Abstract
In order to facilitate efficient and reproducible metagenomic analysis, we introduce Kraken Protocols, an end-to-end pipeline for the classification, quantification, and visualization of metagenomic datasets. Our protocol describes the execution of the Kraken programs, via a sequence of easy to use scripts, in two scenarios: (1) quantification of the species in a metagenomics sample, and (2) detection of a pathogenic agent from a clinical sample taken from a human patient. The protocols can be run by any users who are familiar with the Unix command-line environment.
Introduction
Metagenomics sequencing has greatly improved our understanding of the microscopic world by revealing a vast range of microbial organisms that were previously unobserved, many of which cannot be grown in laboratory cultures 1. Metagenomics experiments take many forms, two of which can be broadly categorized as either microbiome experiments or pathogen identification experiments. In microbiome experiments, researchers evaluate all of the microbial organisms identified in a given sample, often with the goal of describing what is present. In contrast, in a pathogen identification experiment, researchers focus on identifying one or a few pathogenic microbes, with the goal of diagnosing an infection. The suite of tools in the Kraken package were developed to cover many of the bioinformatics needs of both microbiome and pathogen metagenomics experiments. These tools include Kraken 2, KrakenUniq 3, Kraken 24, Kraken2Uniq (based on KrakenUniq), Bracken 5, KrakenTools, and Pavian 6.
Microbiome experiments begin with (1) removing host DNA and then (2) classifying a set of sequencing reads, with each read assigned to a taxonomic category (species, genus, or higher-level taxa), followed by (3) computing the relative abundance of different species in the sample. When computing relative abundance, researchers sometimes focus on a limited number of “marker” genes, classifying only the reads that align within these genes. The most widely used marker gene is ribosomal RNA, but protein-coding genes can also be used, particularly those that are expected to be present in exactly one copy per cell. However, many studies prefer to analyze all of the sequencing reads collected from a sample, which is the strategy we focus on in this protocol. Following characterization of the original data, downstream analyses may include (4) statistical methods for comparing the microbial compositions of different environments or (5) visualization methods for understanding microbial compositions. Kraken 2 and Kraken 24 (an improved version of Kraken) were previously developed by some of us for rapid, accurate classification of sequencing reads. Bracken 5 was developed to work in conjunction with Kraken to compute species abundance using Kraken classification results. Finally, KrakenTools and Pavian 6 provide a comprehensive set of tools for downstream statistical analysis and visualization of the classification and abundance estimation results.
In pathogen identification experiments, researchers and clinicians are interested in identifying pathogenic microbes that might be the cause of a harmful infection. Pathogen identification studies conceptually include the following steps: (1) removal of host DNA from the microbial reads; (2) classification of the remaining microbial reads; (3) comparison of sample reads against control samples; and (4) validation of pathogen classifications. The second and third steps are essentially the same as those done for microbiome experiments, while the first and last steps are needed to ensure the accuracy of any pathogen that is identified. For removal of host DNA, one can use Bowtie 27 (also developed by some of the authors of this protocol) as a fast, sensitive aligner that can compare sequencing reads to the human reference genome. Classification of the remaining reads is then accomplished by using KrakenUniq 3 and Kraken2Uniq. Finally, KrakenTools and Pavian 6 cover the remaining steps of pathogen identification experiments, assisting in the identification and verification of potential pathogens.
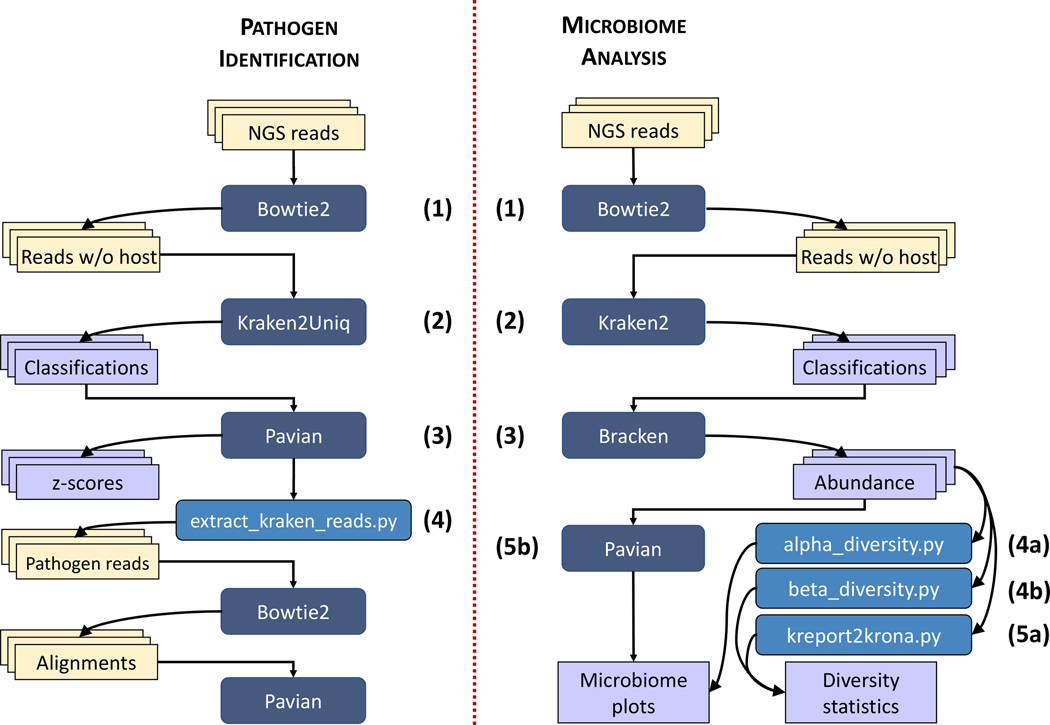
Here, we describe in detail how to use the Kraken suite to analyze metagenomic data for both microbiome and pathogen identification experiments. Figure 1 outlines the protocols for each of the two types of studies.
Fig 1. Protocol workflow.
Overview of two workflows (1) pathogen identification and (2) microbiome analysis. (1) Here we try to detect an infectious agent using NGS reads. For this we start with a sample from the infection site and (ideally) a negative control. As a first step, host DNA is removed by excluding all reads aligning to the host genome using Bowtie 2. This step usually removes a large fraction of the reads. Remaining reads are then classified by Kraken2Uniq against a reference database, and the taxonomic reports are compared using Pavian. Pavian can distinguish large abundance changes between controls and infected samples using z-statistics. For all potential pathogen candidates, reads can be extracted using extract kraken reads.py. In workflow (2) we try to estimate the abundance of species in microbiome samples and compute the diversity changes between them. In the protocol, we start with multiple sets of reads from a microbiome before and after fecal transfer. All samples are classified using Kraken 2. Bracken takes the classified read counts and estimates the abundance of each taxon in the sample. Pavian can be used to explore and visualize this sample to spot the difference. Additionally, alpha diversity.py can be used to quantify the diversity in a sample and beta diversity.py can be used to compare diversity across samples.
Overview of the protocol
The protocols described here focus on two major categories of metagenomic experiments: microbiome analysis and pathogen identification. We detail the protocol steps using DNA samples from the human gut and cornea. However, these protocols may also be applied to RNA data as well as other biomes and species. For microbiome analysis, we consider an experiment describing the change in gut microbiome for a single human patient who underwent a fecal microbiota transplant and antibiotic treatment. For pathogen identification, we consider 8 separate human corneal samples with bacterial, viral, or fungal infections and two non-infectious human corneal samples. For both samples, human reads were already removed prior to analysis in the protocol. However, we include the host removal step in the protocol to demonstrate how to remove host reads using Bowtie 27.
For microbiome analysis, host-filtered microbial reads undergo classification against bacterial, viral, fungal, archaeal, and human genomes using Kraken 2 (Figure 1). The classification report is then used by Bracken for species abundance estimation, which provides estimated reads per species in the sample. Bracken output files are then passed to KrakenTools and Pavian for visualization. Pavian provides Sankey visualization of samples and read count comparisons between samples. For more in-depth downstream analysis, we recently developed KrakenTools (https://github.com/jenniferlu717/KrakenTools) as a set of individual programs for visualizing or transforming Kraken and Bracken output. For microbiome projects, KrakenTools provides functions to compute Krona plots, filter and combine reports, and calculate alpha and beta diversity metrics about each sample. The software is not previously published but is already packaged and available through github or bioconda (https://bioconda.github.io/recipes/krakentools/README.html).
For pathogen identification, we classify 8 infected patient samples and 2 control samples against the same Kraken 2 database used in microbiome analysis (Figure 1). However, the additional feature of unique k-mer counting from KrakenUniq 3 is enabled for accurate pathogen identification. We refer to this as Kraken2Uniq. Following classification by Kraken2Uniq, the Kraken report files are uploaded to the Pavian Shiny App6, which compiles all of the read counts per species and allows between sample visualization and comparison. The Pavian app then calculates z-scores between read counts, with higher z-scores resulting from samples with higher than usual read counts for a given species. We then sort the classified species by z-score, with the highest z-score species for each sample being the most likely pathogen. The filtered table can then be saved as a tab-delimited table that can undergo further visualization. Finally, pathogen identification can be validated using the extract kraken reads.py script from KrakenTools. The KrakenTools script allows users to extract classified reads and confirm potential pathogens using other tools such as BLAST 8 or Bowtie 27.
The target audience for this protocol is biologists and clinicians working in microbiome or metagenomics analysis. This protocol does not require programming expertise, but it does assume familiarity with the Unix command-line interface. Users should be comfortable running programs from the command line in the Unix environment.
Differences between Kraken methods
There are currently four major versions of the Kraken software: (1) Kraken2, (2) KrakenUniq3, (3) Kraken 24 and (4) Kraken2Uniq. All four versions are based on the same classification algorithm which uses exact-matching of read k-mers against a Kraken database of k-mers from existing genomes. However, the methods differ in the ways they count, access and store the k-mer information, with each version improving upon the previous Kraken version.
Kraken and KrakenUniq apply the same k-mer retrieval strategy, and both tools can use the same databases. The reference database needs roughly 12 bytes per k-mer. Building and storing the Kraken index for a thousand distinct bacterial genomes of length 4Mb would result in an index of size ≈ 45GB.
However, the RefSeq 9;10 database already contains ~ 64K bacterial species, and trying to store all k-mers from this ever-growing database became the main bottleneck of Kraken and KrakenUniq. Therefore, Kraken 2 was developed as a much more efficient version of Kraken, reducing the memory usage by 85% over Kraken and KrakenUniq. In addition to the reduction in database size, Kraken 2 also runs about 5 times faster than Kraken/KrakenUniq.
Another distinguishing difference is in how the Kraken methods count reads. While Kraken and Kraken 2 provide cumulative statistics of the total read count per taxa, KrakenUniq and Kraken2Uniq additionally count and report the number of unique k-mers per taxon, using an efficient HyperLogLog implementation. In other words, for each species in the output, KrakenUniq will report how many distinct k-mers from that species were observed in the reads. Unique counts are especially useful for pathogen detection to distinguish real from spurious signals.
Alternative analysis packages
The Kraken/KrakenUniq tools assign every read to a taxon of origin. Other software tools solve this problem, including CLARK 11, Centrifuge 12, and Kaiju 13. Kraken 2 generally exhibits a more advantageous combination of speed, memory footprint, and accuracy compared to those tools 4, but it is still possible to substitute those tools into earlier steps in this protocol. Recent benchmarking studies14, 15 evaluated multiple metagenomic classifiers and found that Kraken, Kraken 2, and Bracken were among the best-performing methods.
Other tools seek to reduce the memory footprint of the index by including only the portions of the genome sequences that can distinguish between taxa, e.g. MetaPhlAn216. This reduces the size of the index, but prevents the tool from classifying many of the reads. So, while these tools can be used in a microbiome analysis like the one described in this protocol, they are not appropriate for pathogen identification.
While approaches based on large-scale machine learning have been shown to have advantages in certain scenarios, e.g. for classifying reads from species that are absent from the database16, these approaches have tended to be computationally outperformed by classifiers that assign reads directly based on sequence content, without the need to first train a model 17;18. For more details on competing paradigms for read classification, see Breitwieser et al. 19.
The Bowtie 2 tool used to filter host reads can be replaced with similar read alignment tools such as BWA-MEM or minimap2.
Limitations
Removal of host reads.
For metagenomic studies, removal of host (usually human) DNA is critical: either as a pre-processing step, using a program such as Bowtie 27 to align all of the reads to the host, or by including the host genome in the Kraken database. The pre-processing strategy may be more effective because external alignment programs such as Bowtie 2 are more sensitive than Kraken, and because pre-processing yields a greatly reduced set of reads, making subsequent steps faster. Bowtie 2 and other alignment programs are suitable for the pre-processing step because they can identify and remove any reads belonging to a known host (e.g., human), while Kraken can then identify the wide variety of unknown microbial species in the remainder of the sample.
However, this pre-processing step may cause sequences of interest to be removed from the sample set, especially in the rare case of viral sequences found to be similar to a human endogenous retrovirus. In this protocol, we include the pre-processing with Bowtie 2 as an optional step that may or may not be included at the discretion of the users of the protocol.
Reference Database.
Kraken 2’s classification sensitivity and specificity highly depend on how (1) complete and (2) accurate the used reference database is. (1) The entire microbial biosphere is still far from being completely sequenced and thus missing or partial genomes may introduce biases and reduce classification performance. Reads without a reference in the database will be labeled as unknown or imprecisely assigned to the next closest taxon. (2) Additionally, genomes can also be contaminated with DNA from other organisms 20;21 introduced during sequencing. Contamination causes Kraken to assign wrong taxonomic labels to reads. While this kind of classification error affects only a small fraction of reads, it can make the detection of weak signals difficult.
For the purpose of this protocol, we use the complete genomes from RefSeq bacteria, archaea, viral, and plasmid libraries along with extensively screened eukaryotic pathogen genomes 22. We use only complete genomes to avoid potential contamination from draft genomes.
For users that may have limited RAM, Kraken 2 provides the ability to generate a minikraken database. Minikraken databases compress the full Kraken 2 database by saving a subset, but representative set of the database k-mers. To allow all users to execute this protocol, we use an 8Gb minikraken database.
Alternatively, Kraken 2 can also classify reads against protein databases using six-frame-translation. Protein-level classification is more sensitive (but less specific) and therefore can help to reduce the missing reference bias. For even longer evolutionary distances, homology detection alignment-based classification methods like DIAMOND 23 or MMseqs224 can be used.
Other issues can arise as reference databases grow and as the distribution of reference genomes per taxon becomes more lopsided. For example, Nasko et al. 25 showed that as the RefSeq database has grown over time, Kraken has tended to assign a greater proportion of reads to higher levels of the taxonomy. In particular, they showed that a greater proportion of reads tend to be assign to the genus level, at the expense of assignments to the species level and below. As public databases of reference genomes continue grow, it will be important to continue to evaluate how classification results are affected by large or lopsided numbers of genomes per taxon.
Long read classification
This protocol is designed for Illumina sequencing reads, which are highly accurate (error rates under 0.5%) but short, usually 100–250bp. However, in some cases, users may want to evaluate longer, higher error-rate reads from Pacific Biosciences or Oxford Nanopore. For longer reads with a different error profile, users might remove host DNA using minimap226, and they might also need to adjust the parameters of the Kraken software. Specifically, higher error rate reads, such as those from Pacific Biosciences or Oxford Nanopore, yield better classification with Kraken databases generated using smaller k-mer and minimizer lengths. Previous analyses have indicated better performance using k-mer and minimizer lengths of 26 (as opposed to the default k-mer length of 35 and minimizer length of 31.
Also the read count measurement might not be as useful for long reads, because these experiments typically generate fewer reads that differ significantly in length. For these experiments, it may make more sense to compare k-mer counts.
Assembly-based approaches
When a species is present in sufficiently high amounts, one might wish to assemble the reads de novo to create large, contiguous sequences from that species. The protocols described here do not perform genome assembly, which requires other software methods.
Materials
Equipment
The Kraken 2 protocol website http://ccb.jhu.edu/data/kraken2_protocol/ summarizes all required software and data, along with details about how to download all required data for the protocol.
Data (the example corneal reads, microbiome fecal reads, and Kraken 2 database for use in this protocol are available from NCBI SRA and Amazon AWS; see Equipment Setup for details)
Kraken 2 software (https://github.com/DerrickWood/kraken2/, version 2.1.1 or later)
Bracken software (https://github.com/jenniferlu717/Bracken, version 2.6.2 or later)
KrakenTools software (https://github.com/jenniferlu717/KrakenTools, version 1.1. or later)
Pavian software (https://github.com/fbreitwieser/pavian, version 1.0 or later)
Bowtie 2 (https://github.com/BenLangmead/bowtie2, version 2.4.4)
samtools (http://www.htslib.org/download/), version 0.1.20 or later)
RStudio (https://www.rstudio.com/)
Hardware (64-bit computer running either Linux or Mac OS X (10.7 Lion or later); 8GB of RAM; see Equipment Setup)
EQUIPMENT SETUP
Required data
The microbiome analysis portion of this protocol is illustrated with a sample microbiome dataset of fecal samples from Xavier et al. (2018) 27. Many samples from the paper are available at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA491657. Here, we use 3 samples from a single patient, T11, who began antibiotic treatment at day 0, underwent stem cell engraftment on day 14, and received autologous fecal microbiota transplantation (auto-FMT) to return the patient’s gut microbiota diversity at day 29.
The infectious disease analysis portion of this protocol is illustrated with selected corneal samples from Li et al. (2018) 28. All 20 samples from the paper are available at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA381365. However, for clarity, we used a select group of 10 of the corneal samples (including 2 controls).
Both protocols require a Kraken 2 database. For this protocol, we use the same pre-built Kraken 2 database, containing RefSeq complete genomes from December 2020 for bacterial, archaeal, and viral genomes, the Human reference genome GRCh38, and the cleaned eukaryotic pathogen genomes for EuPathDB4822 (available at http://ccb.jhu.edu/data/eupathDB/). Additional pre-built Kraken 2 databases are available in the AWS cloud, detailed here: https://benlangmead.github.io/aws-indexes/k2. The following subsection details how to create a Kraken 2 Database if the desired database is not already available.
Kraken 2 Databases
Several pre-built Kraken 2 databases are available at https://benlangmead.github.io/aws-indexes/k2. The most commonly used database is the standard Kraken 2 database (which includes RefSeq archaea, bacteria, viruses, plasmid complete genomes, UniVec Core, and the most recent human reference genome, GRCh38). In addition to the standard database, we provide a database of the cleaned eukaryotic pathogens 22, 16S rRNA databases, and expanded standard databases with RefSeq protozoa, fungi, and plant genomes. All databases are updated monthly to include the most recent genomes.
In this protocol, we use an 8GB minikraken database of the combined standard Kraken 2 database with the cleaned eukaryotic pathogen genomes.
If the desired database is not available, we describe here how to create a custom Kraken 2 database using the kraken2-build script options:
First, download the NCBI taxonomy.
$ kraken2-build --db krakendb --download-taxonomy
Second, download one or more reference libraries. (The full list of available libraries is at https://github.com/DerrickWood/kraken2/wiki/Manual#custom-databases.)
$ kraken2-build --db krakendb --download-library bacteria $ kraken2-build --db krakendb --download-library archaea $ kraken2-build --db krakendb --download-library viral $ kraken2-build --db krakendb --download-library protozoa $ kraken2-build --db krakendb --download-library UniVec_Core
Third, download additional genomes by adding multi-FASTA or single-FASTA files. The FASTA sequence headers must include either 1) NCBI accession numbers or 2) the text kraken:taxid followed by the taxonomy ID for the genome (e.g. >sequence100|kraken:taxid|9606|). If this requirement is met, the following commands will add the sequences to the database:
$ kraken2-build --db krakendb --add-to-library chr1.fa $ kraken2-build --db krakendb --add-to-library chr2.fa
Finally, build the Kraken 2 database and generate the Bracken database files.
$ kraken2-build --db krakendb --build --threads 8 $ bracken-build -d krakendb -t 8 -k 35 -l 100
Downloading and organizing required data
This section details how to organize the database and samples used in this protocol. You should first download the database from https://genome-idx.s3.amazonaws.com/kraken/k2_standard_eupath_20201202.tar.gz and the samples from NCBI SRA. For details on how to download the SRA samples, see https://www.ncbi.nlm.nih.gov/sra/docs/sradownload/. Table 1 lists the SRA accession IDs for each of the samples used in the Kraken Microbiome Analysis Protocol and the Kraken Pathogen Identification Protocol.
Table 1.
NCBI SRA Samples
| Microbiome Analysis | ||||
| Sample | SRA ID | File size | Description | Expected Diversity |
| 1 | SRR14143424 | 5.9Gb | Paired, Day -2 | Normal |
| 2 | SRR14092160 | 3.4Gb | Paired, Day -9 | Normal |
| 3 | SRR14092310 | 2.3Gb | Paired, Day 12 | Low |
| Pathogen Identification | ||||
| Sample | SRA ID | File size | Case ID | Expected Pathogen |
| 1 | SRR12486971 | 178.2Mb | Case 10 | Anncaliia algerae |
| 2 | SRR12486972 | 318.5Mb | Case 9 | Aspergillus fumigatus |
| 3 | SRR12486974 | 141Mb | Case 7 | Candida albicans |
| 4 | SRR12486978 | 110.5Mb | Case 3 | Mycobacteroides chelonae |
| 5 | SRR12486979 | 112.3Mb | Case 20 | Control |
| 6 | SRR12486981 | 311.1Mb | Case 18 | Control |
| 7 | SRR12486983 | 236.5Mb | Case 16 | HSV 1 |
| 8 | SRR12486988 | 351Mb | Case 11 | Acanthamoeba castellanii |
| 9 | SRR12486989 | 228.2Mb | Case 2 | Streptococcus agalactiae |
| 10 | SRR12486990 | 465.7Mb | Case 1 | Staphylococcus aureus |
Create four folders for required data: k2protocol_db, m_samples, p_samples, b_index
Inside the k2protocol_db folder, unpack the database:
$ tar -xzvf k2_standard_eupath_20201202.tar.gz
Inside the b_index folder, download the k2protocol_bowtie2indices.tgz file from
http://ccb.jhu.edu/data/kraken2_protocol/ and unpack the files:
$ tar -xzvf k2protocol_bowtie2indices.tgz
Download the fastq files for SRA samples for the microbiome analysis and for the pathogen identification analysis. Move the three microbiome analysis files into m_samples and move the 10 pathogen identification samples into p_samples.
Downloading and installing software
There are two ways to install the required programs: (1) using conda and (2) downloading the binaries. Conda is an open-source package manager that helps to install software. All the software tools mentioned here are packaged in Bioconda 29, which is a particular repository (“channel”) within conda. (1) To install the software using conda use the following command:
Install all required using Conda
$ conda install -c conda-forge -c bioconda kraken 2 krakentools bracken \ r bowtie2 samtools
Pavian has no conda package. To run the protocol there are two ways:
(a) Install it locally. Open the R console and use the following command:
$ R
if (!require(remotes)) { install.packages (“remotes”)
remotes::install_github(“fbreitwieser/pavian”)
(b) Use the pavian webserver https://fbreitwieser.shinyapps.io/pavian/.
(2) Alternatively all software can be also downloaded as binary using the following commands:
Create a directory to store all of the executable programs used in this protocol (if none already exists):
$ mkdir $HOME/bin
Add the above directory to your PATH environment variable: $ export PATH=$HOME/bin:$PATH
To install Kraken 2, download the latest release from https://github.com/DerrickWood/kraken2/releases, unpack the Kraken 2 zip archive, cd to the unpacked directory, and run the install script.
$ unzip v2.1.1.zip $ cd kraken2–2.1.1/ $ ./install_kraken 2.sh .
To install Bracken, download Bracken version 2.6.2 from https://github.com/JenniferLu717/Bracken, unpack the Bracken zip archive, cd to the unpacked directory and run the install script.
$ unzip v2.6.2.zip $ cd Bracken-2.6.2/ $ sh install_bracken.sh
To install KrakenTools, download Kraken 2 version 2.1.1 from https://github.com/JenniferLu717/KrakenTools and unpack the KrakenTools zip archive.
$ unzip v1.2
Copy the kraken executables to a directory in your PATH and create symlinks for the Bracken and KrakenTools executables.
$ cp kraken2–2.1.1/kraken2 $HOME/bin $ cp kraken2–2.1.1/kraken2 -build $HOME/bin $ cp kraken2–2.1.1/kraken2 -inspect $HOME/bin $ ln -s Bracken-2.6.2/bracken $HOME/bin/bracken $ ln -s Bracken-2.6.2/bracken -build $HOME/bin/bracken-build $ ln -s KrakenTools $HOME/bin/KrakenTools
Download Bowtie 2 version 2.4.4 (x86 64) from https://github.com/BenLangmead/bowtie2/releases/
$ unzip bowtie2 -2.4.4-linux-x86_64.zip $ cp bowtie2–2.4.4-linux-x86_64/bowtie2* $HOME/bin
To install Pavian, open RStudio or R and run the following:
if (!require(remotes)) { install.packages (“remotes”) }
remotes::install_github(“fbreitwieser/pavian”)
Procedure
There are two protocols included: Microbiome Analysis and Pathogen Identification. The Microbiome Analysis portion details how to use the Kraken suite to analyze the overall diversity of microbiome samples while also statistically differentiating between microbiome compositions. We illustrate this protocol using samples of a patient’s fecal microbiome before and after antibiotic treatment, as studied in Taur et al. 27. The Pathogen Identification protocol demonstrates how to identify pathogenic, atypical microbes within the overall microbiome using a set of 10 corneal samples from Li et al. 28, 8 of which contain pathogenic microbes while 2 are control samples.
Procedure 1: Microbiome Analysis
We will explore and visualize the depletion of patient T11’s microbiota diversity due to antibiotic treatment, using our microbiome analysis pipeline. In the study by Taur et al. 27, all of the patients underwent allogeneic hematopoietic stem cell transplantation (allo-HSCT) where antibiotic treatment is essential for optimal clinical outcomes. However, antibiotics deplete a patient’s microbiome diversity. In the paper, the authors showed that auto-FMT treatment restores the microbial diversity to a patient’s gut microbiome. In this example, we explicitly detail three samples from patient T11 collected before and during antibiotic treatment. Samples 1 and 2 were taken before T11 began antibiotic treatment and sample 3 was collected during treatment.
Remove host DNA
1. Depending on the source of the metagenomic sample, users may remove host DNA by aligning reads to the host genome. The provided sample set has already undergone removal of host (human) DNA. We provide these command lines as a guide for how to remove human or host DNA from other samples using Bowtie 27.
$ bowtie2 -x b_index/GRCh38 -p 8 -1 paired_reads_1.fastq -2 paired_reads_2.fastq \ --un-conc nonhuman_reads.fastq -S human_reads.sam
Classify Microbiome Samples using Kraken
Timing ~10 min
2. Run Kraken 24 to classify all reads, providing each read with its taxonomy identification. Kraken 2 examines the k-mers within a query sequence and uses this information to query a database. In normal usage, the only required inputs are the database name, specified with --db, and the input FASTA file (or 2 files if the reads are paired). Below is the simplest example:
$ kraken2 --db $DBNAME seqs.fq
In our protocol, we specify the number of threads to improve the timing; with more threads the program will generally finish faster. With --report we specify the location and name of the k2report file and output the results of the run to a kraken2 file with > path/to/file. The --minimum-hit-groups flag specifies the minimum number of ”hit groups” needed to make a classification call. Hit groups are overlapping k-mers sharing the same minimizer. Kraken 2 uses minimizers to compress the input genomic sequences, thereby reducing storage memory needed and run time. In this example we increase the minimum number of hit groups from the default 2 groups to 3 groups for increased accuracy. Lastly, the --report-minimizer-data flag reports minimizer and distinct minimizer count information in addition to the normal Kraken 2 report.
$ kraken2 --db k2protocol_db --threads 8 --report kreports/SRR14143424.k2report \ --report-minimizer-data --minimum-hit-groups 3 samples/SRR14143424_1.fastq \ samples/SRR14143424_2.fastq > kraken_outputs/SRR14143424.kraken2 $ kraken2 --db k2protocol_db --threads 8 --report kreports/SRR14092160.k2report \ --report-minimizer-data --minimum-hit-groups 3 samples/SRR14092160_1.fastq \ samples/SRR14092160_2.fastq > kraken_outputs/SRR14092160.kraken2 $ kraken2 --db k2protocol_db --threads 8 --report kreports/SRR14092310.k2report \ --report-minimizer-data --minimum-hit-groups 3 samples/SRR14092310_1.fastq \ samples/SRR14092310_2.fastq > kraken_outputs/SRR14092310.kraken2
Kraken 2 has two output files, a standard Kraken 2 output, .kraken2 file, and a Kraken 2 report, .k2report file. The standard Kraken 2 file outputs lines containing five tab-delimited fields; 1) ”C”/”U” indicating classified or unclassified, 2) sequence ID, 3) taxonomy ID (0 if unclassified), 4) length of sequence in base pairs and 5) a space-delimited list indicating the lowest common ancestor mapping of each k-mer. The Kraken report file outputs lines containing 6 tab-delimited fields; 1) percentage of fragments covered by the clade rooted at this taxon, 2) number of fragments covered by the clade rooted at this taxon 3) number of fragments assigned directly to this taxon, 4) rank code such as S for species, 5) NCBI taxonomic ID and 6) indented scientific name. This information and more can be found in the Kraken 2 manual.
Run Bracken for Abundance Estimation of Microbiome Samples
Timing < 1 min
3. Following classification, we use Bracken 5 to estimate species abundance in each sample. Here is a generic Bracken invocation:
$ bracken -d kraken_database-i sample.k2report -r read_length \ -l taxonomic_level -t read_threshold -o sample.bracken -w sample.breport\
Below are the commands used to generate the abundance estimation in our example. Here we set the read length as the average read length in the dataset: 100bp. We set the level for abundance estimation to Species (with -l S), and with the -t 10 we require 10 reads prior to abundance estimation to perform re-estimation. This effectively removes any species with fewer than 10 reads, thereby removing some noise from low abundance species.
$ bracken -d k2protocol_db -i kreports/SRR14143424.k2report -r 100 -l S -t 10 \ -o bracken_outputs/SRR14143424.bracken -w breports/SRR14143424.breport $ bracken -d k2protocol_db -i kreports/SRR14092160.k2report -r 100 -l S -t 10 \ -o bracken_outputs/SRR14092160.bracken -w breports/SRR14092160.breport $ bracken -d k2protocol_db -i kreports/SRR14092310.k2report -r 100 -l S -t 10 \ -o bracken_outputs/SRR14092310.bracken -w breports/SRR14092310.breport
Calculate Alpha Diversity
Timing ~ 35 sec
4. When trying to measure biodiversity, it is useful to quantify differences within and between ecosystems. Whittaker et al 30 defined several forms of alpha diversity as measures that capture the diversity within a particular ecosystem and can be expressed by the number of species in that ecosystem. Using the KrakenTools alpha_diversity.py script, you can calculate Berger Parker’s 31 (BP), Fisher’s 32 (F), Simpson’s 33 (Si), Inverse Simpson’s (ISi) 33, and Shannon’s 34 (Sh) alpha diversity for each sample after running Kraken 2 and Bracken. You can specify which alpha diversity metric you want to output to terminal with the -a flag, as shown below.
$ python KrakenTools/DiversityTools/alpha_diversity.py \ -f bracken_outputs/SRR14143424.bracken -a BP $ python KrakenTools/DiversityTools/alpha_diversity.py \ -f bracken_outputs/SRR14143424.bracken -a Sh $ python KrakenTools/DiversityTools/alpha_diversity.py \ -f bracken_outputs/SRR14143424.bracken -a F $ python KrakenTools/DiversityTools/alpha_diversity.py \ -f bracken_outputs/SRR14143424.bracken -a Si $ python KrakenTools/DiversityTools/alpha_diversity.py \ -f bracken_outputs/SRR14143424.bracken -a ISi $ python KrakenTools/DiversityTools/alpha_diversity.py \ -f bracken_outputs/SRR14092160.bracken -a BP $ python KrakenTools/DiversityTools/alpha_diversity.py \ -f bracken_outputs/SRR14092160.bracken -a Sh $ python KrakenTools/DiversityTools/alpha_diversity.py \ -f bracken_outputs/SRR14092160.bracken -a F $ python KrakenTools/DiversityTools/alpha_diversity.py \ -f bracken_outputs/SRR14092160.bracken -a Si $ python KrakenTools/DiversityTools/alpha_diversity.py \ -f bracken_outputs/SRR14092160.bracken -a ISi $ python KrakenTools/DiversityTools/alpha_diversity.py \ -f bracken_outputs/SRR14092310.bracken -a BP $ python KrakenTools/DiversityTools/alpha_diversity.py \ -f bracken_outputs/SRR14092310.bracken -a Sh $ python KrakenTools/DiversityTools/alpha_diversity.py \ -f bracken_outputs/SRR14092310.bracken -a F $ python KrakenTools/DiversityTools/alpha_diversity.py \ -f bracken_outputs/SRR14092310.bracken -a Si $ python KrakenTools/DiversityTools/alpha_diversity.py \ -f bracken_outputs/SRR14092310.bracken -a ISi
Calculate Beta Diversity
Timing ~5 sec
5. Beta diversity is useful when trying to examine the change in species diversity between two or more ecosystems. Next, we used the beta_diversity.py script to compute the Bray-Curtis 35 dissimilarity matrix, which will contain the pairwise dissimilarities among three microbiome samples. With this metric, an output of 0 means the two samples are exactly the same and 1 means they are maximally divergent.
$ python KrakenTools/DiversityTools/beta_diversity.py -i \ bracken_outputs/SRR14092160.bracken bracken_outputs/SRR14092310.bracken \ bracken_outputs/SRR14143424.bracken --type bracken
Generate Krona Plots
Timing < 1 min
6. Krona plots are multi-layered pie charts frequently used in metagenomic visualization for viewing data in a phylogenetic hierarchy. Using these plots, we can view the hierarchical output from Bracken in a way to better visualize the number of reads coming from different species, genera, etc. as percentages. We run kreport2krona.py to generate Krona plots 36 using the following commands.
$ python KrakenTools/kreport2krona.py -r breports/SRR14143424.breport \ -o b_krona_txt/SRR14143424.b.krona.txt --no-intermediate-ranks $ KronaScripts/ktImportText b_krona_txt/SRR14143424.b.krona.txt \ -o krona_html/SRR14143424.krona.html $ python KrakenTools/kreport2krona.py -r breports/SRR14092160.breport \ -o b_krona_txt/SRR14092160.b.krona.txt --no-intermediate-ranks $ KronaScripts/ktImportText b_krona_txt/SRR14092160.b.krona.txt \ -o krona_html/SRR14092160.krona.html $ python KrakenTools/kreport2krona.py -r breports/SRR14092310.breport \ -o b_krona_txt/SRR14092310.b.krona.txt --no-intermediate-ranks $ KronaScripts/ktImportText b_krona_txt/SRR14092310.b.krona.txt \ -o krona_html/SRR14092310.krona.html
Generate Pavian Plots using the Shiny App
Timing < 1 min
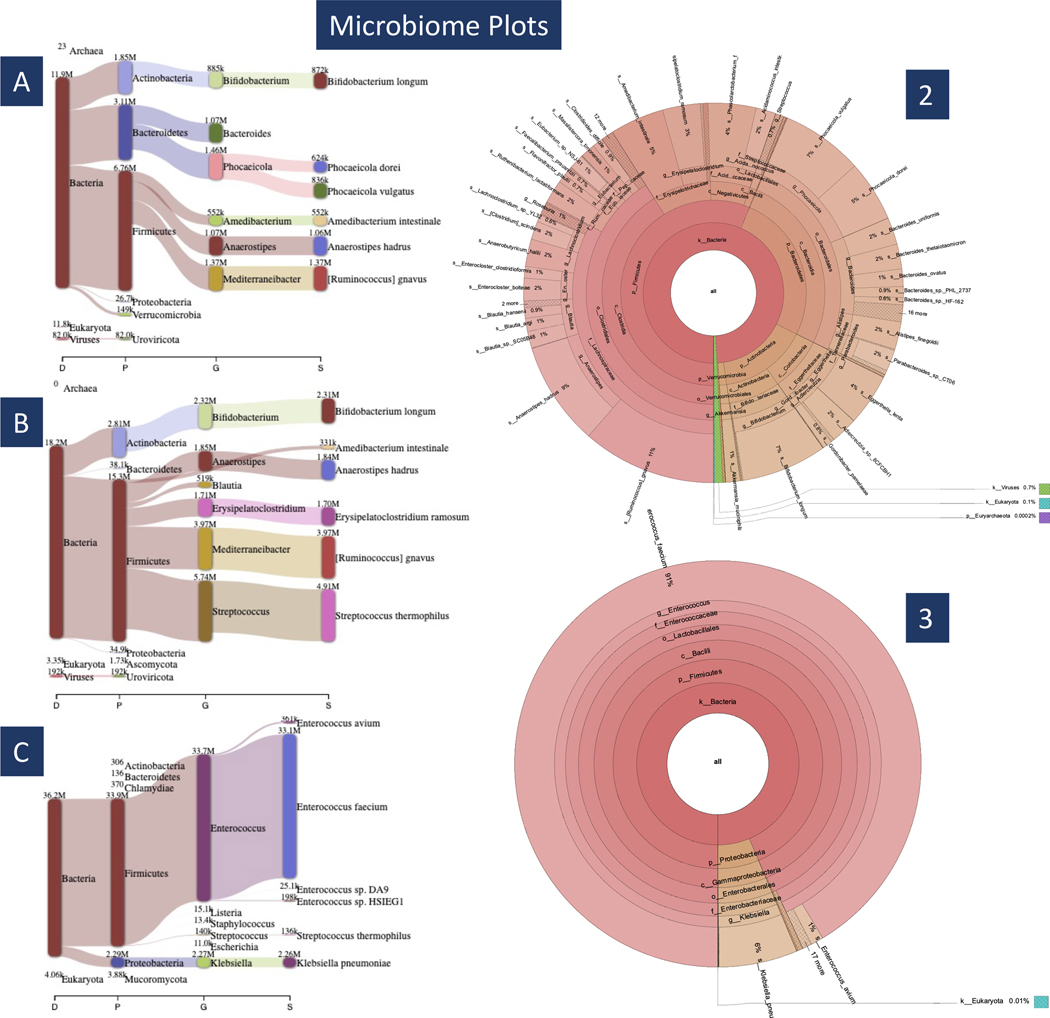
7. Using the .breport files from the Bracken run, you can use Pavian to create the plots shown in Figure 6A, B and C. Figure 2 shows the Pavian interface and the steps for creating classification networks. Open the Pavian shiny app via https://fbreitwieser.shinyapps.io/pavian/.
Fig 6. Microbiome Plots.
In subplots A–C we see Pavian visualization for samples 1–3. Samples 1 and 2 have a similar taxonomic breakdown (A & B), corresponding to T11’s normal microbiome diversity. Subplot C shows that sample 3 is dominated by a few bacteria when T11 is taking antibiotics. On the right are Krona plots generated from samples 2 and 3. The plot on top (2) shows the diversity of patient T11 before receiving any antibiotic treatment (sample 2) and the plot below it (3) shows how depleted his microbiome diversity while he is taking antibiotics (sample 3).
Fig 2. Pavian Output for Hierarchical Visualization.
Upon (1) opening the Pavian app, users should (2) upload the microbiome sample files. (3) Choose ”Sample” to view classification visualization results. (4) Select sample from the drop-down menu. (5) Select plot settings to customize visualization. (6) Save image of network.
8. Click ”Browse...” and Upload the 3 separate SRR*.breport files.
9. Once files are uploaded, click ”Sample” to see the hierarchical classification visualization results.
10. Click on the drop-down list to select the sample that you are viewing.
11. (Optional) Choose ”Configure Sankey” to change what taxonomical ranks to display, number of taxa at each level, etc. there are 10 different settings that can be changed to customize the plots.
12. Save the network by clicking ”Save Network”
Procedure 2: Pathogen Identification
Remove Human DNA using Bowtie 2
1. (Optional) Remove human DNA from the sample set. The provided sample set has already undergone removal of host (human) DNA using the steps shown next, so they do not need to be run again. We provide these command lines as a guide for how to remove human or host DNA from other samples using Bowtie 27.
$ bowtie2 -x bowtie_index/GRCh38 -p 8 -f -1 paired_reads_1.fastq \ -2 paired_reads_2.fastq –-un-conc nonhuman_reads.fa -S human_reads.sam
Classify Reads with Kraken2-Uniq
Timing ~6min, 42 seconds for 10 samples (8 Gb)
2. Following removal of host DNA, the remaining reads undergo classification using Kraken2Uniq. The --report-minimizer-data flag forces Kraken 2 to provide unique k-mer counts per classification. Additionally, we use 8 threads for faster run times, and --minimum-hit-groups 3 for increased classification precision (i.e., fewer false positives).
$ kraken2 --db k2protocol_db --threads 8 --minimum-hit-groups 3 \ --report-minimizer-data --report SRR12486971.k2report \ --paired SRR12486971_1.fastq SRR12486971_2.fastq > SRR12486971.kraken2 $ kraken2 --db k2protocol_db --threads 8 --minimum-hit-groups 3 \ --report-minimizer-data --report SRR12486972.k2report \ --paired SRR12486972_1.fastq SRR12486972_2.fastq > SRR12486972.kraken2 $ kraken2 --db k2protocol_db --threads 8 --minimum-hit-groups 3 \ --report-minimizer-data --report SRR12486974.k2report --paired SRR12486974_1.fastq SRR12486974_2.fastq > SRR12486974.kraken2 $ kraken2 --db k2protocol_db --threads 8 --minimum-hit-groups 3 \ --report-minimizer-data --report SRR12486978.k2report \ --paired SRR12486978_1.fastq SRR12486978_2.fastq > SRR12486978.kraken2 $ kraken2 --db k2protocol_db --threads 8 --minimum-hit-groups 3 \ --report-minimizer-data --report SRR12486979.k2report \ --paired SRR12486979_1.fastq SRR12486979_2.fastq > SRR12486979.kraken2 $ kraken2 --db k2protocol_db --threads 8 --minimum-hit-groups 3 \ --report-minimizer-data --report SRR12486981.k2report \ --paired SRR12486981_1.fastq SRR12486981_2.fastq > SRR12486981.kraken2 $ kraken2 --db k2protocol_db --threads 8 --minimum-hit-groups 3 \ --report-minimizer-data --report SRR12486983.k2report \ --paired SRR12486983_1.fastq SRR12486983_2.fastq > SRR12486983.kraken2 $ kraken2 --db k2protocol_db --threads 8 --minimum-hit-groups 3 \ --report-minimizer-data --report SRR12486988.k2report \ --paired SRR12486988_1.fastq SRR12486988_2.fastq > SRR12486988.kraken2 $ kraken2 --db k2protocol_db --threads 8 --minimum-hit-groups 3 \ --report-minimizer-data --report SRR12486989.k2report \ --paired SRR12486989_1.fastq SRR12486989_2.fastq > SRR12486989.kraken2 $ kraken2 --db k2protocol_db --threads 8 --minimum-hit-groups 3 \ --report-minimizer-data --report SRR12486990.k2report \ --paired SRR12486990_1.fastq SRR12486990_2.fastq > SRR12486990.kraken2
Compare Samples to Controls using Pavian
Timing ~5 min
3. Following classification, we compare samples using Pavian 6 (Figure 3) Open the Pavian shiny app via https://fbreitwieser.shinyapps.io/pavian/.
Fig 3. Pavian Output for Pathogen Identification.
Upon (1) opening the Pavian app, users should (2) upload the pathogen sample files. (3) Choose “Comparison” to view the table of read counts per sample per taxon. (4) Select ‘Species’ and ‘Z-score (reads)’ to filter the table and calculate z-scores. (5). Finally, sort by max z-scores to focus on species that are most likely pathogen candidates.
4. Click ”Browse...” and upload the 10 separate SRR*.kreport2 files. Once files are uploaded, click ”Comparison” to see how read counts vary across samples.f
5. Choose ”Species” and ”Z-score (reads)” to focus on species-level reads and calculate z-scores.
6. Sort by maximum z-scores.
The species with the max z-score for each sample is the most likely pathogen. Note that if two or more pathogens have a high z-score in a given sample, we should consider the possibility that the infection has multiple causes. Species with high read counts (or high z-scores) across all samples are considered background noise or contamination.
Verify Classification
Timing ~30 min
7. For verifying the classification results, use the extract_kraken_reads.py script to extract reads for each of the top z-score species for each sample. This script extracts reads that matched a particular species, identified by the taxonomy ID that is provided with the -t parameter.
$ python extract_kraken_reads.py -k SRR12486971.kraken2 -–include-children \ -s SRR12486971_1.fastq -s2 SRR12486971_2.fastq -t 723287 \ -r SRR12486971.k2report -o SRR12486971_A.algerae.tid723287.1.fa \ -o2 SRR12486971_A.algerae.tid723287.2.fa $ python extract_kraken_reads.py -k SRR12486972.kraken2 –-include-children \ -s SRR12486972_1.fastq -s2 SRR12486972_2.fastq -t 5059 \ -r SRR12486972.k2report -o SRR12486972_A.flavus.tid5059.1.fa \ -o2 SRR12486972_A.flavus.tid5059.2.fa $ python extract_kraken_reads.py -k SRR12486974.kraken2 –-include-children \ -s SRR12486974_1.fastq -s2 SRR12486974_2.fastq -t 5476 \ -r SRR12486974.k2report -o SRR12486974_C.albicans.tid5476.1.fa \ -o2 SRR12486974_C.albicans.tid5476.2.fa $ python extract_kraken_reads.py -k SRR12486978.kraken2 –-include-children \ -s SRR12486978_1.fastq -s2 SRR12486978_2.fastq -t 1774 \ -r SRR12486978.k2report -o SRR12486978_M.chelonae.tid1774.1.fa \ -o2 SRR12486978_M.chelonae.tid1774.2.fa $ python extract_kraken_reads.py -k SRR12486983.kraken2 –-include-children \ -s SRR12486983_1.fastq -s2 SRR12486983_2.fastq -t 10298 \ -r SRR12486983.k2report -o SRR12486983_HSV1.tid10298.1.fa \ -o2 SRR12486983_HSV1.tid10298.2.fa $ python extract_kraken_reads.py -k SRR12486988.kraken2 --include–children \ -s SRR12486988_1.fastq -s2 SRR12486988_2.fastq -t 61605 \ -r SRR12486988.k2report -o SRR12486988_A.lugunensis.tid61605.1.fa \ -o2 SRR12486988_A.lugdunensis.tid61605.2.fa $ python extract_kraken_reads.py -k SRR12486989.kraken2 --include-children \ -s SRR12486989_1.fastq -s2 SRR12486989_2.fastq -t 1311 \ -r SRR12486989.k2report -o SRR12486989_S.agalactiae.tid1311.1.fa \ -o2 SRR12486989_S.agalactiae.tid1311.2.fa $ python extract_kraken_reads.py -k SRR12486990.kraken --include-children \ -s SRR12486990_1.fastq -s2 SRR12486990_2.fastq -t 1280 \ -r SRR12486990.k2report -o SRR12486990_S.aureus.tid1280.1.fa \ -o2 SRR12486990_S.aureus.tid1280.2.fa
8. Align extracted reads to the species genomes using alignment programs such as Bowtie 27 or Minimap226. We use Bowtie 2 for alignment.
$ bowtie2 -x b_index/Aalgerae -f -p 8 \ -1 SRR12486971_A.algerae.tid723287.1.fa \ -2 SRR12486971_A.algerae.tid723287.2.fa \ -S SRR12486971_A.algerae_aligned.sam $ bowtie2 -x b_index/Aflavus -f -p 8 \ -1 SRR12486972_A.flavus.tid5059.1.fa \ -2 SRR12486972_A.flavus.tid5059.2.fa \ -S SRR12486972_A.flavus_aligned.sam $ bowtie2 -x b_index/Calbicans -f -p 8 \ -1 SRR12486974_C.albicans.tid5476.1.fa \ -2 SRR12486974_C.albicans.tid5476.2.fa \ -S SRR12486974_C.albicans_aligned.sam $ bowtie2 -x b_index/Mchelonae -f -p 8 \ -1 SRR12486978_M.chelonae.tid1774.1.fa \ -2 SRR12486978_M.chelonae.tid1774.2.fa \ -S SRR12486978_M.chelonae_aligned.sam $ bowtie2 -x b_index/HSV1 -f -p 8 \ -1 SRR12486983_HSV1.tid10298.1.fa \ -2 SRR12486983_HSV1.tid10298.2.fa \ -S SRR12486983_HSV1_aligned.sam $ bowtie2 -x b_index/Alug -f -p 8 \ -1 SRR12486988_A.lugunensis.tid61605.1.fa \ -2 SRR12486988_A.lugdunensis.tid61605.2.fa \ -S SRR12486988_Alug_aligned.sam $ bowtie2 -x b_index/Sagalactiae -f -p 8 \ -1 SRR12486989_S.agalactiae.tid1311.1.fa \ -2 SRR12486989_S.agalactiae.tid1311.2.fa \ -S SRR12486989_Sagalactiae_aligned.sam $ bowtie2 -x b_index/Saureus -f -p 8 \ -1 SRR12486990_S.aureus.tid1280.1.fa \ -2 SRR12486990_S.aureus.tid1280.2.fa \ -S SRR12486990_Saureus_aligned.sam
9. Following alignment, we use SAMtools 37 to convert the SAM files to sorted BAM files.
$ samtools view -bS -F4 SRR12486971_A.algerae_aligned.sam \ > SRR12486971_A.algerae_aligned.bam $ samtools view -bS -F4 SRR12486972_A.flavus_aligned.sam \ > SRR12486972_A.flavus_aligned.bam $ samtools view -bS -F4 SRR12486974_C.albicans_aligned.sam \ > SRR12486974_C.albicans_aligned.bam $ samtools view -bS -F4 SRR12486978_M.chelonae_aligned.sam \ > SRR12486978_M.chelonae_aligned.bam $ samtools view -bS -F4 SRR12486983_HSV1_aligned.sam \ > SRR12486983_HSV1_aligned.bam $ samtools view -bS -F4 SRR12486988_Alug_aligned.sam \ > SRR12486988_Alug_aligned.bam $ samtools view -bS -F4 SRR12486989_Sagalactiae_aligned.sam \ > SRR12486989_Sagalactiae_aligned.bam $ samtools view -bS -F4 SRR12486990_Saureus_aligned.sam \ > SRR12486990_Saureus_aligned.bam $ samtools sort SRR12486971_A.algerae_aligned.bam \ -o SRR12486971_A.algerae_sorted.bam $ samtools sort SRR12486972_A.flavus_aligned.bam \ -o SRR12486972_A.flavus_sorted.bam $ samtools sort SRR12486974_C.albicans_aligned.bam \ -o SRR12486974_C.albicans_sorted.bam $ samtools sort SRR12486978_M.chelonae_aligned.bam \ -o SRR12486978_M.chelonae_sorted.bam $ samtools sort SRR12486983_HSV1_aligned.bam \ -o SRR12486983_HSV1_sorted.bam $ samtools sort SRR12486988_Alug_aligned.bam \ -o SRR12486988_Alug_sorted.bam $ samtools sort SRR12486989_Sagalactiae_aligned.bam \ -o SRR12486989_Sagalactiae_sorted.bam $ samtools sort SRR12486990_Saureus_aligned.bam \ -o SRR12486990_Saureus_sorted.bam $ samtools index SRR12486971_A.algerae_sorted.bam $ samtools index SRR12486972_A.flavus_sorted.bam $ samtools index SRR12486974_C.albicans_sorted.bam $ samtools index SRR12486978_M.chelonae_sorted.bam $ samtools index SRR12486983_HSV1_sorted.bam $ samtools index SRR12486988_Alug_sorted.bam $ samtools index SRR12486989_Sagalactiae_sorted.bam $ samtools index SRR12486990_Saureus_sorted.bam
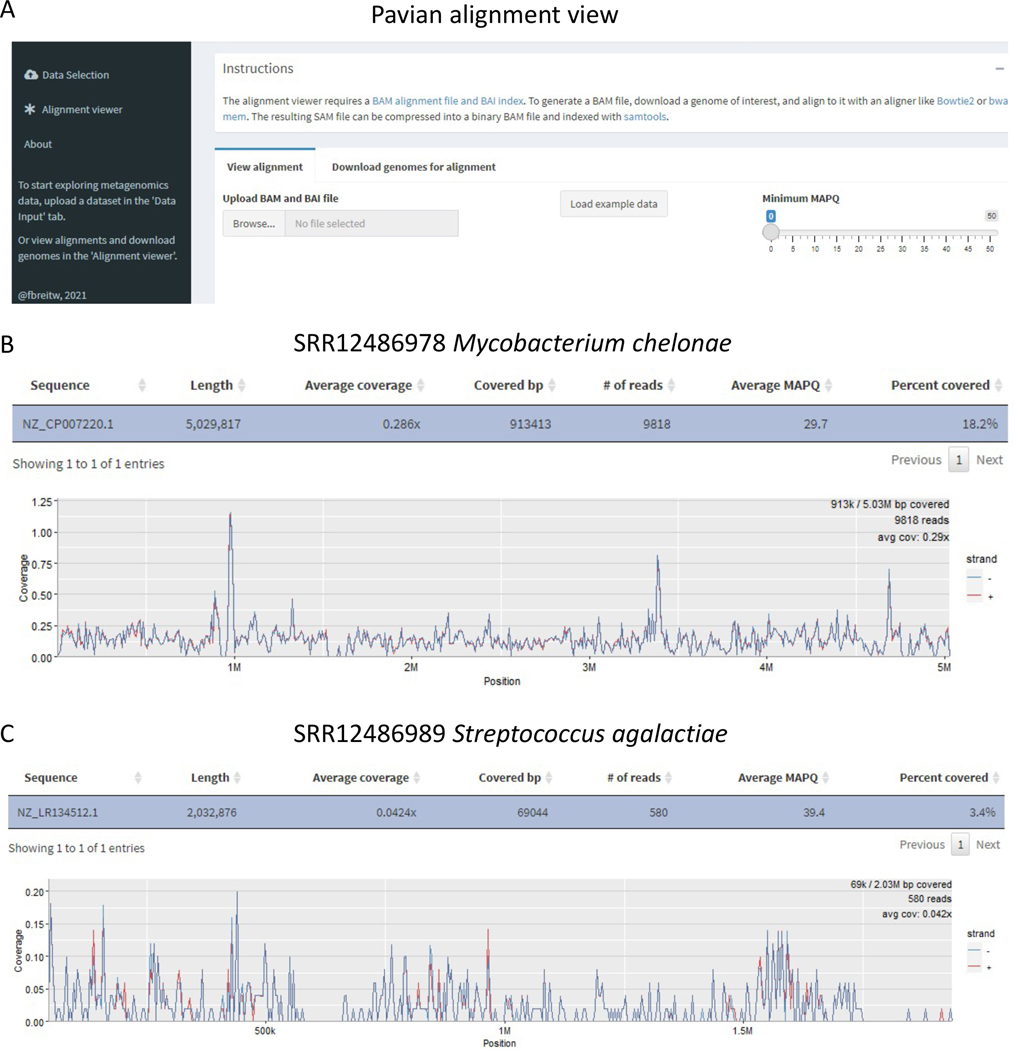
10. Finally, upload the sorted BAM file and the index of the sorted BAM file (.bam and .bai files) to Pavian’s Alignment Viewer for the coverage plot and statistics. Figure 4 shows example coverage plots for the SRR12486978 reads aligned to the Mycobacterium chelonae genome and the SRR12486989 reads aligned to the Streptococcus agalactiae genome.
Fig 4. Pavian Alignment Viewer.
A) shows the graphical interface of Pavian’s alignment viewer. Users should upload the .bam and .bai files to the alignment viewer. B) and C) show two example coverage plots for the pathogen identification samples. Pavian displays the coverage plot along with summarizing coverage statistics.
Timing
Troubleshooting
Step 2. Run Kraken 2 results in large numbers of unclassified reads
If your Kraken 2 output files have too many unclassified reads, it probably means there are organisms missing from your working Kraken 2 database. To classify more reads, you should download additional genomes and add them to the database.
Steps 3–6. After Kraken 2 and Bracken calls
If there are any errors during these steps, you should check that your kraken2 and bracken files were successfully made. For example, if the output from kraken2 looks like this, there was a problem in the kraken2 call:
Loading database information … done. 0 sequences (0.00 Mbp) processed in 0.001s (0.0 Kseq/m, 0.00 Mbp/m). 0 sequences classified (-nan %) 0 sequences unclassified (-nan %)
The command used to create the erroneous output shown above is as follows:
for sample in SRR14092160 SRR14092310; do kraken2 --db k2protocol_db --threads 8 --report kreports/$sample.k2report \ --report-minimizer-data --minimum-hit-groups 3 samples/$sample_1.fasta \ samples/$sample_2.fasta > kraken_outputs/$sample.kraken2 done
The problem in this example is the bash syntax. Here, the input filenames are not interpolated correctly because the “$sample” variable is not protected against trailing underscores in the filenames. Therefore, bash is then attempting to reference a nonexistent variable. In this example, the commands that follow will throw an error because 0 sequences were classified and the .kraken2 file is empty. To fix this, you should check that you are referencing the files correctly. The output of a successful Kraken 2 run for sample SRR14092160 is as follows (you can specify as many samples as you want, separated by spaces):
Loading database information … done. 73021500 sequences (8907.00 Mbp) processed in 154.050 s (28440.6 Kseq/m, 3469 Mbp/m). 11996432 sequences classified (16.43%) 61025068 sequences unclassified (83.57%)
There should be non-zero values for the number of sequences processed and for the number of reads classified. This can be done by protecting the code via some form of quoting. The following command, with double quotes around the filenames, does this:
for sample in SRR14092160 SRR14092310; do
kraken2 --db k2protocol_db --threads 8 --report “ kreports/${sample}.k2report” \
--report-minimizer-data --minimum-hit-groups 3 “samples/${sample}_1.fasta” \
“samples/${sample}_2.fasta” > “kraken_outputs/${sample}.kraken2”
done
Anticipated results
The accuracy of classification and abundance estimation is dependent on the quality and composition of the genomes used in the Kraken and Bracken databases. If the database is missing genomes present in the sequenced samples, there might be a high proportion of unclassified reads when using Kraken. If the database contains contaminated genomes (e.g. bacteria that have bits of human sequence in them20;21), this will lead to false positives, where sequencing reads will be identified as incorrect species. Unclassified reads and mis-classified reads will lead to additional problems downstream for Bracken, which will only report abundances for species identified by Kraken. To ensure a higher classification percentage, we recommend using a comprehensive Kraken database containing, at minimum, the human genome and bacterial, viral, archaeal, vector, and eukaryotic pathogen genomes. For a low rate of false positives, we recommend using only complete or quality-controlled genomes in the building of the Kraken database.
In this protocol, we use a ”MiniKraken” version of this comprehensive database that includes the human genome, complete bacterial, viral, and archaeal genomes from RefSeq, and a filtered eukaryotic pathogen database. The MiniKraken version of the comprehensive database contains sequences from all of the aforementioned genomes, but it down-samples the k-mers from each genome sufficiently to allow Kraken to run with only 8GB of RAM. Kraken should still detect accurate bacterial, viral, archaeal, and eukaryotic microbial signatures, but as we limit the number of sequences, we also expect a significant fraction of unclassified reads.
Microbiome Analysis
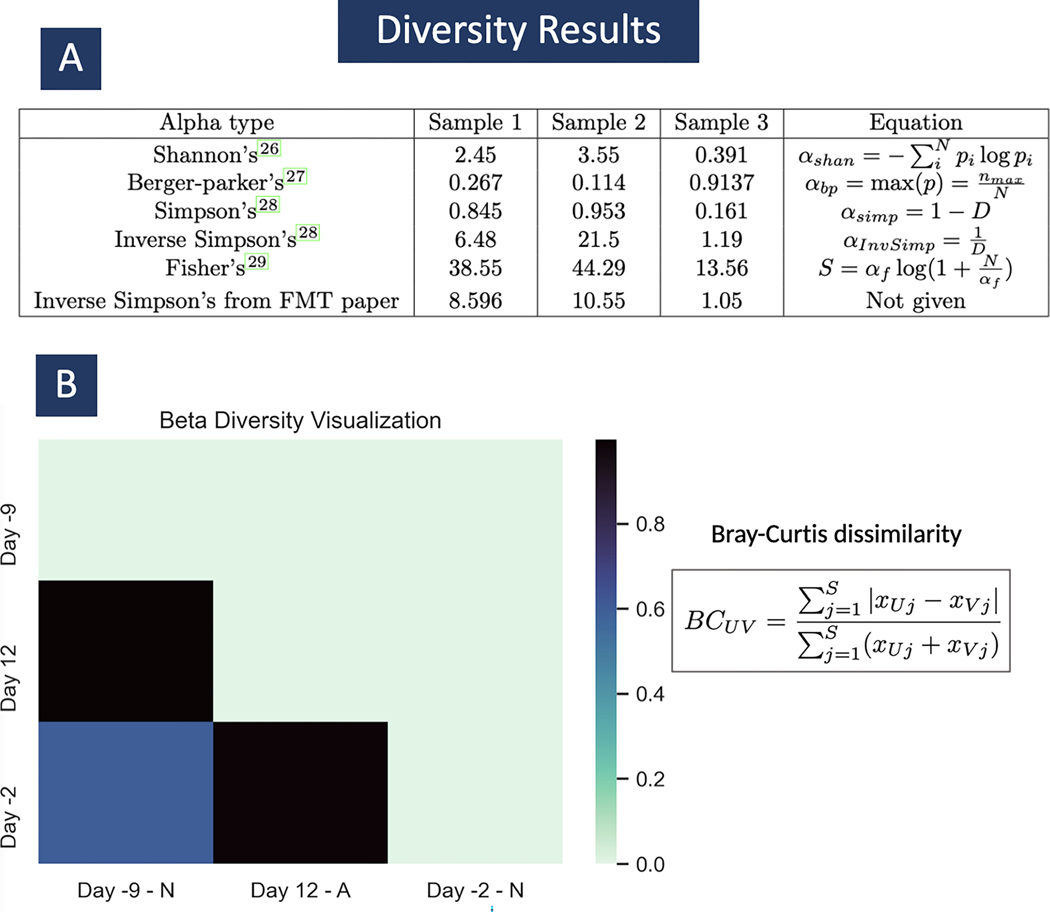
The three metagenomic sequencing samples that we use in this protocol illustrate the normal state of patient T11’s microbiome prior to treatment and its depleted diversity state after antibiotic treatment. (The original study also measured diversity after a fecal microbial transplant restored much of the microbiome.) Supplemental Table 1 lists the read counts per species in the final Bracken analysis of the three samples. In Figure 5, we can find the computed alpha diversity outputs for each sample, for each alpha diversity type currently available. We calculate these values using the abundance estimation calculation from Bracken using KrakenTools’ alpha diversity script. In samples 1 and 2, both taken when patient T11 had normal microbiome diversity, we expect to see a higher value for alpha diversity than in sample 3, which was taken during antibiotic treatment. In Figure 5 A, we see exactly that across the board for all the alpha diversity types. In the original study, they published the alpha diversity values shown in the last row of table A. They specified that it is Inverse Simpson’s alpha diversity however they did not include the exact tool or equations they used to find these values. The Inverse Simpson’s alpha value on row 4 of table A is the value we found using Bracken’s abundance estimation and using the equations shown. For sample 2, we found a significant discrepancy in the alpha diversity published in the original paper (10.55) and our calculation (21.5). However, this can be explained by the large percentage of unclassified reads for this sample (about 80%) and a difference in formulation. Since the Kraken 2 report has many unclassified reads, the Bracken abundance estimation is not being calculated for the entire sample but only for the 20% of reads that are classified. In order to have more classified reads, a larger database needs to be used.
Fig 5. Alpha and Beta diversity results.
In subplot A, we can see the computed results for alpha diversity. In the equations p is a vector of pis where for all i species i.e. . And . In subplot B, we can see a heatmap of the 3 samples from 3 different time points in patient T11’s treatment. The sample taken on day 12, was taken while T11 was taking antibiotics, marked with an ‘A’. The samples taken from days -9 and -2, were taken before the commencement of antibiotic treatment, marked with an ‘N’. Here we use beta_diversity.py to compare diversity across samples. This is the Bray-Curtis dissimilarity matrix. We see that the two samples taken before commencement of treatment are more similar to each other than either sample compared to sample A, from day 12.
Figure 5 also shows the output of the beta diversity calculation based on the Bray-Curtis dissimilarity matrix shown in section B. The heat map shows that when comparing the same two samples, for example Day -2 (sample 1) with itself, the beta diversity value is 0 because the two samples being compared are exactly the same. When comparing Day -2 with Day 12 (sample 3) the beta diversity is close to 1 because the diversity levels in each are very different from each other. Also, as expected, we see that samples 1 and 2 (day -2 and -9) are somewhat similar with a beta diversity value of 0.6.
In Figure 6, we have the Pavian and two of the Krona plots we created. First, focusing on the left-side, subplots A-C are the Pavian plots for samples 1–3 respectively. Here we see that plot C is dominated by a single species of bacteria, Enterococcus faecium, showing patient T11 only had one species of bacteria in his microbiome while undergoing antibiotic treatment. In plots A and B, we see several species of bacteria showing T11’s normal variation in microbiome diversity. On the right-side of figure 6, we have two Krona plots made using samples 2 and 3, top to bottom. Here we see the top Krona plot is divided into several different sections which means represent the different species present in the sample. In the bottom plot, we see much fewer partitions showing again how E. faecium dominates the sample.
Using the MiniKraken database, there will be a large number of unclassified reads, as many as 42,899,387 in sample 1. For plotting purposes, we simply discarded all unclassified reads from the kreport2krona.py output when creating the plots shown in Figure 6. All of the visualizations methods used, Krona plot, Pavian plot, and Beta diversity heatmap, show the great difference in microbiome diversity between patient T11 before and during antibiotic treatment.
Pathogen Detection
Because we are using the MiniKraken version of our comprehensive database, we expect a high proportion of unclassified reads across all infectious disease samples. Table 3 lists the total read counts per sample and the number of unclassified and classified reads per sample. However, because the MiniKraken database was selected from a comprehensive set of genomes, we still expect to see classifications across all major taxonomic clades (e.g. bacteria, archaea, viruses, fungi, etc.). Table 4 lists the read counts for major clades, showing 80K–2.7M bacterial reads per sample, along with varying numbers of reads from archaea, fungi, and amoebae. Full species read counts for each sample are listed in Supplemental Table 2.
Table 3.
Classification read counts from corneal samples using the MiniKraken database
| Sample | Total Reads | Unclassified Reads | Unclassified % | Classified Reads | Classified % |
|---|---|---|---|---|---|
|
| |||||
| SRR12486971 | 3,664,512 | 2,899,189 | 79.1% | 765,323 | 20.9% |
| SRR12486972 | 7,594,644 | 7,285,624 | 95.9% | 309,020 | 4.1% |
| SRR12486974 | 3,335,998 | 3,162,788 | 94.8% | 173,210 | 5.2% |
| SRR12486978 | 2,625,249 | 2,496,107 | 95.1% | 129,142 | 4.9% |
| SRR12486979 | 2,371,302 | 2,023,600 | 85.3% | 347,702 | 14.7% |
| SRR12486981 | 6,730,160 | 6,093,775 | 90.5% | 636,385 | 9.5% |
| SRR12486983 | 4,819,760 | 3,234,956 | 67.1% | 1,584,804 | 32.9% |
| SRR12486988 | 8,369,736 | 8,122,882 | 97.1% | 246,854 | 2.9% |
| SRR12486989 | 5,440,369 | 5,252,849 | 96.6% | 187,520 | 3.4% |
| SRR12486990 | 9,402,750 | 6,619,669 | 70.4% | 2,783,081 | 29.6% |
Table 4. Cornea samples per-clade read counts.
This table lists the per-clade read counts for the major clades in these samples. Only select major clades are listed with read counts for clarity.
| Sample | Bacteria | Archaea | Virus | Fungi | Amoeba |
|---|---|---|---|---|---|
|
| |||||
| SRR12486971 | 649,685 | 34 | 228 | 85,396 | 45 |
| SRR12486972 | 150,302 | 10 | 50 | 31,602 | 54 |
| SRR12486974 | 85,361 | 7 | 14 | 36,833 | 18 |
| SRR12486978 | 91,448 | 2 | 763 | 87 | 3 |
| SRR12486979 | 317,804 | 21 | 64 | 885 | 36 |
| SRR12486981 | 465,581 | 51 | 157 | 2,082 | 110 |
| SRR12486983 | 888,749 | 44 | 646,017 | 1,496 | 54 |
| SRR12486988 | 80,095 | 12 | 698 | 325 | 5587 |
| SRR12486989 | 97,912 | 7 | 37 | 256 | 83 |
| SRR12486990 | 2,674,036 | 152 | 1,109 | 4,449 | 166 |
Despite the many clades detected in the corneal samples, the diagnostic challenge is determining the most likely pathogen(s) per sample. Samples SRR12486979 and SRR12486981 are control samples, providing a baseline for read counts from species are likely to be contaminants or possibly non-infectious components of the samples. For each of the remaining patient samples, we are interested in any species with higher than average read counts as compared to the control samples or other corneal samples. For example, sample SRR12486971 has 84,000+ Anncaliia algerae reads. All other samples have 12 A. algerae reads or fewer, making it appear that A. algerae is a likely pathogen in that sample (which indeed it is, as was confirmed in the original study).
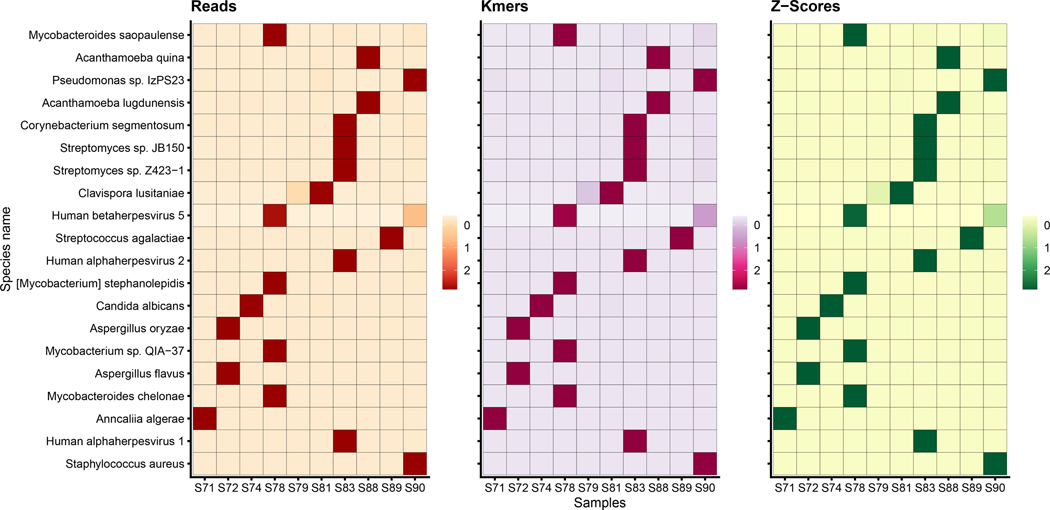
The easiest method for finding the most likely pathogen is by uploading a set of similar samples to Pavian 6, as described in Step 3 of our Pathogen Identification protocol. Using the comparison tab, Pavian can calculate and sort the species by z-scores, a metric used to determine whether a species (or any other taxonomic clade) has significantly higher read counts in one sample as compared to the remaining other samples. Table 5 lists the species with the highest z-score in each sample. As shown in the table, the highest z-score correctly identified the true pathogen for each of the non-control samples.
Table 5. Species with the highest Z-scores in cornea samples.
This table lists the species with the highest z-scores for each of non-control samples.
| Sample | True Infection | Z-score Species | Taxid | Reads | Z-score |
|---|---|---|---|---|---|
|
| |||||
| SRR12486971 | Anncaliia algerae | Anncaliia algerae | 723287 | 84,409 | 56930 |
| SRR12486972 | Aspergillus flavus | Aspergillus flavus | 5059 | 3,814 | 3814 |
| SRR12486974 | Candida albicans | Candida albicans | 5476 | 36,609 | 1452 |
| SRR12486978 | Mycobacterium chelonae | Mycobacterium chelonae | 1774 | 11,320 | 3817 |
| SRR12486979 | Control | - | - | - | - |
| SRR12486981 | Control | - | - | - | - |
| SRR12486983 | HSV 1 | HSV 1 | 10298 | 635,691 | 65960 |
| SRR12486988 | Acanthamoeba | Acanthamoeba lugdunensis | 61605 | 1,004 | 338 |
| SRR12486989 | Streptococcus agalactiae | Streptococcus agalactiae | 1311 | 797 | 797 |
| SRR12486990 | Staphylococcus aureus | Staphylococcus aureus | 1280 | 1,414,661 | 76330 |
The highest z-score species can be further checked by using the k-mer-counting feature in KrakenUniq3 (which is also available as a parameter in Kraken 2). As described in the KrakenUniq paper, for most species in a metagenomics sample, each read will be a random sample from the genome, which means that each read will tend to contribute approximately r-k distinct k-mers to the k-mer count, where r is the read length. However, if a bacterial genome is contaminated with a small amount of human sequence (for example), then large numbers of human reads will incorrectly match a small part of that genome. In these cases, the number of distinct k-mers that are observed from a genome will be very small compared to the number of reads. Thus, if the k-mer counts listed in the Kraken 2 report are relatively small, the species is probably not present but instead is a false positive caused by a contaminated genome in the database. Figure 7 summarizes the pathogen identification results for each individual sample, displaying a heat map of reads, k-mers, and z-scores across all samples.
Fig 7. Pathogen Identification Results.
The above plot summarizes the Kraken2Uniq results across the 10 corneal samples. The number of reads, number of k-mers, and z-scores reveals the most likely pathogen for each sample. For example, Acanthamoeba quina has high read and k-mer counts in S88 alone, Staphylococcus aureus is prevalent in S90, and Human alphaherpesvirus 1 is likely to infect S83. For each sample, the true pathogen is the pathogen with the highest z-score for that particular sample.
Finally, we validate the classification results by extracting the classified reads using KrakenTools and aligning the reads against the suspected pathogen genomes using Bowtie 27. We upload the alignment BAM files to Pavian 6 for a clear visualization of the read coverage across the genomes, confirming that the reads are derived from the full pathogen’s genome.
As a result of this protocol, we have a set of potential infectious agents discovered in the samples. The infectious agents should be presented to pathologists and clinicians for further verification with other independent methods.
Supplementary Material
Table 2.
Estimated time required for the microbiome analysis steps
| Step | Description | Timing |
|---|---|---|
|
| ||
| 1 | Download and Setup | 26 min |
| 2 | Classify/Estimate abundance | 10 min |
| 3 | Alpha Diversity | 15 sec |
| 4 | Beta Diversity | 5 sec |
| 5 | Generate Krona | 5 sec |
| 6 | Run Pavian | 10 sec |
|
| ||
| Total | 37 min | |
Acknowledgments
Indexes for tools in the Kraken suite, including the indexes used in this protocol, are made freely available on Amazon Web Services thanks to the AWS Public Dataset Program.
BL was supported by NIH/NIGMS grant R35GM139602 to BL. MS acknowledges support from the National Research Foundation of Korea grant [2019R1A6A1A10073437, 2020M3A9G7103933, 2021R1C1C102065, 2021M3A9I4021220]; New Faculty Startup Fund; and the Creative-Pioneering Researchers Program through Seoul National University.
Footnotes
Competing interests
The authors declare no competing financial interest
Availability
The following website details and links all software and databases used in this protocol: http://ccb.jhu.edu/data/kraken2_protocol/. We also provide easy-to-use jupyter notebooks for both workflows, which can be executed in the browser using Google Collab: https://github.com/martin-steinegger/kraken-protocol/
References
- 1.Rappé MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–394. [DOI] [PubMed] [Google Scholar]
- 2.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014. Mar;15(3):R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breitwieser FP, Baker DN, Salzberg SL. KrakenUniq: confident and fast metagenomics classification using unique k-mer counts. Genome Biol. 2018. Nov;19(1):198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biology. 2019. Nov:762302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu J, Breitwieser FP, Thielen P, Salzberg SL. Bracken: estimating species abundance in metagenomics data. PeerJ Comput Sci. 2017. Jan;3:e104. [Google Scholar]
- 6.Breitwieser FP, Salzberg SL. Pavian: Interactive analysis of metagenomics data for microbiome studies and pathogen identification. Bioinformatics. 2019. Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012. Mar;9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990. Oct;215(3):403–410. [DOI] [PubMed] [Google Scholar]
- 9.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007. Jan;35(Database issue):D61–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016. Jan;44(D1):D733–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ounit R, Wanamaker S, Close TJ, Lonardi S. CLARK: fast and accurate classification of metagenomic and genomic sequences using discriminative k -mers. BMC Genomics. 2015. Mar;16(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim D, Song L, Breitwieser FP, Salzberg SL. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res. 2016. Dec;26(12):1721–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menzel P, Ng KL, Krogh A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun. 2016. Apr;7:11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye SH, Siddle KJ, Park DJ, Sabeti PC. Benchmarking Metagenomics Tools for Taxonomic Classification. Cell. 2019. Aug;178(4):779–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seppey M, Manni M, Zdobnov M. LEMMI: a continuous benchmarking platform for metagenomics classifiers. Genome Research. 2020. Jul; 30: 1208–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012. Jun;9(8):811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vervier K, Mahé P, Tournoud M, Veyrieras JB, Vert JP. Large-scale machine learning for metagenomics sequence classification. Bioinformatics. 2016. Apr;32(7):1023–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo Y, Yu YW, Zeng J, Berger B, Peng J. Metagenomic binning through low-density hashing. Bioinformatics. 2019. Jan;35(2):219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Breitwieser FP, Lu J, Salzberg SL. A review of methods and databases for metagenomic classification and assembly. Brief Bioinform. 2017. Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breitwieser FP, Pertea M, Zimin AV, Salzberg SL. Human contamination in bacterial genomes has created thousands of spurious proteins. Genome Res. 2019. Jun;29(6):954–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steinegger M, Salzberg SL. Terminating contamination: large-scale search identifies more than 2,000,000 contaminated entries in GenBank. Genome Biol. 2020. May;21(1):115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu J, Salzberg SL. Removing contaminants from databases of draft genomes. PLoS Comput Biol. 2018. Jun;14(6):e1006277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015. Jan;12(1):59–60. [DOI] [PubMed] [Google Scholar]
- 24.Mirdita M, Steinegger M, Breitwieser F, Söding J, Levy Karin E. Fast and sensitive taxonomic assignment to metagenomic contigs. Bioinformatics. 2021. Mar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nasko DJ, Koren S, Phillippy AM, Treangen TJ. RefSeq database growth influences the accuracy of k-mer-based lowest common ancestor species identification. Genome Biol. 2018. Oct;19(1):165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018. Sep;34(18):3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taur Y, Coyte K, Schluter J, Robilotti E, Figueroa C, Gjonbalaj M, et al. Reconstitution of the gut microbiota of antibiotic-treated patients by autologous fecal microbiota transplant. Sci Transl Med. 2018. Sep;10(460). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Z, Breitwieser FP, Lu J, Jun AS, Asnaghi L, Salzberg SL, et al. Identifying Corneal Infections in Formalin-Fixed Specimens Using Next Generation Sequencing. Invest Ophthalmol Vis Sci. 2018. Jan;59(1):280–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grüning B, Dale R, Sjödin A, Chapman BA, Rowe J, Tomkins-Tinch CH, et al. Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat Methods. 2018. Jul;15(7):475–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whittaker RH. Evolution and Measurement of Species Diversity. Taxon. 1972;21(2/3):213–251. [Google Scholar]
- 31.Berger WH, Parker FL. Diversity of Planktonic Foraminifera in Deep-Sea Sediments. Science. 1970. Jun;168(3937):1345–1347. [DOI] [PubMed] [Google Scholar]
- 32.Fisher RA, Corbet AS, Williams CB. The Relation Between the Number of Species and the Number of Individuals in a Random Sample of an Animal Population. J Anim Ecol. 1943;12(1):42–58. [Google Scholar]
- 33.Simpson EH. Measurement of Diversity. Nature. 1949. Apr;163(4148):688–688. [Google Scholar]
- 34.Shannon CE. A mathematical theory of communication. The Bell System Technical Journal. 1948. Jul;27(3):379–423. [Google Scholar]
- 35.Bray JR, Curtis JT. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecological Monographs. 1957;27(4):325–349. Available from: https://esajournals.onlinelibrary.wiley.com/doi/abs/10.2307/1942268. [Google Scholar]
- 36.Ondov BD, Bergman NH, Phillippy AM. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics. 2011. Sep;12:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021. Feb;10(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following website details and links all software and databases used in this protocol: http://ccb.jhu.edu/data/kraken2_protocol/. We also provide easy-to-use jupyter notebooks for both workflows, which can be executed in the browser using Google Collab: https://github.com/martin-steinegger/kraken-protocol/