site for O-PS on the β-linked Glc side branch on GlcII gives a structural arrangement in the R1 core OS (Fig. 1) rather different from that in the R2 core OS and the classical example of the S. enterica serovar Typhimurium core OS (16).
While this paper was in the final stages of preparation for submission, another group reported a correlation between the presence of the R3 core type and VT production in isolates of E. coli representing serotypes O157, O111, O86, and O26, based on reactivity with an R3 MAb (6). Our results with VTEC isolates both confirm and extend their observations. The finding of a single core OS type (R3) in VTEC isolates of serogroups O157 and O55 and in “traditional” EPEC serogroups such as O111 is explained by the phylogenetic relationships among these isolates. EHEC (36, 51) and EPEC (32, 36) isolates reflect relatively homogeneous groups of organisms. They are only distantly related to extraintestinal pathogenic E. coli isolates. The occurrence of a single core OS type (R3) among VTEC O157:H7 and O55:H7 isolates is consistent with the observation that they form a closely related and recently emergent clone and with the proposal that O157:H7 arose from an O55:H7 progenitor (53). In one MLEE study, O157:H7 isolates were found to cluster with ECOR37 and ECOR42 (36). ECOR37 has an R3 core OS (Fig. 3). On reexamination of ECOR37 isolate, we found it was a representative of serogroup O55:H7 but was VT negative (Ziebell et al., unpublished data). The ECOR42 strain was serotyped as O87:H26.
VTEC O157:H7 isolates are only distantly related to other VT-positive serogroups (51), but we found that the R3 core type was distributed in VT-positive isolates representing a variety of different serogroups (Table 2). Currie and Poxton (6) speculated that the correlation between the R3 core OS and VTEC isolates might reflect a role for the R3 core as a receptor for lysogenic phages carrying the VT genes. However, the R3 core OS alone is unlikely to form the VT phage receptor since E. coli C600 (a K-12 strain) has been used as a recipient for such phages (29, 44, 46, 48). Also, receptor data for two Shiga toxin 2 (VT) phages clearly implicate outer membrane proteins FadL and LamB as phage receptors (48). It has been reported that isolates with R-LPS are easier to lysogenize with VT phages (44), and it is certainly possible that lysogenization is eased by some characteristic of S-LPS-containing R3-type cores (e.g., reduced O-antigen capping), as has been suggested previously (6). In a broader analysis of VTEC isolates, we detected the R1 core in isolates of O113:H21 (three isolates), O145:H8 (one isolate), O145:H− (three isolates), and O6:H34 (one isolate); the R4 core type in an O112:H2 isolate; and the K-12 core in single isolates of O85:H− and O145:H− (Table 2). It is conceivable that processes other than lysogenization mediate the spread of VT genes to the isolates with other core OSs. Indeed, conjugation is reported to be an alternative method for transfer of VT genes (44).
In contrast to the high frequency of the R3 core type in VTEC isolates, the R1 core type predominated in the 20 VT-negative isolates included in the study. The overall representation of R1 in VT-negative isolates was lower than that seen among the septicemic and UTI isolates and more in line with values determined for multiclonal human commensals (10) and for phylogenetic group A (Table 3). Core-type analysis also revealed diversity within the O157 serogroup, with R1, R2, and R3 core OSs represented in VT-negative isolates belonging to serotype E. coli O157:H− and in O157 isolates with flagellar antigens other than H7. Testing for EHEC virulence factors suggested that the two VT-negative O157:H7 isolates were VTEC isolates that had lost their toxin genes (21). Their R3 core OS type is therefore understandable. Overall, our findings for the O157 serogroup correlate well with results of MLEE studies (53), which indicate that members of serogroup O157 are genetically diverse with no strong linkage between O157:H7 and other members of the O157 serogroup.
Data from this and other studies indicate that there is no formal link between core OS type and O serotypes. It has been established that the O serotype does not provide a reliable assessment of clonal structure in most clinical isolates due to horizontal transfer and genetic recombination (42). As a result, isolates from a given serotype can be distributed in distinct and sometimes distantly related clones. The LPS core OS is a much more conserved structure, and, based on the results for groups B1, B2, and D and for the most common EHEC serogroups, it appears to be a more stable genetic character.
ACKNOWLEDGMENTS
This work was supported by a funding from the Canadian Bacterial Diseases Network (NCE program) awarded to C.W. and by Health Canada. E.F. was supported by a Natural Sciences and Engineering Research Council postgraduate scholarship, and D.E.H. was the recipient of a postdoctoral fellowship from the Medical Research Council.
We thank B. Allen, B. J. Appelmelk, H. Brade, H. Ochman, and F. Scheutz for generously supplying bacterial isolates and Irene Yong for assistance in serotyping.
REFERENCES
- 1.Aleksic S. WHO report on Shiga-like toxin producing Escherichia coli (SLTEC), with emphasis on zoonotic aspects. Geneva, Switzerland: World Health Organization; 1995. [Google Scholar]
- 2.Appelmelk B J, An Y-Q, Hekker T A M, Thijs L G, MacLaren D M, deGraaf J. Frequencies of lipopolysaccharide core types in Escherichia coli strains from bacteraemic patients. Microbiology. 1994;140:1119–1124. doi: 10.1099/13500872-140-5-1119. [DOI] [PubMed] [Google Scholar]
- 3.Bergthorsson U, Ochman H. Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol Biol Evol. 1998;15:6–16. doi: 10.1093/oxfordjournals.molbev.a025847. [DOI] [PubMed] [Google Scholar]
- 4.Bergthorsson U, Ochman H. Heterogeneity of genome sizes among natural isolates of Escherichia coli. J Bacteriol. 1995;177:5784–5789. doi: 10.1128/jb.177.20.5784-5789.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd E F, Hartl D L. Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution. J Bacteriol. 1998;180:1159–1165. doi: 10.1128/jb.180.5.1159-1165.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Currie C G, Poxton I R. The lipopolysaccharide core type of Escherichia coli O157:H7 and other non-O157 verotoxin-producing E. coli. FEMS Immunol Med Microbiol. 1999;24:57–62. doi: 10.1111/j.1574-695X.1999.tb01265.x. [DOI] [PubMed] [Google Scholar]
- 7.Czeczulin J R, Whittam T S, Henderson I R, Navarro-Garcia F, Nataro J P. Phylogenetic analysis of enteroaggregative and diffusely adherent Escherichia coli. Infect Immun. 1999;67:2692–2699. doi: 10.1128/iai.67.6.2692-2699.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ewing W H. Edwards and Ewing's identification of Enterobacteriaceae. 4th ed. New York, N.Y: Elsevier Science Publishing, Inc.; 1996. [Google Scholar]
- 9.Gamian A, Romanowska E. The core structure of Shigella sonnei lipopolysaccharide and the linkage between O-specific polysaccharide and the core region. Eur J Biochem. 1982;129:105–109. doi: 10.1111/j.1432-1033.1982.tb07027.x. [DOI] [PubMed] [Google Scholar]
- 10.Gibb A R, Barclay G R, Poxton I R, di Padova F. Frequencies of lipopolysaccharide core types among clinical isolates of Escherichia coli defined with monoclonal antibodies. J Infect Dis. 1992;166:1051–1057. doi: 10.1093/infdis/166.5.1051. [DOI] [PubMed] [Google Scholar]
- 11.Gyles C, Johnson R, Gao A, Ziebell K, Pierard D, Aleksic S, Boerlin P. Association of enterohemorrhagic Escherichia coli hemolysin with serotypes of Shiga-like toxin-producing Escherichia coli of human and bovine origins. Appl Environ Microbiol. 1998;64:4134–4141. doi: 10.1128/aem.64.11.4134-4141.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haishima Y, Holst O, Brade H. Structural investigation of the lipopolysaccharide of Escherichia coli rough mutant F653 representing the R3 core type. Eur J Biochem. 1992;203:127–134. doi: 10.1111/j.1432-1033.1992.tb19837.x. [DOI] [PubMed] [Google Scholar]
- 13.Hammerling G, Luderitz O, Westphal O, Makela P H. Structural investigations on the core polysaccharide of Escherichia coli O100. Eur J Biochem. 1971;22:331–344. doi: 10.1111/j.1432-1033.1971.tb01549.x. [DOI] [PubMed] [Google Scholar]
- 14.Heinrichs D E, Monteiro M A, Perry M B, Whitfield C. The assembly system for the lipopolysaccharide R2 core-type of Escherichia coli is a hybrid of those found in Escherichia coli K-12 and Salmonella enterica. Structure and function of the R2 WaaK and WaaL homologs. J Biol Chem. 1998;273:8849–8859. doi: 10.1074/jbc.273.15.8849. [DOI] [PubMed] [Google Scholar]
- 15.Heinrichs D E, Yethon J A, Amor P A, Whitfield C. The assembly system for the outer core portion of R1 and R4-type lipopolysaccharides of Escherichia coli. The R1 core-specific β-glucosyltransferase provides a novel attachment site for O polysaccharides. J Biol Chem. 1998;273:29497–29505. doi: 10.1074/jbc.273.45.29497. [DOI] [PubMed] [Google Scholar]
- 16.Heinrichs D E, Yethon J A, Whitfield C. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol Microbiol. 1998;30:221–232. doi: 10.1046/j.1365-2958.1998.01063.x. [DOI] [PubMed] [Google Scholar]
- 17.Herzer P J, Inouye S, Inouye M, Whittam T S. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J Bacteriol. 1990;172:6175–6181. doi: 10.1128/jb.172.11.6175-6181.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holst O. Chemical structure of the core region of lipopolysaccharides. In: Brade H, Opal S M, Vogel S N, Morrison D C, editors. Endotoxin in health and disease. New York, N.Y: Marcel Dekker Inc.; 1999. pp. 305–330. [Google Scholar]
- 19.Hull S. Escherichia coli lipopolysaccharide in pathogenesis and virulence. In: Sussman M, editor. Escherichia coli: mechanisms of virulence. Cambridge, United Kingdom: Cambridge University Press; 1997. pp. 145–167. [Google Scholar]
- 20.Johnson R P, Clarke R C, Wilson J B, Read S C, Rahn K, Renwick S A, Sandhu K A, Alves D, Karmali M A, Lior H, McEwen S A, Spika J, Gyles C L. Growing concerns and recent outbreaks involving non-O157 serotypes of verotoxigenic Escherichia coli. J Food Protect. 1996;59:1112–1122. doi: 10.4315/0362-028X-59.10.1112. [DOI] [PubMed] [Google Scholar]
- 21.Karch H, Meyer T, Russmann H, Heesemann J. Frequent loss of Shiga-like toxin genes in clinical isolates of Escherichia coli upon subcultivation. Infect Immun. 1992;60:3464–3467. doi: 10.1128/iai.60.8.3464-3467.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katzenellenbogen E, Romanowska E. Structural studies on Shigella flexneri serotype 6 core region. Eur J Biochem. 1980;113:205–211. doi: 10.1111/j.1432-1033.1980.tb06157.x. [DOI] [PubMed] [Google Scholar]
- 23.Levine M M. Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic, and enteroadherent. J Infect Dis. 1987;155:377–389. doi: 10.1093/infdis/155.3.377. [DOI] [PubMed] [Google Scholar]
- 24.Liu D, Reeves P R. Escherichia coli K12 regains its O antigen. Microbiology. 1994;140:49–57. doi: 10.1099/13500872-140-1-49. [DOI] [PubMed] [Google Scholar]
- 25.Lugowski C, Kulakowska M, Romanowska E. Characterization and diagnostic application of a lipopolysaccharide core oligosaccharide-protein conjugate. J Immunol Methods. 1986;95:187–194. doi: 10.1016/0022-1759(86)90405-9. [DOI] [PubMed] [Google Scholar]
- 26.Mariani-Kurkdjian P, Denamur E, Milon A, Picard B, Cave H, Lambert-Zechovsky N, Loirat C, Goullet P, Sansonetti P J, Elion J. Identification of a clone of Escherichia coli O103:H2 as a potential agent of hemolytic-uremic syndrome in France. J Clin Microbiol. 1993;31:296–301. doi: 10.1128/jcm.31.2.296-301.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maslow J N, Whittam T S, Gilks C F, Wilson R A, Mulligan M E, Adams K S, Arbeit R D. Clonal relationships among bloodstream isolates of Escherichia coli. Infect Immun. 1995;63:2409–2417. doi: 10.1128/iai.63.7.2409-2417.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Müller-Loennies S, Holst O, Brade H. Chemical structure of the core region of Escherichia coli J-5 lipopolysaccharide. Eur J Biochem. 1994;224:751–760. doi: 10.1111/j.1432-1033.1994.t01-1-00751.x. [DOI] [PubMed] [Google Scholar]
- 29.O'Brien A D, Newland J W, Miller S F, Holmes R K, Smith H W, Formal S B. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science. 1984;226:694–696. doi: 10.1126/science.6387911. [DOI] [PubMed] [Google Scholar]
- 30.Ochman H, Selander R K. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984;157:690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olsthoorn M M, Petersen B O, Schlecht S, Haverkamp J, Bock K, Thomas-Oates J E, Holst O. Identification of a novel core type in Salmonella lipopolysaccharide. Complete structural analysis of the core region of the lipopolysaccharide from Salmonella enterica sv. Arizonae O62. J Biol Chem. 1998;273:3817–3829. doi: 10.1074/jbc.273.7.3817. [DOI] [PubMed] [Google Scholar]
- 32.Ørskov F, Whittam T S, Cravioto A, Ørskov I. Clonal relationships among classic enteropathogenic Escherichia coli (EPEC) belong to different O groups. J Infect Dis. 1990;162:76–81. doi: 10.1093/infdis/162.1.76. [DOI] [PubMed] [Google Scholar]
- 33.Paton A W, Paton J C. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J Clin Microbiol. 1998;36:598–602. doi: 10.1128/jcm.36.2.598-602.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Picard B, Garcia J S, Gouriou S, Duriez P, Brahimi N, Bingen E, Elion J, Denamur E. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun. 1999;67:546–553. doi: 10.1128/iai.67.2.546-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pierard D, Stevens D, Moriau L, Lior H, Lauwers S. Isolation and virulence factors of verocytotoxin-producing Escherichia coli in human stool samples. Clin Microb Infect. 1997;3:531–540. doi: 10.1111/j.1469-0691.1997.tb00303.x. [DOI] [PubMed] [Google Scholar]
- 36.Pupo G M, Karaolis D K, Lan R, Reeves P R. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect Immun. 1997;65:2685–2692. doi: 10.1128/iai.65.7.2685-2692.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raetz C R H. Bacterial lipopolysaccharides: a remarkable family of bioactive macroamphiphiles. In: Niedhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1035–1063. [Google Scholar]
- 38.Read S C, Clarke R C, Martin A, De Grandis S A, Hii J, McEwen S A, Gyles C L. Polymerase chain reaction for detection of verocytotoxigenic Escherichia coli isolated from animal and food sources. Mol Cell Probes. 1992;6:153–161. doi: 10.1016/0890-8508(92)90060-b. [DOI] [PubMed] [Google Scholar]
- 39.Rode C K, Melkerson-Watson L J, Johnson A T, Bloch C A. Type-specific contributions to chromosome size differences in Escherichia coli. Infect Immun. 1999;67:230–236. doi: 10.1128/iai.67.1.230-236.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmidt G, Jann B, Jann K. Genetic and immunochemical studies on Escherichia coli O14:K7:H−. Eur J Biochem. 1974;42:303–309. doi: 10.1111/j.1432-1033.1974.tb03340.x. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt G, Jann B, Jann K. Immunochemistry of R lipopolysaccharides of Escherichia coli. Different core regions in the lipopolysaccharides of O group 8. Eur J Biochem. 1969;10:501–510. doi: 10.1111/j.1432-1033.1969.tb00717.x. [DOI] [PubMed] [Google Scholar]
- 42.Selander R K, Caugant D A, Whittam T S. Genetic structure and variation in natural populations of Escherichia coli. In: Niedhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 1625–1648. [Google Scholar]
- 43.Selander R K, Korhonen T K, Vaisanen-Rhen V, Williams P H, Pattison P E, Caugant D A. Genetic relationships and clonal structure of strains of Escherichia coli causing neonatal septicemia and meningitis. Infect Immun. 1986;52:213–222. doi: 10.1128/iai.52.1.213-222.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith H W, Green P, Parsell Z. Vero cell toxins in Escherichia coli and related bacteria: transfer by phage and conjugation and toxic action in laboratory animals, chickens and pigs. J Gen Microbiol. 1983;129:3121–3137. doi: 10.1099/00221287-129-10-3121. [DOI] [PubMed] [Google Scholar]
- 45.Stevenson G, Neal B, Liu D, Hobbs M, Packer N H, Batley M, Redmond J W, Lindquist L, Reeves P R. Structure of the O antigen of Escherichia coli K-12 and the sequence of its rfb cluster. J Bacteriol. 1994;176:4144–4156. doi: 10.1128/jb.176.13.4144-4156.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strockbine N A, Marques L R, Newland J W, Smith H W, Holmes R K, O'Brien A D. Two toxin-converting phages from Escherichia coli O157:H7 strain 933 encode antigenically distinct toxins with similar biologic activities. Infect Immun. 1986;53:135–140. doi: 10.1128/iai.53.1.135-140.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsang R S, Chan K H, Chau P Y, Wan K C, Ng M H, Schlecht S. A murine monoclonal antibody specific for the outer core oligosaccharide of Salmonella lipopolysaccharide. Infect Immun. 1987;55:211–216. doi: 10.1128/iai.55.1.211-216.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watarai M, Sato T, Kobayashi M, Shimizu T, Yamasaki S, Tobe T, Sasakawa C, Takeda Y. Identification and characterization of a newly isolated Shiga toxin 2-converting phage from Shiga toxin-producing Escherichia coli. Infect Immun. 1998;66:4100–4107. doi: 10.1128/iai.66.9.4100-4107.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whittam T S. Clonal dynamics of Escherichia coli in its natural habitat. Antonie Leeuwenhoek. 1989;55:23–32. doi: 10.1007/BF02309616. [DOI] [PubMed] [Google Scholar]
- 50.Whittam T S. Genetic variation and evolutionary processes in natural populations of Escherichia coli. In: Niedhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: ASM Press; 1996. pp. 2708–2720. [Google Scholar]
- 51.Whittam T S, Wachsmuth I K, Wilson R A. Genetic evidence of clonal descent of Escherichia coli O157:H7 associated with hemorrhagic colitis and hemolytic uremic syndrome. J Infect Dis. 1988;157:1124–1133. doi: 10.1093/infdis/157.6.1124. [DOI] [PubMed] [Google Scholar]
- 52.Whittam T S, Wilson R A. Genetic relationships among pathogenic Escherichia coli of serogroup O157. Infect Immun. 1988;56:2467–2473. doi: 10.1128/iai.56.9.2467-2473.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whittam T S, Wolfe M L, Wachsmuth I K, Ørskov F, Ørskov I, Wilson R A. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect Immun. 1993;61:1619–1629. doi: 10.1128/iai.61.5.1619-1629.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]