Abstract
Covid-19 has become a worldwide epidemic which has caused the death of millions in a very short time. This disease, which is transmitted rapidly, has mutated and different variations have emerged. Early diagnosis is important to prevent the spread of this disease. In this study, a new deep learning-based architecture is proposed for rapid detection of Covid-19 and other symptoms using CT and X-ray chest images. This method, called CovidDWNet, is based on a structure based on feature reuse residual block (FRB) and depthwise dilated convolutions (DDC) units. The FRB and DDC units efficiently acquired various features in the chest scan images and it was seen that the proposed architecture significantly improved its performance. In addition, the feature maps obtained with the CovidDWNet architecture were estimated with the Gradient boosting (GB) algorithm. With the CovidDWNet+GB architecture, which is a combination of CovidDWNet and GB, a performance increase of approximately 7% in CT images and between 3% and 4% in X-ray images has been achieved. The CovidDWNet+GB architecture achieved the highest success compared to other architectures, with 99.84% and 100% accuracy rates, respectively, on different datasets containing binary class (Covid-19 and Normal) CT images. Similarly, the proposed architecture showed the highest success with 96.81% accuracy in multi-class (Covid-19, Lung Opacity, Normal and Viral Pneumonia) X-ray images and 96.32% accuracy in the dataset containing X-ray and CT images. When the time to predict the disease in CT or X-ray images is examined, it is possible to say that it has a high speed because the CovidDWNet+GB method predicts thousands of images within seconds.
Keywords: Covid-19 diagnosis, Deep learning, Depthwise dilated convolutions, Feature reuse residual block, Gradient boosting
Code metadata
Permanent link to reproducible Capsule: https://doi.org/10.24433/CO.2183919.v1.
1. Introduction
Coronavirus (Covid-19) is a disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-COV-2). After emerging in Wuhan, China in December 2019, it soon spread around the world and became a global pandemic [1]. According to the data of the World Health Organization (WHO), it has been determined that more than 410 million cases have been seen so far, and close to 6 million people have died [2]. WHO declared the coronavirus infection as a Covid-19 pandemic in March 2020 due to the increasing number of deaths and cases. Due to the increasing cases and deaths, many states have had to close their borders to prevent the spread of the pandemic. In addition, many countries have imposed curfews for a certain period as a precaution [3].
This disease usually affects the respiratory system, such as the lungs, and also appears to cause pneumonia-like symptoms [4]. Patients commonly experience symptoms such as fever, cough, sneezing, and shortness of breath. It spreads rapidly through respiratory droplets produced by the cough or sneeze of an infected person. Elderly people and people with chronic illnesses appear to be more prone to Covid-19 infection [5].
One of the most common methods used to diagnose Covid-19 is reverse transcription-polymerase chain reaction (RT-PCR) tests. These tests are performed to determine whether individuals have been infected with SARS-COV-2, the virus that causes Covid-19 disease, momentarily or in the past. The disadvantages of these tests are that test results take time, the number of available RT-PCR test kits is low, and the risk of health personnel contracting the disease during the test is high [6]. It is also costly in that special equipment, materials, and tools are often required for RT-PCR examinations. Therefore, many countries have difficulties in procuring test kits due to budgetary and technical constraints [7]. At the same time, the sensitivity of the RT-PCR test is a cause for concern because of sample and laboratory errors that may occur [8], [9]. Liu et al. [10] have expressed their opinion on the poor performance of RT-PCR in its sensitivity. Similarly, in a study conducted by Drame et al. [11], they expressed their reservations about the use of RT-PCR to determine the viral load in the diagnosis of 2019 coronavirus disease (Covid-19). In addition, it was stated in another study that the sensitivity of these tests could be as low as 38% [12].
Covid-19, which manifests itself as a lung infection, computed tomography (CT) and chest X-ray (X-ray) images are other methods used for the detection of this disease [5]. Typical radiographic features can be reliably detected in patients with pneumonia caused by this disease with CT imaging. Although these methods have some advantages over RT-PCR testing in terms of early detection of Covid-19, specialist physicians are needed to understand and make sense of images. Considering the disadvantages of RT-PCR tests, CT and X-ray imaging techniques used in the diagnosis of Covid-19, Artificial Intelligence (AI), and Deep Learning (DL) based methods are seen as alternative methods. AI and DL methods can help the early diagnosis of this disease and make the treatment process faster by leading experts to reach a fast and accurate diagnosis through CT and X-ray images in the detection process of Covid-19 [13], [14], [15].
Artificial intelligence and deep learning methods are widely used by researchers for the detection of Covid-19 infection from X-ray and CT images. Due to the improved performance of deep learning methods, they are widely used compared to traditional methods. One of the most important reasons that led researchers to this field is that, unlike machine learning and traditional methods, in deep learning architectures, there is no need for feature extraction in the data during the preprocessing stage. Deep learning architectures can be trained with the help of the hyperparameters of the convolutional neural network (CNN) architecture to learn the best features according to the dataset used [3]. Researchers used deep learning methods in many areas classification of white blood cells [16], segmentation of brain MRI images [17], synthetic image generation [18], generating images from EEG signals [19], skin cancer classification [20], fundus image segmentation [21], diagnose different types of Otitis media [22], breast cancer detection [23], breast lymph node segmentation [24], brain disease classification [25], lung segmentation [26], [27], detection of arrhythmia [28], [29], [30] and detecting pneumonia from chest X-ray images [31]. With the pandemic, the use of deep learning methods for the detection of coronavirus symptoms from X-ray and CT images has increased significantly [3].
In literature, it can be seen that many deep learning-based studies have been carried out for the diagnosis of Covid-19 with the help of radiological images [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42].
In the study by Leracitano et al. [32], The authors proposed a fuzzy logic-based deep learning approach to differentiate X-ray images of patients with Covid-19 pneumonia and non-Covid-19-related interstitial pneumonia. The model developed here, called CovNNet, uses the blurry edge detection algorithm together with the blurry images to extract some relevant features from the X-ray images. This study [32] for the detection of Covid-19 from binary class (Covid, Non-Covid) X-ray images, performed poorly compared to many other studies in the literature, with an accuracy rate of 81%. Ahamed et al. [33] proposed a deep learning-based Covid-19 case detection model trained with a dataset of chest CT scans and X-ray images. A modified ResNet50V2 architecture is used as a deep learning architecture in the proposed model. High performance was achieved in this study using two, three, and four classes CT and X-ray images. However, a complex architecture with high processing power was used. Verma et al. [34] proposed different models such as vanilla (vanilla) LSTM, stacked LSTM, ED_LSTM, BiLSTM, CNN, and hybrid CNN+LSTM model to capture the complex trend of the COVID-19 outbreak and perform the Covid-19 prediction. In another study by Khan et al. [35], two new deep learning-based models named deep hybrid learning (DHL) and deep boosted hybrid learning (DBHL) are proposed for effective Covid-19 detection in X-ray datasets. In the proposed DHL architecture, the representation learning capability of the two developed COVID-RENet-1 & 2 models and a machine learning classifier is used separately. In the Covid-RENet model, region and edge-based attention mechanisms were applied to extract boundary features and learn region homogeneity. In addition, the transfer learning method was used in chest X-rays in the proposed architectures. In this study, in which two-class (Covid, Non-Covid) X-ray images are used, it is seen that it has an accuracy of 98.53%. In this study, performance evaluation with only binary class X-ray images is seen as a disadvantage in terms of the performance of the architectures. Because it is important to use different data sets for Covid-19 detection. The success of CNN architectures may vary according to the number of classes and image type.
In the study by Loey et al. [36], a bayesian optimization-based CNN model was proposed for the classification of chest X-ray images. In the proposed model, CNN architecture is used to extract and learn deep features. In addition, CNN hyperparameters are adjusted according to an objective function using a Bayes-based optimizer method. In another study by Lahsaini et al. [37], they used a dataset of Covid and non-Covid CT images validated by RT-PCR tests at Tlemcen hospital in Algeria. A comparative study was carried out on Inception, Resnet-V2, VGG16, VGG19, DenseNet121, DenseNet201, ImageNet, and Xception deep models using the transfer learning method. Also, a model based on DenseNet201 architecture and the GradCam algorithm is proposed. In another study by Toğaçar et al. [38], images were preprocessed using the fuzzy color technique to classify X-ray images. Then, the features obtained with MobileNetV2, and SqueezeNet models were processed with the help of the social mimic optimization method. The productive features obtained were classified using support vector machines (SVM). The DarkCovidNet method developed by Ozturk et al. [39] was used as a classifier for the YOLO real-time object detection system. By applying seventeen convolution layers and adding different filtering to each layer. As in previous studies for the detection of Covid-19, only CT in [37] and only X-ray images were used in [38], [39].
In addition, when the studies were examined, different models were developed by the researchers, defined by the names CoroNet [40], CovidXrayNet [41], and CovXNet [42]. The CoroNet [40] model, which is proposed as a deep CNN model, is based on the Xception architecture pre-trained on the ImageNet dataset. The CovidXrayNet [41] method, based on the EfficientNet-B0 model and based on the optimization method, is proposed. In this study [41], the data augmentation method is used to increase accuracy and CNN hyperparameters are optimized. In the CovXNet [42] technique, deep CNN-based architecture and a model that uses depthwise convolution and varying dilation rates to extract features efficiently are proposed. In the proposed method, different forms of CovXNets are designed and trained with X-ray images of various resolutions. In addition, a stacking algorithm was used to increase the performance rate, and abnormal regions of X-ray images were distinguished by integrating a gradient-based discriminative localization. Looking at the time complexity of the CovXNets architecture (Fig. 10), it was seen that it had a higher time complexity compared to the other architectures studied. This shows that the CovXNets architecture has a complex structure.
Fig. 10.
The time complexity of architectures: (a) Time complexity based on training times (in hours), (b) Time complexity based on test times (in seconds).
When the studies for the detection of Covid-19 disease with deep learning methods are examined, it is seen that researchers generally use X-ray [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53] or CT [54], [55], [56], [57], [58] images, but few studies use both X-ray and CT images [59], [60], [61], [62]. At the same time, it has been seen that the researchers only examined the performance of their architectures on the dataset used or compared them only with traditional architectures. In addition, it has been determined that no performance evaluation has been made according to the training and test times in the literature reviews. In our study, contrary to these studies, CT and X-ray images were used, and a performance evaluation was carried out on the same dataset, including traditional architectures as well as different current architectures. In addition, performance evaluation was made by considering the training and test times of the architectures. The proposed model was developed to reduce the workload of specialist physicians by providing effective, efficient, and rapid detection of Covid-19 and similar cases. With the thought that the CovidDWNet+GB architecture will guide different studies, it has been opened to everyone’s access on the Github page (https://github.com/GaffariCelik/Covid-19).
Our main contributions to this work are listed as follows:
-
•
A new deep learning-based model (CovidDWNet) has been proposed for the detection of Covid-19 and other pneumonia cases.
-
•
The performance of the CovidDWNet architecture has been increased by using multiple feature reuse residual blocks and depthwise dilated convolutions neural networks. In addition, the success rate has been increased by performing the disease prediction process with the Gradient boosting algorithm of the feature vectors obtained with the CovidDWNet architecture.
-
•
By using different CT and X-ray datasets, a real performance evaluation was made among the current architectures in the literature.
2. Material
In this study, three datasets were used: Covid-CT [63] and Sars-Cov-2 [64] datasets containing CT images, and Dataset-X-ray [65] dataset containing X-ray images. These datasets have been made publicly available for researchers to carry out their work. Example images in datasets are given in Fig. 1.
Fig. 1.
Example images are included in the datasets.
The Covid-CT [63] consists of 812 CT images, 349 of which are Covid-19 and 463 normal, taken from 216 patients. This dataset has been validated by a senior radiologist at Tongji Hospital in Wuhan, China, who diagnosed and treated a large number of Covid-19 patients at the time of emergence of this disease between January and April 2019. The Sars-Cov-2 [64] dataset contains a total of 2482 CT chest scan images, of which 1252 are Covid-19 and 1230 are normal. This dataset was obtained from different hospitals in Sao Paulo, Brazil. The Database-X-ray [65] dataset was created for COVID-19 positive cases with the collaboration of a team of researchers from the University of Qatar, Dhaka University, Bangladesh, and medical doctors in Pakistan and Malaysia. This dataset includes X-ray images of Covid-19, normal and other lung infection diseases. It consists of 21165 X-ray images in total, including 3616 Covid-19, 10192 normal, 6012 lung opacity (Non-Covid lung infection), and 1345 Viral pneumonia.
3. Method
As a method, a CNN-based architecture has been proposed for the detection of Covid-19 and other pneumonia symptoms. This architecture is a method based on feature reuse residual block (FRB) and depthwise dilated convolutions (DDC) units.
Convolutional Neural Networks (CNNs) are models that provide high classification performance in multi-class problems and have self-learning capabilities. CNNs are coordinated combinations of multilayer perceptrons, in which every neuron in one layer is associated with all neurons in the next. A convolutional network consists of a convolutional layer and a rectified linear unit (ReLu). Convolution layers form the basic structure of CNN models. Inputs are convolutionalized and applied with nuclei across the entire visual field with convolution filters. Thus, simpler, small patterns are obtained with more complex, detailed patterns. In this way, the hierarchical network structure enables the extraction of the highest feature maps, enhanced generalization capability, and reduced computational complexity [15], [53], [66], [67]. The basic convolution operation can be written mathematically as [33]:
| (1) |
Here, represents the input matrix (may be an image), filter size, and the two-dimensional filter.
3.1. Feature reuse residual block (FRB)
The feature reuse block (FRB) is a widely used technique in computer vision. In this method, feature maps of previous layers are given as input to all subsequent layers. Thus, the performance of the network is highly increased by reusing the features of the previous layers in all subsequent layers [68], [69]. The mathematical formulation of the FRB technique used in this study is defined as follows:
| (2) |
Here, and represent the inputs and outputs of the considered layers, weights, and the combination of those features. The architecture of the FRB technique is presented in Fig. 2(a).
Fig. 2.
Feature reuse residual block architecture-(a) and depthwise dilated convolutions architecture-(b), which constitute the basic structure of the proposed architecture.
3.2. Depthwise dilated convolutions (DDC)
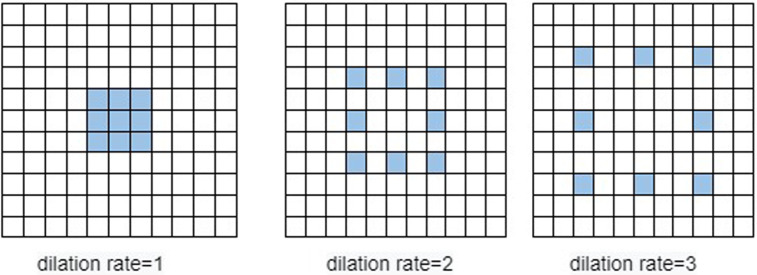
As shown in Fig. 3, dilated convolutions can be expanded in the scope of the convolution kernel by changing the dilation ratio compared to standard convolution. By expanding the scope of the convolution kernel, multi-scale features (information) can be obtained. Choosing a dilation rate of one captures the same properties as standard convolution. However, in dilated convolution, when the dilation ratio is selected as greater than one, more detailed features can be obtained than in standard convolution [70], [71], [72].
Fig. 3.
Dilated Convolution, covering different areas for different dilation ratios when the kernel size () is selected.
In depthwise dilated convolutions operation, the convolution operation is applied to each input channel separately. With point convolution (conventional convolution with 1 × 1 window), inter-channel features are projected into a new space. More efficient features are obtained by using a combination of 1 × 1 convolution and 3 × 3 deep convolution instead of 3 × 3 standard convolution. Therefore, various spatial information is extracted from local information to broader generalized information. In this way, different features extracted with varying dilation ratios with different convolution operations will result in greater diversity in the feature extraction process [42], [73], [74]. The mathematical formulation of depthwise convolution is as follows:
| (3) |
Here, represents layer, , and layer size, and layer index, and , and filter respectively. denotes a learnable convolution filter and an element-wise multiplication operator.
In addition, if the pneumonia disease has spread over a larger area rather than just one region in X-ray images, it may be necessary to combine features from different observation levels. Therefore, the depthwise dilated convolution technique can be used effectively in the diagnosis of pneumonia [42] (see Fig. 4).
Fig. 4.
(a) Normal convolution and (b) depthwise convolutions operations. In depthwise convolutions, the number of filters is equal to the number of channels of the input [75].
3.3. Gradient boosting (GB)
Gradient boosting (GB) is a machine learning algorithm used for classification and regression problems [76]. GB aims to combine strong learner models to obtain a weak learner with high prediction accuracy [77], [78]. The GB method tries to minimize the cost function to find an additive model. Therefore, the GB algorithm iteratively adds weak learners (a new decision tree) to the model, reducing the cost function at the highest rate at each step [77]. The steps of the GB algorithm are given below mathematically [76], [77]:
-
1.
Input variable () and target variable () are determined. The cost function () is defined.
-
2.
A simple decision tree () is initialized that establishes the relationship between and . Here, it is aimed to minimize the cost function ( = ).
-
3.
A pseudo-residue is defined to obtain a new target variable. The defined pseudo-residue is used as the new target variable ().
-
4.
A new decision tree () suitable for the pseudo-residual is developed. By including in the model, is updated (( = ).
-
5.
Step 3 and step 4 are repeated for the specified number of cycles.
-
6.
Finally, all decision trees are combined and the result of the GB model is obtained ( = ).
3.4. Proposed architecture
In this study, a new architecture named CovidDWNet+GB is proposed. This architecture consists of four blocks as shown in detail in Fig. 5. First, the input image is represented in the larger area of the input information by two successive convolution operations. In the second stage, new features are extracted after the obtained information FRB and then DDC operations. The same operations were repeated four times with different filter (f) numbers and dilation ratios (maxd) to increase the depth of the mesh. The depth of the DDC unit was determined by reducing the dilation rate by five at the beginning and by one in the next steps. Then, the obtained feature map is applied to the global average pooling layer (GAP) and three fully connected layers, respectively. In addition, after each convolution operation, Relu was used as the activation function and BatchNormalization was used as the normalization operation. Finally, after the CovidDWNet architecture was trained, the feature vectors obtained from the second fully connected (FC(64, R)) layer were estimated using Gradient Boosting (GB) machines. The proposed architecture is detailed in Fig. 5 and its methodology is shown in Eq. (4)–(5).
| (4) |
| (5) |
Fig. 5.
Proposed architecture (CovidDWNet+GB).
Here, stands for convolution layer, global average pooling, fully connected, softmax activation function, and gradient boosting classifier. represents the number of repetitions of the operation (> = 1).
Blocks in the proposed architecture include DDC, FRB units, and Convolution layer. The FRB unit (Fig. 2(a)) consists of four interconnected Convolution layers. The features obtained in the last step are combined with the input features. In this way, it adds depth to the architecture by reusing previous features. The FRB unit is mathematically shown in Eq. (6).
| (6) |
Here, weights represent outputs. * indicates the convolution operation. means applying the Relu and BatchNormalization (BN) operations of the layer output, respectively. [ ] denotes the merge operation. refers to the FRB operation of the th block.
The DDC unit, detailed in Fig. 2(b), is expanded with varying dilatation rates and the receptive area of X-ray and CT images. In this way, distinctive features are obtained effectively and more diversity is provided in the feature extraction process. The mathematical notation of the DDC unit used in the proposed architecture is given in Eq. (7).
| (7) |
Here, shows weights, outputs, and operation. means the join operation. refers to the DDC operation of the block.
Finally, the features obtained by FRB and DCC operations in blocks are given to the Convolution layer. In this way, important properties are obtained. The mathematical expression required for this operation is given in Eq. (8).
| (8) |
Here weights, * denotes the convolution operation.
3.5. Training and optimization of the proposed architecture
The CovidDWNet architecture is trained with a backpropagation algorithm using Cross-Entropy Eq. (9) for training multi-class datasets and binary cross-entropy Eq. (10) cost functions for training two-class datasets. These cost functions can be expressed mathematically as:
| (9) |
| (10) |
Here, is the number of samples, is the actual value, and is the predicted value.
Adam optimization [42], [76] algorithm is used as the optimization algorithm for updating the weights in the architecture. Adam optimization algorithm with learning coefficient at time :
| (11) |
| (12) |
| (13) |
Here stands for weights, hyperparameters and time t learning rate coefficient. represents the gradient at time and represent the exponential mean of gradients and squares of gradients along, . In the proposed architecture, the Relu activation function used after each convolution operation is given in Eq. (14) [34].
| (14) |
Fully connected layers (FC) form a fundamental part of CNN architectures, where all neurons in the previous layer connect to all neurons in the next layer and calculate how much each value matches the class. As the last layer, the output of the FC is combined with the activation functions of sigmoid, SVM, softmax, etc. for class prediction. Softmax activation function used for classification in this study, a probability distribution of number of output categories is calculated according to Eq. (15) [33], [79].
| (15) |
Here, is the input vector, is the number of classes, up to, , and is the output vector. The sum of all values is equal to 1.
4. Experimental results and discussion
For the detection of Covid-19, some literature studies based on deep learning using X-ray and CT images are presented in Table 1. It can be said that there are differences in success rates according to the datasets that researchers use by developing different architectures. In general, it is seen that the success rates of studies with two classes are higher than those with multiple classes. Marques et al. [80] performed binary and triple classification on X-ray images with the CNN-based architecture they developed using the EfficientNet architecture. This method has shown the highest success in the classification of binary class (Covid-19, normal) X-ray images with an accuracy rate of 99.6% compared to other architectures. On four-class X-ray datasets, Umer et al. [81] showed the lowest performance with 85.0% accuracy, while the proposed architecture achieved the highest performance with 96.8% accuracy. In addition, when studies using two-class CT images containing Covid-19 and Normal images were examined, Gifani et al. [82] showed the lowest performance with 85% accuracy using CNNs-based architecture, while the recommended architecture showed the highest performance with 100% accuracy. However, when the results here are examined, it is seen that the datasets used by the researchers affect the success rates of the methods. At the same time, the number of samples and the number of classes in the datasets are the factors affecting the success of the architectures.
Table 1.
Some deep learning approaches and success results for Covid-19 diagnosis from X-ray and CT images.
| Study | Architecture | Class | Scanning | Accuracy (%) |
|---|---|---|---|---|
| Sethy et al. [83] | ResNet50 plus | 2-class (Covid-19, noncovid-19) | CT | 95.38 |
| Li et al. [84] | Stacked-autoencoder | 2-class (Covid-19, Pneumonia, normal) | CT | 94.7 |
| Gifani et al. [82] | CNNs models | 2-class( Covid-19, noncovid-19) | CT | 85.0 |
| Xu et al. [85] | ResNet + Loc-ation Attention | 3-class (Influenza-A, Normal, covid-19) | CT | 86.7 |
| Heidarian et al. [86] | COVID-FACT | 3-class (Covid-19, Pneumonia, normal) | CT | 90.82 |
| Mukherjee et al. [87] | Tailored Deep NN | 2-class (Covid-19, noncovid-19) | CT | 95.83 |
| Mukherjee et al. [87] | Tailored Deep NN | 2-class (Covid-19, noncovid) | X-ray | 96.13 |
| Wang et al. [88] | COVID-Net | 3-class (Covid-19, pneumonia, normal) | X-ray | 93.3 |
| Heidari et al. [89] | VGG16-based CNN | 3-class (Covid-19, pneumonia, normal) | X-ray | 94.5 |
| Chakraborty [90] | Corona-Nidaan | 3-class (Covid-19, normal, pneumonia,) | X-ray | 95.0 |
| Umer et al. [81] | COVINet | 2-class (Covid-19, normal) | X-ray | 97.0 |
| Umer et al. [81] | COVINet | 3-class (Covid-19, normal, virus pneumonia) | X-ray | 90.0 |
| Umer et al. [81] | COVINet | 4-class (Covid-19, normal, virus pneumonia, bacterial pneumonia) | X-ray | 85.0 |
| Babukarthik et al. [91] | Genetic deep CNN | 2- class (Covid-19, normal) | X-ray | 98.8 |
| Apostolopoulos et al. [92] |
Pretrained CNNs | 3-class (Covid-19, nonCovid-19 pneumonia, normal) | X-ray | 96.7 |
| Ismael et al [93] | Deep CNNs | 2- class (Covid-19, normal) | X-ray | 92.6 |
| Oh et al. [94] | ResNet-18 | 4-class (Covid-19+viral pneumonia, bacterial pneumonia, tuberculosis, normal) | X-ray | 91.9 |
| Ezzat et al. [95] | GSA-DenseNet121 | 2-class (Covid-19, pneumonia) | X-ray | 93.4 |
| Marques et al. [80] | CNN + EfficientNet | 2- class (Covid-19, normal) | X-ray | 99.6 |
| Marques et al. [80] | CNN + EfficientNet | 3- class (Covid-19, nonCovid-19 pneunomia, normal) | X-ray | 96.7 |
| Hussain et al. [96] | CoroDet | 2- class (Covid-19, normal) | X-ray | 99.1 |
| Hussain et al. [96] | CoroDet | 3- class (Covid-19, normal, pneumonia) | X-ray | 94.2 |
| Hussain et al. [96] | CoroDet | 4- class (Covid-19, normal, non-Covid-19 pneumonia, non-Covid-19 bacterial pneumonia) | X-ray | 91.2 |
| Proposed | CovidDWNet | 2- class (Covid-19, normal) | CT | 100.0 |
| proposed | CovidDWNet | 4-class (Covid-19, Lung Opacity, Normal, Viral pneumonia) | X-ray | 96.8 |
To evaluate the performance of the architectures developed by the researchers more fairly, it is important to conduct experimental studies using common datasets. Therefore, in this study, four different experimental applications were carried out using three different datasets containing CT and X-ray images for the diagnosis of Covid-19. To objectively evaluate the performance of current architectures mentioned in the literature with CovidDWNet+GB (Our model), training was carried out on the same dataset by keeping certain parameters the same. The results obtained according to different metrics by training each model 200 epochs are presented in Table 4, 5, 6, and 7. Commonly used metrics accuracy, precision, recall, F1-Score, specificity, and AUC were used to evaluate the results. These metrics are:
| (16) |
| (17) |
| (18) |
| (19) |
| (20) |
Table 4.
The success of the models in detecting Covid-19 on the Sars-Cov-2 dataset containing CT images.
| Model | Accuracy (%) | Precision | Recall | F1-Score | Specificity(%) | AUC (%) |
|---|---|---|---|---|---|---|
| DenseNet | 97.37 | 0.97 | 0.97 | 0.97 | 98.57 | 97.37 |
| AlexNet | 94.14 | 0.94 | 0.94 | 0.94 | 94.12 | 94.12 |
| ResNet | 95.96 | 0.96 | 0.96 | 0.96 | 95.97 | 95.97 |
| CspNet [98] | 95.15 | 0.95 | 0.95 | 0.95 | 95.14 | 95.14 |
| VGG16 | 50.51 | 0.50 | 0.34 | 0.25 | 50.00 | 50.00 |
| VGG19 | 50.51 | 0.50 | 0.34 | 0.25 | 50.00 | 50.00 |
| CovXNet [42] | 98.18 | 0.98 | 0.98 | 0.98 | 98.18 | 98.18 |
| CoroNet [40] | 98.59 | 0.99 | 0.99 | 0.99 | 98.57 | 98.57 |
| CovidXrayNet [41] | 97.97 | 0.98 | 0.98 | 0.98 | 98.00 | 98.00 |
| DarkCovidNet [39] | 95.35 | 0.95 | 0.95 | 0.95 | 95.35 | 95.35 |
| Proposed (No DDC) | 98.38 | 0.98 | 0.98 | 0.98 | 98.39 | 98.39 |
| Proposed+ DataAug. | 97.78 | 0.98 | 0.98 | 0.98 | 97.78 | 97.78 |
| Proposed (No GB) | 98.59 | 0.99 | 0.99 | 0.99 | 98.58 | 98.58 |
| Proposed(CovidDWNet+GB) | 100.0 | 1.00 | 1.00 | 1.00 | 100.0 | 100.0 |
Table 5.
Success rates of models according to different metrics in detecting Covid-19 on the Covid-CT and Sars-Cov-2 datasets containing CT images.
| Model | Accuracy | Precision | Recall | F1-Score | Specificity | AUC (%) |
|---|---|---|---|---|---|---|
| DenseNet | 92.09 | 0.92 | 0.92 | 0.92 | 92.09 | 92.09 |
| AlexNet | 86.51 | 0.87 | 0.86 | 0.87 | 86.49 | 86.49 |
| ResNet | 87.44 | 0.88 | 0.97 | 0.87 | 87.47 | 87.47 |
| CspNet [98] | 85.58 | 0.86 | 0.86 | 0.86 | 85.53 | 85.53 |
| VGG16 | 50.39 | 0.25 | 0.50 | 0.34 | 50.00 | 50.00 |
| VGG19 | 50.39 | 0.25 | 0.50 | 0.34 | 50.00 | 50.00 |
| CovXNet [42] | 88.99 | 0.89 | 0.89 | 0.89 | 89.03 | 89.03 |
| CoroNet [40] | 92.25 | 0.92 | 0.92 | 0.92 | 92.26 | 92.26 |
| CovidXrayNet [41] | 91.16 | 0.92 | 0.91 | 0.91 | 91.12 | 91.12 |
| DarkCovidNet [39] | 88.92 | 0.88 | 0.87 | 0.87 | 88.92 | 88.92 |
| Proposed (No DDC) | 91.63 | 0.92 | 0.92 | 0.92 | 91.62 | 91.62 |
| Proposed+ DataAug. | 86.36 | 0.86 | 0.86 | 0.86 | 86.33 | 86.33 |
| Proposed (No GB) | 93.33 | 0.93 | 0.93 | 0.93 | 93.31 | 93.31 |
| Proposed(CovidDWNet+GB) | 99.84 | 1.0 | 1.00 | 1.00 | 99.85 | 99.85 |
Table 6.
Results of architectures for Covid-19 detection according to different metrics on the Dataset-X-ray dataset containing X-ray images.
| Model | Accuracy | Precision | Recall | F1-Score | Specificity | AUC (%) |
|---|---|---|---|---|---|---|
| DenseNet | 87.10 | 0.88 | 0.89 | 0.88 | 87.66 | 92.12 |
| AlexNet | 90.62 | 0.91 | 0.91 | 0.91 | 88.12 | 93.72 |
| ResNet | 92.70 | 0.94 | 0.93 | 0.94 | 90.79 | 95.14 |
| CspNet [98] | 82.63 | 0.79 | 0.84 | 0.81 | 80.09 | 88.84 |
| VGG16 | 91.09 | 0.92 | 0.91 | 0.92 | 87.59 | 93.88 |
| VGG19 | 91.33 | 0.93 | 0.92 | 0.92 | 87.55 | 93.96 |
| CovXNet [42] | 92.44 | 0.95 | 0.89 | 0.91 | 89.28 | 92.76 |
| CoroNet [40] | 92.11 | 0.93 | 0.92 | 0.92 | 90.52 | 94.40 |
| CovidXrayNet [41] | 95.39 | 0.96 | 0.96 | 0.96 | 94.33 | 96.88 |
| DarkCovidNet [39] | 94.33 | 0.96 | 0.94 | 0.95 | 92.17 | 95.67 |
| Proposed (No DDC) | 93.30 | 0.94 | 0.94 | 0.94 | 90.75 | 95.50 |
| Proposed+ DataAug. | 93.19 | 0.95 | 0.92 | 0.94 | 89.72 | 94.54 |
| Proposed (No GB) | 93.76 | 0.95 | 0.94 | 0.94 | 90.91 | 95.67 |
| Proposed(CovidDWNet+GB) | 96.81 | 0.98 | 0.97 | 0.98 | 95.54 | 97.98 |
Table 7.
Performance of Architectures for Covid-19 detection by different metrics on all datasets containing X-ray and CT images.
| Model | Accuracy | Precision | Recall | F1-Score | Specificity | AUC (%) |
|---|---|---|---|---|---|---|
| DenseNet | 93.05 | 0.94 | 0.93 | 0.94 | 90.41 | 95.11 |
| AlexNet | 90.98 | 0.92 | 0.91 | 0.91 | 86.91 | 93.66 |
| ResNet | 92.02 | 0.93 | 0.93 | 0.93 | 90.30 | 94.70 |
| CspNet [98] | 90.71 | 0.92 | 0.91 | 0.91 | 87.14 | 93.67 |
| VGG16 | 89.65 | 0.91 | 0.90 | 0.90 | 84.90 | 92.88 |
| VGG19 | 89.24 | 0.91 | 0.89 | 0.90 | 83.64 | 92.27 |
| CovXNet [42] | 92.45 | 0.94 | 0.91 | 0.93 | 88.85 | 93.86 |
| CoroNet [40] | 92.23 | 0.94 | 0.92 | 0.93 | 87.28 | 94.43 |
| CovidXrayNet [41] | 95.30 | 0.96 | 0.96 | 0.96 | 92.93 | 96.85 |
| DarkCovidNet [39] | 90.59 | 0.92 | 0.91 | 0.91 | 88.05 | 93.06 |
| Proposed (No DDC) | 92.19 | 0.93 | 0.92 | 0.93 | 87.69 | 94.40 |
| Proposed+ DataAug. | 91.92 | 0.94 | 0.91 | 0.90 | 89.90 | 93.10 |
| Proposed (No GB) | 93.36 | 0.94 | 0.94 | 0.94 | 91.73 | 95.50 |
| Proposed(CovidDWNet+GB) | 96.32 | 0.97 | 0.97 | 0.97 | 95.17 | 97.67 |
Here, (True Positives) denotes correctly classified diseased cases, (True Negatives) correctly defined healthy cases, (False Positives) misclassified diseased cases, (False Negatives) misclassified healthy cases [3], [97]. The receiver operating characteristic (ROC) curve is used in classification problems to evaluate the performance of models by plotting the true positive rate (TPR) versus the false positive rate (FPR). The area under the curve (AUC) indicates the area under the ROC, which is a probability curve [3].
| (21) |
| (22) |
The hyperparameters of the architectures during the training phase are presented in Table 2. CovidDWNet architecture, developed on Keras/Tensorflow platform, takes images as input by scaling 128 × 128; Adam (Learning_rate = 0.001) optimization function and batch size value 32 are given.
Table 2.
Hyperparameters of architectures for Covid-19 detection.
| Model | Data augmentation | Software | Input size | Optimizer | Learning rate | Batch size |
|---|---|---|---|---|---|---|
| DenseNet | No | Keras, TensorFlow | 224 × 224 | Adam | 0.0001 | 32 |
| AlexNet | No | Keras, TensorFlow | 224 × 224 | Adam | 0.0001 | 32 |
| ResNet | No | Keras, TensorFlow | 224 × 224 | Adam | 0.0001 | 32 |
| CspNet [98] | No | Keras, TensorFlow | 224 × 224 | Adam | 0.0001 | 32 |
| VGG16 | No | Keras, TensorFlow | 224 × 224 | Adam | 0.0001 | 32 |
| VGG19 | No | Keras, TensorFlow | 224 × 224 | Adam | 0.0001 | 32 |
| CovXNet [42] | Yes | Keras, TensorFlow | 128 × 128 | Adam | 0.001 | 16 |
| CoroNet [40] | Yes | Keras, TensorFlow | 150 × 150 | Adam | 0.0001 | 10 |
| CovidXrayNet [41] | Yes | Fastai, PyTorch | 256 × 256 | Adam | – | 32 |
| DarkCovidNet [39] | No | Fastai, PyTorch | 256 × 256 | Adam | 0.003 | 32 |
| Proposed (No DDC) | No | Keras, TensorFlow | 128 × 128 | Adam | 0.001 | 32 |
| Proposed | No | Keras, TensorFlow | 128 × 128 | Adam | 0.001 | 32 |
The image distribution of the datasets used in experimental applications for the detection of Covid-19 and other pneumonia diseases according to training and test sets is given in Table 3 in detail. At the same time, the number of images according to the types of diseases in each application is presented in this table. Datasets are reserved for approximately 80% training and 20% testing. In the first application, the Sars-Cov-2 [64] dataset was used. This dataset is divided into two datasets, training and testing. The training dataset contains a total of 1986 images, 1002 Covid, and 984 Normal. The test dataset contains 495 images, 250 of which are Covid and 245 are normal. Similarly, a second application was performed using the Covid-CT [63] and Sars-Cov-2 [64] datasets containing CT images. In this application, there are 2589 images (1288 Covid, 1301 Normal) in the training dataset and 645 images (320 Covid, 325 Normal) in the test dataset. In the third application, Dataset-X-ray [65] dataset containing X-ray images was used. In this application, there are 16933 images (2893 Covid, 8154 Normal, 4810 Lung Opacity, and 1079 Viral Pneumonia) in the training dataset and 4232 images (723 Covid, 2038 Normal, 1202 Lung Opacity, and 269 Viral Pneumonia) in the test dataset. The fourth application was performed by combining all datasets containing X-ray and CT images. In this application, a total of 19515 images, including 4174 Covid, 9455 Normal, 4810 Lung Opacity, and 1079 Viral Pneumonia, in the training dataset; In the test dataset, there are a total of 4877 images, including 1043 Covid, 2363 Normal, 1202 Lung Opacity and 269 Viral Pneumonia.
Table 3.
Number of records in datasets used in applications.
| Application (s) | Data set(s) | Image (s) | Train/Test | Covid | Normal | Lung Opacity | Viral Pneumonia | Total |
|---|---|---|---|---|---|---|---|---|
| Application1 | Sars-Cov-2 [64] | CT | Train Set | 1002 | 984 | – | – | 1986 |
| Test Set | 250 | 245 | – | – | 495 | |||
| Application2 | Covid-CT [63] and Sars-Cov-2 [64] |
CT | Train Set | 1288 | 1301 | 2589 | ||
| Test Set | 320 | 325 | 645 | |||||
| Application3 | Dataset-X-ray [65] | X-ray | Train Set | 2893 | 8154 | 4810 | 1076 | 16933 |
| Test Set | 723 | 2038 | 1202 | 269 | 4232 | |||
| Application4 | All datasets | X-ray + CT | Train Set | 4174 | 9455 | 4810 | 1076 | 19515 |
| Test Set | 1043 | 2363 | 1202 | 269 | 4877 |
The results of the experimental study with the SARS-COV-2 [64] dataset containing CT images for Covid-19 detection are given in Table 4. The success rate has been increased by adding a DDC module to the proposed architecture. In addition, high performance has been achieved by adding GB classifier to the proposed architecture. However, when the data augmentation method is applied to the proposed architecture, it is seen that the success decreases. When the results obtained in this application are examined in a general way, we can say that our model exhibits the highest performance in all metrics with a 100% success rate. Also, the confusion matrix results of the proposed architecture for this application are given in Fig. 6(a). When the results are examined, it is seen that the proposed architecture correctly predicts Covid-19 patients and non-patients with 100% high performance.
Fig. 6.
Performance results of the proposed architecture in binary and multiple classes. (a) The first application, (b) second application, (c) third application, (d) fourth application results.
In the second application, an experimental study was performed by combining the Covid-CT [63] and Sars-Cov-2 [64] datasets containing Covid-19 and Normal CT images, and the results are presented in Table 5. When the results are examined, the proposed architecture (CovidDWNet+GB) showed the highest success with 99.84% according to the accuracy metric and 100% (1.00) according to the precision, recall, and F1-Score metrics. Similarly, the CovidDWNet architecture achieved the highest success with 99.85% performance according to specificity and AUC metrics. In addition, when the confusion matrix results are examined in Fig. 6(b), It is seen that the CovidDWNet+GB architecture detects Covid-19 patients and normal people who are not sick with 100% accuracy.
In the third experimental study for Covid-19 detection, the four-class Dataset-X-ray [65] dataset was used. The results of the experimental study are shown in Table 6. When the results of the application are examined, our model (CovidDWNet+GB) achieved the highest performance with 96.81% accuracy, 0.98 precision, 0.97 recall, 0.98 F1-Score, 95.54% specificity, and 97.98% AUC. At the same time, when the training and testing times of the third application (Application3) are examined (in Table 8), it is seen that the proposed architecture is faster than the CovidXrayNet and DarkCovidNet architectures, which are the closest to the success rate. In addition, when the success distribution of the CovidDWNet+GB architecture according to classes is analyzed in Fig. 6(c), we can say that it correctly predicts Covid-19 patients 99%, Lung Opacity patients 92%, people who are not sick 98% and Viral Pneumonia patients 100%.
Table 8.
Training and testing times of architectures. Training times in hours and minutes; Test times are shown in seconds.
| Model | Application1 |
Application2 |
Application3 |
Application4 |
||||
|---|---|---|---|---|---|---|---|---|
| Train time (hr.min.) | Test time (s.) | Train time (hr.min.) | Test time (s.) | Train time (hr.min.) | Test time (s.) | Train time (hr.min.) | Test time (s.) | |
| DenseNet | 0.53 | 1 | 1.18 | 2 | 5.00 | 6 | 6.10 | 7 |
| AlexNet | 0.53 | 1 | 1.16 | 1 | 4.50 | 3 | 6.04 | 4 |
| ResNet | 1.10 | 2 | 1.50 | 3 | 6.56 | 8 | 7.36 | 10 |
| CspNet [80] | 1.30 | 2 | 1.46 | 3 | 6.00 | 7 | 8.20 | 9 |
| VGG16 | 1.07 | 3 | 1.30 | 3 | 7.50 | 18 | 9.13 | 19 |
| VGG19 | 1.14 | 3 | 1.33 | 4 | 8.16 | 19 | 9.23 | 21 |
| CovXNet [42] | 5.05 | 7 | 6.05 | 8 | 15.33 | 26 | 18.33 | 29 |
| CoroNet [40] | 1.20 | 2 | 1.43 | 2 | 6.33 | 5 | 7.50 | 7 |
| CovidXrayNet [41] | 1.10 | 2 | 1.43 | 3 | 8.33 | 12 | 9.13 | 15 |
| DarkCovidNet [39] | 1.18 | 2 | 1.48 | 2 | 7.54 | 13 | 8.43 | 15 |
| Proposed (CovidDWNet) | 1.16 | 2 | 1.45 | 3 | 7.24 | 8 | 8.32 | 11 |
In the last experimental study for Covid-19 detection, an application was performed by combining all datasets (Covid-CT, Sars-Cov-2, and Dataset-X-ray). The results obtained according to different metrics are given in Table 7. When the results are examined, we can say that CovidDWNet+GB, 96.32% accuracy, 0.97 precision, 0.97 recall, 0.97 F1-Score, 95.17% specificity, and 97.67% AUC showed the highest success. Also, the confusion matrix results for the CovidDWNet+GB architecture of this application are given in Fig. 6(d). When the results are examined, it is seen that he predicted Covid 19 patients at 97%, Lung Opacity patients at 93%, non-sick people at 97%, and Viral Pneumonia patients at 100% correct.
According to the experimental application results, the class performances (confusion matrix) of the proposed architecture (CovidDWNet+GB) for Covid-19 detection are given in Fig. 6. It shows the results of the binary classification in (a) and (b), and multi-classification in (c) and (d). (a) gives the results of the first application, (b) the second application, (c) the third application, and (d) the fourth application. In applications containing the proposed architectural CT images (Fig. 6 (a–b)), it appears to predict Covid-19 and Normal images extremely successfully with 100% success rates.
Similarly, multiple classification performances of the CovidDWNet architecture are given in Fig. 6-(c) and (d). In the third experimental study including X-ray images, it is seen that the proposed architecture, Covid-19, Lung Opacity, Normal and Viral Pneumonia images were estimated with success rates of 99%, 92%, 98%, and 100%, respectively. In addition, in the fourth application containing all datasets, it was observed that he predicted Covid-19 images with 97%, Lung Opacity images with 93%, Normal images with 97%, and Viral pneumonia images with a rate of 100%. It can be said that it performs extremely satisfactorily in classes other than the Lung Opacity class. It is thought that its lower success in images containing Lung Opacity is due to its overlapping features with other classes.
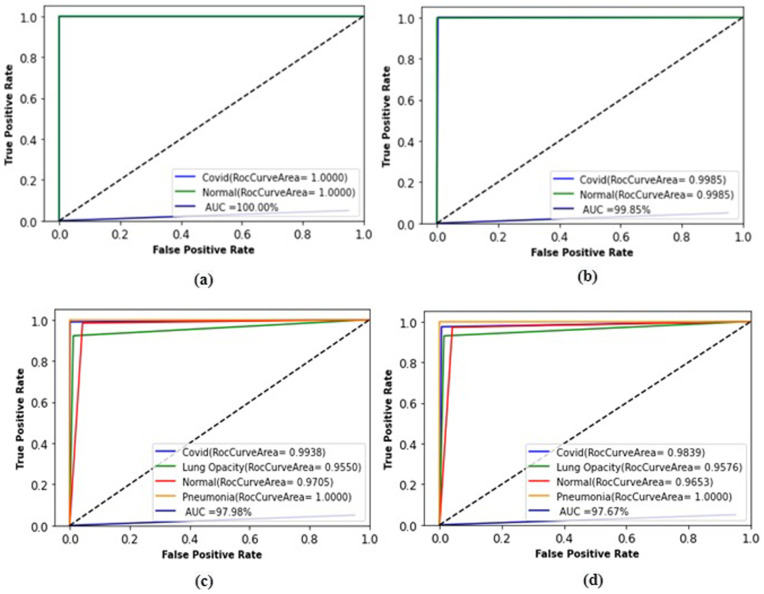
The ROC curve of our proposed model is shown in Fig. 7. The ROC curve is a graphical representation of the classification performance of the network. The closer the curve is to its upper left limit, the higher the performance. Fig. 7 (a–b) shows the results of the CT images, (c) the results of the X-ray images, and (d) the results of the X-ray and CT images. In CT images, it is seen that AUC values of 99.85% and 100% results are obtained. We can say that AUC values of 97.98% and 97.67% were obtained in X-ray and all images, respectively.
Fig. 7.
ROC analysis of the proposed model. (a) First application, (b) second application, (c) third application, (d) fourth application results.
Gradient-based class activation mapping (Grad-CAM) algorithm [99] is used to highlight important points on X-ray and CT images that affect the performance of CNN architectures. The main purpose of this algorithm was developed to create stronger deep networks. The last convolutional layer is considered to be the stage where the best balance is achieved between important spatial information and the highest semantics [100]. Grad-CAM generates heatmap heat zones to highlight key locations from features derived from the final convolution layer. This information indicates which regions the algorithm pays more attention to. In Fig. 8, heatmap and Grad-CAM images obtained for sample Covid-19 images with the Grad-CAM algorithm are given. Green and yellow areas on heatmaps highlight key regions where the CovidDWNet architecture is concentrated. Regions with dark yellow in heatmaps and red in Grad-CAM indicate important regions with high distinctiveness.
Fig. 8.
From CovidDWNet architecture using Grad-CAM resulting sample heatmaps and Covid-19 visuals.
CNNs are used for classification and recognition problems by making use of fully connected layers of feature maps obtained as a result of the convolution process [101]. Feature maps are obtained with filters defined by convolution operations on the input image. Feature maps obtained for a particular input image are used to understand which features of the input are detected or preserved. It is expected to detect small or fine details from the image given as input to the models. However, the models will capture more general feature maps close to the output [102]. In Fig. 9, an example of tens of feature maps obtained from the images given as an introduction to the CovidDWNet architecture is given. It is seen that the different features of the images are emphasized in the first two convolution layers. These images appear to be understandable images. We can say that the feature maps obtained from the last convolution layer of the next blocks (Block1-4) capture more fine details. These attributes are meaningful features that are not understood by humans but can be understood by CNN models. At the same time, it is possible to say that the feature maps show fewer and fewer details as they go deeper and that these details are meaningful features in the decision-making process by CNN models.
Fig. 9.
Feature maps were obtained from sample CT and X-ray images.
Also, the training and test times of the applications are given in Table 8. Training times in hours and minutes; Test times are shown in seconds. Training times, architectures 200 epoch training time; test times represent the time elapsed during the estimation of all samples in the test dataset. When the training and testing times are examined, we can say that the AlexNet architecture has a higher speed compared to other architectures. However, when the overall success of the AlexNet architecture is examined in the experimental applications, it has been observed that it exhibits a low performance.
The time complexity of the architectures according to the training and test times is given in Fig. 10. When the time complexity diagram is carefully examined, it is seen that the AlexNet architecture has the smallest time complexity. We can also say that the CovXNet architecture has the highest time complexity. It is possible to say that the proposed architecture has moderate time complexity.
When the results of experimental studies are examined in general, it is seen that it predicts X-ray and CT images with high performance. A higher success was achieved with CT images compared to X-ray images. We can say that this is due to the more sensitive and finer detailed structures of CT images [13], [42].
In addition, a higher performance has been achieved by integrating the DDC module into the CovidDWNet architecture, providing different expansion rates and deepening the feature map with depthwise convolution. However, when the data augmentation method is applied to the proposed architecture, it has been observed that it affects success negatively. The hyperparameters and values of the applied data augmentation method are given in Table 9.
Table 9.
Data augmentation hyperparameters for the proposed architecture.
| Parameters | Value |
|---|---|
| Width shift | 0.2 |
| Height shift | 0.2 |
| Shear | 0.25 |
| Zoom | 0.2 |
| Rotation | 30 |
| Horizontal flip | True |
| Vertical flip | True |
5. Conclusion
Covid-19 pandemic cases are increasing day by day and cause the death of many people. It has caused millions of cases and the death of millions of people so far. This disease, which brings with it different health problems, poses serious threats to human health with the emergence of new variations. Many states are taking many measures to prevent the spread of the disease and reduce deaths. RT-PCR tests are generally used to detect this disease. However, considering the inadequacy of RT-PCR tests, the risk of transmission to healthcare personnel, pain to patients, and cost, it brings with it many problems. In this sense, different researches are carried out and different solutions are offered. Deep learning architectures with high performance are one of these studies. When the literature is examined, it is possible to see many studies with deep learning. In these studies, it is seen that only one of the CT or X-ray datasets is used mostly. At the same time, it was seen that the performance evaluation of the studies was limited in themselves.
In this study for the detection of Covid-19 and similar symptoms, datasets containing CT or X-ray images were used. A new architecture is proposed, called CovidDWNet, based on feature reuse residual block (FRB) and Depthwise dilated convolutions (DDC) units. High performance has been achieved by providing the combination of the proposed architecture and the Gradient boosting (GB) algorithm (CovidDWNet+GB). In addition, the current architectures in the literature were examined, the architectures were trained on the same data sets and performance evaluation was made accordingly.
It has been observed that CovidDWNet+GB exhibits the highest success with 99.84% and 100% accuracy rates in applications performed on CT datasets with two classes (Covid-19, and non Covid-19). In addition, it has been observed that it provides the highest success according to precision, recall, F1-Score, specificity, and AUC metrics. The proposed architecture showed the highest success in the application using four classes (Covid-19, Lung Opacity, Normal and Viral Pneumonia) X-ray images, with 96.81% accuracy, 0.98 precision, 0.97 recall, 0.98 F1-Score, 95.54% specificity, and 97.98% AUC. Similarly, we can say that the CovidDWNet+GB architecture showed the highest success in the experimental study using X-ray and CT images, with 96.32% accuracy, 0.97 precision, 0.97 recall, 0.97 F1-Score, 95.17% specificity, and 97.67% AUC. Also, it has been observed that the proposed architecture predicts 4877 images in the test dataset with a high speed of 11 s.
As a result, when the performances of different architectures are examined by keeping certain parameters constant on the same datasets, it is possible to say that the proposed architecture exhibits a respectable success in the literature and shows a remarkable performance among current architectures.
CRediT authorship contribution statement
Gaffari Celik: Conceptualization, Methodology, Software, Formal analysis, Data curation, Writing – original draft, Writing – review & editing, Visualization, Investigation, Validation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
The code (and data) in this article has been certified as Reproducible by Code Ocean: (https://codeocean.com/). More information on the Reproducibility Badge Initiative is available at https://www.elsevier.com/physical-sciences-and-engineering/computer-science/journals.
Data availability
No data was used for the research described in the article.
References
- 1.Wu F., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronavirus disease (COVID-19) pandemic. https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
- 3.Subramanian N., Elharrouss O., Al-Maadeed S., Chowdhury M. A review of deep learning-based detection methods for COVID-19. Comput. Biol. Med. 2022;143 doi: 10.1016/j.compbiomed.2022.105233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubin G.D., et al. The role of chest imaging in patient management during the COVID-19 pandemic. Chest. 2020;158(1):106–116. doi: 10.1016/j.chest.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh R., et al. Corona virus (COVID-19) symptoms prevention and treatment: A short review. J. Drug Deliv. Ther. 2021;11(2-S):118–120. doi: 10.22270/jddt.v11i2-S.4644. [DOI] [Google Scholar]
- 6.R S., et al. An efficient hardware architecture based on an ensemble of deep learning models for COVID -19 prediction. Sustain. Cities Soc. 2022 doi: 10.1016/j.scs.2022.103713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heidari A., Jafari Navimipour N., Unal M., Toumaj S. The COVID-19 epidemic analysis and diagnosis using deep learning: A systematic literature review and future directions. Comput. Biol. Med. 2022;141(2021) doi: 10.1016/j.compbiomed.2021.105141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharfstein J.M., Becker S.J., Mello M.M. Diagnostic testing for the novel coronavirus. JAMA. 2020;323(15):1437. doi: 10.1001/jama.2020.3864. [DOI] [PubMed] [Google Scholar]
- 9.Stephanie S., et al. Determinants of chest radiography sensitivity for COVID-19: A multi-institutional study in the United States. Radiol. Cardiothorac. Imaging. 2020;2(5) doi: 10.1148/ryct.2020200337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu R., et al. Clinica Chimica Acta positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to 2020. Clin. Chim. Acta. 2020;505(March):172–175. doi: 10.1016/j.cca.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dramé M., et al. Should RT-PCR be considered a gold standard in the diagnosis of COVID-19? J. Med. Virol. 2020;92(11):2312–2313. doi: 10.1002/jmv.25996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie J., et al. Characteristics of patients with coronavirus disease (COVID-19) confirmed using an IgM-IgG antibody test. J. Med. Virol. 2020;92(10):2004–2010. doi: 10.1002/jmv.25930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hassan H., et al. Supervised and weakly supervised deep learning models for COVID-19 CT diagnosis: A systematic review. Comput. Methods Programs Biomed. 2022;218 doi: 10.1016/j.cmpb.2022.106731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaur P., Malaviya V., Gupta A., Bhatia G., Pachori R.B., Sharma D. COVID-19 disease identification from chest CT images using empirical wavelet transformation and transfer learning. Biomed. Signal Process. Control. 2022;71(PA) doi: 10.1016/j.bspc.2021.103076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ucar F., Korkmaz D. COVIDiagnosis-Net: Deep Bayes-SqueezeNet based diagnosis of the coronavirus disease, 2019 (COVID-19) from X-ray images. Med. Hypotheses. 2020;140(April) doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Başaran E. Classification of white blood cells with SVM by selecting SqueezeNet and LIME properties by mRMR method. Signal, Image Video Process. 2022 doi: 10.1007/s11760-022-02141-2. [DOI] [Google Scholar]
- 17.Çelik G., Talu M.F. A new 3D MRI segmentation method based on generative adversarial network and atrous convolution. Biomed. Signal Process. Control. 2022;71(PA) doi: 10.1016/j.bspc.2021.103155. [DOI] [Google Scholar]
- 18.Goodfellow I., et al. Generative adversarial networks. Commun. ACM. 2020;63(11):139–144. doi: 10.1145/3422622. [DOI] [Google Scholar]
- 19.Çelik G., Talu M.F. Generating the image viewed from EEG signals. Pamukkale Univ. J. Eng. Sci. 2021;27(2):129–138. doi: 10.5505/pajes.2020.76399. [DOI] [Google Scholar]
- 20.Esteva A., et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan J.H., et al. Automated segmentation of exudates, haemorrhages, microaneurysms using single convolutional neural network. Inf. Sci. (Ny) 2017;420:66–76. doi: 10.1016/j.ins.2017.08.050. [DOI] [Google Scholar]
- 22.Başaran E., Cömert Z., Çelik Y. Neighbourhood component analysis and deep feature-based diagnosis model for middle ear otoscope images. Neural Comput. Appl. 2022 doi: 10.1007/s00521-021-06810-0. [DOI] [Google Scholar]
- 23.Celik Y., Talo M., Yildirim O., Karabatak M., Acharya U.R. Automated invasive ductal carcinoma detection based using deep transfer learning with whole-slide images. Pattern Recognit. Lett. 2020;133:232–239. doi: 10.1016/j.patrec.2020.03.011. [DOI] [Google Scholar]
- 24.Bozdag Z., Talu F.M. Pyramidal nonlocal network for histopathological image of breast lymph node segmentation. Int. J. Comput. Intell. Syst. 2021;14(1):122–131. doi: 10.2991/ijcis.d.201030.001. [DOI] [Google Scholar]
- 25.Talo M., Yildirim O., Baloglu U.B., Aydin G., Acharya U.R. Convolutional neural networks for multi-class brain disease detection using MRI images. Comput. Med. Imaging Graph. 2019;78 doi: 10.1016/j.compmedimag.2019.101673. [DOI] [PubMed] [Google Scholar]
- 26.Gaál G., Maga B., Lukács A. Attention U-net based adversarial architectures for chest X-ray lung segmentation. CEUR Workshop Proc. 2020;2692:1–7. [Google Scholar]
- 27.Souza J.C., Bandeira Diniz J.O., Ferreira J.L., França da Silva G.L., Corrêa Silva A., de Paiva A.C. An automatic method for lung segmentation and reconstruction in chest X-ray using deep neural networks. Comput. Methods Programs Biomed. 2019;177:285–296. doi: 10.1016/j.cmpb.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Yıldırım Ö., Pławiak P., Tan R.S., Acharya U.R. Arrhythmia detection using deep convolutional neural network with long duration ECG signals. Comput. Biol. Med. 2018;102(September):411–420. doi: 10.1016/j.compbiomed.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 29.Hannun A.Y., et al. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nature Med. 2019;25(1):65–69. doi: 10.1038/s41591-018-0268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Acharya U.R., et al. A deep convolutional neural network model to classify heartbeats. Comput. Biol. Med. 2017;89(August):389–396. doi: 10.1016/j.compbiomed.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 31.Rajpurkar P., et al. 2017. CheXNet: Radiologist-level pneumonia detection on chest X-rays with deep learning; pp. 3–9. [Online]. Available: http://arxiv.org/abs/1711.05225. [Google Scholar]
- 32.Ieracitano C., et al. A fuzzy-enhanced deep learning approach for early detection of Covid-19 pneumonia from portable chest X-ray images. Neurocomputing. 2022;481:202–215. doi: 10.1016/j.neucom.2022.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahamed K.U., et al. A deep learning approach using effective preprocessing techniques to detect COVID-19 from chest CT-scan and X-ray images. Comput. Biol. Med. 2021;139(October) doi: 10.1016/j.compbiomed.2021.105014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verma H., Mandal S., Gupta A. Temporal deep learning architecture for prediction of COVID-19 cases in India. Expert Syst. Appl. 2021;195(January) doi: 10.1016/j.eswa.2022.116611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khan S.H., et al. COVID-19 detection in chest X-ray images using deep boosted hybrid learning. Comput. Biol. Med. 2021;137(August) doi: 10.1016/j.compbiomed.2021.104816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loey M., El-Sappagh S., Mirjalili S. Bayesian-based optimized deep learning model to detect COVID-19 patients using chest X-ray image data. Comput. Biol. Med. 2022;142(January) doi: 10.1016/j.compbiomed.2022.105213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lahsaini I., El Habib Daho M., Chikh M.A. Deep transfer learning based classification model for COVID-19 using chest CT-scans. Pattern Recognit. Lett. 2021;152:122–128. doi: 10.1016/j.patrec.2021.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toğaçar M., Ergen B., Cömert Z. COVID-19 detection using deep learning models to exploit social mimic optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput. Biol. Med. 2020;121(March) doi: 10.1016/j.compbiomed.2020.103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Rajendra Acharya U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020;121(April) doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khan A.I., Shah J.L., Bhat M.M. CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput. Methods Programs Biomed. 2020;196 doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monshi M.M.A., Poon J., Chung V., Monshi F.M. CovidXrayNet: Optimizing data augmentation and CNN hyperparameters for improved COVID-19 detection from CXR. Comput. Biol. Med. 2021;133(March) doi: 10.1016/j.compbiomed.2021.104375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahmud T., Rahman M.A., Fattah S.A. CovXNet: A multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Comput. Biol. Med. 2020;122(June) doi: 10.1016/j.compbiomed.2020.103869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calderon-Ramirez S., Yang S., Elizondo D., Moemeni A. Dealing with distribution mismatch in semi-supervised deep learning for COVID-19 detection using chest X-ray images: A novel approach using feature densities. Appl. Soft Comput. 2022;123 doi: 10.1016/j.asoc.2022.108983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta A., Anjum, Gupta S., Katarya R. InstaCovNet-19: A deep learning classification model for the detection of COVID-19 patients using Chest X-ray. Appl. Soft Comput. 2021;99 doi: 10.1016/j.asoc.2020.106859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feki I., Ammar S., Kessentini Y., Muhammad K. Federated learning for COVID-19 screening from Chest X-ray images. Appl. Soft Comput. 2021;106 doi: 10.1016/j.asoc.2021.107330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Moura J., Novo J., Ortega M. Fully automatic deep convolutional approaches for the analysis of COVID-19 using chest X-ray images. Appl. Soft Comput. 2022;115 doi: 10.1016/j.asoc.2021.108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shankar K., et al. An optimal cascaded recurrent neural network for intelligent COVID-19 detection using Chest X-ray images. Appl. Soft Comput. 2021;113 doi: 10.1016/j.asoc.2021.107878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Albahli S., Ayub N., Shiraz M. Coronavirus disease (COVID-19) detection using X-ray images and enhanced DenseNet. Appl. Soft Comput. 2021;110 doi: 10.1016/j.asoc.2021.107645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elazab A., Elfattah M.A., Zhang Y. Novel multi-site graph convolutional network with supervision mechanism for COVID-19 diagnosis from X-ray radiographs. Appl. Soft Comput. 2022;114 doi: 10.1016/j.asoc.2021.108041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ozcan T. A new composite approach for COVID-19 detection in X-ray images using deep features. Appl. Soft Comput. 2021;111 doi: 10.1016/j.asoc.2021.107669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Calderon-Ramirez S., et al. Correcting data imbalance for semi-supervised COVID-19 detection using X-ray chest images. Appl. Soft Comput. 2021;111 doi: 10.1016/j.asoc.2021.107692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karthik R., Menaka R., Hariharan M. Learning distinctive filters for COVID-19 detection from chest X-ray using shuffled residual CNN. Appl. Soft Comput. 2021;99 doi: 10.1016/j.asoc.2020.106744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Demir F. DeepCoroNet: A deep LSTM approach for automated detection of COVID-19 cases from chest X-ray images. Appl. Soft Comput. 2021;103 doi: 10.1016/j.asoc.2021.107160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou T., Lu H., Yang Z., Qiu S., Huo B., Dong Y. The ensemble deep learning model for novel COVID-19 on CT images. Appl. Soft Comput. 2021;98 doi: 10.1016/j.asoc.2020.106885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bandyopadhyay R., Basu A., Cuevas E., Sarkar R. Harris Hawks optimisation with simulated annealing as a deep feature selection method for screening of COVID-19 CT-scans. Appl. Soft Comput. 2021;111 doi: 10.1016/j.asoc.2021.107698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ye Q., et al. Robust weakly supervised learning for COVID-19 recognition using multi-center CT images. Appl. Soft Comput. 2022;116 doi: 10.1016/j.asoc.2021.108291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song L., et al. A deep fuzzy model for diagnosis of COVID-19 from CT images. Appl. Soft Comput. 2022;122 doi: 10.1016/j.asoc.2022.108883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liang S., Nie R., Cao J., Wang X., Zhang G. FCF: Feature complement fusion network for detecting COVID-19 through CT scan images. Appl. Soft Comput. 2022;125 doi: 10.1016/j.asoc.2022.109111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saygılı A. A new approach for computer-aided detection of coronavirus (COVID-19) from CT and X-ray images using machine learning methods. Appl. Soft Comput. 2021;105 doi: 10.1016/j.asoc.2021.107323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Naeem H., Bin-Salem A.A. A CNN-LSTM network with multi-level feature extraction-based approach for automated detection of coronavirus from CT scan and X-ray images. Appl. Soft Comput. 2021;113 doi: 10.1016/j.asoc.2021.107918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vinod D.N., Jeyavadhanam B.R., Zungeru A.M., Prabaharan S.R.S. Fully automated unified prognosis of Covid-19 chest X-ray/CT scan images using Deep Covix-Net model. Comput. Biol. Med. 2021;136(August) doi: 10.1016/j.compbiomed.2021.104729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li J., et al. Multi-task contrastive learning for automatic CT and X-ray diagnosis of COVID-19. Pattern Recognit. 2021;114 doi: 10.1016/j.patcog.2021.107848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang X., He X., Zhao J., Zhang Y., Zhang S., Xie P. 2020. COVID-CT-dataset: A CT scan dataset about COVID-19; pp. 1–14. [Online]. Available: http://arxiv.org/abs/2003.13865. [Google Scholar]
- 64.Soares E., Angelov P., Biaso S., Froes M.H., Abe D.K. 2020. SARS-CoV-2 CT-scan dataset: A large dataset of real patients CT scans for SARS-CoV-2 identification. medRxiv, p. 2020.04.24.20078584, [Online]. Available: https://www.medrxiv.org/content/10.1101/2020.04.24.20078584v3%0Ahttps://www.medrxiv.org/content/10.1101/2020.04.24.20078584v3.abstract. [Google Scholar]
- 65.Chowdhury M.E.H., et al. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. 2020;8(July):132665–132676. doi: 10.1109/ACCESS.2020.3010287. [DOI] [Google Scholar]
- 66.Gu J., et al. Recent advances in convolutional neural networks. Pattern Recognit. 2018;77:354–377. doi: 10.1016/j.patcog.2017.10.013. [DOI] [Google Scholar]
- 67.Budak Ü., Cömert Z., Çıbuk M., Şengür A. DCCMED-Net: Densely connected and concatenated multi Encoder-Decoder CNNs for retinal vessel extraction from fundus images. Med. Hypotheses. 2019;134:2020. doi: 10.1016/j.mehy.2019.109426. [DOI] [PubMed] [Google Scholar]
- 68.Ren F., Liu W., Wu G. Feature reuse residual networks for insect pest recognition. IEEE Access. 2019;7:122758–122768. doi: 10.1109/ACCESS.2019.2938194. [DOI] [Google Scholar]
- 69.He K., Zhang X., Ren S., Sun J. 2015. Deep residual learning for image recognition. [Online]. Available: http://arxiv.org/abs/1512.03385. [Google Scholar]
- 70.Kim S., Park I., Kwon S., Han J. Motion retargetting based on dilated convolutions and skeleton-specific loss functions. Comput. Graph. Forum. 2020;39(2):497–507. doi: 10.1111/cgf.13947. [DOI] [Google Scholar]
- 71.Sooksatra S., Kondo T., Bunnun P., Yoshitaka A. Redesigned skip-network for crowd counting with dilated convolution and backward connection. J. Imaging. 2020;6(5) doi: 10.3390/JIMAGING6050028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li X., Zhai M., Sun J. DDCNNC: Dilated and depthwise separable convolutional neural network for diagnosis COVID-19 via chest X-ray images. Int. J. Cogn. Comput. Eng. 2021;2(April):71–82. doi: 10.1016/j.ijcce.2021.04.001. [DOI] [Google Scholar]
- 73.Chollet F. Proc. - 30th IEEE Conf. Comput. Vis. Pattern Recognition, CVPR 2017, 2017-Janua. 2017. Xception: Deep learning with depthwise separable convolutions; pp. 1800–1807. [DOI] [Google Scholar]
- 74.Ma Y., Wang C. SdcNet for object recognition. Comput. Vis. Image Underst. 2020;215 doi: 10.1016/j.cviu.2021.103332. 2022. [DOI] [Google Scholar]
- 75.Wang C.-F. 2018. A basic introduction to separable convolutions. https://l24.im/hrH8qwp. (Accessed 22 Nov. 2021) [Google Scholar]
- 76.Friedman J.H. Greedy function approximation: A gradient boosting machine. Ann. Statist. 2001;29(5):1189–1232. doi: 10.1214/aos/1013203451. [DOI] [Google Scholar]
- 77.Chen H., Shen Z., Wang L., Qi C., Tian Y. Prediction of undrained failure envelopes of skirted circular foundations using gradient boosting machine algorithm. Ocean Eng. 2022;258(May) doi: 10.1016/j.oceaneng.2022.111767. [DOI] [Google Scholar]
- 78.Touzani S., Granderson J., Fernandes S. Gradient boosting machine for modeling the energy consumption of commercial buildings. Energy Build. 2018;158:1533–1543. doi: 10.1016/j.enbuild.2017.11.039. [DOI] [Google Scholar]
- 79.Gao B., Pavel L. 2017. On the properties of the softmax function with application in game theory and reinforcement learning; pp. 1–10. [Online]. Available: http://arxiv.org/abs/1704.00805. [Google Scholar]
- 80.Marques G., Agarwal D., de la Torre Díez I. Automated medical diagnosis of COVID-19 through EfficientNet convolutional neural network. Appl. Soft Comput. 2020;96 doi: 10.1016/j.asoc.2020.106691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Umer M., Ashraf I., Ullah S., Mehmood A., Choi G.S. COVINet: a convolutional neural network approach for predicting COVID-19 from chest X-ray images. J. Ambient Intell. Humaniz. Comput. 2022;13(1):535–547. doi: 10.1007/s12652-021-02917-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.gifani P., Shalbaf A., Vafaeezadeh M. Automated detection of COVID-19 using ensemble of transfer learning with deep convolutional neural network based on CT scans. Int. J. Comput. Assist. Radiol. Surg. 2021;16(1):115–123. doi: 10.1007/s11548-020-02286-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sethy P.K., Behera S.K., Ratha P.K., Biswas P. Detection of coronavirus disease (COVID-19) based on deep features. Int. J. Math. Eng. Manag. Sci. 2020;5(4):643–651. doi: 10.20944/preprints202003.0300.v1. [DOI] [Google Scholar]
- 84.Li D., Fu Z., Xu J. Stacked-autoencoder-based model for COVID-19 diagnosis on CT images. Appl. Intell. 2021;51(5):2805–2817. doi: 10.1007/s10489-020-02002-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu X., et al. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. 2020;6(10):1122–1129. doi: 10.1016/j.eng.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Heidarian S., et al. COVID-FACT: A fully-automated capsule network-based framework for identification of COVID-19 cases from chest CT scans. Front. Artif. Intell. 2021;4(May):1–13. doi: 10.3389/frai.2021.598932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mukherjee H., Ghosh S., Dhar A., Obaidullah S.M., Santosh K.C., Roy K. Deep neural network to detect COVID-19: one architecture for both CT scans and chest X-rays. Appl. Intell. 2021;51(5):2777–2789. doi: 10.1007/s10489-020-01943-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang L., Lin Z.Q., Wong A. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci. Rep. 2020;10(1):19549. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Heidari M., Mirniaharikandehei S., Khuzani A.Z., Danala G., Qiu Y., Zheng B. Improving the performance of CNN to predict the likelihood of COVID-19 using chest X-ray images with preprocessing algorithms. Int. J. Med. Inform. 2020;144(June) doi: 10.1016/j.ijmedinf.2020.104284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chakraborty M., Dhavale S.V., Ingole J. Corona-Nidaan: lightweight deep convolutional neural network for chest X-ray based COVID-19 infection detection. Appl. Intell. 2021;51(5):3026–3043. doi: 10.1007/s10489-020-01978-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Babukarthik R.G., Ananth Krishna Adiga V., Sambasivam G., Chandramohan D., Amudhavel A.J. Prediction of COVID-19 using genetic deep learning convolutional neural network (GDCNN) IEEE Access. 2020;8:177647–177666. doi: 10.1109/ACCESS.2020.3025164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Apostolopoulos I.D., Mpesiana T.A. COVID-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020;43(2):635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ismael A.M., Şengür A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst. Appl. 2020;164:2021. doi: 10.1016/j.eswa.2020.114054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Oh Y., Park S., Ye J.C. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans. Med. Imaging. 2020;39(8):2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- 95.Ezzat D., Hassanien A.E., Ella H.A. An optimized deep learning architecture for the diagnosis of COVID-19 disease based on gravitational search optimization. Appl. Soft Comput. 2021;98 doi: 10.1016/j.asoc.2020.106742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hussain E., Hasan M., Rahman M.A., Lee I., Tamanna T., Parvez M.Z. CoroDet: A deep learning based classification for COVID-19 detection using chest X-ray images. Chaos Solitons Fractals. 2021;142 doi: 10.1016/j.chaos.2020.110495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Başaran E., Cömert Z., Çelik Y. Convolutional neural network approach for automatic tympanic membrane detection and classification. Biomed. Signal Process. Control. 2020;56 doi: 10.1016/j.bspc.2019.101734. [DOI] [Google Scholar]
- 98.Wang C.Y., Mark Liao H.Y., Wu Y.H., Chen P.Y., Hsieh J.W., Yeh I.H. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit. Work., 2020-June. 2020. CSPNet: A new backbone that can enhance learning capability of CNN; pp. 1571–1580. [DOI] [Google Scholar]
- 99.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. Proc. IEEE Int. Conf. Comput. Vis., 2017-Octob. 2017. Grad-CAM: Visual explanations from deep networks via gradient-based localization; pp. 618–626. [DOI] [Google Scholar]
- 100.Abbasniya M.R., Sheikholeslamzadeh S.A., Nasiri H., Emami S. 2022. Classification of Breast Tumours Based on Histopathology Images using Deep Features and Ensemble of Gradient Boosting Methods, Vol. 103. arXiv Prepr. arXiv2209.01380. June. 108382. [DOI] [Google Scholar]
- 101.Kim H., Jung W.K., Park Y.C., Lee J.W., Ahn S.H. Broken stitch detection method for sewing operation using CNN feature map and image-processing techniques. Expert Syst. Appl. 2022;188 doi: 10.1016/j.eswa.2021.116014. [DOI] [Google Scholar]
- 102.Brownlee J. 2019. How to visualize filters and feature maps in convolutional neural networks. https://3c5.com/w7im4. (Accessed 25 Sep. 2022) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.