Abstract
Pseudomonas putida rarely results in infection, primarily in patients undergoing invasive procedures or immunocompromised hosts. We aimed to investigate the characteristics of Pseudomonas putida infections. This is a retrospectively designed cross-sectional observational study. We retrospectively scanned the data from our hospital for the 10 years before February 15, 2022. The patients with Pseudomonas putida growth in the microbiological cultures and with antibiotic susceptibility tests were included in the study. We recorded culture isolates types, age, gender, comorbidities, immunosuppressive factors, symptoms, invasive medical procedures, length of hospital stay, and radiological findings. The mean age of the patients was 66.2 ± 14.5 years, and the male patients predominated (76.3%, n = 58/76). There was growth in bronchial lavage in 33 patients, sputum in 28, pleural effusion fluid in 12, and tracheal aspirate in 3 patients. The rate of antibiotic-resistant strains was 56.6% (n = 43). All strains were sensitive to colistin (100%), and carbapenem, amikacin, and gentamicin sensitivity rates were high. We observed that the risk of antibiotic resistance increased 4.29 times in the patients in the intensive care unit (Cl:1.27–14.47, P = .01). The patients with Diabetes Mellitus had a higher risk (OR 4.33, Cl:1.11–16.77, P = .03), and in cancer cases, the risk was 3.31 times higher (Cl:1.06–10.32, P = .03). The risk of Pseudomonas putida infection should be considered, particularly in patients with comorbid disorders causing immunosuppression, including Diabetes Mellitus and Cancer.
Keywords: antibiotic resistance, immunosuppression, invasive procedures, Pseudomonas putida
Highlights.
-
•
This study was performed in a Chest and Respiratory Diseases Hospital and on the bacterial culture-positive cases of P. putida. All samples were pulmonary in origin. To our knowledge, this study contains the largest number of P. putida cases in the literature.
-
•
This study reports the antibiotic resistance profile, risk factors, symptoms, and pulmonary radiological features of pulmonary P. putida cases.
1. Introduction
Pseudomonas putida (P. putida) belongs to the fluorescent group of opportunistic Pseudomonas species. It is commonly found on inanimate surfaces in hospitals and humid places due to its strong tolerance to harsh living conditions and causes nosocomial infections.[1,2]
It has been reported that the pathogenicity of P. putida is less compared to other Pseudomonas species, and it is sensitive to many antimicrobial agents. Therefore, P. putida infections are rarely seen in clinics.[3]
In recent years, the isolation rate of re P. putida has increased, and the emergence of multi-drug resistant and even extensively drug-resistant strains has been a cause for concern. Although this microbial agent causes infections in cancer and immunosuppressed patients and is less virulent than Pseudomonas aeruginosa, fatal diseases have been reported.[4,5]
P. putida infections used to be known to have a good prognosis. However, recent studies reported a high mortality rate (40%), particularly in patients with comorbid conditions,[5,6] attracting the attention of clinicians.[7]
Our study investigated the clinical features, risk factors, and antimicrobial resistance rates in patients with positive P. putida cultures.
2. Materials and Methods
This study is a retrospectively designed cross-sectional observational study. We retrospectively scanned the data of our hospital for the 10 years before February 15, 2022, and 76 patients (58 inpatients, 18 outpatients) with P. putida growth in the microbiological cultures and with antibiotic susceptibility tests were included in the study. Four patients with missing information in their files were excluded. Resistance to any antibiotic was considered antibiotic resistance.
We recorded localizations of bacterial isolation (sputum, bronchial lavage, pleural effusion, tracheal aspirate), age, gender, comorbidities, factors causing immunosuppression (cancer type in patients with lung cancer [LC], chemoradiotherapy), symptoms, invasive medical procedures (catheter, pleural drainage, tube thoracostomy, intubation), length of hospital stay, microbiology culture results, antibiotic resistance, and the radiological findings on thorax computed tomography (T-CT) including bronchial obstruction, bronchiectasis, pleural effusion, and parenchyma findings.
2.1. Study group
We retrospectively scanned the data from our hospital for the 10 years before February 15, 2022. The patients with P. putida growth in the microbiological cultures and with antibiotic susceptibility tests were included in the study.
2.2. Bacterial identification and antimicrobial susceptibility test
Bacterial culture and antibiogram procedures were studied according to “the European committee on antimicrobial susceptibility testing (EUCAST)” Clinical breakpoints-bacteria (v 12.0) standards. We used the BD Phoenix™ M50 automated identification and susceptibility tester to identify and antibiogram P. putida strains. In addition to the BD Phoenix ™ M50 device, we used conventional methods such as disc diffusion and gradient tests for sensitivity tests. Diagnostics MIC-COL (Diagnostics I.n.c., Galanta, Slovakia) microtube dilution method was used for colistin within the framework of EUCAST standards. Pseudomonas aeruginosa ATCC 27853 was used as a reference strain for quality control. Inhibition zone diameters and MIC values for the Gradient test and Microtube dilution test were measured and interpreted according to EUCAST standards. We showed the final results as susceptible (S), high dose susceptible (I), and resistant (R). The antimicrobial agents involved were: colistin, meropenem, ciprofloxacin, gentamicin, amikacin, imipenem, ceftazidime, aztreonam, levofloxacin, piperacillin/tasomeric acid – sensitivity disk provided by OX-OID Company.
2.3. Statistical analysis
We analyzed the data of the patients with statistical package for the social sciences (SPSS, IBM Corp., Armonk, New York) for Windows 23.0 software. Mann-Whitney U test was used to analyze the descriptive statistical parameters (mean, standard deviation) that did not conform to a normal distribution. We performed binary logistic regression analysis to determine the effects of demographic data, computed tomography findings, and the presence of comorbid disorders on the likelihood of having antibiotic-resistant P. putida isolate. We evaluated results at a 95% confidence interval and a significance level of P < .05.
2.4. Limitations
This study had some limitations. Since our hospital where this cross-sectional study was conducted is a Chest and Respiratory Diseases Hospital, the number of patients diagnosed with chronic obstructive pulmonary disease (COPD) was high. The isolates were only acquired from the respiratory tract, which might have affected the symptom diversity. We may obtain different results in a hospital where general patients are hospitalized. In patients with lung disorders, we may relate the findings to the primary disease, which should be kept in mind. Data on antimicrobial treatments administered before culture results and changes in the treatment after obtaining the culture results were available in a limited number of cases. We did not include the management of the patients in the study. As this is a retrospective study, electronic data records were adhered to.
3. Results
3.1. Sample source and distribution of p. Putida
The mean age of 76 patients included in the study was 66.2 ± 14.5 years. Male patients were predominant (76.3%, n = 58/76). The length of hospital stay was 12.6 ± 10.2 days (76.3%, n = 58). The distribution of samples to clinics was determined as 44.7% (n = 34) from the chest diseases clinic, 26.3% (n = 20) from the intensive care unit (ICU), and 28.9% (n = 22) from the thoracic surgery clinic. The bacterial growth was observed in the bronchial lavage in 43.4% (n = 33), in sputum in 36.8% (n = 28), in pleural effusion in 15.8% (n = 12), and in tracheal aspirate in 3.9% (n = 3) of the patients.
3.2. Risk factors
Nineteen patients had a history of invasive procedures, including tube drainage (n = 12), tracheostomy (n = 4), and intubation (n = 3). The most common comorbidities were COPD in 53.9% (n = 41), hypertension in 42.1% (n = 32), and cardiovascular disorders in 39.5% (n = 27) of the patients. Non-small cell lung cancer (NSCLC) was the most frequent LC (n = 21), and squamous cell cancer was the most frequent NSCLC (n = 8/21) (See Table 1).
Table 1.
The demographic characteristics of patients [n = 76 (%)].
| Demographic data | Computed Tomography Findings | 71 (93.4) | |
|---|---|---|---|
| Age (yrs) | 66.2 ± 14.5 | Consolidation | 31 (40.7) |
| Gender (M) | 58 (76.3) | Pleural effusion | 19 (25.0) |
| Smoking | 26 (34.2) | Central bronchiectasis | 14 (18.4) |
| Hospitalized patients | 58 (76.3) | Mass lesion | 12 (14.4) |
| Mean hospital stay | 12.6 ± 10.2 | Ground glass | 7 (9.2) |
| The clinic that requested the culture | Emphysema | 7 (9.2) | |
| Chest Diseases Clinic | 34 (44.7) | Cavitation | 6 (7.8) |
| Thoracic Surgery Clinic | 22 (28.9) | Bronchial obstruction | 6 (7.8) |
| Intensive Care Unit | 20 (26.3) | Pneumothorax | 2 (2.6) |
| Site of sampling | Comorbidity | 63 (82.9) | |
| Bronchial lavage | 33 (43.4) | COPD | 41 (53.9) |
| Sputum | 28 (36.8) | Hypertension | 32 (42.1) |
| Pleural effusion | 12 (15.8) | Cardiovascular disorder | 27 (39.5) |
| Tracheal aspirate | 3 (3.9) | Lung cancer | 21 (27.6) |
| No symptoms | 8 (10.5) | Squamous cell | 8 (10.5) |
| Presence of invasive Procedures | 19 (25) | Adenocancer | 7 (9.2) |
| Tube drain | 12 (15.7) | Small Cell | 4 (5.3) |
| Tracheostomy | 4 (5.2) | Other | 2 (2.6) |
| Intubation | 3 (3.9) | Diabetes mellitus | 16 (21.1) |
| Polymicrobial growth | 26 (34.2) | Tuberculosis History | 8 (10.5) |
| One co-pathogen | 23 (30.3) | Cerebrovascular disease | 5 (6.6) |
| Two co-pathogens | 3 (3.9) | Chronic kidney failure | 3 (3.9) |
| Single drug resistance | 7 (9.2) | Mortality within 30 d | 11 (14.4) |
| Multidrug resistance | 36 (47.4) | Geriatric patient (>65 yrs) | 9 (11.8) |
| Coinfection | 7 (9.2) | ||
| Invasive procedure | 7 (9.2) | ||
| Multidrug resistance | 4 (5.3) | ||
| Lung cancer | 1 (1.3) | ||
COPD = chronic obstructive pulmonary disease.
3.3. Clinical and radiological findings
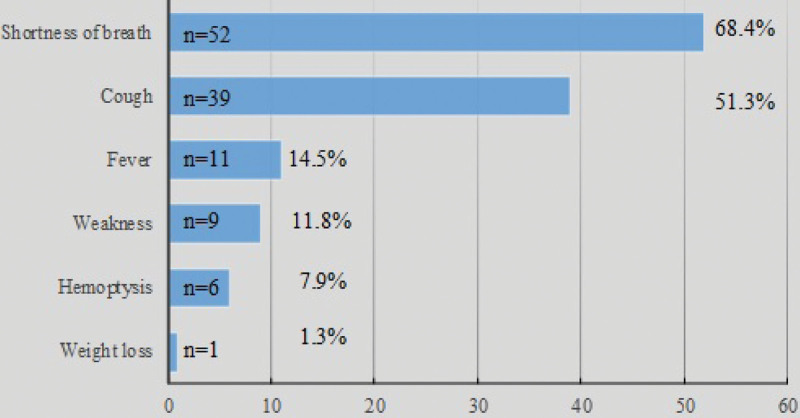
As the symptoms were considered, there was dyspnea in 68.4% (n = 52), cough in 51.3% (n = 39), and fever in 14.5% (n = 11) of the patients. Since our study was conducted in a Chest and Respiratory Diseases Hospital, it was inevitable that respiratory symptoms were predominant in the patients (See Fig. 1).
Figure 1.
Distribution of symptoms.
T-CT was obtained in 93.4% (n = 71/76) of the patients. The most common radiological findings were consolidation in 40.7% (n = 31), pleural effusion in 25.0% (n = 19) and central bronchiectasis in 18.4% (n = 14) of the cases.
All-cause mortality within the 30 days after the positive microbiological culture was seen in 11 patients (14.4%). Nine of them were over 65 years, 7 of them had coinfection with other microorganisms and 7 of them had invasive procedures before P. putida growth (See Table 1).
3.4. The prevalence of p. Putida resistance and polymicrobial growth
There were resistant strains in 56.6% (n = 43) of the patients. We detected single antibiotic resistance in 9.2% (n = 7); multi-antibiotic resistance was seen in 43.4% (n = 33) of the patients. One co-pathogen growth was seen in 30.3% (n:23), and 2 or more co-pathogen growth was seen in 3.9% (n = 3).
While all P. putida isolates were sensitive to colistin, their sensitivity to gentamicin, amikacin, and meropenem was high. In the resistant group, resistance rates were high for aztreonam (62.5%), levofloxacin, and ciprofloxacin (46.1%) (See Table 2).
Table 2.
Antibiotic resistance of P. Putida isolates.
| Antibiotic | n | Sensitive n = 33 (43.4%) | Resistant n = 43 (56.6%) |
|---|---|---|---|
| Colistin | 76 | 76 (100) | - |
| Meropenem | 76 | 63 (82.9) | 13 (17.1) |
| Amikacin | 76 | 73 (96.1) | 3 (3.9) |
| Ceftazidime | 76 | 62 (81.6) | 14 (18.4) |
| Piperacillin-Tazobactam | 76 | 50 (65.3) | 26 (34.2) |
| Gentamicin | 51 | 45 (88.2) | 6 (11.8) |
| İmipenem | 50 | 43 (86) | 7 (14) |
| Levofloxacin | 76 | 41 (53.9) | 35 (46.1) |
| Ciprofloxacin | 76 | 41 (53.9) | 35 (46.1) |
| Aztreonam | 24 | 9 (35.5) | 15 (62.5) |
| (%) row percentage |
Binary logistic regression analysis was employed to examine the correlation of the findings with antibiotic resistance. It was determined that age (OR 1.00, Cl: 0.97–1.03, P = .92), gender (OR 1.74, Cl: 0.57–5.27, P = .32), and being symptomatic (OR 1.34, Cl: 0.31-5.83, P = .69) did not increased the risk for antibiotic resistance.
The risk of antibiotic resistance was 3.52 (Cl:1.15–10.76, P = .02) times higher in hospitalized patients than in outpatients, and the risk was 4.29 (CI:1.27–14.47, P = .01) times higher in the patients in ICU compared to the 1s who were not in ICU.
Invasive procedures (OR 0.60, Cl:0.21–1.73, P = .35) or smoking history (OR 0.84, Cl:0.32–2.19, P = .72) did not significantly increase the risk for resistance.
When the radiological findings were examined, it was determined that 39 patients had T-CTs in the antibiotic resistance group. Central bronchiectasis was strongly correlated with antibiotic resistance (OR 6.66, Cl:1.36–32.51, P = .01). We found that the presence of consolidation was associated or correlated to resistant strain 2.84 times (Cl:1.06–7.58, P = .03). Although the presence of a cavitary lesion seemed to be associated or correlated to resistant strain 4.55 times but the result was not statistically significant (Cl: 0.50–41.20, P = .17). The presence of emphysema (OR 0.29, Cl: 0.06–1.62, P = .15) and bronchial obstruction (OR 0.80, Cl: 0.15–4.29, P = .80) seemed to be negatively correlated. However, the result was not statistically significant.
There were comorbid disorders in 33 patients in the risk group for antibiotic resistance. COPD, the most common comorbid condition in the patient group, increased the risk 2.87 times (Cl:1.12–7.34, P = .03). It was determined that the comorbid disorder which increased the risk for antibiotic resistance the most was diabetes mellitus (DM) (OR 4.33, Cl:1.11–16.77, P = .03). The risk was 3.31 times higher in patients with LC (Cl:1.06–10.32, P = .03). Other comorbidities were not correlated with an increased risk for antibiotic resistance (See Table 3).
Table 3.
Evaluation of the risk for antibiotic resistance.
| Resistant group | OR (95% CI) for antibiotic-resistant | p | |
|---|---|---|---|
| Demographic data | n = 43 | ||
| Age | 66.4 ± 14.6 | 1.00 (0.97–1.03) | .92 |
| Gender (F) | 12 (27.9) | 1.74 (0.57–5.27) | .32 |
| Smoking | 14 (32.6) | 0.84 (0.32–2.19) | .72 |
| Hospitalized patients | 37 (86.0) | 3.52 (1.15–10.76) | .02 * |
| Hospitalization in the intensive care unit | 16 (37.2) | 4.29 (1.27–14.47) | .01 * |
| Presence of symptoms | 39 (90.7) | 1.34 (0.31–5.83) | .69 |
| Invasive procedures | 9 (20.9) | 0.60 (0.21–1.73) | .35 |
| Computed Tomography Findings | |||
| Consolidation | 22 (56.4) | 2.84 (1.06–7.58) | .03 * |
| Pleural effusion | 11 (28.2) | 1.17 (0.40–3.40) | .76 |
| Central bronchiectasis | 12 (30.7) | 6.66 (1.36–32.51) | .01 * |
| Mass lesion | 7 (17.9) | 1.18 (0.33–4.15) | .79 |
| Ground glass appearance | 6 (15.3) | 2.07 (0.21–19.67) | .52 |
| Emphysema | 2 (5.1) | 0.29 (0.06–1.62) | .15 |
| Cavitary lesion | 5 (12.8) | 4.55 (0.50–41.20) | .17 |
| Bronchial obstruction | 3 (7.6) | 0.80 (0.15–4.29) | .80 |
| Presence of comorbid disorder | n = 33 | ||
| COPD | 28 (84.8) | 2.87 (1.12–7.34) | .03 * |
| Hypertension | 18 (54.5) | 0.97 (0.39–2.44) | .96 |
| Cardiovascular disease | 14 (42.4) | 1.50 (0.54–4.18) | .43 |
| Lung cancer | 16 (48.4) | 3.31 (1.06–10.32) | .03 * |
| Diabetes mellitus | 13 (39.3) | 4.33 (1.11–16.77) | .03 * |
| Tuberculosis History | 6 (18.2) | 2.51 (0.47–13.35) | .27 |
| Cerebrovascular disease | 2 (6.1) | 0.48 (0.07–3.10) | .44 |
| Chronic kidney failure | 1 (3) | 0.36 (0.03–4.25) | .42 |
(%) = column percentages, COPD = chronic obstructive pulmonary disease, OR = 1 implies the probability of antibiotic resistance.
P value = 0.05 significant.
4. Discussion
In the 1980s, P. putida bacteremia was first reported in cancer patients[8,9] and later in patients with catheter-related bloodstream infections, pneumonia, acute cholecystitis, cholangitis, skin, and soft tissue infections[6,10,11]
P. putida strains are rarely isolated from clinical specimens, suggesting that they survive in a hospital setting and sometimes cause nosocomial infections in seriously ill or immunocompromised patients.[12–14]
In multidisciplinary studies, P. putida was isolated most frequently from the patients hospitalized in surgery clinics, including neurosurgery (15.6%), organ transplantation (9.4%), orthopedics (9.4%), as well as chest diseases clinic (9.4%).[7,14]
Our study was performed in the clinics of a Chest and Respiratory Diseases Hospital, and 44.7% of P. putida growth was observed in the samples sent from the chest diseases clinic, 28.9% were grown from the samples sent from the thoracic surgery clinic, and 26.3% were grown in the samples sent from ICU. In binary logistic regression analysis, the risk of antibiotic resistance was 4.29 times higher in the intensive care unit patients (Cl:1.27–14.47, P = .01). In addition, it was 3.52 times higher in hospitalized patients than in outpatients (Cl:1.15–10.76, P = .02).
Urine and blood are in the first place for culture-positive P. putida samples in general hospitals, and P. putida growth rate in sputum was reported as 12.5%.[7,15] Our study was performed only with respiratory tract samples, and P. putida growth in the cultures was most frequent in bronchial lavage (43.4%) and sputum (36.8%) and the least frequent in tracheal aspirate samples (3.9%).
In the studies, the mortality reates were inconsistent. One of the study reported that the mortality rate was 7.9% (1/28)[6] and another study reported that the case fatality rate was 29% (12/41).[15] Our cohort had all-cause mortality within the 30 days after the positive microbiological culture was 14.4% (11/76).
In studies conducted in multidisciplinary centers, invasive surgical procedures, drainage tubes, urinary catheters, and femoral venous catheters have been reported in the 1st place in parallel with the fact that surgical services are in the first place in terms of samples positive for P. putida growth.[7] In another study, they reported that P. putida was colonized in the tissues of immunocompromised patients with bile drainage tubes.[6] In our study, the most common invasive procedure was tube drainage (15.7%), which is consistent with the literature. Despite the general knowledge that the presence of an invasive procedure increases the risk of P. putida infection, our study found that the presence of an invasive procedure did not significantly increase the risk of antibiotic resistance (OR 0.60, Cl:0.21–1.73, P = .35).
In addition to the study which reported solid tumors, hematological malignancy, and immunosuppressive conditions as the most common underlying disorders in patients with P. putida growth,[5] another study reported that 85.7% of 28 cases with P. putida growth were immunosuppressed, and opportunistic P. putida infections were observed mainly in the patients with immunosuppression, low physical performance, and comorbid disorders.[7,9,16,17]
In our study on patients with pulmonary disorders, COPD may be regarded as a disorder causing poor physical performance, which was present in 53.9% of the patients. In the antibiotic resistance risk analysis, we found that COPD increased the risk 2.87 times (Cl:1.12–7.34, P = .03). In the immunosuppressed group, the rate of lung cancer patients receiving chemoradiotherapy was 27.6%. The most common lung cancer subtype was NSCLC (Squamous cell, 10.5%) and the risk for antibiotic resistance was 3.31 times higher in cancer patients (CI:1.06–10.32, P = .03).
The most common extrapulmonary disorders were hypertension (42.1%), cardiovascular disorders (39.5%), and DM (21.1%). In evaluating antibiotic resistance risk with binary logistic regression analysis, DM was determined as the comorbid disorder that increased the risk the most (OR 4.33, Cl:1.11–16.77, P = .03).
In this study which we conducted in a Chest and Respiratory Diseases Hospital and only with respiratory tract samples, the most common T-CT findings were consolidation (40.7%), pleural effusion (25.0%), central bronchiectasis (18.4%), and more rarely mass, ground glass appearance, emphysema, cavitary lesion, bronchial obstruction, and pneumothorax. T-CT was performed on 39 patients in the antibiotic-resistant group. We found that central bronchiectasis strongly correlated with antibiotic resistance (OR 6.66, Cl:1.36–32.51, P = .01), while pneumonic consolidation increased the risk 2.84 times (CI:1.06–7.58, P = .03). Although the presence of a cavitary lesion seemed to increase the risk 4.55 times (Cl: 0.50–41.20, P = .17), the result was not statistically significant.
Fever was reported as the most common symptom in the previous studies that included multidisciplinary clinical data.[7] In our study, the most common symptom was dyspnea (68.4%) (cough 51.3%, fever 14.5%), but it was observed that being symptomatic or not was not correlated with the risk of antibiotic resistance (OR 1.34, CI: 0.31–5.83, P = .69).
In our Chest and Respiratory Diseases Hospital, it was inevitable that respiratory symptoms were predominant in the patients, both comorbid disorders and the factors causing immunosuppression were pulmonary in origin, and our study was conducted only with respiratory tract samples.
4.1. Antibiotic resistance in p. Putida hospital isolates
P. putida has generally been considered low virulence and has little clinical significance. In the literature, it has been reported that the prognosis of P. putida bacteremia is good, with an improvement rate of 93%.[6,18]
In the earlier studies, Fass et al found that clinical isolates of P. putida were susceptible to various antibiotics (100% sensitivity to ciprofloxacin and tobramycin, 87% sensitivity to imipenem and piperacillin/tazobactam).[19] Another recent study reported sensitivity to ciprofloxacin (54.5%), amikacin (86.4%), and gentamicin (56.8%).[7]
Our study found the sensitivity rates for tobramycin, ciprofloxacin, imipenem, piperacillin/tazobactam, amikacin, and gentamicin to 96.0%, 53.9%, 86.0%, 65.3%, 96.1%, and 88.2%, respectively. The low sensitivity to ciprofloxacin may be attributed to the widespread use of this agent over the years. We conducted this study with data from a tertiary medical center, and the frequent use of antibiotics in hospitalized patients may be the reason for the low sensitivity to piperacillin/tazobactam. Likewise, our study’s lowest preference for gentamicin in our chest diseases hospital might have resulted in high sensitivity to gentamicin. In addition, Sader et al reported amikacin sensitivity as 79.8%.[20] Our amikacin sensitivity rate was similar to the literature.
Studies conducted in the following years have reported an increasing number of carbapenem-resistant P. putida isolates.[21,22] Within the scope of the Korean nationwide surveillance of antimicrobial resistance, 8 (67%) isolates were identified as P. putida in the analysis of carbapenem-resistant (12 imipenem-resistant isolates) of Pseudomonas species other than P. aeruginosa.[23]
In a study conducted with 18 P. putida isolates, 22% were resistant to imipenem, and 28% were resistant to meropenem. The same study compared carbapenem resistance rates with P. aeruginosa and found no significant difference. The aztreonam resistance rate of P. putida (72%) was higher than P. aeruginosa. With these findings, it was reported that multi-drug and carbapenem resistance is common not only in P. aeruginosa but also in P. putida isolates.[5] In another study, it was sensitive to aztreonam (88.6%), imipenem (62.8%), and meropenem (45.5%).[7]
We found the aztreonam resistance rate (62.5%) to be compatible with some studies in the literature and inconsistent with others. We found carbapenem resistance rates to be lower than other studies in the literature (meropenem 17.1%, imipenem 14.0%). These low resistance rates may be due to the controlled use of carbapenems or their usage as the last step in our tertiary medical center.
In our study, the prevalence of antibiotic resistance was 9.2% for a single antibiotic and 43.4% for more-than-1 antibiotic multi-drug resistant, which was lower than the literature data (75%).[7] On the other hand, the polymicrobial growth rate was 3.9% for 2 or more pathogens and 30.3% for 1 pathogen, and lower than the literature data (>2 pathogens 18.8%, 1 pathogen 59.4%).[7]
5. Conclusion
High rates of antibiotic resistance may be observed in patients with lung cancer, DM as comorbidity, hospitalized in the ICU, and patients with radiological bronchiectasis. The susceptibility rates of P. putida for imipenem, meropenem, gentamicin, and amikacin are still high. These agents may be used as the first choice antibiotics in P. putida infections for successful treatment of the condition.
Author contributions
Conceptualization: Hüsnü Baykal, Deniz Çelik, A. Füsun Ülger, Sedat Vezir, M. Ömür Güngör.
Data curation: Hüsnü Baykal, Deniz Çelik, A. Füsun Ülger, Sedat Vezir, M. Ömür Güngör.
Formal analysis: M. Ömür Güngör.
Investigation: Deniz Çelik, A. Füsun Ülger, Sedat Vezir, M. Ömür Güngör.
Methodology: Hüsnü Baykal, Deniz Çelik, A. Füsun Ülger, Sedat Vezir, M. Ömür Güngör.
Project administration: Hüsnü Baykal.
Supervision: Hüsnü Baykal, Deniz Çelik, A. Füsun Ülger, Sedat Vezir, M. Ömür Güngör.
Validation: Hüsnü Baykal, A. Füsun Ülger, Sedat Vezir, M. Ömür Güngör.
Visualization: Hüsnü Baykal, Sedat Vezir, M. Ömür Güngör.
Writing – original draft: Hüsnü Baykal, M. Ömür Güngör.
Writing – review & editing: Deniz Çelik, A. Füsun Ülger, Sedat Vezir, M. Ömür Güngör.
Abbreviations:
- COPD =
- chronic obstructive pulmonary disease
- DM =
- diabetes mellitus
- EUCAST =
- the European committee on antimicrobial susceptibility testing
- ICU =
- the intensive care unit
- LC =
- lung cancer
- NSCLC =
- non-small cell lung cancer
- P. Putida =
- Pseudomonas putida
- T-CT =
- thorax computed tomography
The ethics committee approval obtained from Health Sciences University Ankara Atatürk Sanatorium Training and Research Hospital, Clinical Studies Ethic Committee (2012-KAEK-15/2481) in 22.02.2022.
The authors have no funding and conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
How to cite this article: Baykal H, Çelik D, Ülger AF, Vezir S, Güngör MÖ. Clinical features, risk factors, and antimicrobial resistance of pseudomonas putida isolates. Medicine 2022;101:48(e32145).
Contributor Information
Hüsnü Baykal, Email: drhusnubaykal@hotmail.com.
A. Füsun Ülger, Email: fusunulger67@gmail.com.
Sedat Vezir, Email: sedatvezir@gmail.com.
M. Ömür Güngör, Email: omurgungor70@hotmail.com.
References
- [1].Devarajan N, Kohler T, Sivalingam P, et al. Antibiotic-resistant Pseudomonas spp. in the aquatic environment: a prevalence study under tropical and temperate climate conditions. Water Res. 2017;115:256–65. [DOI] [PubMed] [Google Scholar]
- [2].Rodriguez A, Escobar S, Gomez E, et al. Behavior of several Pseudomonas putida strains growth under different agitation and oxygen supply conditions. Biotechnol Prog. 2018;34:900–9. [DOI] [PubMed] [Google Scholar]
- [3].Cho CH, Lee SB. Comparison of clinical characteristics and antibiotic susceptibility between Pseudomonas aeruginosa and P. putida keratitis at a tertiary referral center: a retrospective study. BMC Ophthalmol. 2018;18:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Usta Atmaca H, Akbas F. A Extensively drug-resistant Pseudomonas putida bacteremia that was resolved spontaneously. J Infect Dev Ctries. 2019;13:577–80. [DOI] [PubMed] [Google Scholar]
- [5].Kim SE, Park SH, Park HB, et al. Nosocomial Pseudomonas putida bacteremia: high rates of carbapenem resistance and mortality. Chonnam Med J. 2012;48:91–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yoshino Y, Kitazawa T, Kamimura M, et al. Pseudomonas putida bacteremia in adult patients: five case reports and a review of the literature. J Infect Chemother. 2011;17:278–82. [DOI] [PubMed] [Google Scholar]
- [7].Genmei T, Yang X, Peihong Y, et al. Risk factors and antimicrobial resistance profiles of Pseudomonas putida infection in central China, 2010-2017. Medicine (Baltim). 2019;98:e17812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Taylor M, Keane CT, Falkiner FR. Pseudomonas putida in transfused blood. Lancet. 1984;2:107. [DOI] [PubMed] [Google Scholar]
- [9].Anaissie E, Fainstein V, Miller P, et al. Pseudomonas putida. Newly recognized pathogen in patients with cancer. Am J Med. 1987;82:1191–4. [DOI] [PubMed] [Google Scholar]
- [10].Ladhani S, Bhutta ZA. Neonatal Pseudomonas putida infection presenting as staphylococcal scalded skin syndrome. Eur J Clin Microbiol Infect Dis. 1998;17:642–4. [DOI] [PubMed] [Google Scholar]
- [11].Thomas BS, Okamoto K, Bankowski MJ, et al. A lethal case of Pseudomonas putida bacteremia due to soft tissue infection. Infect Dis Clin Pract (Baltim Md). 2013;21:147–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Carpenter RJ, Hartzell JD, Forsberg JA, et al. Pseudomonas putida war wound infection in a US Marine: a case report and review of the literature. J Infect. 2008;56:234–40. [DOI] [PubMed] [Google Scholar]
- [13].Treviño M, Moldes L, Hernández M, et al. Nosocomial infection by VIM-2 metallo-beta-lactamase-producing Pseudomonas putida. J Med Microbiol. 2010;59:853–5. [DOI] [PubMed] [Google Scholar]
- [14].Liu H, Zhao J, Xing Y, et al. Nosocomial infection in adult admissions with hematological malignancies originating from different lineages: a prospective observational study. PLoS One. 2014;9:e113506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yang CH, Young T, Peng MY, et al. Clinical spectrum of Pseudomonas putida infection. J Formos Med Assoc. 1996;95:754–61. [PubMed] [Google Scholar]
- [16].Molina L, Udaondo Z, Duque E, et al. Specific gene loci of clinical Pseudomonas putida isolates. PLoS One. 2016;11:e0147478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Franzetti F, Cernuschi M, Esposito R, et al. Pseudomonas infections in patients with AIDS and AIDS-related complex. J Intern Med. 1992;231:437–43. [DOI] [PubMed] [Google Scholar]
- [18].Blazevic DJ, Koepcke MH, Matsen JM. Incidence and identification of Pseudomonas fluorescens and Pseudomonas putida in the clinical laboratory. Appl Microbiol. 1973;25:107–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fass RJ, Barnishan J, Solomon MC, et al. In vitro activities of quinolones, beta-lactams, tobramycin, and trimethoprim-sulfamethoxazole against nonfermentative gram-negative bacilli. Antimicrob Agents Chemother. 1996;40:1412–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sader HS, Jones RN. Nadiren izole edilen enterik olmayan Gram negatif basillerin antimikrobiyal duyarliliği. Int J Antimicrob Ajanlari. 2005;25:95–109. [Google Scholar]
- [21].Almuzara M, Radice M, de Gárate N, et al. VIM-2-producing Pseudomonas putida, Buenos Aires. Emerg Infect Dis. 2007;13:668–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bogaerts P, Huang TD, Rodriguez-Villalobos H, et al. Nosocomial infections caused by multidrug-resistant Pseudomonas putida isolates producing VIM-2 and VIM-4 metallo-beta-lactamases. J Antimicrob Chemother. 2008;61:749–51. [DOI] [PubMed] [Google Scholar]
- [23].Lee K, Park AJ, Kim MY, et al. Metallo-beta-lactamase-producing Pseudomonas spp. in Korea: high prevalence of isolates with VIM-2 type and emergence of isolates with IMP-1 type. Yonsei Med J. 2009;50:335–9. [DOI] [PMC free article] [PubMed] [Google Scholar]